Abstract
By using oligonucleotide-directed saturation mutagenesis, we collected 366 different single amino acid substitutions in a 109-aa segment (residues 95–203) in the fingers and palm subdomains of the HIV-1 reverse transcriptase (RT), the enzyme that replicates the viral genome. After expression in Escherichia coli, two phenotypic assays were performed. The first assay tested for RNA-dependent DNA polymerase activity. The other assay used Western blot analysis to estimate the stability of each mutant protein by measuring the processing of the RT into its mature heterodimeric form, consisting of a 66-kDa subunit and a 51-kDa subunit. The resulting phenotypic data provided a “genetic” means to identify amino acid side chains that are important for protein function or stability, as well as side chains located on the protein surface. Several HIV-1 RT crystal structures were used to evaluate the mutational analysis. Our genetic map correlates well with the crystal structures. Combining our phenotype data with crystallographic data allowed us to study the genetically defined critical residues. The important functional residues are found near the enzyme active site. Many residues important for the stability of the RT participate in potential hydrogen bonding or hydrophobic interactions in the protein interior. In addition to providing a better understanding of the HIV-1 RT, this work demonstrates the utility of saturation mutagenesis to study the function, structure, and stability of proteins in general. This strategy should be useful for studying proteins for which no crystallographic data are available.
The development of oligonucleotide-directed mutagenesis (1) enhanced the power of genetic analysis by providing a method to introduce a mutation at a specific position in a DNA sequence. This technique may be used to make a specific amino acid substitution in a protein by altering codons. An adaptation of site-specific mutagenesis led to saturation mutagenesis (reviewed in ref. 2), which provides an approach to make libraries containing a collection of mutations at every base in a DNA element (3) or at every amino acid in a protein (4).
X-ray crystallography is a powerful tool for studying protein function and structure. However, the use of this method is limited to proteins that crystallize. With the constant discovery of new proteins with biological and medical importance, additional methods that provide functional and structural information are extremely valuable. In this study, we assess the potential of saturation mutagenesis for the study of function and stability of proteins for which structural data are difficult to obtain, and as a means to offer additional insight into proteins whose structures are known. By using saturation mutagenesis we collected 366 unique single missense mutants in a 109-aa region (residues P95 through E203) in the fingers and palm subdomains of the HIV-1 reverse transcriptase (RT).
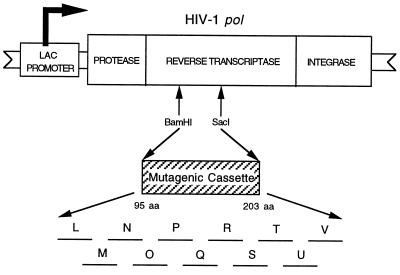
The HIV-1 RT is encoded by the viral pol gene, which contains the coding sequences for the protease, RT (with both DNA polymerase and ribonuclease H activities), and integrase. We previously developed a bacterial expression system for the HIV-1 pol gene (5, 6) (Fig. 1). In this system the pol gene products are expressed initially as a 120-kDa polypeptide that is processed by the viral protease to produce the mature protease (11 kDa), RT (66-kDa and 51-kDa heterodimer), and integrase (34 kDa). This process appears indistinguishable from protease processing in the virus.
Figure 1.
pE66M HIV-1 RT expression vector. The HIV-1 RT expression vector pE66M contains the entire HIV-1 pol gene cloned into the phagemid pIBI20 (IBI). The pol gene is under control of the inducible lac promoter. The 11 mutant libraries (L through V) are contained in the BamHI–SacI cassette. This figure is not drawn to scale.
The wild-type RT expressed in our system is catalytically active. We have two biological assays to test the effect of RT mutations. The first assay is an in vitro RNA-dependent DNA polymerase assay to test for the enzymatic activity of the RT. Mutations at amino acid residues involved either directly or indirectly in enzyme catalysis will result in loss of RT activity. Alternatively, loss of RT activity may be caused by a mutation at a residue involved in the protein stability of the RT. In our second phenotype assay, we used Western blot analysis to estimate the stability of each mutant by measuring the processing of the RT into its mature heterodimeric form. There is an equilibrium between the catalytically inactive denatured state and the native state where the protein is correctly folded in its biologically active conformation (7). Destabilizing mutations shift the equilibrium toward the unfolded state. Unstable proteins sometimes aggregate and become insoluble, which can interfere with normal processing. Furthermore, an unstable protein may become accessible to cellular proteases and be degraded. Our Western blot analysis shows the amounts of correctly processed protein, as well as unprocessed precursor or improperly processed products. We also used Western blots to determine the relative amount of soluble protein.
Combining the RT activity and Western blot phenotype data for all our mutants, we constructed a genetic map of the HIV-1 RT that predicts the residues involved in protein function and stability, as well as which side chains are on the protein surface. We then compared our predictions with crystallographic data to demonstrate the utility of saturation mutagenesis as a method for gaining insight into both protein function and structure.
MATERIALS AND METHODS
RT Expression System.
The HIV-1 RT expression plasmid is pART1E66M (pE66M) (8). In pE66M (Fig. 1) the pol gene is under the control of the inducible lac promoter. This vector is a phagemid, with origins of replication to allow propagation as a plasmid or as a single-stranded DNA phage.
Mutant Library Construction.
By using an improved oligonucleotide-directed saturation mutagenesis strategy, a 326-bp region of the HIV-1 RT spanning amino acid residues P95 through E203 was randomly mutagenized (8, 9). This process was accomplished through the production of 11 mutant libraries (labeled L, M, N, O, P, Q, R, S, T, U, and V) (Fig. 1). These libraries were stored as phagemid stocks. An additional 21 mutants were made by using site-directed mutagenesis to fill gaps. We made nine more site-specific mutants (T107S, T107V, V118T, L120A, L120T, T128V, Y146F, S191T, and H198Q) to study the role of certain residues in more detail.
Initial Genotype Screening.
Escherichia coli strain JM101 was infected with each phagemid library and plated onto 2× YT (1.0% tryptone/1.0% yeast extract/0.5% NaCl) ampicillin plates. Individual clones were randomly picked from the plates and inoculated into 2× YT liquid medium [supplemented with 200 μg/ml of ampicillin, 10 μg/ml of thiamine, 0.2% glucose, and M13KO7 helper phage (10) or VCSM13 helper phage (Stratagene)]. The cultures were grown for 12–18 hr at 37°C, then pelleted by centrifugation and two aliquots of the phage supernatant were removed: an 800-μl aliquot (for single-stranded DNA isolation) and a 150-μl aliquot (a phage stock for long-term storage of each mutant). The single-stranded DNA was isolated from the phage particles by using a NaI method (11), a QIAprep Spin M13 Kit (Qiagen), or as described previously (6). The region spanning the site of mutagenesis in each clone was sequenced according to the manufacturer’s protocol for the Sequenase version 2.0 DNA sequencing kit (United States Biochemical) or with the Klenow fragment of E. coli DNA polymerase. The resulting clones contained 0, 1, or more nucleotide substitutions. Clones with a nucleotide substitution(s) resulting in a single missense mutation were subjected to phenotype assays.
Preparing Lysates for Phenotype Analysis.
RT mutant phagemid clones were infected into E. coli strain JM101 and plated onto glucose-minimal-ampicillin plates. Individual colonies were inoculated into two separate cultures: 2× YT (supplemented as described above) to confirm the mutant’s genotype and M9 minimal medium (without glucose and supplemented with 0.4% casamino acids, 0.001% thiamine, and 200 μg/ml of ampicillin) to determine the phenotype. The genotype cultures were processed as described above. The phenotype cultures were grown at 37°C to an optical density of 0.2–0.4 at 600 nm, adjusted to equal concentrations of cells, and induced with 1 mM or 5 mM isopropyl-β-d-thiogalactopyranoside at 37°C for 2 hr. Each culture then was used to prepare lysates for the assay of RT activity (8) and Western analysis (6). Lysates of the soluble fraction to be used for Western blot analysis were prepared by adding 15 μl of 4× sample buffer (63 mM Tris, pH 6.8/2% SDS/10% glycerol/5% β-mercaptoethanol/1 mg/ml bromophenol blue) to 30 μl of the lysate made for the RT activity assay.
Phenotype Assays.
The RT activity was assayed by using a poly(rC)-oligo(dG) template-primer as previously described (8). RT activity for each mutant was expressed as a percentage of RT activity of the wild-type clone (pE66M). Western blot analysis of total cell lysates and soluble lysates was performed as described previously (6), with the exception that for mutants of residues P95 through M164 the blots were stained with the NEA-9304 mouse monoclonal anti-HIV-1 RT antibody (DuPont) instead of a polyclonal anti-HIV-1 RT antibody.
Computer Analysis of Crystal Structures.
The crystal structure viewing program Kinimage version 4.2 (12) was used to analyze the mutant phenotypes, build models, and find inter-atomic distances. The computer program ribbons (13) was used to create the crystal structure figures. The coordinates of the crystal structures used in the analyses were obtained from the Brookhaven Protein Data Bank [1har (14), 1rtj (15), 1hmi (16), and 1mml (17)].
Sequence Variability Calculation.
An alignment of 29 retroviral RT sequences (18) was used to calculate the variability of each residue position (P95 through E203 in HIV-1 RT). Variability is defined at each position as the number of different residues found at that position divided by the frequency of the most common residue (19). Variability is calculated as v = d/(m/s) = ds/m, where d is the number of different residues found at that position, m is the number of occurrences of the most frequent residue at that position, and s is 29, the number of sequences in the alignment. At a few positions where gaps were introduced into some sequences to produce the best alignment, the gap has been treated as one type of residue for the purposes of the calculation. For example, an invariant residue among all 29 retroviral RTs would have a variability of one.
Molecular Surface Area (MSA) Calculation.
An MSA analysis was performed on the HIV-1 RT crystal structure 1rtj (15) by using the program gepol (20, 21) to determine the exposed MSA for every atom in the HIV-1 RT heterodimer. The sum of the MSA of side-chain atoms for each residue was calculated to give the exposed MSA for amino acids in the fingers and palm subdomains for the 66-kDa subunit (22). The buried MSA was calculated by subtracting the exposed MSA for each residue from the MSA of the amino acid side chain in the second position in an alanine-X-alanine tripeptide by using MSAs compiled in ref. 23.
RESULTS AND DISCUSSION
Collection of Single Amino Acid Substitutions.
By sequencing individual clones from our 11 mutant libraries spanning HIV-1 RT residues P95 through E203, we identified 336 unique single missense mutations. We constructed an additional 30 mutations by using site-directed mutagenesis. A survey of the literature revealed 258 unique single amino acid replacements isolated by others in the entire HIV-1 RT (22). Of these replacements, 99 mutations representing 48 different sites are in the region of our cassette, but only 33 are common to our collection. These mutations are found in either HIV-1 RT clones or in viral isolates.
Interpretation of Phenotype Data.
RT assay.We tested each mutant for RNA-dependent DNA polymerase activity by performing an in vitro RNA-dependent DNA polymerase assay using a crude lysate of bacteria expressing each mutant. RT activity is expressed as a percentage of the activity obtained from the wild-type clone pE66M. The results are divided into four categories: negative (<3% of wild-type activity), low intermediate (≥3% but <20% of wild-type activity), high intermediate (≥20% but <50% of wild-type activity), and wild-type or positive (≥50% of wild-type activity).
Western blot analysis.
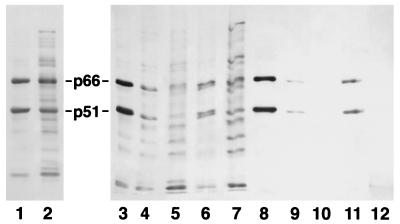
Because our system expresses the entire HIV-1 pol gene, we are able to use Western blot analysis to measure protease processing of the RT into its mature heterodimeric form. On a Western blot of wild-type RT, we detect a 66-kDa band (p66) and a 51-kDa band (p51), representing the two subunits of the active RT. Abnormal processing is assumed to result from protein stability changes, because the processing cleavage sites are outside the mutagenic region. Mutations affecting processing could result in the aggregation of the mutant protein (leading to incomplete processing) or proteolytic degradation (causing loss of signal). Examples of Western blot phenotypes of HIV-1 RT mutants are shown in Fig. 2.
Figure 2.
Examples of Western blot analysis of HIV-1 RT mutants. Western blot analysis demonstrating the four phenotypes for total cell lysates (wild type, US, UL, and Lt) and the four phenotypes for soluble lysates (wild type, faint, very faint, and no bands). The Western phenotypes are described in Results and Discussion. The Western blot is stained with a monoclonal anti-HIV-1 RT antibody. p66 and p51 are the subunits of the mature heterodimeric HIV-1 RT. Samples in lanes 1–7 are total cell lysates: 1) V111G (wild-type phenotype); 2) Y144F (US phenotype); 3) pE66M (a wild-type HIV-1 RT clone); 4) P133A (Lt phenotype); 5) L149R (Lt phenotype); 6) A129G (Lt phenotype); 7) Y146D (UL phenotype). Samples in lanes 8–12 are soluble lysates: 8) pE66M (a wild-type HIV-1 RT clone); 9) P133A (very faint phenotype); 10) L149R (no bands phenotype); 11) A129G (faint phenotype); 12) Y146D (no bands phenotype).
We obtained Western blots for both soluble fraction lysates and total cell lysates. In interpreting the Western blot data, we classified the phenotype for individual mutants as either positive or negative for protein stability. A specific mutant was considered positive for protein stability if either the total cell lysate or the soluble lysate gave a wild-type phenotype. A specific mutant was considered to be negative for protein stability if the total cell lysate gave a non-wild-type phenotype and, if determined, a non-wild-type phenotype for soluble lysates.
The results for the Western blot analysis of the soluble lysates are classified as “wild-type” for the presence of wild-type levels of the p66 and p51 forms of the mature RT, “faint” for reduced levels of p66 and p51, “very faint” for a further reduction of p66 and p51 when compared with those in the faint category, and “no bands” for the complete absence of p66 and p51.
The results for the total cell lysates for libraries S, T, U, and V (M164–E203) were described previously (8). The phenotypes for mutants in these libraries were classified as wild type or positive for efficient processing to produce stable products, as indicated by the presence of wild-type levels of p66 and p51 forms of the mature RT, negative for inefficient processing, indicated by the near absence of p51, along with a much reduced level of p66, with some high and low molecular weight products when compared with a wild-type clone, and intermediate for a variety of phenotypes, generally indicated by a reduced level of the RT heterodimer.
The phenotypes as measured by Western blot analysis for mutants in libraries L, M, N, O, P, Q, and R (residues P95–M164) for total cell lysates are classified as wild type for the presence of wild-type levels of the p66 and p51 forms of the mature RT with no unprocessed high molecular weight bands; upper strong (US) for the presence of wild-type levels (strong) of the p66 and p51 forms of the mature RT with unprocessed high molecular weight (upper) bands; upper light (UL) for the presence of reduced levels (light) of the p66 and p51 forms of the mature RT with unprocessed high molecular weight (upper) bands; and light (Lt) for the presence of reduced levels (light) of p66 and p51 forms of the mature RT with no unprocessed high molecular weight bands. These unprocessed high molecular weight bands are insoluble, because they are absent in Western blots of the soluble lysates.
Genetic Classification of HIV-1 RT Residues.
By using the collection of RT activity and Western blot data for the 366 mutants, we produced a genetic map of critical residues within the HIV-1 RT (Fig. 3). Fig. 4 describes our classification of individual amino acid residues with respect to their roles in the protein. This approach works well with a few mutations at a particular residue, although our classification system is not absolute. Occasionally, when there are many substitutions at a particular residue, exceptions are observed and the site is classified according to the preponderance of the data (see discussion below).
Figure 3.
Summary and analysis of HIV-1 RT mutant phenotypes. Phenotypes of single missense mutations in a 109-aa region of the HIV-1 RT. Mutants were isolated from 11 overlapping random substitution libraries designated L, M, N, O, P, Q, R, S, T, U, and V. The region mutagenized in each library is indicated by a horizontal dashed line. The secondary structural elements indicated are from the 3hvt HIV-1 RT crystal structure (24) and not from the 1rtj crystal structure (15), which is shown in Fig. 5. There are minor differences between them with respect to the end points assigned for α helices and β strands. The conservative amino acids are grouped as follows: (A, S, T, G, P); (V, L, I, M); (R, K, H); (D, E, N, Q); (F, Y, W); (C) (25). RT phenotypes are expressed as follows: uppercase = wild-type or positive; italicized uppercase = high intermediate; italicized lowercase = low intermediate; lowercase = negative. In the line labeled “RT sensitivity pattern,” those residues that are mutationally insensitive for RT activity are indicated by X; those residues that have extreme mutational sensitivity for RT activity are indicated by O; those residues that have extreme mutational sensitivity for RT activity with nonconservative substitutions, but not with conservative substitutions, are indicated by Z. The RT and Western sensitivity patterns are defined in Fig. 4. The phenotypes from Western blots of total cell lysates for residues P95 to M164 are expressed as follows: uppercase = wild type; italicized uppercase = US; italicized lowercase = UL; lowercase = Lt. The phenotypes from Western blots of total cell lysates for residues M164 to E203 are expressed as follows: uppercase = wild type; italicized uppercase = intermediate; lowercase = negative. The phenotypes from Western blots of soluble lysates are expressed as follows: red = wild type; green = faint; purple = very faint; cyan = no bands; black = not determined. In the line labeled “Western sensitivity pattern,” those residues that are mutationally insensitive for protein stability are indicated by X; those residues that have extreme mutational sensitivity for protein stability are indicated by O; those residues that have extreme mutational sensitivity for protein stability with nonconservative substitutions, but not with conservative substitutions, are indicated by Z. Roles of key residues are designated as follows: F = functionally important; C = catalytically important; S = important for protein stability (conservative interpretation); s = additional residues important for protein stability from a liberal interpretation; E = external. The roles indicated were those assigned before we made additional substitutions to study residues critical for stability in more detail. These mutants (T107S, T107V, V118T, L120A, L120T, T128V, Y146F, S191T, and H198Q) are underlined, and their phenotypes were not used to establish the sensitivity patterns. A table listing the data used to assemble this figure can be viewed at www.unc.edu/∼clyde. This table includes the actual percent RT activity for each HIV-1 RT mutant.
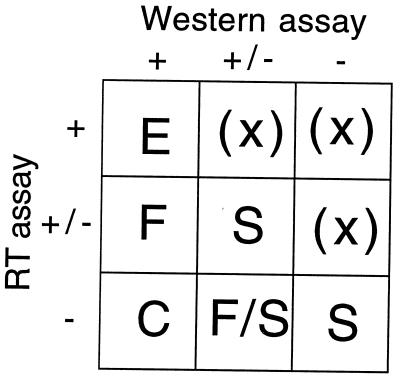
Figure 4.
Significance of mutational sensitivity patterns. Positions that show theoretical mutational patterns of extreme sensitivity (−), sensitivity to nonconservative changes (+/−), or insensitivity (+), for the two assays identify different classes of significant residues. Extreme sensitivity for RT assay data occurs when all mutations, both conservative and nonconservative, result in RT activity less than 3%. Sensitivity to nonconservative changes for RT assay data occurs when all nonconservative changes, but not all conservative changes, have less than 3% RT activity. Insensitivity for RT assay data occurs when all mutations, both conservative and nonconservative, result in wild-type levels of RT activity. Extreme sensitivity from Western blot analysis occurs when all mutations, both conservative and nonconservative, are negative [where the total cell lysate has a non-wild-type phenotype (US, UL, Lt, intermediate, or negative phenotype) and, if determined, a non-wild-type phenotype (faint, very faint, or no bands) for soluble lysates]. Sensitivity to nonconservative changes from Western blot analysis occurs when all nonconservative changes, but not all conservative changes, have a negative phenotype. Insensitivity from Western blot analysis is where all mutations, both conservative and nonconservative, are positive (where either the total cell lysate or the soluble lysate has a wild-type phenotype). E indicates the pattern expected for external residues. S indicates residues important for protein stability. C and F designate residues of catalytic and functional importance. Classes indicated by (X) are expected to be empty.
Positions at which all mutations have wild-type characteristics for both RT activity and protein stability as measured by Western blot analysis are classified as “external.” External side chains are not in contact with other critical residues or substrate and are expected generally to be insensitive to substitutions. In our scheme, mutations resulting in wild-type RT activity, along with wild-type levels of mature RT in both total cell and soluble lysates, and with only small amounts of unprocessed product in total cell lysate, are classified as wild type for stability (e.g., K103T).
Catalytic residues are defined as those at positions where all substitutions, both conservative and nonconservative, result in proteins that are negative for RT activity and have wild-type Western phenotypes. Functional residues are defined as those in which all nonconservative changes result in mutant proteins that are negative for RT activity, but where conservative changes are not all negative for RT activity. All changes in functional residues have wild-type phenotypes for protein stability. For residues with four or more substitutions, we allowed one mutation to deviate from the strict rules for functional/catalytic classification by one phenotype category (i.e., low intermediate category for RT activity or intermediate or US category for Western blot analysis). These residues include: D185 (classified as catalytic although D185V gives an intermediate Western phenotype), M184 (classified as functional although M184V gives an intermediate Western phenotype), Y183 (classified as functional although Y183C has 5% of wild-type RT activity), and G99 (classified as functional although G99L gave a US Western phenotype).
Residues important for stability are defined as those in which every nonconservative change (and in some cases some of the conservative substitutions) resulted in RTs with non-wild-type Westerns (for both total cell lysates and soluble lysates) and negative RT activity less than 3% (conservative interpretation) and low intermediate RT activity less than 20% (liberal interpretation). After initial identification of residues potentially involved in stability, we made additional site-directed substitutions at some of these positions (T107S, T107V, V118T, L120A, L120T, T128V, Y146F, S191T, and H198Q) to study their role in stability in more detail. Data from these new mutations sometimes altered our initial classification.
Evaluation of the Genetic Map of Critical Residues in HIV-1 RT.
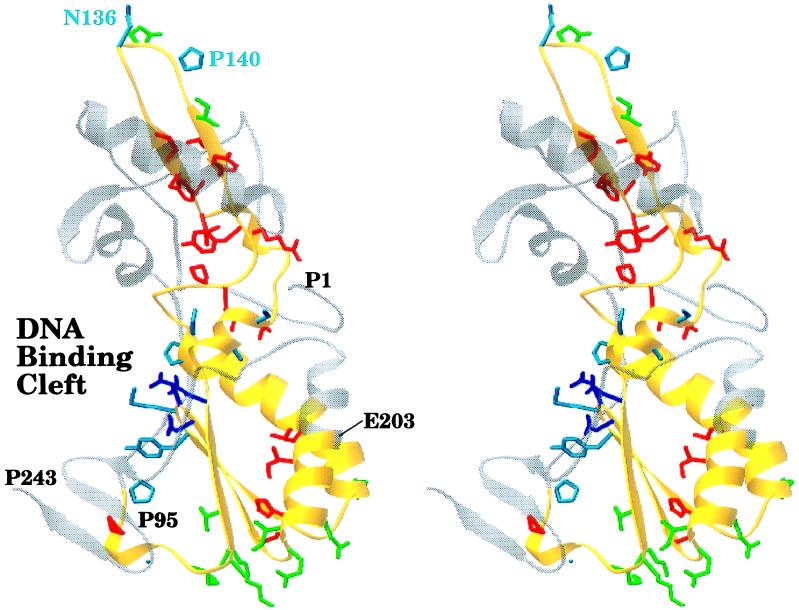
Visual inspection of crystal structures. Because the HIV-1 RT is a heterodimer consisting of 66-kDa and 51-kDa subunits resulting from different processing of the same polypeptide sequence, a mutation made at a specific amino acid residue will be present in both subunits. The observed phenotype from a particular amino acid substitution will result from the change in the 66-kDa subunit, the 51-kDa subunit, or both. Fig. 5 shows the placement of the functional/catalytic, stability, and external residue side chains on the fingers and palm subdomains of the 66-kDa subunit in the HIV-1 RT crystal structure 1rtj (15). The 66-kDa subunit plays the major role in enzyme catalysis. As expected, most of the functional/catalytic residues cluster around the active site in the DNA binding cleft. Generally, the protein stability side chains face the interior of the protein, and the external side chains point away from the surface. We also analyzed the placement of important residues by using the crystal structure 1hmi (16) complexed with DNA. The functional/catalytic residues are located near the DNA template-primer (data not shown). Because this structure lacks side chains, we are unable to analyze the interaction of the functional/catalytic residues with the DNA template-primer.
Figure 5.
Stereo view of critical residues of the HIV-1 RT. The polypeptide backbone of the fingers and palm subdomains of the 66-kDa subunit of the HIV-1 RT from the 1rtj crystal structure (15) is shown. The polypeptide backbone of the region we mutagenized (residues P95 through E203) is colored in yellow. Side chains for critical residues are colored as follows: catalytic (D110, D185, and D186) in purple, functional in cyan, stability (from a conservative interpretation of the mutational data) in red, and external in green. This figure was generated by using the program ribbons (13).
Residue variability among other retroviral RTs.
Residues involved in protein function/catalysis and stability are expected to be conserved among other retroviral RTs. On the other hand, external residues are expected to be highly variable. To test this hypothesis, by using a sequence alignment of 29 retroviral RTs (18) we calculated the variability for HIV-1 RT residues P95 through E203. As expected most of the external residues are highly variable, whereas the functional/catalytic and stability residues are conserved (22).
MSA analysis.
Stability residue side chains should be buried, whereas external residue side chains should be exposed on the surface. Functional/catalytic side chains also should be exposed to be in a position to interact with substrate. To test this hypothesis, we calculated the actual MSA buried and the percent MSA buried for amino acids in the fingers and palm subdomains of the 66-kDa subunit and the 51-kDa subunit. Sorting the residues in order of percent MSA buried shows that, in general, the stability residues have higher percentages of MSA buried compared with the functional/catalytic and external residues (22).
Variability versus MSA.
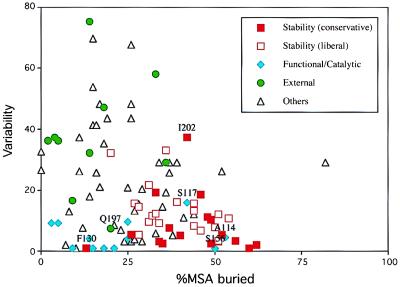
A scatter graph plotting variability versus percent MSA buried was used to further evaluate the genetic map for the 66-kDa subunit (Fig. 6). In theory, the external residues should fall in the upper-left portion of the graph (high variability and low side-chain burial), the catalytic/functional residues should fall in the lower-left portion of the graph (low variability and low side-chain burial), and the residues important for protein stability should fall in the lower-right portion of the graph (low variability and high side-chain burial). As seen in the graph, our genetic classification of the residues fits these predictions quite well for the 66-kDa subunit. Similar results were obtained for the 51-kDa subunit, with the exception that most of the catalytic/functional residues were more buried (22).
Figure 6.
RT sequence variability vs. percent MSA buried for residues P95 through E203 of the HIV-1 RT 66-kDa subunit. This figure is a scatter graph plotting RT sequence variability (y axis) against percent MSA buried (x axis) for HIV-1 RT residues P95 through E203. The percent MSA buried is for residues in the 66-kDa subunit. Glycine residues are not included. Residues assigned a role from an interpretation of the mutational data are designated as follows: red ▪ (stability), red □ (additional stability residues from a more liberal interpretation of the mutational data), cyan ⧫ (functional/catalytic), and green • (external). Unclassified residues are indicated with black ▵. Residues that fall outside of their predicted region are labeled with their sequence position (functional residues A114, S117, and S156, stability residues F130 and I202, and the external residue Q197).
A few of the residues do not fall within their predicted region in the variability versus percent MSA buried scatter graph. These residues are indicated on the scatter graph for the 66-kDa subunit in Fig. 6. Functional residues that play an indirect role by maintaining the local conformation for other functional residues that interact directly with substrate are expected to be conserved and buried and appear in the lower-right portion of the graph. S156, a functional residue that is conserved but buried, plays an indirect role in RT function. We propose that S156 helps determine the conformation of the β8-αE loop in the 66-kDa subunit. A nonconservative substitution (S156L) here may alter the local conformation of the loop and thus interfere with RT function, but does not appear to disrupt the overall stability. This loop interacts with substrate (17, 26). Another functional residue that is conserved but buried, A114, lies in a region believed to be involved in template-primer binding (27). A valine substitution at residue A114, has negative RT activity, without affecting stability. It is possible that the larger and more bulky hydrophobic valine substitution interferes indirectly with template-primer binding. Although less buried and less conserved than A114, S117 (another functional residue exception) also is located in this template-primer binding region.
Two residues important for protein stability (F130 and I202) fall outside of the predicted region in the plot of variability versus percent MSA buried (Fig. 6). The side chain of I202 has a greater variability but a percent molecular surface area buried similar to other stability residues. From inspection of the crystal structure 1rtj for the HIV-1 RT (15), I202 appears to be involved in a stabilizing van der Waals interaction (unpublished observation). Likewise, an inspection of the crystal structure 1mml for the murine leukemia virus RT (17) reveals that T240 (the equivalent of I202 in HIV-1 RT) may be involved in a stabilizing hydrogen-bonding interaction (unpublished observation). The higher variability of residue I202 appears to result from the ability of a residue at that position to stabilize the protein by at least two different mechanisms. In contrast to I202, F130 is invariant in the sequence alignment of 29 retroviral RTs (18), but is more exposed than other residues involved in protein stability. Although more exposed, all of the side-chain atoms of F130, except for carbons Cβ and Cγ, are in van der Waals contacts with carbon atoms of other hydrophobic side chains in the 66-kDa subunit. These van der Waals interactions most likely are important for protein stability. Furthermore, F130 is more buried in the 51-kDa subunit and all of its side-chain atoms are in van der Waals contacts with carbon atoms of other hydrophobic side chains (22).
The variability calculated for one external residue, Q197, is unusually low. However, the value of molecular surface area buried for this residue is typical of external residues. Only nine of the 29 aligned RT sequences have an amino acid at this position in the alignment (20 are gapped) (18). The available sequences probably do not adequately sample the potential variability at position 197, and the variability calculation used here (19) is not designed to deal with this situation.
External residues.
External residues are expected to be hydrophilic to interact with solvent water. Consistent with this expectation, most (7 of 10) of the external residues are hydrophilic with the exception of V106, I142, and I195. The sequence variability and MSA analyses support our genetic classification of the two hydrophobic residues I142 and I195 as external. I142 has a high variability and a percent MSA buried of 5% in the 66-kDa subunit and 23% in the 51-kDa subunit, whereas I195 also has a high sequence variability and a percent MSA buried of 2% in the 66-kDa subunit and 8% in the 51-kDa subunit. V106 also has a high sequence variability, although it is more buried than the other external residues. One interesting observation is that mutations at four of the external residues (K101, K103, V106, and E138) result in HIV-1 RT resistance to various non-nucleoside inhibitors (28, 29).
Functional residues.
We were able to identify D110, D185, and D186 as catalytically important. These three residues comprise the evolutionarily conserved aspartic acid triad of the active site of RT. All of the functional/catalytic residues in the 66-kDa subunit of the 1hmi crystal structure are located near the template-primer substrate, except for N136 and P140, which both are part of the β-β loop (shown in Fig. 5 for the 1rtj crystal structure). N136 and P140 in the 51-kDa subunit are located near the DNA substrate, suggesting their functional role is realized through the smaller subunit.
Protein stability residues.
Several regions are important for stability: F124–P133, R143–P150, I167–E169, and L187–S191. Both hydrophobic and hydrophilic residues are classified as important for protein stability. We have combined crystal structure analysis with our mutational data to propose hydrogen bonding and hydrophobic stabilizing interactions for many of the residues important for protein stability (unpublished work). An example of this approach is described in the next section.
T107/S191/H198 interaction.
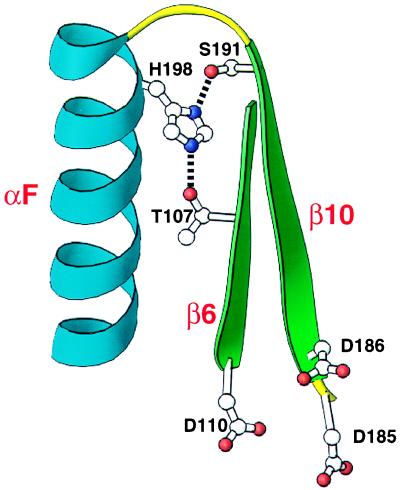
Three hydrophilic stability residues (T107, S191, and H198) are within hydrogen bonding distances of one another. In the two highest resolution HIV-1 RT structures, 1har (2.2 Å) (14) and 1rtj (2.35 Å) (15), the side-chain oxygen of S191 is within hydrogen bonding distance of the Nδ1 nitrogen of H198, whereas the side-chain oxygen of another residue T107 is within hydrogen bonding distance of the Nɛ2 nitrogen of H198. In the 1har structure, the distance between S191 Oγ and H198 Nδ1 is 2.7 Å, and the distance between T107 Oγ1 and H198 Nɛ2 is 2.8 Å. In this model, H198 forms two hydrogen bonds, one with S191 and another with T107. These potential hydrogen bonds appear significant because they bring together αF, β6, and β10. The aspartic acids of the catalytic triad (D110, D185, and D186) are in β6, β10, and the loop joining β10 and β9. This interaction can be viewed in Fig. 7.
Figure 7.
T107/S191/H198 hydrogen bonding interaction. Potential hydrogen bonding interactions between T107 and H198 and between S191 and H198 are shown. The polypeptide backbone is shown for amino acid residues S105–D110 and D185–W212 from HIV-1 RT crystal structure 1har (14). Secondary structural elements shown are β6 (green), β10 (green), and αF (blue). Amino acid chains (T107, D110, D185, D186, S191, and H198) are shown. Side-chain nitrogen atoms are represented as purple dots, and side-chain oxygens as red dots. In the crystal structure 1har, the atomic distance between T107 Oγ1 and H198 Nɛ2 is 2.8 Å and between S191 Oγ and H198 Nδ1 is 2.7 Å. Side chains D110, D185, and D186 (catalytic aspartic acid triad of the active site) are included as a reference point. This structural representation is approximately inverted compared with the one in Fig. 5. This figure was generated by using the program ribbons (13).
Mutations at H198 and S191 clearly establish the importance of hydrogen bonding in protein stability. Replacing S191 with nonpolar side chains (alanine and phenylalanine) that cannot hydrogen-bond disrupts protein stability. An alanine side chain is identical to that of serine, except for the presence of a hydroxyl group on the serine with hydrogen bonding potential. Although a tyrosine substitution also abolishes stability, the tyrosine side chain may be too bulky to properly hydrogen-bond with H198. A threonine replacement at S191 has wild-type stability. Threonine has a small polar side chain like serine and differs only in the addition of a methyl group. Substituting a proline at S191 most likely destabilizes the protein by altering the configuration of the backbone. The role of H198 seems to be quite specific. Replacing H198 with another basic residue (arginine) disrupts stability. Other changes with the polar side chains tyrosine and asparagine, as well as a proline, also destroy protein stability. A glutamine substitution partially restores stability. Glutamine is similar in size to histidine and has two side-chain atoms that can form hydrogen bonds. Some of the low-resolution HIV-1 RT crystal structures do not have the S191 and H198 side chains positioned within hydrogen bonding distance in either subunit, thus showing the usefulness of our mutational approach as a method to evaluate the accuracy of certain crystal structures.
On the other hand, the T107-H198 hydrogen bond is not essential for stability. Replacing T107 with an alanine, which cannot form a hydrogen bond, does not affect stability. Because T107 is within hydrogen bonding distance of H198 it is likely that they hydrogen-bond, but that this hydrogen bond is not essential to protein stability. These hydrogen bonds (S191-H198 and T107-H198) illustrate the use of mutational analysis to differentiate between an interaction that is critical for protein stability and one that is not.
Utility of Saturation Mutagenesis in the Study of Proteins.
This study demonstrates the usefulness of large-scale saturation mutagenesis as a method for understanding protein function and structure. Independent of crystallographic data, we identified residues that are involved in protein function and stability, in addition to residues with external side chains. Subsequent analysis using the available crystal structures reveals that our genetic map of critical residues within the RT is quite accurate. This analysis illustrates the potential power of saturation mutagenesis in the study of proteins for which no crystal structure is available.
Even when structural information is available, as in the case of the HIV-1 RT, it is difficult to infer the roles of specific residues in protein function or stability directly from the structure. In such cases saturation mutagenesis complements the crystallographic data by providing biological evidence concerning the roles of specific residues. In certain cases mutational analysis may be used to test specific features of a structural model or distinguish between alternative structures.
Acknowledgments
We thank Susan Elmore for synthesis of oligonucleotides, Dave Lado for technical assistance, David Cohen and Donald Doyle for running the gepol computer program, and Michael Batalia and Edward Collins for preparing the crystal structure figures. We also thank Thomas Kunkel and Jack Griffith for helpful comments on the manuscript. This work was supported by National Institutes of Health Grants AI08998, GM21313, AI25321, and GM42501.
ABBREVIATIONS
- RT
reverse transcriptase
- MSA
molecular surface area
- US
upper strong
- UL
upper light
- Lt
light
References
- 1.Hutchison C A, III, Phillips S, Edgell M H, Gillam S, Jahnke P, Smith M. J Biol Chem. 1978;253:6551–6560. [PubMed] [Google Scholar]
- 2.Hutchison C A, III, Swanstrom R, Loeb D D. Methods Enzymol. 1991;202:356–390. doi: 10.1016/0076-6879(91)02019-6. [DOI] [PubMed] [Google Scholar]
- 3.Hutchison C A, III, Nordeen S K, Vogt K, Edgell M H. Proc Natl Acad Sci USA. 1986;83:710–714. doi: 10.1073/pnas.83.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loeb D D, Swanstrom R, Everitt L, Manchester M, Stamper S E, Hutchison C A., III Nature (London) 1989;340:397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- 5.Farmerie W G, Loeb D D, Casavant N C, Hutchison C A, III, Edgell M H, Swanstrom R. Science. 1987;236:305–308. doi: 10.1126/science.2436298. [DOI] [PubMed] [Google Scholar]
- 6.Loeb D D, Hutchison C A, III, Edgell M H, Farmerie W G, Swanstrom R. J Virol. 1989;63:111–121. doi: 10.1128/jvi.63.1.111-121.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lattman E E, Rose G D. Proc Natl Acad Sci USA. 1993;90:439–441. doi: 10.1073/pnas.90.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao S F, Chan V L, Juranka P, Kaplan A H, Swanstrom R, Hutchison C A., III Nucleic Acids Res. 1995;23:803–810. doi: 10.1093/nar/23.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao S F. Ph.D. thesis. Chapel Hill: The University of North Carolina; 1992. [Google Scholar]
- 10.Vieira J, Messing J. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 11.Wilson R K. Biotechniques. 1993;15:414–422. [PubMed] [Google Scholar]
- 12.Richardson D C, Richardson J S. Trends Biochem Sci. 1994;19:135–138. doi: 10.1016/0968-0004(94)90207-0. [DOI] [PubMed] [Google Scholar]
- 13.Carson M. J Mol Graphics. 1987;5:103–106. [Google Scholar]
- 14.Unge T, Knight S, Bhikhabhai R, Lövgren S, Dauter Z, Wilson K, Strandberg B. Structure (London) 1994;2:953–961. doi: 10.1016/s0969-2126(94)00097-2. [DOI] [PubMed] [Google Scholar]
- 15.Esnouf R, Ren J, Ross C, Jones Y, Stammers D, Stuart D. Nat Struct Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 16.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgiadis M M, Jessen S M, Ogata C M, Telesnitsky A, Goff S P, Hendrickson W A. Structure (London) 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 18.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu T T, Kabat E A. J Exp Med. 1970;132:211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silla E, Tuñón I, Pascual-Ahuir J L. J Comput Chem. 1991;12:1077–1088. [Google Scholar]
- 21.Pascual-Ahuir J L, Silla E, Tuñón I. gepol 92. Department of Chemistry, Indiana University: Quantum Chemistry Program Exchange; 1992. , Program 554. [Google Scholar]
- 22.Wrobel J A. Ph.D. thesis. Chapel Hill: The University of North Carolina; 1996. [Google Scholar]
- 23.Doyle D F. Ph.D. thesis. Chapel Hill: The University of North Carolina; 1996. [Google Scholar]
- 24.Wang J, Smerdon S J, Jäger J, Kohlstaedt L A, Rice P A, Friedman J M, Steitz T A. Proc Natl Acad Sci USA. 1994;91:7242–7246. doi: 10.1073/pnas.91.15.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dayhoff M O, Schwartz R M, Orcutt B C. In: Atlas of Protein Sequence and Structure. Dayhoff M O, editor. Vol. 5. Silver Spring, MD: Natl. Biomed. Res. Found.; 1978. pp. 345–352. [Google Scholar]
- 26.Sarafianos S G, Pandey V N, Kaushik N, Modak M J. Biochemistry. 1995;34:7207–7216. doi: 10.1021/bi00021a036. [DOI] [PubMed] [Google Scholar]
- 27.Boyer P L, Ferris A L, Clark P, Whitmer J, Frank P, Tantillo C, Arnold E, Hughes S H. J Mol Biol. 1994;243:472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]
- 28.Byrnes V W, Sardana V V, Schleif W A, Condra J H, Waterbury J A, Wolfgang J A, Long W J, Schneider C L, Schlabach A J, Wolanski B S, Graham D J, Gotlib L, Rhodes A, Titus D L, Roth E, Blahy O M, Quintero J C, Staszewski S, Emini E A. Antimicrob Agents Chemother. 1993;37:1576–1579. doi: 10.1128/aac.37.8.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyer P L, Ding J, Arnold E, Hughes S H. Antimicrob Agents Chemother. 1994;38:1909–1914. doi: 10.1128/aac.38.9.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]