Abstract
While recent work has implicated Tbx20 in myocardial maturation and proliferation, the role of Tbx20 in heart valve development remains relatively unknown. Tbx20 expression was manipulated in primary avian endocardial cells in order to elucidate its function in developing endocardial cushions. Tbx20 gain of function was achieved with a Tbx20-adenovirus, and endogenous Tbx20 expression was inhibited with Tbx20-specific siRNA in cultured endocardial cushion cells. With Tbx20 gain of function, the expression of chondroitin sulfate proteoglycans (CSPG), including aggrecan and versican, was decreased, while the expression of the matrix metalloproteinases (MMP) mmp9 and mmp13 was increased. Consistent results were observed with Tbx20 loss of function where the expression of CSPG genes increased and MMP genes decreased. In addition, cushion mesenchyme proliferation increased with infection of a Tbx20-adenovirus and decreased with transfection of Tbx20-specfic siRNA. Furthermore, BMP2 treatment resulted in increased Tbx20 expression in endocardial cushion cells, and loss of Tbx20 led to increased Tbx2 and decreased N-myc gene expression. Taken together, these data support a role for Tbx20 in repressing extracellular matrix remodeling and promoting cell proliferation in mesenchymal valve precursor populations in endocardial cushions during embryonic development.
Keywords: Tbx20, endocardial cushion development, aggrecan, versican, mmp9, mmp13, cell proliferation, siRNA, chicken
Introduction
Heart valve development is a complex process that is essential to normal heart function. Congenital valve defects can result in valve dysfunction (Rabkin-Aikawa et al., 2005) and there is mounting evidence that early developmental defects in valvulogenesis can lead to valve disease later in life (Cripe et al., 2004; Garg et al., 2005). Much is known about the early events in valve formation, however relatively little is known about how primitive endocardial cushions remodel into mature valves. T-box transcription factors function in various aspects of cardiogenesis, including cardiac lineage determination, chamber specification, epicardial development, and specialization of the conduction system (Plageman and Yutzey, 2005; Stennard and Harvey, 2005). Tbx20 has been implicated in cardiac muscle maturation and is expressed in developing heart valves, however its specific role in valves and their precursor cell populations has not been identified.
Recently, the role of Tbx20 in mammalian heart development was investigated using targeted mutagenesis and RNAi strategies in mice. In these studies, loss of Tbx20 function resulted in decreased or delayed cardiac gene expression, hypoplasia of the myocardium, and decreased chamber maturation (Cai et al., 2005; Singh et al., 2005; Stennard et al., 2005; Takeuchi et al., 2005). In addition, a specific role for Tbx20 in myocardial proliferation was proposed through increased N-myc gene expression (Cai et al., 2005). In each case, mutant mice lacking Tbx20 gene expression were embryonic lethal due to failure of cardiac muscle maturation at approximately embryonic day (E) 9, prior to valve development (Cai et al., 2005; Stennard et al., 2005; Takeuchi et al., 2005). Further evidence for Tbx20’s role in heart muscle development comes from loss of function studies in other animal model systems. In zebrafish and Xenopus embryos, the loss of Tbx20 results in heart looping and chamber maturation defects (Brown et al., 2003; Stennard et al., 2003; Szeto et al., 2002). Taken together, these studies have defined a role for Tbx20 in early cardiac muscle development where it promotes primitive, proliferative myocardium. Later in development, Tbx20 is expressed in the myocardium as well as in the atrioventricular (AV) canal and outflow tract (OFT) endocardial cushions and in the mitral and tricuspid valves (Plageman and Yutzey, 2004; Stennard et al., 2003; Yamagishi et al., 2004). While the loss of function studies were very informative regarding Tbx20’s role in regulating myocardial proliferation and maturation, the role of Tbx20 in valve development is still unclear.
Heart valvulogenesis is initiated in the AV canal and OFT by signaling events originating in the myocardium that cause cells in the endocardium to undergo an epithelial to mesenchymal transformation (EMT) and migrate into the intervening cardiac jelly. The result of EMT is the formation of endocardial cushions composed of highly proliferative, undifferentiated, mesenchymal valve progenitor cells embedded in an unorganized extracellular matrix (ECM) (Armstrong and Bischoff, 2004; Hinton et al., 2006; Lincoln et al., 2006b; Person et al., 2005; Schroeder et al., 2003). These cushions will ultimately elongate and undergo ECM remodeling in order to form functionally mature valves (Hinton et al., 2006; Lincoln et al., 2004). One hallmark of the transition from endocardial cushion to remodeling valve is a decrease in endocardial cushion cell proliferation (Hinton et al., 2006; Lincoln et al., 2004). As previously reported, murine endocardial cushion cells are approximately 6 times more proliferative than cells in the remodeling valve leaflet (Hinton et al., 2006) and decreased proliferation is also a feature of valve remodeling in avian embryos (Lincoln et al., 2004). In addition, valve remodeling is characterized by increased organization and complexity of the ECM. During endocardial cushion formation, proteoglycans are diffusely and variably expressed throughout the cushion (Hinton et al., 2006). However, beginning in the remodeling stage of valve development and continuing into postnatal time points, the ECM becomes stratified into three distinct layers: the elastin rich atrialis, the proteoglycan rich spongiosa, and the collagen rich fibrosa (Flanagan and Pandit, 2003; Hinton et al., 2006; Lincoln et al., 2006b). While recent progress has been made to describe this process histologically, the molecular mechanisms governing this transition have not been elucidated.
The process by which endocardial cushions develop into mature valve leaflets is marked by the expression of specific ECM proteins and remodeling enzymes (Lincoln et al., 2006b). Aggrecan (agg) and versican (vers) are major proteoglycan constituents of the spongiosa layer in avian valves. Both are large chondroitin sulfate proteoglycans (CSPG) that aggregate with hyaluronan to form hydrated compressible ECM (Arciniegas et al., 2004; Luo et al., 2000). These CSPGs are not only found in developing valves, but also in articular cartilage, where they function to provide resistance against compressive forces (Arciniegas et al., 2004; Pirok et al., 1997). The proper distribution and organization of ECM in remodeling valves is important for normal valve function (Hinton et al., 2006; Rabkin et al., 2001) and is dependent on a coordinated deposition and degradation of individual matrix proteins. As in other tissues, matrix metalloproteinases (MMP) can mediate the degradation and remodeling of ECM components in the heart (Coker et al., 1998; Rabkin et al., 2001; Sternlicht and Werb, 2001). Family members, including MMP9 (Gelatinase B) and MMP13 (Collagenase-3), are involved in the degradation and reorganization of collagens, elastin, and proteoglycans (Passi et al., 1999; Rabkin-Aikawa et al., 2005; Sternlicht and Werb, 2001). In addition, increased expression of MMP9 and MMP13 was observed in diseased human mitral and aortic valves with disorganized ECM (Rabkin et al., 2001; Soini et al., 2001). Taken together, normal valve maturation requires regulated ECM organization and remodeling, however the mechanisms controlling this process are still unclear.
To investigate the role of Tbx20 in endocardial cushion maturation and valve development, the expression of Tbx20, CSPGs, and MMPs in the endocardial cushions and remodeling valves of avian embryos was examined. Tbx20 and MMP genes are expressed at higher levels in endocardial cushions relative to remodeling valves, while CSPG genes are expressed at higher levels in remodeling valves relative to endocardial cushions. In addition, a primary chicken endocardial cushion culture system was used for Tbx20 gain and loss of function studies. The effects of altered Tbx20 expression on CSPG and MMP gene expression as well as the ability of Tbx20 to affect endocardial cushion cell proliferation were examined. In these studies, increased Tbx20 expression led to repression of CSPG genes and increased expression of MMP genes, whereas the opposite was observed with loss of Tbx20 function. Furthermore, increased Tbx20 function led to increased proliferation in endocardial cushion cells, while loss of Tbx20 led to decreased proliferation. These findings coincide with high levels of Tbx20 promoting proliferation in endocardial cushions while lower levels of Tbx20 in remodeling valves corresponds to decreased rates of proliferation. Taken together, these studies are consistent with a role for Tbx20 in antagonizing the transition from endocardial cushion to remodeling valve.
Materials and methods
Chicken embryo collection
Fertilized white leghorn chicken eggs (CBT Farms, MD) were incubated at 38°C under high humidity. Embryos were collected at Hamburger Hamilton (HH) stages 25, 26, 34, and 36, corresponding to embryonic days 4.5, 5, 8, and 10, respectively (Hamburger and Hamilton, 1951). For histology, hearts were dissected in 1× phosphate-buffered saline (PBS) and fixed for 2 h in 4% paraformaldehyde/PBS. After fixation, embryonic tissue was dehydrated in a graded ethanol/PBS series (25%, 50%, 75%, 95%, 100%) and washed in xylene before being embedded in paraplast (Sigma) for further processing. All animal procedures were approved and performed in accordance with institutional guidelines.
In situ hybridizations
The chicken Tbx20 sequence (Genbank accession number AB070544) was amplified from HH stage 20 heart cDNA using previously reported degenerate primers 5′-TGCTGRAAGTARTGRTG-3′ and 5′-GTGGAYAAYAAGAGATA-3′ where R represents purine and Y represents pyrimidine (Iio et al., 2001; Plageman and Yutzey, 2004). The chicken aggrecan sequence (Genbank accession number U78555) was amplified from HH stage 34 wing cDNA using the previously reported primers 5′-CTGCGTTCCCTGAGATTAC-3′ and 5′-TTGCCAGGTCGATCTCAC-3′ (Li et al., 1996; Lincoln et al., 2006a). The chicken versican sequence (Genbank accession number NM_204787) was amplified from HH stage 36 wing cDNA using the primers 5′-CAAGGCGCTGAGTGCTAAATG-3′ and 5′-AGGGGCTAATACTGCTCTGG-3′ (Shinomura et al., 1993). The chicken mmp9 sequence (Genbank accession number AF222690) was amplified from HH stage 30 heart cDNA using the primers 5′-GCTGCCACTTCCCCTTCATCTTTG-3′ and 5′-CGGGGGCCCACTGCGTTCTTG-3′ (Hahn-Dantona et al., 2000). The chicken mmp13 sequence (Genbank accession number AF070478) was amplified from HH stage 30 cardiac outflow tract cDNA using the primers 5′-TGATGCCATAACAAAACTTCGTG-3′ and 5′-AGATGCTAGATTGCTGGGACTTA-3′ (Lei et al., 1999). All sequences were amplified by reverse transcriptase polymerase chain reaction (RT-PCR), subcloned into pGEM T-vector (Promega), and confirmed by sequencing. For each sequence, antisense RNA probes were generated as previously reported (Ehrman and Yutzey, 1999) with the following modifications. The Tbx20 probe was synthesized with T3 polymerase from a plasmid linearized with Xho I. The aggrecan and mmp13 probes were synthesized with Sp6 polymerase from plasmids linearized with Nco I. The versican and mmp9 probes were synthesized using T7 polymerase from plasmids linearized with Not I.
In situ hybridization of tissue sections was performed as previously described (Somi et al., 2004) with the following modifications. Paraformaldehyde-fixed chicken hearts were embedded in paraffin, and 14μm sections were cut and mounted onto Superfrost Plus microscope slides (Fisher Scientific). Sections were then deparaffinized in xylene, rehydrated through an ethanol/distilled water series (100%, 95%, 75%, 50%), and then rinsed in 1×PBS. Sections were treated with 20μg/ml proteinase K/PBS for 6 min at 37°C. Hybridizations using 170μl of 0.5μg/ml DIG labeled riboprobe were carried out in Coverwell Incubation Chambers (Grace Biolabs). Color reactions using nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP; Roche) were allowed to develop from 3-48 h. Slides were then rinsed in 1×PBS/0.1%Tween 20, dehydrated in a graded ethanol series, rinsed in xylene, and cover slipped using Cytoseal (Electron Microscopy Sciences).
Endocardial cushion cell culture
Embryonic chicken hearts were collected at HH stage 25, and pre-fused endocardial cushions were dissected using tungsten needles. Cushion cells free of myocardial contamination were cultured as previously reported (Lincoln et al., 2006a) with the following modifications. Dissected cushions from 12 embryos were plated onto a 0.01% collagen coated two-well chamber slide (Labtek) with 1ml media (10% fetal bovine serum, 1% penicillin/streptomycin, 1×M199 (Invitrogen)). Endocardial cushion cultures were incubated for a total of 4 days. In some cases, recombinant human BMP2 or recombinant human Noggin (R&D Systems) was added to the culture media, with a final concentration of 200ng/ml at the time of plating and replenishment after 48 h. Cells were collected after 4 days in culture for RNA isolation or fixed in 4% paraformaldehyde/PBS for immunohistochemistry.
Recombinant adenovirus
A recombinant adenovirus containing the full length coding region of murine Tbx20 (AdTbx20) was generated from a pAC-CMV-Tbx20 expression plasmid (Plageman and Yutzey, 2004) using previously described methods (Gomez-Foix et al., 1992). For Tbx20 gain of function studies, endocardial cushion cells cultured for 24 h were infected with 108 plaque forming units of the AdTbx20 virus or a β-galactosidase (Adβ-gal) control virus in serum free media (1 ×M199 (Invitrogen)). Cells were incubated with the infection media for 6 h, which was then replaced with new supplemented culture media. After 48 h, RNA was isolated or cells were fixed in 4% paraformaldehyde/PBS for immunohistochemistry. Infection efficiency was determined to be greater than 90% in Adβ-gal infected cultures as measured by staining with 1mg/ml X-gal (Amresco) using previously a reported method (Liberatore et al., 2002). High expression of the murine Tbx20 viral transcript was confirmed using RT-PCR and ectopic Tbx20 protein expression was confirmed by immunohistochemistry with a rabbit polyclonal antibody directed against Tbx20 (Orbigen).
Tbx20 siRNA
A 19 nucleotide RNA duplex corresponding to the chicken Tbx20 sequence (Genbank accession number AB070544) was designed using BLOCK-iT™ RNAi Designer (Invitrogen). The Tbx20 siRNA with sequence 5′-GCAUCCAUUGCUACACCUAdTdT -3′ and 5′-UAGGUGUAGCAAUGGAUGCdTdT-3′ and a scrambled control siRNA with sequence 5′-CCGGUAAUGACACCCAAUUdTdT-3′ and 5′-AAUUGGGUGUCAUUACCGGdTdT-3′ were obtained from Invitrogen. A final siRNA concentration of 100nM was used with Lipofectamine 2000 (Invitrogen) to transfect cultured endocardial cushion cells as described by the manufacture’s protocol. Transfected cells were incubated 48 h before RNA isolation or fixation for immunohistochemistry. The transfection efficiency was determined by co-transfection of the FITC-labeled (λex=494nm) BLOCK-iT™ Fluorescent Oligo (Invitrogen) along with siRNA oligos. Co-transfected cells were cultured and fixed in 4% paraformaldehyde/PBS for 15 min and then counterstained with a 1:1000 dilution of TO-PRO-3 iodide (λex=642) (Molecular Probes) in 1×PBS. Fluorescence was detected using a Nikon PCM 2000 confocal microscope and images were obtained using Simple PCI software. The percent of positively transfected cells was calculated by dividing the number of FITC/TO-PRO-3 iodide double-labeled nuclei by the number of total TO-PRO-3 iodide-labeled nuclei per microscopic field. In three independent experiments, a total of 10 fields containing at least 75 cells per field were counted for each treatment group. The level of Tbx20 mRNA expression was determined using real time RT-PCR, and Tbx20 protein expression was detected by immunohistochemistry on Tbx20 siRNA and scrambled control transfected cultures using a rabbit polyclonal antibody directed against Tbx20 (Orbigen).
Immunohistochemistry
Endocardial cushion cell cultures were fixed with 4% paraformaldehyde/PBS for 30 min, washed three times in PBS/0.1%Tween 20, and treated with 3% hydrogen peroxide/PBS for 30 min. Immunohistochemistry was performed using an ABC peroxidase staining kit (Pierce) according to the manufacture’s protocol. A rabbit polyclonal antibody directed against Tbx20 (Orbigen) was used at a 1:200 dilution in blocking solution. Mouse monoclonal antibodies directed against aggrecan (Abcam) and versican (Developmental Studies Hybridoma Bank) were used at a 1:200 dilution in blocking solution. All primary antibodies were incubated overnight at 4°C. Detection of antibody reactivity was visualized using DAB substrate (Pierce).
BrdU incorporation and quantification
BrdU positive nuclei were identified via immunohistochemistry using a BrdU detection kit (Zymed). BrdU labeling reagent (Zymed) was diluted 1:100 in culture media and incubated with endocardial cushion cell cultures for 1.5 h prior to fixation with cold 70% ethanol for 15 min at 4°C. Cells were treated with 3% hydrogen peroxide/methanol for 10 min, washed three times with 1×PBS, and incubated in blocking solution for 10 min. A biotinylated mouse anti-BrdU primary antibody was used followed by a streptavadin-peroxidase conjugated secondary antibody. A colorimetric reaction was carried out using DAB followed by counterstaining with hematoxylin. The percent of proliferating cells was calculated by dividing the number of BrdU labeled nuclei by the number of total nuclei per microscopic field. In three independent experiments, a total of 10 fields containing at least 75 cells per field were counted for each treatment group. Statistical significance of observed differences was determined by Student’s t-test.
RT-PCR analysis of gene expression
Total RNA was isolated from cultures of 12 endocardial cushions per experimental group using 200μl Trizol reagent (Invitrogen) and cDNA was generated from the entire RNA sample from each group using SuperScript II (Invitrogen), as described by the manufacture’s protocol. Total RNA was also isolated from 6 chicken AV canals at HH stages 25 and 36 using 800μl Trizol reagent and cDNA was generated from 5μg of each RNA sample using SuperScript II as described by the manufacture’s protocol. 1μl cDNA was used for analysis by semi-quantitative RT-PCR or quantitative real time RT-PCR (MJ Research Opticon 2). RT-PCR reactions were performed at 35 cycles using 20 pmol of the following primers: Tbx20 5′-CAGGCAACGCAAAGCAGAG-3′ and 5′-TTGGCATGTGGAAAGAAGG-3′, aggrecan 5′-CCTGCCTGACCTCTTTGC-3′ and 5′-TGGGGAGGAGGGCAACAT-3′, versican 5′-CCTCACTGGTAAGCCCACAT-3′ and 5′-TGATTCTTCTTGGCCCATTC-3′, mmp9 5′-GCCACTTCCCCTTCATCT-3′ and 5′-GTTGCCACCATTGGTGTA-3′, mmp13 5′-TGATGCCATAACAAAACTTCGTG-3′ and 5′-AGATGCTAGATTGCTGGGACTTA-3′, N-myc 5′-ACCACTTTTCCATCGGTCAG-3′ and 5′-TTGGTTGGATCATGGGTTTT-3′, Tbx2 5′-AACACGGCTTTACCATCCTG-3′ and 5′-TTCAGCTGCGTGATCTTGTC-3′, β-actin 5′-ATCACAGGGGTGTGGGTGTT-3′ and 5′-AATGAGAGGTTCAGGTGCCC-3′. Gene expression levels determined by quantitative real time RT-PCR were calculated as previously reported (Lincoln et al., 2006a) with the following modifications. A standard curve was generated for each primer set using HH stage 34 whole heart cDNA and all values were normalized to GAPDH expression. Consistent GAPDH expression in all experimental groups was confirmed by normalizing GAPDH values to β-actin expression. Real time RT-PCR results represent at least three independent experiments (n=3–6) with reactions performed in duplicate. For developmental studies, expression is represented as arbitrary units of fluorescence intensity for data generated with equivalent RNA input and normalized to GAPDH. For endocardial cushion cultures, expression was calculated as fold increase or percent decrease determined by dividing the experimental value by the control value. The control value was then set to 1 or 100%, respectively. Statistical significance of observed differences was determined by Student’s t-test.
Results
CSPG and MMP genes are differentially expressed in endocardial cushions and remodeling valves
Tbx20 is expressed in chicken and mouse endocardial cushions and in the remodeling valves (Plageman and Yutzey, 2004; Stennard et al., 2003; Yamagishi et al., 2004). In addition, CSPG genes, including aggrecan and versican, and MMP genes, including MMP9 and MMP13, are expressed in mature and diseased valves. In situ hybridizations were performed on sectioned embryonic chicken hearts in order to localize the expression of aggrecan (agg), versican (vers), mmp9, and mmp13 in relation to Tbx20 during the stages of endocardial cushion formation (HH stage 25) and valve remodeling (HH stage 34) in vivo. At HH stage 25, Tbx20 is expressed at high levels throughout the entire AV endocardial cushion (Fig. 1A). In contrast, agg expression is restricted to the subatrial region (arrow) of the endocardial cushion and is excluded from the core (star) of the cushion (Fig. 1C). vers is also expressed in the subatrial region of the endocardial cushion and excluded from the core region (Fig.1E). In contrast, mmp9 is expressed throughout the entire cushion including the subatrial and core regions, and expression was also observed in the subepicardial space (open arrowhead) at this stage (Fig.1G). Similarly, mmp13 is expressed throughout the subatrial and core regions of the endocardial cushion at HH stage 25 (Fig.1I).
Figure 1. CSPG and MMP genes are differentially expressed in endocardial cushions and remodeling valves.
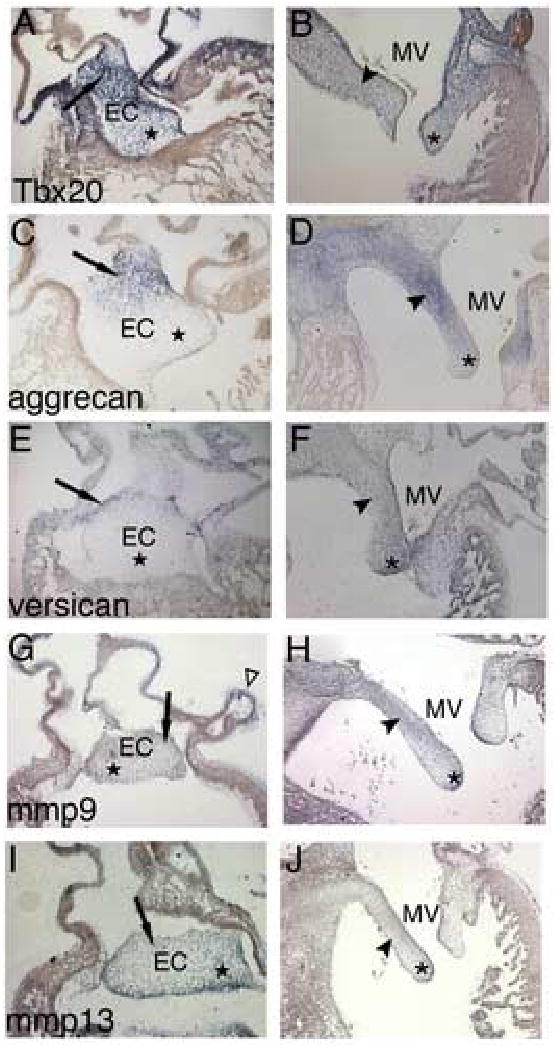
Expression of Tbx20, aggrecan, versican, mmp9, and mmp13 was examined in sectioned HH stage 25 and HH stage 34 chicken hearts. In situ hybridizations show Tbx20 (A), mmp9 (G), and mmp13 (I) are expressed throughout the entire AV endocardial cushion including the subatrial region (arrow) and cushion core (star). In addition, expression of mmp9 was also detected in the subepicardial space (open arrowhead in G). In contrast, aggrecan (C) and versican (E) are expressed only in the subatrial region of the cushion (arrows). In remodeling mitral valves, Tbx20 (B) is expressed in the valve leaflet (arrowhead) and distal tips (asterisk). At HH stage 34, aggrecan (D) is expressed at high levels in the valve leaflet, but is excluded from the distal tips, while versican (F) is expressed throughout the leaflet, with increased expression at the distal tips. In remodeling valves, mmp9 (H) is expressed throughout the leaflet with concentrated expression in the atrial aspect of the leaflet and in the distal tips, while mmp13 (J) is expressed in the ventricular aspect of the leaflet and in the distal tips. EC, endocardial cushion; MV, mitral valve.
In HH stage 34 remodeling valves, Tbx20 is expressed at more diffuse levels in the leaflets (arrowhead) and tips (asterisks) of the remodeling mitral valve (Fig.1B). In contrast, agg becomes highly expressed throughout the valve leaflet but is expressed at lower levels at the most distal tips (Fig.1D). vers is also expressed throughout the remodeling valve but is predominant at the distal tips of the valve leaflets (Fig.1F). By HH stage 34, mmp9 is expressed at low levels in the remodeling mitral valve, with expression being concentrated at the atrial aspect of the valve leaflet and the distal tips (Fig.1H), while mmp13 is expressed at low levels in the ventricular aspect of the valve leaflet and at the distal tips (Fig.1J). Similar expression of Tbx20, agg, vers, mmp9, and mmp13 was found in the remodeling tricuspid valve leaflets with the exception of the muscular portion of the mural leaflet (data not shown). Together these expression studies show that Tbx20, mmp9, and mmp13 are expressed throughout the core of the endocardial cushion while the expression of agg and vers is restricted to the subatrial region. Later in remodeling valves, the expression of agg and vers is expanded throughout the valve leaflet while Tbx20, mmp9, and mmp13 expression is more diffuse and compartmentalized compared to the expression in endocardial cushions.
Tbx20, CSPG and MMP expression was further quantified using RNA isolated from AV canals of HH stage 25 and HH stage 36 chicken embryos. The expression levels of Tbx20, agg, vers, mmp9, and mmp13 were measured using quantitative real time RT-PCR (Fig.2). At HH stage 25 (endocardial cushion stage), the expression of Tbx20 is approximately 1.5 fold higher than later at HH stage 36 (valve remodeling stage). Similarly, the expression of mmp9 and mmp13 is more than 2 fold higher at HH stage 25 than later at HH stage 36. In contrast, the expression of agg and vers is more than 4 fold higher in remodeling valves (HH stage 36) than in endocardial cushions (HH stage 25). These results indicate that the expression of Tbx20, mmp9, and mmp13 is relatively higher in endocardial cushions than in remodeling valves. In contrast, expression of agg and vers is increased in remodeling valves relative to undifferentiated endocardial cushions.
Figure 2. Differential expression of Tbx20, CSPG, and MMP genes during endocardial cushion and remodeling valve stages.
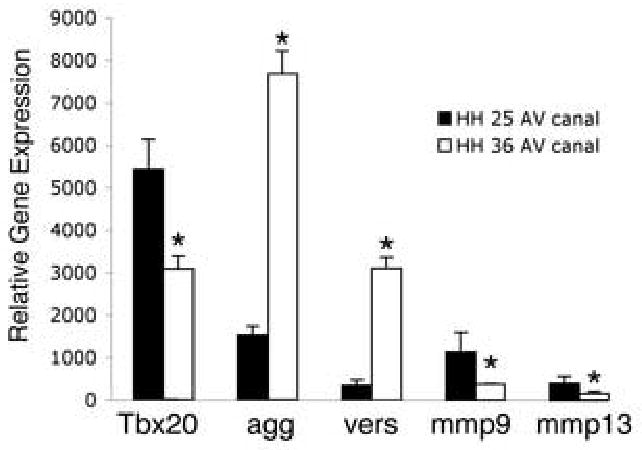
Expression of Tbx20, aggrecan, versican, mmp9, and mmp13 was examined in isolated AV canals from HH stage 25 or HH stage 36 chicken embryos using real time RT-PCR. The expression of Tbx20, mmp9, and mmp13 was decreased at HH stage 36 compared to the expression at HH stage 25. In contrast the expression of aggrecan and versican at HH stage 36 was increased compared to the expression at HH stage 25. Values on the y-axis represent arbitrary units of fluorescence intensity for data obtained with equivalent RNA input normalized to GAPDH. These data are representative of 3 independent real time RT-PCR experiments performed in duplicate (n=3). Statistical significance of observed differences between gene expression levels at HH stage 25 and HH stage 36 is indicated by an asterisk (P<0.05), and error bars represent standard error of the mean.
Tbx20 gain of function results in decreased CSPG expression and increased MMP expression in cultured endocardial cushion cells
The ability of Tbx20 to affect CSPG and MMP expression in undifferentiated endocardial cushions was examined. Primary cultures derived from unfused endocardial cushions removed from the AV canals of HH stage 25 chicken embryos were infected with an adenovirus that expresses murine Tbx20 (AdTbx20) or a control adenovirus that expresses β-gal (Adβ-gal). Endocardial cushion cells were infected with greater than 90% efficiency, as determined by X-gal staining of Adβ-gal infected cultures (data not shown). High levels of expression of the Tbx20 viral transcript were determined by real time RT-PCR and increased Tbx20 protein was detectable by immunohistochemistry (data not shown). The effect of increased Tbx20 on the expression of CSPG genes was determined by real time RT-PCR. Endocardial cushion cultures infected with AdTbx20 had an approximately 60% reduction in agg (Fig. 3E) and vers (Fig.3F) mRNA levels relative to Adβ-gal infected controls. This reduction was confirmed by immunohistochemistry using antibodies specifically directed against Agg and Vers. Expression of Agg and Vers protein was apparent in cells infected with Adβ-gal (Fig.3A,C), but not in cells infected with AdTbx20 (Fig.3B,D). In addition, the expression of mmp9 and mmp13 mRNA was measured by real time RT-PCR in cells infected with Adβ-gal or AdTbx20. In cells infected with AdTbx20, the expression of mmp9 was increased 3 fold (Fig.3G), and the expression of mmp13 was increased 5.5 fold (Fig.3H). These studies demonstrate that Tbx20 gain of function results in decreased agg and vers expression and increased mmp9 and mmp13 expression in endocardial cushion cell cultures.
Figure 3. Tbx20 gain of function results in decreased expression of CSPGs and increased expression of MMPs.
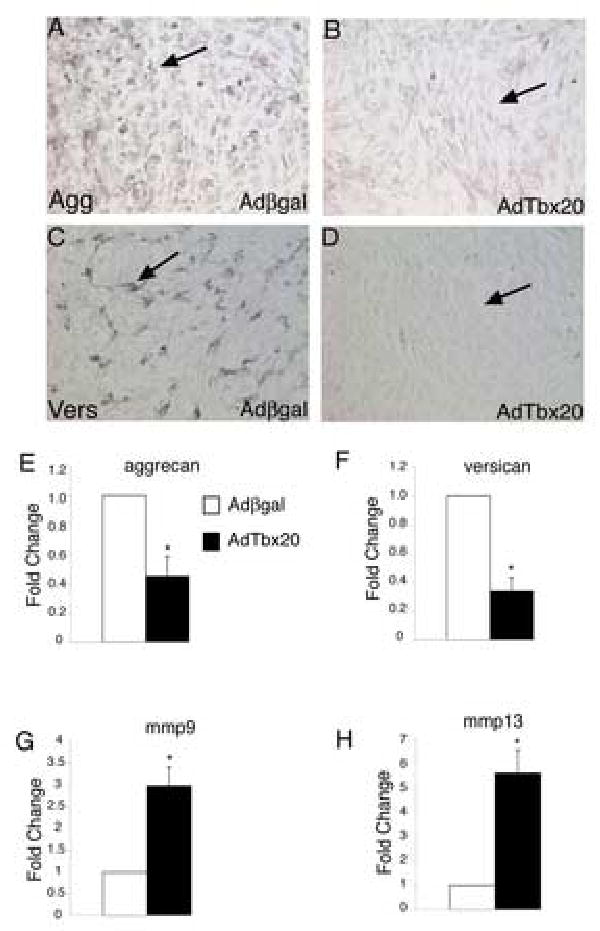
Adenoviruses expressing β-gal (Ad β-gal) or Tbx20 (AdTbx20) were used to infect primary avian HH stage 25 endocardial cushion cells. Immunohistochemistry with antibodies specific for Aggrecan (A,B) or Versican (C,D) was used to measure aggrecan and versican protein levels following infection. Cells infected with AdTbx20 had less immunoreactivity (B,D arrows indicate lack of staining) than cells infected with Adβ-gal (A,C arrows indicate staining). These results were confirmed by real time RT-PCR. Cells infected with AdTbx20 had reduced expression of aggrecan (E) and versican (F) (n=6, P<0.01). The expression of mmp9 and mmp13 was also measured in infected cells using real time PT-PCR. In cells infected with AdTbx20, mmp9 (G) and mmp13 (H) expression was increased relative to expression in cells infected with Adβ-gal (n=6, P<0.05). Fold changes were determined by dividing experimental values by control values with control values set to 1. Asterisks indicate statistically significant differences from control values and error bars represent standard error of the mean.
Tbx20 loss of function results in increased CSPG expression and decreased MMP expression
In order to achieve endogenous Tbx20 loss of function, a Tbx20-specific siRNA was transfected into primary chicken endocardial cushion cells. Scrambled control siRNA was also transfected in parallel experiments. To determine the efficiency of transfection, a fluorescently labeled oligonucleotide was co-transfected with the siRNA, and cells were counterstained with TO-PRO-3 iodide to visualize the nuclei (Fig.4A,C,E). Primary endocardial cushion cells were transfected with greater than 70% efficiency in the presence or absence of co-transfected siRNA (Fig.4G). Knockdown of Tbx20 protein expression was evident using immunohistochemistry with a Tbx20-specific antibody (Fig.4B,D,F). Cells transfected with Tbx20-specific siRNA had significantly reduced Tbx20 expression in the nucleus (Fig.4B) relative to scrambled siRNA(Fig.4D) or untransfected controls (Fig.4F). In addition, transfection with Tbx20-specific siRNA resulted in an approximately 85% reduction of Tbx20 mRNA levels (Fig.4H) as determined by real time RT-PCR. There was no detectable change in Tbx20 gene expression following transfection of the scrambled siRNA control (Fig.4H). These data indicate that primary chicken endocardial cushion cells can be efficiently transfected with sequence-specific siRNA in order to produce a significant loss of Tbx20 function.
Figure 4. Tbx20-specific siRNA transfected into primary chicken endocardial cushion cells results in decreased Tbx20 mRNA and protein expression.
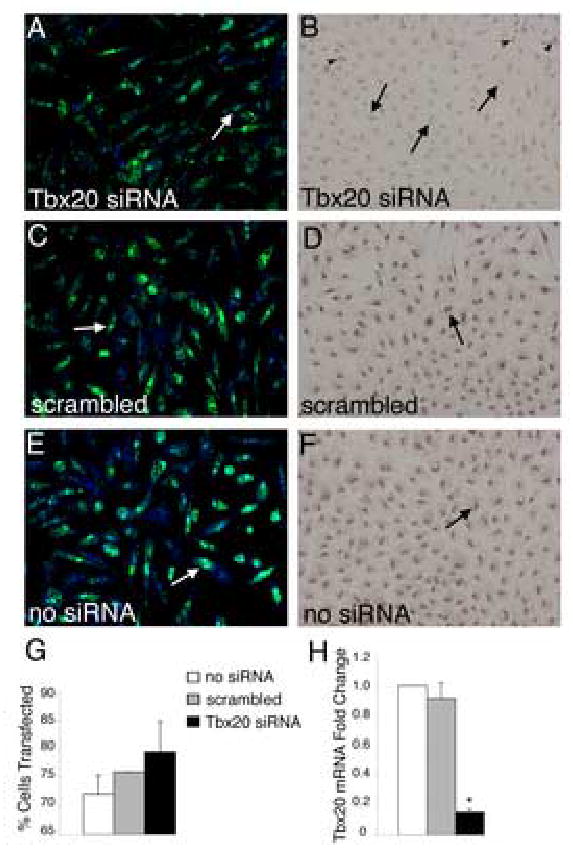
(A,C,E) Primary chicken endocardial cushion cells were co-transfected with siRNA and FITC-labeled fluorescent oligos (green) to visualize transfection efficiency. Cells were counterstained with TO-PRO-3 iodide (blue) to visualize the nuclei. Examples of positively transfected cells are indicated by white arrows. Transfection efficiency was determined as the number of cells with incorporation of FITC-labeled oligonucleotide/total TO-PRO-3 positive nuclei (G). These analyses demonstrated that cultures were consistently transfected at 70–80% efficiency. (B,D,F) Tbx20 protein expression was examined by immunohistochemistry with an antibody specific for Tbx20 in parallel cultures of siRNA-transfected cells. Cells transfected with Tbx20-specific siRNA (B) had significantly reduced nuclear staining (arrows indicate cells with reduced Tbx20 immunoreactivity) compared to controls (D,F, arrows indicate Tbx20 immunopositive cells). In Tbx20-specific siRNA transfected cultures (B), a subset of cells remained Tbx20 immunopositive (arrowheads). Tbx20 mRNA expression was quantified using real time RT-PCR (H). Cells transfected with Tbx20-specific siRNA had an 80% reduction in Tbx20 mRNA relative to untransfected and scrambled siRNA controls. Asterisks indicate statistically significant differences from control values and error bars represent standard error of the mean (n=3, P<0.01).
The regulatory effects of Tbx20 knockdown on CSPG and MMP expression were examined in primary endocardial cushion cells removed from HH stage 25 AV canals. In cells transfected with Tbx20-specific siRNA, agg expression increased approximately 4 fold (Fig.5A) and vers expression increased approximately 6 fold (Fig.5B). In contrast, mmp9 expression decreased approximately 80% (Fig.5C) and mmp13 expression decreased approximately 50% (Fig.5D) as measured by real time RT-PCR. Taken together, loss of Tbx20 function leads to increased expression of CSPG genes and decreased expression of MMP genes. These results from loss of Tbx20 function are consistent with the adenoviral gain of Tbx20 function data and support a role for Tbx20 in repressing CSPG expression and promoting MMP expression in primitive endocardial cushion cells.
Figure 5. Tbx20 loss of function results in increased CSPG expression and decreased MMP expression.
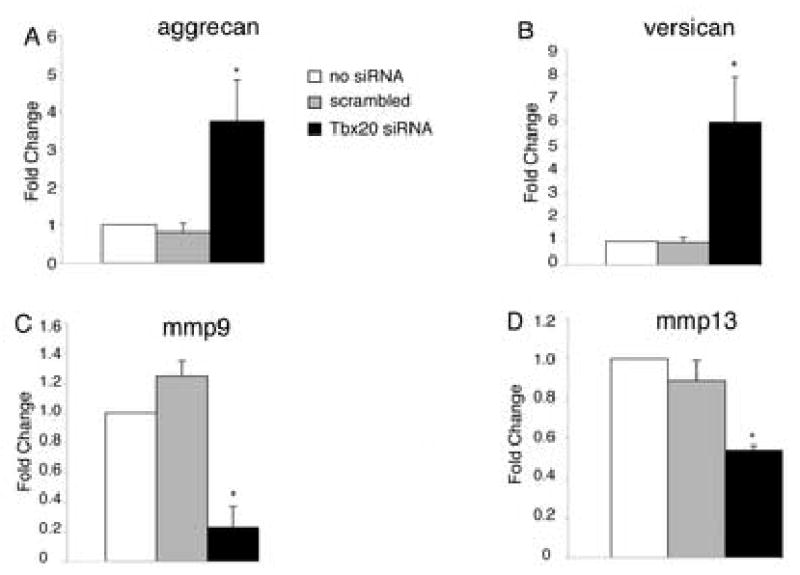
Primary endocardial cushion cells were transfected with Tbx20-specific or scrambled control siRNA. Cells transfected with Tbx20-specific siRNA had increased expression of aggrecan (A) and versican (B) and decreased expression of mmp9 (C) and mmp13 (D), relative to untransfected and scrambled siRNA controls as measured by real time RT-PCR. Asterisks indicate statistically significant differences from control values and error bars represent standard error of the mean (n=4, P<0.05).
Tbx20 promotes endocardial cushion cell proliferation
Heart valve development is characterized by high proliferation in endocardial cushions and decreased proliferation in remodeling valves (Hinton et al., 2006; Lincoln et al., 2004). Targeted mutagenesis of Tbx20 in mice suggests Tbx20 promotes myocardial cell proliferation in the primitive heart tube (Cai et al., 2005). To determine if Tbx20 has a similar role in endocardial cushion cells, Tbx20 gain and loss of function studies were performed and cell proliferation was assessed. Endocardial cushion cultures were infected with AdTbx20 or Adβ-gal for gain of function studies or transfected with Tbx20-specific siRNA or scrambled control siRNA for loss of function studies. The number of cells in S-phase of the cell cycle was measured by BrdU incorporation and used as an indication of proliferation. A proliferation index was determined by dividing the number of BrdU labeled nuclei by the number of total nuclei per microscopic field. The normal proliferation index of endocardial cushion cells infected with Adβ-gal (Fig.6A) or transfected with scrambled siRNA (Fig.6C) was approximately 20% (Fig.6E). Cells that were infected with AdTbx20 (Fig.6B) were significantly more proliferative, with a proliferation index of over 40% (Fig.6E). In contrast, cells transfected with Tbx20-specific siRNA (Fig.6D) were significantly less proliferative, with a proliferation index of less than 10% (Fig.6E). These data are consistent with a role for Tbx20 in promoting proliferation in endocardial cushions.
Figure 6. Tbx20 promotes endocardial cushion cell proliferation.
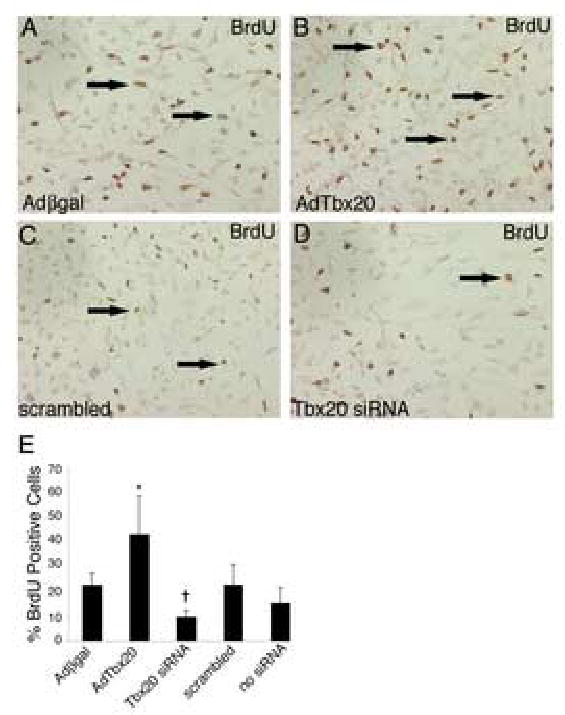
Primary endocardial cushion cells were infected with adenoviruses expressing β-gal (Ad β-gal) or Tbx20 (AdTbx20) for gain of function or transfected with Tbx20-specific or scrambled control siRNA for loss of function. Proliferation was measured using immunohistochemistry for BrdU incorporation. Cells infected with AdTbx20 (B) exhibit increased BrdU incorporation (arrows indicate positively stained cells) relative to cells infected with Adβ-gal (A). Cells transfected with Tbx20-specific siRNA (D) are less proliferative than cells transfected with scrambled siRNA (C). The percent of BrdU positive cells relative to total nuclei in each group is quantified (E). The asterisk represents a statistically significant difference compared to the Adβ-gal control group while the cross represents a statistically significant difference compared to the no siRNA control group (n=3, P<0.01). Error bars represent standard error of the mean.
BMP2 induces Tbx20 and altered Tbx20 function affects the expression of Tbx2 and N-myc in endocardial cushion cells
BMP2 induces Tbx20 expression in cardiac primordia prior to cardiomyogenic differentiation and its expression in AV canal myocardium is required for endocardial cushion formation (Ma et al., 2005; Plageman and Yutzey, 2004; Rivera-Feliciano and Tabin, 2006). The ability of BMP2 to induce Tbx20 expression in endocardial cushion cells was investigated. AV endocardial cushions were removed from the surrounding myocardium of HH stage 25 chicken embryos and cultured with or without the addition of recombinant human BMP2 or Noggin, a BMP inhibitor. RNA was isolated and Tbx20 expression levels were measured using real time RT-PCR. Addition of BMP2 induced Tbx20 expression while addition of Noggin repressed endogenous Tbx20 expression below levels in untreated controls (Fig. 7A). These data demonstrate that BMP2 can induce Tbx20 in endocardial cushion cells and that inhibition of endogenous BMP signaling by Noggin decreases Tbx20 gene expression levels. Bmp2 is required for endocardial cushion formation (Ma et al., 2005; Rivera-Feliciano and Tabin, 2006; Sugi et al., 2004), and the ability of BMP2 to induce Tbx20 in endocardial cushion cells is consistent with Tbx20 regulation by BMP signaling in the early stages of endocardial cushion development.
Figure 7. BMP2 induces Tbx20 and Tbx20 regulates the expression of Tbx2 and N-myc in endocardial cushion cells.
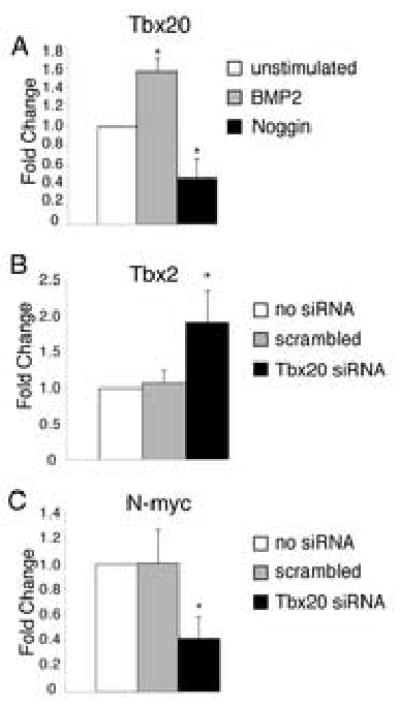
Endocardial cushion cells were cultured with or without the addition of soluble recombinant BMP2 or Noggin, a BMP inhibitor. Addition of BMP2 resulted in increased Tbx20 expression relative to untreated controls, while addition of Noggin attenuated that response (A), as measured by real time RT-PCR. The expression of Tbx2 and N-myc, genes associated with Tbx20 regulated proliferation, was measured in siRNA transfected endocardial cushion cells using real time RT-PCR. In cells transfected with Tbx20-specific siRNA, Tbx2 expression is increased (B) while N-myc expression is decreased relative to untransfected or scrambled siRNA controls (C). Asterisks indicate statistically significant differences from control values and error bars represent standard error of the mean (n=5, P<0.01).
It was previously reported that Tbx20 affects myocardial proliferation by directly repressing Tbx2, and thus relieving the direct repression of Tbx2 on N-myc expression (Cai et al., 2005). To determine if a similar mechanism occurs in endocardial cushion cells, cultured cells were transfected with Tbx20-specific siRNA or scrambled control siRNA. Changes in Tbx2 and N-myc mRNA levels were measured by real time RT-PCR. In cells transfected with Tbx20-specific siRNA, Tbx2 expression increased by approximately 2 fold (Fig.7B), while N-myc expression decreased by approximately 60% (Fig.7C). These studies provide evidence that Tbx20 similarly regulates endocardial cushion and myocardial cell proliferation through Tbx2 and N-myc gene expression. Taken together, these results are consistent with a role for Tbx20 in maintaining the endocardial cushions as primitive, undifferentiated, and proliferative.
Discussion
Gain and loss of function strategies were used to examine the role of Tbx20 in developing avian endocardial cushions. During normal valve development Tbx20, mmp9, and mmp13 are expressed at higher levels in endocardial cushions relative to remodeling valves, while agg and vers are expressed at higher levels in remodeling valves relative to endocardial cushions. Additionally, endocardial cushion cells are more proliferative than cells in remodeling valves (Hinton et al., 2006). In these experiments, increased Tbx20 leads to decreased CSPG expression and increased MMP expression whereas the opposite trend occurs with loss of Tbx20. Furthermore, increased Tbx20 induces cell proliferation in endocardial cushion cultures, while proliferation decreases with loss of Tbx20. These results are consistent with Tbx20 expression in highly proliferative, unremodeled valve precursors and suggest that Tbx20 functions to maintain valve precursors in an immature, highly proliferative state, as has also been noted for Tbx20 function in primitive myocardium. Taken together, a model for the functions of Tbx20 in valve development (Fig.8) can be generated in which high levels of Tbx20 early in cushion mesenchyme maintain primitive, proliferative, unremodeled, endocardial cushions, while lower levels of Tbx20 later in development occur in more highly differentiated, less proliferative, remodeling valves.
Figure 8. Model for Tbx20 function in endocardial cushions and remodeling valves.
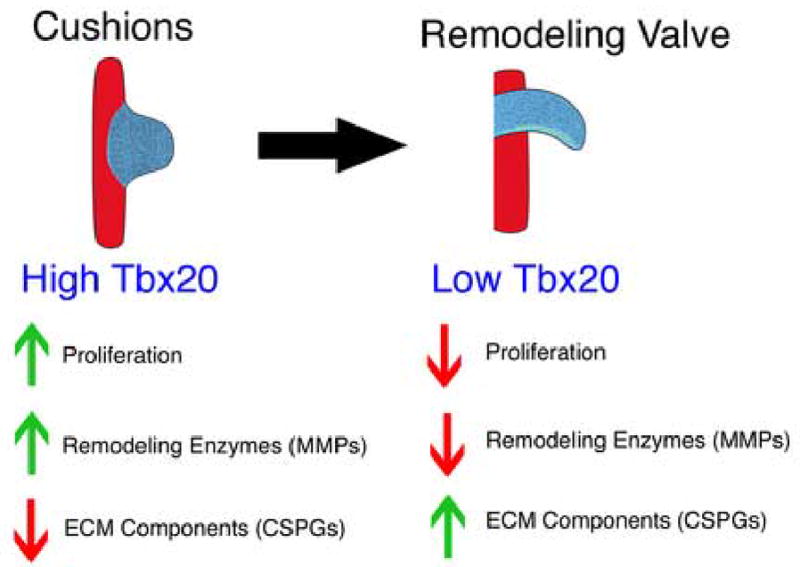
Endocardial cushions are characterized by highly proliferative mesenchymal cells and unorganized extracellular matrix, while remodeling valves are characterized by less proliferative interstitial cells and organized extracellular matrix. These molecular and cellular characteristics are consistent with a model in which high levels of Tbx20 in endocardial cushions induce proliferation while maintaining an unremodeled, unorganized extracellular matrix. In contrast, lower levels of Tbx20 in remodeling valves allow for a less proliferative cell population and a more remodeled and organized extracellular matrix.
Increased Tbx20 expression promotes proliferation in endocardial cushion cells. In this report and in a previously published study, altered Tbx20 function affects the expression of N-myc and Tbx2 (Cai et al., 2005). In studies of primitive mouse myocardium lacking Tbx20 it was suggested that Tbx20 acts to increase N-myc expression by repressing Tbx2, a transcriptional repressor (Cai et al., 2005). However it is also possible that Tbx20 directly activates N-myc gene expression since Tbx20 can have activator or repressor function depending on transcriptional partners or cellular context (Plageman and Yutzey, 2004; Stennard et al., 2003). The relevance of N-myc to normal heart development was demonstrated directly by targeted mutagenesis in mice. Mutant mice lacking N-myc have complex cardiac abnormalities including a lack of endocardial cushions, atrial and ventricular septal defects, and hypoplasia of the compact myocardium (Charron et al., 1992; Hurlin, 2005; Moens et al., 1993). Extensive cell culture studies have demonstrated that N-myc promotes cell cycle progression at the G1/S-phase, and studies of Tbx5 in Xenopus embryos support a role for this T-box family member in the G1 to S-phase transition in cardiac myocytes (Goetz et al., 2006; Hooker and Hurlin, 2006). In addition, TBX2 and TBX3 expression is increased and promotes proliferation in a subset of mammary carcinomas and breast cancer cell lines (Carlson et al., 2002; Fan et al., 2004; Prince et al., 2004; Vance et al., 2005). The ability of Tbx20 to promote N-myc expression in the developing heart may be indicative of an important role in regulation of cell cycle progression at the G1/S-phase in both myocardial and endocardial cushion cell populations.
Bmp2 is expressed in the myocardium of the AV canal and is required for the initial stages of endocardial cushion formation (Ma et al., 2005; Rivera-Feliciano and Tabin, 2006; Yamada et al., 2000). In this study, we report BMP2 induces Tbx20 in endocardial cushion cells and that loss of Tbx20 function in cushion cells results in increased Tbx2 expression. In murine atrial and ventricular chamber myocardium, Tbx20 represses Tbx2 expression, thereby allowing chamber-specific differentiation and myocardial proliferation to occur (Cai et al., 2005; Singh et al., 2005; Stennard et al., 2005). A similar regulatory relationship may occur during endocardial cushion maturation. BMP2 can induce expression of both Tbx20 and Tbx2 in the primary heart field and AV canal (Plageman and Yutzey, 2004; Rivera-Feliciano and Tabin, 2006; Yamada et al., 2000). In addition, Tbx20 may exert positive feedback on Bmp2 expression (Cai et al., 2005) although, decreased Tbx20 expression has also been observed to result in ectopic Bmp2 expression, suggesting that Tbx20 can negatively feedback on Bmp2 (Singh et al., 2005). In light of all of these studies, the mechanism that regulates the precise coordination of Bmp2, Tbx20, and Tbx2 expression still remains unclear. In addition, future experiments are needed to assess independent functions of Tbx20 and Tbx2 in chamber myocardium and endocardial cushions.
Increased Tbx20 function leads to decreased expression of CSPGs and increased expression of MMPs in endocardial cushion cells. This is consistent with a role for Tbx20 in ECM remodeling during valve development. While altered Tbx20 function affects the expression of ECM remodeling genes, it is not known whether these interactions are direct or indirect. In previous studies a limited number of direct downstream targets of Tbx20 have been identified, including Nppa and Tbx2 (Cai et al., 2005; Stennard et al., 2003; Takeuchi et al., 2005). To determine if Tbx20 interacts directly or indirectly with CSPG and MMP regulatory elements, promoter regions of murine Agg and Mmp13 were analyzed for T-box binding sites (TBE). While several conserved TBE sites were found in addition to binding sites for common T-box cofactors, Tbx20 alone was not sufficient to induce Agg or Mmp13 reporter constructs and induction was not observed in co-transfection experiments (data not shown). However, these studies do not rule out the possibility that Tbx20 directly regulates CSPG and MMP gene expression through other regulatory elements or with additional co-factors. Direct transcriptional regulators ofAgg and Mmp13 have been identified in other cell types. Sox9 and Scleraxis can directly activate Agg expression in chondrocytic and osteoblastic cell lines respectively (Liu et al., 1997; Sekiya et al., 2000), while Tgf-β and core-binding factor 1 (CBFA1) can directly activate Mmp13 expression in human fibroblasts and chondrocytic and osteoblastic cell lines (Jimenez et al., 1999; Sternlicht and Werb, 2001; Uria et al., 1998). It is possible that the effect of Tbx20 on CSPG and MMP expression is indirect through the expression of genes such as Sox9, scleraxis, Tgf-β, or CBFA1, which are also present in endocardial cushions and valve primordia (Lincoln et al.,2006a; Yutzey, unpublished). With the data currently available, the effect of Tbx20 as a direct or indirect transcriptional regulator of CSPG or MMP gene expression cannot be determined.
In general, the ECM is important for many cellular processes including cell shape, movement, growth, differentiation, and survival. Similarly, MMPs that degrade and remodel ECM components can also affect all of these processes by altering the structure and composition of the ECM (Sternlicht and Werb, 2001). During angiogenesis and bone development, MMPs function to create a permissive environment for cell migration (Sternlicht and Werb, 2001). Therefore, the expression of MMPs in endocardial cushions composed of a loosely arranged ECM is consistent with migratory, proliferative, mesenchymal cells. The observed decrease in mmp9 and mmp13 expression in remodeling valves may be required for organization of the ECM and compartmentalization of interstitial cells (Hinton et al., 2006). Correct expression and assembly of CSPGs is also necessary for normal valve development. Transgenic mice containing an insertional mutation in the versican gene fail to form endocardial cushions (Kern et al., 2006; Mjaatvedt et al., 1998) and proteoglycans are integral components of the mature valve leaflet (Hinton et al., 2006). In this report, the observed differential expression of CSPGs and MMPs during endocardial cushion development and valve remodeling stages highlights the importance of establishing a balance between deposition and degradation of ECM components in order for normal valve development to occur.
Human heart valve disease is associated with abnormal organization and increased deposition of ECM constituents. In a recent study of pediatric aortic valve disease, ECM layers were unorganized and disproportionate and interstitial cell compartmentalization was abnormal (Hinton et al., 2006). Similarly, patients with congenital polyvalvular disease had AV and semilunar valves composed of abnormal ECM and disrupted valve architecture (Bartram et al., 2001). Furthermore, in cases of myxomatous and calcified adult aortic valves, disorganized ECM was accompanied by elevated levels of MMPs including MMP9 and MMP13 (Akhtar et al., 1999; Bailey et al., 2004; Rabkin et al., 2001; Soini et al., 2001). These features of valve disease, including unorganized ECM and interstitial cells with elevated MMP levels, are all characteristic of endocardial cushions. While features characteristic of valve disease have been described, relatively little is known about the molecular causes of valve pathogenesis. As shown in this study, altered Tbx20 function affects valve progenitor cell proliferation as well as expression of ECM components and remodeling enzymes in AV valve precursor cells. Therefore, it is possible that that misregulation of Tbx20 could lead to some of the characteristic features of valve disease. Further experiments examining the expression of Tbx20 in diseased valves may give insights into molecular mechanisms of valve disease.
Acknowledgments
The AdTbx20 adenovirus was generated by Timothy F. Plageman, Jr. In addition, we thank Heather Evans-Anderson for technical support and scientific advice. Artwork was generated with the help of Andreas Lange. This work was supported by an AHA Ohio Valley Affiliate pre-doctoral fellowship 0515153B to ELS, NIH grant HL082716 to KEY, and NIH/NHLBI SCCOR in Pediatric Heart Development and Disease P50 HL074728.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhtar S, Meek KM, James V. Ultrastructure abnormalities in proteoglycans, collagen fibrils, and elastic fibers in normal and myxomatous mitral valve chordae tendineae. Cardiovasc Pathol. 1999;8:191–201. doi: 10.1016/s1054-8807(99)00004-6. [DOI] [PubMed] [Google Scholar]
- Arciniegas E, Neves CY, Candelle D, Parada D. Differential versican isoforms and aggrecan expression in the chicken embryo aorta. Anat Rec A Discov Mol Cell Evol Biol. 2004;279:592–600. doi: 10.1002/ar.a.20042. [DOI] [PubMed] [Google Scholar]
- Armstrong EJ, Bischoff J. Heart valve development: endothelial cell signaling and differentiation. Circ Res. 2004;95:459–70. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M, Pillarisetti S, Jones P, Xiao H, Simionescu D, Vyavahare N. Involvement of matrix metalloproteinases and tenascin-C in elastin calcification. Cardiovasc Pathol. 2004;13:146–55. doi: 10.1016/S1054-8807(04)00009-2. [DOI] [PubMed] [Google Scholar]
- Bartram U, Bartelings MM, Kramer HH, Gittenberger-de Groot AC. Congenital polyvalvular disease: a review. Pediatr Cardiol. 2001;22:93–101. doi: 10.1007/s002460010169. [DOI] [PubMed] [Google Scholar]
- Brown DD, Binder O, Pagratis M, Parr BA, Conlon FL. Developmental expression of the Xenopus laevis Tbx20 orthologue. Dev Genes Evol. 2003;212:604–7. doi: 10.1007/s00427-002-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, Li X, Rosenfeld MG, Chen J, Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–87. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson H, Ota S, Song Y, Chen Y, Hurlin PJ. Tbx3 impinges on the p53 pathway to suppress apoptosis, facilitate cell transformation and block myogenic differentiation. Oncogene. 2002;21:3827–35. doi: 10.1038/sj.onc.1205476. [DOI] [PubMed] [Google Scholar]
- Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, Robertson EJ, Alt FW. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992;6:2248–57. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- Coker ML, Thomas CV, Clair MJ, Hendrick JW, Krombach RS, Galis ZS, Spinale FG. Myocardial matrix metalloproteinase activity and abundance with congestive heart failure. Am J Physiol. 1998;274:H1516–23. doi: 10.1152/ajpheart.1998.274.5.H1516. [DOI] [PubMed] [Google Scholar]
- Cripe L, Andelfinger G, Martin LJ, Shooner K, Benson DW. Bicuspid aortic valve is heritable. J Am Coll Cardiol. 2004;44:138–43. doi: 10.1016/j.jacc.2004.03.050. [DOI] [PubMed] [Google Scholar]
- Ehrman LA, Yutzey KE. Lack of regulation in the heart forming region of avian embryos. Dev Biol. 1999;207:163–75. doi: 10.1006/dbio.1998.9167. [DOI] [PubMed] [Google Scholar]
- Fan W, Huang X, Chen C, Gray J, Huang T. TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer Res. 2004;64:5132–9. doi: 10.1158/0008-5472.CAN-04-0615. [DOI] [PubMed] [Google Scholar]
- Flanagan TC, Pandit A. Living artificial heart valve alternatives: a review. Eur Cell Mater. 2003;6:28–45. doi: 10.22203/ecm.v006a04. discussion 45. [DOI] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–4. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Goetz SC, Brown DD, Conlon FL. TBX5 is required for embryonic cardiac cell cycle progression. Development. 2006;133:2575–84. doi: 10.1242/dev.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Foix AM, Coats WS, Baque S, Alam T, Gerard RD, Newgard CB. Adenovirus-mediated transfer of the muscle glycogen phosphorylase gene into hepatocytes confers altered regulation of glycogen metabolism. J Biol Chem. 1992;267:25129–34. [PubMed] [Google Scholar]
- Hahn-Dantona EA, Aimes RT, Quigley JP. The isolation, characterization, and molecular cloning of a 75-kDa gelatinase B-like enzyme, a member of the matrix metalloproteinase (MMP) family. An avian enzyme that is MMP-9-like in its cell expression pattern but diverges from mammalian gelatinase B in sequence and biochemical properties. J Biol Chem. 2000;275:40827–38. doi: 10.1074/jbc.M006234200. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. Dev Dyn. 1951;195:231–72. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Hinton RB, Jr, Lincoln J, Deutsch GH, Osinska H, Manning PB, Benson DW, Yutzey KE. Extracellular matrix remodeling and organization in developing and diseased aortic valves. Circ Res. 2006;98:1431–8. doi: 10.1161/01.RES.0000224114.65109.4e. [DOI] [PubMed] [Google Scholar]
- Hooker CW, Hurlin PJ. Of Myc and Mnt. J Cell Sci. 2006;119:208–16. doi: 10.1242/jcs.02815. [DOI] [PubMed] [Google Scholar]
- Hurlin PJ. N-Myc functions in transcription and development. Birth Defects Res C Embryo Today. 2005;75:340–52. doi: 10.1002/bdrc.20059. [DOI] [PubMed] [Google Scholar]
- Iio A, Koide M, Hidaka K, Morisaki T. Expression pattern of novel chick T-box gene, Tbx20. Dev Genes Evol. 2001;211:559–62. doi: 10.1007/s00427-001-0187-y. [DOI] [PubMed] [Google Scholar]
- Jimenez MJ, Balbin M, Lopez JM, Alvarez J, Komori T, Lopez-Otin C. Collagenase 3 is a target of Cbfa1, a transcription factor of the runt gene family involved in bone formation. Mol Cell Biol. 1999;19:4431–42. doi: 10.1128/mcb.19.6.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern CB, Twal WO, Mjaatvedt CH, Fairey SE, Toole BP, Iruela-Arispe ML, Argraves WS. Proteolytic cleavage of versican during cardiac cushion morphogenesis. Dev Dyn. 2006;235:2238–2247. doi: 10.1002/dvdy.20838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Furth EE, Kalluri R, Wakenell P, Kallen CB, Jeffrey JJ, Leboy PS, Strauss JF., 3rd Induction of matrix metalloproteinases and collagenolysis in chick embryonic membranes before hatching. Biol Reprod. 1999;60:183–9. doi: 10.1095/biolreprod60.1.183. [DOI] [PubMed] [Google Scholar]
- Li H, Domowicz M, Hennig A, Schwartz NB. S103L reactive chondroitin sulfate proteoglycan (aggrecan) mRNA expressed in developing chick brain and cartilage is encoded by a single gene. Brain Res Mol Brain Res. 1996;36:309–21. doi: 10.1016/0169-328x(95)00269-x. [DOI] [PubMed] [Google Scholar]
- Liberatore CM, Searcy-Schrick RD, Vincent EB, Yutzey KE. Nkx-2.5 gene induction in mice is mediated by a Smad consensus regulatory region. Dev Biol. 2002;244:243–56. doi: 10.1006/dbio.2002.0604. [DOI] [PubMed] [Google Scholar]
- Lincoln J, Alfieri CM, Yutzey KE. Development of heart valve leaflets and supporting apparatus in chicken and mouse embryos. Dev Dyn. 2004;230:239–50. doi: 10.1002/dvdy.20051. [DOI] [PubMed] [Google Scholar]
- Lincoln J, Alfieri CM, Yutzey KE. BMP and FGF regulatory pathways control cell lineage diversification of heart valve precursor cells. Dev Biol. 2006a;292:292–302. doi: 10.1016/j.ydbio.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Lincoln J, Lange AW, Yutzey KE. Hearts and bones: shared regulatory mechanisms in heart valve, cartilage, tendon, and bone development. Dev Biol. 2006b;294:292–302. doi: 10.1016/j.ydbio.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Liu Y, Watanabe H, Nifuji A, Yamada Y, Olson EN, Noda M. Overexpression of a single helix-loop-helix-type transcription factor, scleraxis, enhances aggrecan gene expression in osteoblastic osteosarcoma ROS17/2.8 cells. J Biol Chem. 1997;272:29880–5. doi: 10.1074/jbc.272.47.29880. [DOI] [PubMed] [Google Scholar]
- Luo W, Guo C, Zheng J, Chen TL, Wang PY, Vertel BM, Tanzer ML. Aggrecan from start to finish. J Bone Miner Metab. 2000;18:51–6. doi: 10.1007/s007740050011. [DOI] [PubMed] [Google Scholar]
- Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–11. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Mjaatvedt CH, Yamamura H, Capehart AA, Turner D, Markwald RR. The Cspg2 gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation. Dev Biol. 1998;202:56–66. doi: 10.1006/dbio.1998.9001. [DOI] [PubMed] [Google Scholar]
- Moens CB, Stanton BR, Parada LF, Rossant J. Defects in heart and lung development in compound heterozygotes for two different targeted mutations at the N-myc locus. Development. 1993;119:485–99. doi: 10.1242/dev.119.2.485. [DOI] [PubMed] [Google Scholar]
- Passi A, Negrini D, Albertini R, Miserocchi G, De Luca G. The sensitivity of versican from rabbit lung to gelatinase A (MMP-2) and B (MMP-9) and its involvement in the development of hydraulic lung edema. FEBS Lett. 1999;456:93–6. doi: 10.1016/s0014-5793(99)00929-1. [DOI] [PubMed] [Google Scholar]
- Person AD, Klewer SE, Runyan RB. Cell biology of cardiac cushion development. Int Rev Cytol. 2005;243:287–335. doi: 10.1016/S0074-7696(05)43005-3. [DOI] [PubMed] [Google Scholar]
- Pirok EW, 3rd, Li H, Mensch JR, Jr, Henry J, Schwartz NB. Structural and functional analysis of the chick chondroitin sulfate proteoglycan (aggrecan) promoter and enhancer region. J Biol Chem. 1997;272:11566–74. doi: 10.1074/jbc.272.17.11566. [DOI] [PubMed] [Google Scholar]
- Plageman TF, Jr, Yutzey KE. Differential expression and function of Tbx5 and Tbx20 in cardiac development. J Biol Chem. 2004;279:19026–34. doi: 10.1074/jbc.M314041200. [DOI] [PubMed] [Google Scholar]
- Plageman TF, Jr, Yutzey KE. T-box genes and heart development: putting the "T" in heart. Dev Dyn. 2005;232:11–20. doi: 10.1002/dvdy.20201. [DOI] [PubMed] [Google Scholar]
- Prince S, Carreira S, Vance KW, Abrahams A, Goding CR. Tbx2 directly represses the expression of the p21(WAF1) cyclin-dependent kinase inhibitor. Cancer Res. 2004;64:1669–74. doi: 10.1158/0008-5472.can-03-3286. [DOI] [PubMed] [Google Scholar]
- Rabkin E, Aikawa M, Stone JR, Fukumoto Y, Libby P, Schoen FJ. Activated interstitial myofibroblasts express catabolic enzymes and mediate matrix remodeling in myxomatous heart valves. Circulation. 2001;104:2525–32. doi: 10.1161/hc4601.099489. [DOI] [PubMed] [Google Scholar]
- Rabkin-Aikawa E, Mayer JE, Jr, Schoen FJ. Heart valve regeneration. Adv Biochem Eng Biotechnol. 2005;94:141–79. doi: 10.1007/b100003. [DOI] [PubMed] [Google Scholar]
- Rivera-Feliciano J, Tabin CJ. Bmp2 instructs cardiac progenitors to form the heart-valve-inducing field. Dev Biol. 2006;295:580–8. doi: 10.1016/j.ydbio.2006.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JA, Jackson LF, Lee DC, Camenisch TD. Form and function of developing heart valves: coordination by extracellular matrix and growth factor signaling. J Mol Med. 2003;81:392–403. doi: 10.1007/s00109-003-0456-5. [DOI] [PubMed] [Google Scholar]
- Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, Nifuji A, Noda M. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J Biol Chem. 2000;275:10738–44. doi: 10.1074/jbc.275.15.10738. [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nishida Y, Ito K, Kimata K. cDNA cloning of PG-M, a large chondroitin sulfate proteoglycan expressed during chondrogenesis in chick limb buds. Alternative spliced multiforms of PG-M and their relationships to versican. J Biol Chem. 1993;268:14461–9. [PubMed] [Google Scholar]
- Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M, Schuster-Gossler K, Burger A, Ericson J, Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132:2697–707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- Soini Y, Satta J, Maatta M, Autio-Harmainen H. Expression of MMP2, MMP9, MT1-MMP, TIMP1, and TIMP2 mRNA in valvular lesions of the heart. J Pathol. 2001;194:225–31. doi: 10.1002/path.850. [DOI] [PubMed] [Google Scholar]
- Somi S, Buffing AA, Moorman AF, Van Den Hoff MJ. Dynamic patterns of expression of BMP isoforms 2, 4, 5, 6, and 7 during chicken heart development. Anat Rec A Discov Mol Cell Evol Biol. 2004;279:636–51. doi: 10.1002/ar.a.20031. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Elliott DA, Rankin S, Haast SJ, Lai D, McDonald LP, Niederreither K, Dolle P, Bruneau BG, Zorn AM, Harvey RP. Cardiac T-box factor Tbx20 directly interacts with Nkx2–5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262:206–24. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, Elliott DE, Prall OW, Black BL, Fatkin D, Harvey RP. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–62. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Harvey RP. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005;132:4897–910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugi Y, Yamamura H, Okagawa H, Markwald RR. Bone morphogenetic protein-2 can mediate myocardial regulation of atrioventricular cushion mesenchymal cell formation in mice. Dev Biol. 2004;269:505–18. doi: 10.1016/j.ydbio.2004.01.045. [DOI] [PubMed] [Google Scholar]
- Szeto DP, Griffin KJ, Kimelman D. HrT is required for cardiovascular development in zebrafish. Development. 2002;129:5093–101. doi: 10.1242/dev.129.21.5093. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R, Hui CC, Henkelman RM, Nemer M, Black BL, Nagy A, Bruneau BG. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–74. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- Uria JA, Jimenez MG, Balbin M, Freije JM, Lopez-Otin C. Differential effects of transforming growth factor-beta on the expression of collagenase-1 and collagenase-3 in human fibroblasts. J Biol Chem. 1998;273:9769–77. doi: 10.1074/jbc.273.16.9769. [DOI] [PubMed] [Google Scholar]
- Vance KW, Carreira S, Brosch G, Goding CR. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 2005;65:2260–8. doi: 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- Yamada M, Revelli JP, Eichele G, Barron M, Schwartz RJ. Expression of chick Tbx-2, Tbx-3, and Tbx-5 genes during early heart development: evidence for BMP2 induction of Tbx2. Dev Biol. 2000;228:95–105. doi: 10.1006/dbio.2000.9927. [DOI] [PubMed] [Google Scholar]
- Yamagishi T, Nakajima Y, Nishimatsu S, Nohno T, Ando K, Nakamura H. Expression of tbx20 RNA during chick heart development. Dev Dyn. 2004;230:576–80. doi: 10.1002/dvdy.20076. [DOI] [PubMed] [Google Scholar]


