Abstract
In recent years, as a knowledge-based discipline, bioinformatics has been made more computationally amenable. After its beginnings as a technology advocated by computer scientists to overcome problems of heterogeneity, ontology has been taken up by biologists themselves as a means to consistently annotate features from genotype to phenotype. In medical informatics, artifacts called ontologies have been used for a longer period of time to produce controlled lexicons for coding schemes. In this article, we review the current position in ontologies and how they have become institutionalized within biomedicine. As the field has matured, the much older philosophical aspects of ontology have come into play. With this and the institutionalization of ontology has come greater formality. We review this trend and what benefits it might bring to ontologies and their use within biomedicine.
Keywords: bio-ontology, medical ontology, annotation, knowledge, knowledge representation, history
INTRODUCTION
In this briefing, we explore the current state and future prospects of the use of ontologies within bioinformatics and medical informatics. Since an earlier Briefing in 2000 [1], the role of ontologies within bioinformatics has changed markedly. It has moved from a niche activity to one that is, in all respects, a mainstream activity. It is useful, however, to remind ourselves why this interest is so large before we move on to review the current state and future prospects of biomedical ontologies.
Biology is unlike physics and much of chemistry in that—although it contains many laws and models—few of these are reduced to a mathematical form. It is not possible to take a protein’s sequence of amino acids, apply some formula, and derive a set of characteristics such as accurate three-dimensional shape, functionality, forms of modification, etc. Instead of mathematical laws, biomedical scientists use what they understand about characterized entities to make inferences about uncharacterized entities. This is, for example, the basis of the similarity search—similarity between biological sequences is made mathematically, but any inference about that similarity is made by a biologist reading annotations. What we are using to make these inferences is what we know about the entities being compared. This is our knowledge about those entities.
Instead of the convenience of mathematical forms, biomedical scientists collect facts, often recording them in natural language, and then use that knowledge to make inferences about yet uncharacterized observations. Yet, this knowledge is highly heterogeneous. While it is easy to compare, for instance, nucleic acid or polypeptide sequences between bioinformatics resources, the knowledge component of these resources is very difficult to compare, both for humans and computers, because the knowledge is represented in a wide variety of lexical forms [2, 3].
In computer science, ontologies are a technique or technology used to represent and share knowledge about a domain by modeling the things in that domain and the relationships between those things [4]. These relationships describe the properties of those things; in essence, what it is to be one of those things in the domain being modeled. An ontology represents a conceptualization of reality or simply reality (This philosophical aspect of the ontological discipline is beyond the scope of this article). The labels used for the things and their properties in an ontological model can provide a language for a community to talk about their domain. By agreeing on a particular ontological representation, a common vocabulary can be used to describe and ultimately analyze data.
Such sharing has obvious benefits for humans using facts to help make inferences about a domain of study. Those facts, the knowledge about the domain, become much easier to handle as the same things are referred to in the same manner across the resources in which those facts are stored. Ultimately, we would like to be able to handle knowledge computationally in a manner comparable to that in which we handle numeric data. What is more, as will be described later in the ‘New formalisms and tools for representing bio-ontologies’ section, given a well-defined semantics for the knowledge representation language, machines can make inferences about the facts expressed in that language.
This article will show how this basic idea has become a central theme within biomedical research to the stage where it now has a national center in the US (see section titled ‘Institutionalization of bio-ontologies’). The Section titled ‘Timeline and recent additions’ shows how ontologies have a long history in the biomedical domain and, particularly, in biology and now represent a broad spectrum of important biological knowledge. Later in the article, the future direction of these current trends will be explored. It is not possible in such an article to do justice to all the resources available. Our aim, however, is to give a ‘briefing’ as to what exists. Electronic references to the ontological resources are available in Appendix 1.
Appendix.
Ontological and terminological resources mentioned in this article (all URLs are valid as of 9 July, 2006).
| BFO | Basic Formal Ontology
http://ontology.buffalo.edu/bfo/ |
| ChEBI | Chemical Entities of Biological Interest
http://www.ebi.ac.uk/chebi/ |
| CPT | Current Procedural Terminology
http://www.ama-assn.org/ama/pub/category/3113.html |
| CTV3 | Clinical Terms Version 3 (formerly known as the Read Codes)
http://www.nhsia.nhs.uk/terms/pages/readcodes_intro.asp |
| DM&D | Dictionary of Medicines and Devices
http://www.dmd.nhs.uk/ |
| DOLCE | Descriptive Ontology for Linguistic and Cognitive Engineering
http://www.loa-cnr.it/DOLCE.html |
| EMTREE | EMTREE is Elsevier’s Life Science Thesaurus
http://www.info.embase.com/emtree/about/ |
| FMA | Foundational Model of Anatomy
http://fma.biostr.washington.edu/ |
| GALEN | Generalized Architecture for Languages, Encyclopedias and Nomenclatures in medicine
http://www.opengalen.org/ |
| GO | Gene Ontology
http://www.geneontology.org/ |
| ICD | International Classification of Diseases
http://www.who.int/classifications/icd/en/ |
| ICPC | International Classification of Primary Care
http://www.globalfamilydoctor.com/wicc/ |
| MeSH | Medical Subject Headings
http://www.nlm.nih.gov/mesh/ |
| MGED | Microarray Gene Expression Data Ontology
http://mged.sourceforge.net/ontologies/MGEDontology.php |
| NCI Thesaurus. | NCI Thesaurus
http://cancer.gov/cancerinfo/terminologyresources/ |
| OBO | Open Biomedical Ontologies
http://obo.sourceforge.net/ |
| OPCS | Office of Population Censuses and Surveys Intervention Classification
http://www.connectingforhealth.nhs.uk/interventionclassification |
| SNOMED | Systematized Nomenclature of Medicine
http://www.snomed.org/ |
| SNOP | Systematized Nomenclature of Pathology (superseded by SNOMED) |
| SUMO | Suggested Upper Merged Ontology
http://ontology.teknowledge.com/ |
| UMLS | Unified Medical Language System
http://umlsks.nlm.nih.gov/ (cost-free license required) |
TIMELINE AND RECENT ADDITIONS
From Linnaeus to Ashburner
Human beings like to put the things (instances) they see around them into categories. What is more, categories can have subcategories. We see classification throughout human activities: we do it to people, library books, Web pages, etc. Biomedical scientists are no different. Biologists have long classified the phenomena they observe in the world around them. After mediaeval bestiaries, a classic starting point for talking about classification in biology is the Linnaean classification of species [5]. This classification is all pervasive and species taxonomies still form a backbone of how we talk about biological data, especially in the realm of evolution.
Ontology and classification are, however, not the same. Classification might be a component of ontology, but the latter adds something more. An ontology attempts to describe what we understand exists in our domain and to try and capture what it is to belong to one of the classes, categories or types in that model. An ontology, more formally, is a set of logic axioms that form a model of a portion of (a conceptualization) of reality (after [6]). There are many artifacts that are called ontology. One’s bias usually depends on purpose for modeling, representation used for modeling and philosophical viewpoint [5]. What computer scientists call ontologies are not really ontologies; they are knowledge structures or conceptual models, but the term has now been established. So, in this article, we are very inclusive in what we call ‘ontology’.
This article is not the place for a deep discussion of what counts as a real ontology in the true philosophical sense of the discipline. It is not that such a debate is wasted, but, for the large part, what we call ontologies are being built to perform a job of sharing what we understand about the world of biomedicine. The spectrum of ontology-like structures will range from controlled vocabularies, thesauri, directed acyclic graphs and frame-based systems to rich logical axioms encapsulating our knowledge [6]. In this article, almost anything along this spectrum will be included, but the further away from the right-hand end of the spectrum, the more ‘ontology-like’ (from a computer science perspective) the artifact becomes.
The use of the word ontology within biology is relatively recent. Figure 1 shows a timeline for the appearance of what we might call ontologies or ontology-like artifacts within bioinformatics. In the early phase, computer scientists had a technique for knowledge representation (from which they build what they call ontologies). They recognized in biological data a domain in which such techniques were needed in order to overcome the massive semantic heterogeneity in the domain [2, 3]. Rich, high-fidelity models of biology, such as can be provided by ontologies, are also seen as a way of providing a means of forming knowledge bases such as EcoCyc [7], RiboWeb [8] and PharmGKB [9]. In TAMBIS (Transparent Access to Multiple Bioinformatics Information Sources), we also see the use of ontologies to form a global schema over multiple heterogeneous resources [10]. Here, ontology forms a mechanism for building queries by using a common ontological form that is mapped to each of the underlying resources. Finally, in this phase, we see the use of ontology as a reference model of what exists in biology. The Molecular Biology Ontology (MBO) [11] was an early attempt to begin to define the entities in the domain to promote consistent interpretation across resources.
Figure 1.
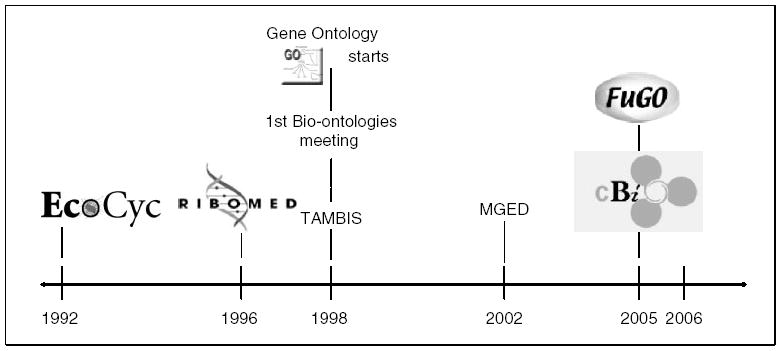
Bio-ontology timeline.
A second phase saw the adoption of ontology by the biological community itself. Pre-eminent among these is the Gene Ontology (GO) [12]. Biologists recognized that, as whole genomes became available, nucleic acid and polypeptide sequence data allowed easy comparative studies. The problem, however, was that, while sequence comparison was easy, comparing functional annotation of those data was hard. In order to address this problem, the mouse, yeast and fly gene communities came together to develop the Gene Ontology (GO). The GO has three aspects or separate ontologies:
Molecular function
Biological process
Cellular component
Together these capture three of the major aspects that biologists wish to describe about the gene products they place in databases. As genome database providers commit to the GO (that is, they agree with its view of the world) and adopt the terminology delivered by the GO, then each resource describes its gene products in a common form. This sharing, together with the structure provided by the relationships between terms in the GO (Figure 2), makes querying of within and between resources possible (Figure 3).
Figure 2.

Representation of the molecular function ‘hexokinase activity’ in the Gene Ontology.
Figure 3.
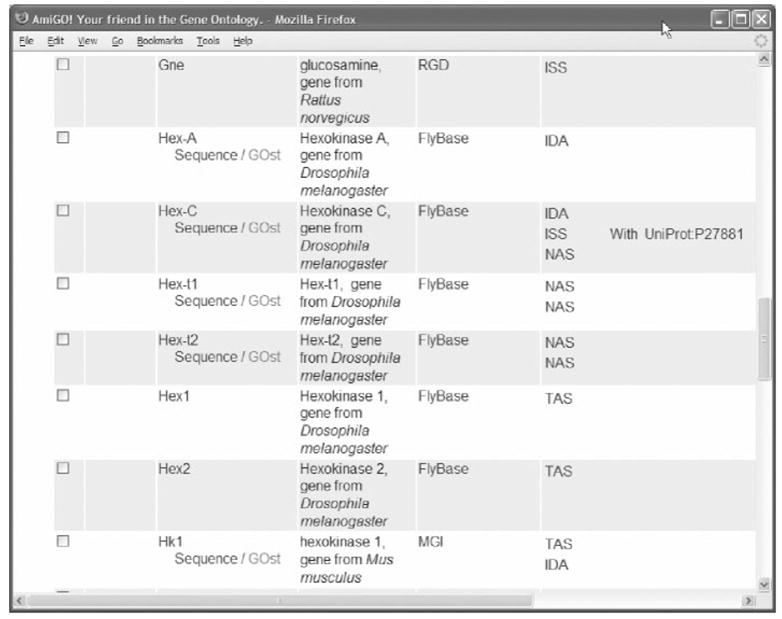
Example of gene products in rat, mouse and fruit fly annotated with the Gene Ontology term ‘hexokinase activity’.
From its start with some 3500 terms in 1998, covering three databases, GO now holds some 20,000 terms and is adopted by about 20 databases. These are largely species-specific genome databases, but also include cross-community resources such as UniProt and InterPro.
The Gene Ontology phenomenon
The Gene Ontology (GO) has been phenomenally successful and it is useful to examine why this has been so. The Gene Ontology has put its success down to the following points [13]:
Community involvement: The development of the GO is a very open process. Response is welcomed from the community that it seeks to serve. It is built by and for biologists. Groups join GO because it suits their needs; this would be less likely to happen with a dictated, unresponsive organization.
Clear goals: The GO had the specific aim of promoting consistent annotation for gene products for the three major functional attributes. While GO has been used for many other purposes, this narrow, clear goal enabled focus to be maintained.
Limited scope: It is obvious that an ontology for the whole of biology would be useful. It is also very impractical. A limited, but very useful scope was able to demonstrate utility. The broadening range of Open Biomedical Ontologies (OBO) is a validation of this approach.
Simple structure: The GO’s use of a simple directed acyclic graph (DAG) was sufficient to its purposes. The OBO language [14] has increased its expressivity over time. Too much too soon was, however, more likely to hamper rather than encourage progress.
Continuous evolution: Our understanding of biology changes and expands. Part of the community engagement is to respond to change and put in place the apparatus to cope with change.
Active curation: As well as the community input, the continuous evolution and necessary maintenance necessitate curators to implement changes.
Early use: As soon as the GO was useful, it was used. Even a relatively small number of gene products with consistent annotation are useful. Again, the spread of use is a validation of this process.
After GO: the ‘OBOization’ of bio-ontologies
The success of the GO in meeting its objectives, its wide uptake by other databases for attributing gene product functionality and finally the use of the GO outside its original use has led to many other groups starting to develop ontologies for database annotation. In order to provide some coordination to these efforts, the OBO consortium was established.
OBO is guided by a set of principles that are used to give coherence to wider ontological efforts across the community:
Openness: All the OBO ontologies are freely available to the community, with appropriate attribution. This encourages usage and community buy-in and effort.
Common representation: This in either the OBO format (http://www.geneontology.org/GO.format.shtml#oboflat) or the Web Ontology Language (OWL) (See http://www.w3.org/TR/owl-features/). This provides common access via open tools. Although not mentioned as part of the criteria, it offers common semantics for knowledge representation. (For more information about representation formalisms, see the ‘New formalisms and tools representing bio-ontologies’ section).
Independence: Lack of replication across separate ontologies encourages combinatorial re-use of ontologies and the interlinking of ontologies via relationships.
Identifiers: Each term should have a semantic-free identifier, the first part of which refers to the originating ontology. This promotes easy management.
Natural language definitions: Terms themselves are often ambiguous, even in the context of their ontology, and definition helps ensure appropriate interpretation. It is usual that arguments over terms are bitter and long, while arguments over definitions are shorter and useful.
Through these simple criteria, the ontology community is attempting not to repeat the errors most of their ontologies have been developed to resolve. That is, the massive syntactic and semantic heterogeneity extant in bioinformatics resources. There are many resources under the OBO umbrella, and most of these are shown in Figure 4, in which OBO have been roughly arranged along a spectrum of genotype to phenotype.
Figure 4.
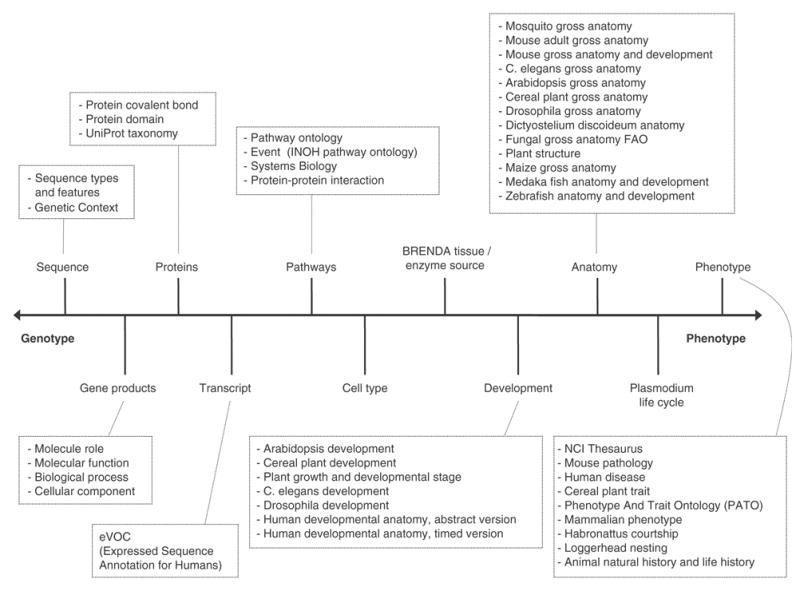
The OBO ontologies arranged on a spectrum of genotype to phenotype, according to their main topic.
The two most significant OBO are the GO [12] and the Sequence Ontology [15]. The former is used to annotate the principle attributes of gene products and the latter provides a vocabulary to describe the features of biological sequences. A common language to describe parts (regions) on nucleic acid and protein sequences across many resources has a potentially huge impact on not only querying but the computational analysis of biological sequence data.
Moving along the spectrum toward phenotype, we see increasing numbers of species ontologies on the same subject: development and anatomy. While the description of sequence features and major attributes of gene products might be core to molecular biology, these descriptions need to be placed in a context. At what stages of development are these sequence features and these gene products important? In what organ, tissue or other anatomical parts are these gene products important? Obviously, each species has its own development and anatomy, but an interesting trend over the coming years will be efforts to explore what different groupings of organisms have in common. In a sense, all explorations of molecular biology are a search for mechanisms that produce a phenotype. As a consequence, we are seeing a general trend towards descriptions of phenotype.
Other OBO ontologies include some that describe experiments that generate biological data. Foremost amongst these is the Microarray Gene Expression Data (MGED) ontology [16]. This ontology provides a vocabulary for describing a biological sample used in an experiment, the treatment that the sample receives in the experiment and the microarray chip technology used in the experiment. This basic information will aid researchers exploring third-party data to validate comparisons between data and help confirm interpretations of data. It is, after all, necessary to know how an experiment was performed in order to interpret findings and make comparison between interpretations. As more high-throughput experimental techniques come into play across the domain, each needing vocabularies, the Functional Genomics Ontology (FUGO) (http://www.fugo.org) has been conceived in order to bring coherence to these ontological developments.
Clinical ontologies
Use of clinical terminologies has a much longer history in medicine. Being able to predict disease outbreak is predicated upon reliable aggregation of statistics on those diseases. Yet, if different communities use different terminologies for the diseases being monitored, then those statistics and hence predictions become unreliable. As long ago as the early 17th century, the authorities in London drew up a list of ‘ways in which people died’. For example, the term ‘French pox’ was used for the same cause of death in each London parish, and consequently more reliable statistics were gathered. The London Bills of Mortality remained in use for many years and not just in London. In the late 1880s, the International Classification of Diseases (ICD) was published. This brought the old Bill of Mortality’s terminology up to date and provided mankind with some 200 ways of dying (what conveniently fitted on two sides of paper). ICD is now in its 10th edition and now has some 13 000 rubrics.
This need for coding is central to the use of terminology in medicine. Originally created for epidemiology purposes, ICD now plays a major role in billing within hospitals. To make this task more complex, several vocabularies have been developed for similar purposes; exactly the problem that the Open Biomedical Ontologies Consortium wishes to avoid. Figure 5 shows the timeline for the appearance of these terminologies. The need for such common, shared means of referred to phenomena of interest has a longer history in medicine, perhaps reflecting its more immediate practical benefit (not dying, for instance). Classification of what we know about the world, the putting of things into categories, is such a natural human activity that no domain can claim its use first. The use of the word ontology, in its computer science usage to denote a means of capturing and sharing a common representation of knowledge, is fairly recent and dates back less than 20 years in both fields.
Figure 5.
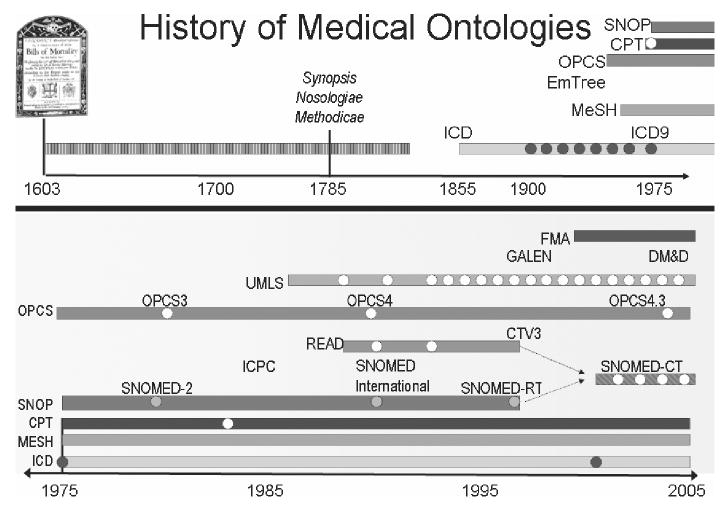
The history of the major players in medical ontologies.
For many years, the ICD was the only medical terminology. In medicine as in biology, the increasing use of information technology and increasing quantities of data have highlighted the need to be able to talk about medicine in a common manner for both humans and machines. It does not take long to think of the consequences of prescribing drugs if inconsistent and confusing terminology is used for drugs, prescribing regimes and side effects. An attempt to make those vocabularies ‘interoperable’ is represented by the Unified Medical Language System® (UMLS®), a terminology integration system comprising over 130 biomedical vocabularies [17]. There is a debate about whether these artifacts are ontology. This is not the forum for that debate, but suffice it to say that these artifacts are structured representations of things in the biomedicine domain.
Figure 6 shows these medical terminologies arranged according to ‘phenome’, or space of observable characteristics and along the ‘prescriptome’ or space of treatments. This movement from left to right transitions from anatomy, physiology and biochemistry (how the normative human organism, or common variants of it, are supposed to work and how they respond to stressors), through symptoms that suggest one or more diseases and further investigations to filter that list, to treatment options with goals and outcomes on the far right.
Figure 6.

The gross subject areas of ontology-like artifacts in medicine arranged in a space from the phenome to the prescriptome.
INSTITUTIONALIZATION OF BIO-ONTOLOGIES
Often referred to as a ‘cottage industry’ by Mark Musen, ontology development was indeed characterized, until recently, by individual researchers modeling knowledge for particular applications, without sophisticated tools or formalisms and independently of existing ontologies. As a result, the ontologies of this era were only minimally sharable and reusable. More recently, the equivalent of an industrial revolution for ontology was marked by the apparition of both new technologies (see section titled ‘New formalisms and tools representing bio-ontologies’) and institutions. It is beyond the scope of this article to give an exhaustive list of ontology centers, even in biomedicine. The institutions presented below were selected because of their impact on the community at large.
IFOMIS
The Institute for Formal Ontology and Medical Information Science (http://www.ifomis.uni-saarland.de/) (IFOMIS) was founded in 2002 with a grant from a German nonprofit foundation, the Alexander von Humboldt Foundation. Directed by Barry Smith, a philosopher, IFOMIS is an interdisciplinary research group, with members from philosophy, computer and information science, logic, medicine, and medical informatics. Over the past years, IFOMIS has contributed to applying formal ontology to biomedicine (e.g. [18]) and has developed collaborations with developers of biomedical ontologies such as the Gene Ontology Consortium and the Structural Informatics Group at the University of Washington.
National Center for Biomedical Ontology
Created as part of the National Centers for Biomedical Computing in 2006 and funded by the National institutes of Health, the National Center for Biomedical Ontology (http://bioontology.org/) (NCBO), led by Mark Musen and Suzanna Lewis, defines itself as ‘a consortium of leading biologists, clinicians, informaticians, and ontologists who develop innovative technology and methods that allow scientists to create, disseminate, and manage biomedical information and knowledge in machine processable form.’ NCBO is now involved in the development of ontologies from the OBO family. The Center draws on the experience of long-time contributors to the field of biomedical ontology, both on the side of the content (with several core members of the Gene Ontology and OBO Consortia—see the section titled ‘The Gene Ontology Phenomenon’) and on the side of the of the tools (with key contributors to the ontology editor and knowledge acquisition system Protégé—see the section titled ‘Protégé’). NCBO is doing much to draw together activity within the biomedical ontology field and to maintain and encourage coherence and perceived best practice in ontology development.
Other ontology centers have been created recently, both in Europe and the US, with a focus on ontological research, but not limited to biomedicine in their applications. The National Center for Ontological Research (http://ncor.us/) (NCOR) was established in 2005 and is codirected by Barry Smith and Mark Musen. The European Center for Ontological Research (http://www.ecor.uni-saarland.de/) (ECOR) was founded in 2004 and is currently directed by Nicola Guarino.
W3C Health Care and Life Sciences Interest Group
Over the past couple of years, the interest of the Semantic Web community (http://www.w3.org/2001/sw/) has shifted in part toward the healthcare and life sciences community [19]. One year after a successful workshop bringing together over 100 biologists, computer scientists and other researchers, the World Wide Web Consortium (W3C) announced the creation of the Health Care and Life Sciences Interest Group (http://www.w3.org/2001/sw/hcls/) in November 2005, ‘‘to develop and support the use of Semantic Web technologies to improve collaboration, research and development, and innovation adoption in the of Health Care and Life Science domains.’’ Several task forces currently address key areas necessary for implementation of a Semantic Web for healthcare and life sciences, for example, the conversion of existing resources into the Semantic Web formalisms RDF (Resource Description Framework) and OWL (Web Ontology Language). Semantic Web technologies are presented in more detail in section 4.3 below.
Bio-ontologies in conferences, journals and books
In the past ten years, bio-ontologies have become ‘mainstream’ in biomedical conferences and the literature. The pioneering workshop in the field was created in 1998 at the Intelligent Systems for Molecular Biology (ISMB) conference (http://www.iscb.org/), and is held annually since. There is now an ontology track at ISMB. A successful session on ‘Biomedical ontologies’ was organized at the Pacific Symposium on Biocomputing (http://psb.stanford.edu/) (PSB) for three years (2003–2005). Similarly, the number of presentations on ontology has regularly increased at medical informatics conferences such as the American Medical Informatics Association (http://www.amia.org/) (AMIA) Annual Symposium, the Medical Informatics Europe (MIE) organized by the European Federation for Medical Informatics (http://www.efmi.org/) (EFMI), and Medinfo, organized by the International Medical Informatics Association (http://www.imia.org/) (IMIA).
As shown in Figure 7, the number of articles on ontology has grown exponentially in PubMed/ Medline, from less than 10 in 1996 to almost 500 in 2005. Noticeably, over half of the growth is attributable to the GO. Bio-ontologies appear in the literature through permanent sections and special issues. For example, the leading journal Bioinformatics has an ontology section. Recently, two major medical informatics journals have devoted a special issue to bio-ontologies. Issues 7–8 of Computers in Biology and Medicine (Volumes 36, 200, July–August 2006) present 14 papers on various aspects of biomedical ontology [20–33], ranging from ontology development, evaluation and mapping to the use of ontologies for ontology integration, semantic similarity computation and task modeling. Also presented are ontologies for specialized domains including public health, colon carcinoma, adverse drug reactions and heart failure. Issue 3 of the Journal of Biomedical Informatics (Volume 39, June 2006) is a collection of 10 papers presented at the 2005 meeting of the International Medical Informatics Association Working Group 6 [5, 34–43]. This series of articles offers a more formal perspective on biomedical ontologies, discussing issues such as reality, granularity, mereology and reference ontologies. Together, these two journal issues provide a panorama of bio-ontologies, with foundational issues and practical aspects.
Figure 7.
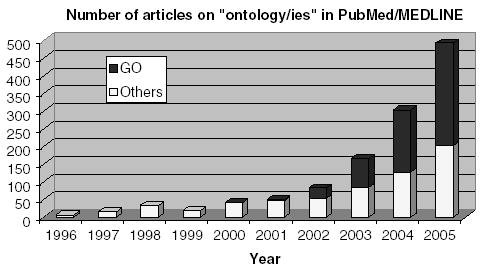
Growth of ontology papers in PubMed/ Medline.
The book Ontologies for Bioinformatics [44] published in 2005 provides a good technological overview of bio-ontologies in the context of the Semantic Web. The introduction to ontologies puts a strong emphasis on the Semantic Web technologies (see the section titled ‘Semantic web technologies’), with examples from bioinformatics. The chapters devoted to ‘Building and using ontologies’ also present query languages and transformation methods based on XML. The last part of the book is an introduction to Bayesian networks. As this summary suggests, this book takes an extremely broad view of ontology, even including XML schema. Also of interest to bioinformaticians is the Handbook on Ontologies [45], presenting ontology from the perspective of computer science rather than bioinformatics. Beside the expected chapters on ontology languages and ontology engineering, the Handbook is also relevant to our community with chapters on building ontologies from medical thesauri [46] and ontologies in bioinformatics [47]. Finally, Ontologies in Medicine [48] is a collection of nine papers reporting on issues in and applications of ontologies in the medical domain.
NEW FORMALISMS AND TOOLS FOR REPRESENTING BIO-ONTOLOGIES
Biomedical terminologies are typically large, covering tens to hundreds of thousands of entities (e.g. about 20 000 for the GO and 300 000 for SNOMED Clinical Terms). Until recently, no widely used ontology development environments (as opposed to ontology editors, to use a software development analogy) were available and ontologies were developed essentially ‘by hand’ or with rudimentary tools such as file-system-like tree editors. In the past 15 years, Protégé has emerged as the leading ontology editor across disciplines. At the same time, description logics (DL) have superseded frame-based languages to become the leading formalism for representing ontologies. Finally, Semantic Web technologies are playing an increasing role in knowledge representation. This cross-discipline view is in contrast to that in bioinformatics and medical informatics. Within bio-ontology, in-house tools have been developed by the Gene Ontology Consortium in the form of DAG-Edit and latterly OBO-Edit. Medical informatics has used a variety of tools, either proprietary or open-source. In this section, we briefly review some knowledge representations and ontology development tools.
Protégé
Developed by the Stanford Medical Informatics group with funding from various US Government agencies in the past 15 years (and now a core technology of the National Center for Biomedical Ontology), Protégé (http://protege.stanford.edu/) is the leading ontology editor across disciplines, with a community of about 50 000 users, representing research and industrial projects in more than 100 countries. Originally developed for representing frame-based ontologies, in accordance with the Open Knowledge Base Connectivity (OKBC) protocol, Protégé has evolved, in collaboration with the University of Manchester, to represent ontologies in the OWL, based on description logics. Many large biomedical ontologies have adopted Protégé for their representation, including the Foundational Model of Anatomy (frame-based) and the NCI Thesaurus (DL-based), though Protégé is not used for the majority of OBO ontologies. Beside the support of OWL, recent changes for Protégé include support for exporting Protégé ontologies into a variety of formats (e.g. RDF/S, OWL and XML Schema—see the section titled ‘semantic web technologies’). Based on an open architecture, Protégé can be extended through plug-in components, some of which are contributed by users. Examples of services provided through the 69 plug-ins currently available for Protégé include ontology visualization (OntoViz), ontology alignment (PROMPT) and interfaces with rule engines (e.g., Jess http://www.jessrules.com/jess/index.shtml) and formalisms (e.g. SWRL—the Semantic Web Rule Language (http://www.w3.org/Submission/SWRL/)).
Description logics
It is beyond the scope of this article to give a detailed introduction to description logics. (The interested reader is referred to [49] for more information). Instead, we will show why they have emerged as a popular ontology language in biomedicine and other domains. Intuitively, highly expressive knowledge representation formalisms such as first-order logic (FOL) could be thought of as ideal for ontologies. In practice, however, FOL is also intractable, or, more simply, too complex to be computed. Description logics represent a family of languages defined as a trade-off between expressivity and tractability. The aforementioned OWL can be used to illustrate this trade-off. OWL actually comes in three varieties of decreasing expressivity but of increasing tractability: OWL Full, OWL DL and OWL Lite [50].
DLs are usually considered sufficiently expressive to represent most biomedical ontologies. The first large biomedical ontology developed with description logics was GALEN—the Generalized Architecture for Languages, Encyclopedias and Nomenclatures in medicine. The development of GALEN started in the early 1990s—before the times of the Semantic Web—and its authors started by designing a DL-based language for representing medical knowledge: GRAIL, the GALEN Representation And Integration Language [51]. Another important milestone in the use of DLs for developing biomedical terminologies is the creation of SNOMED Clinical Terms (SNOMED CT). Not only did SNOMED CT result from merging two major clinical terminologies—SNOMED Reference Terminology (SNOMED RT) and Clinical Terms Version 3 (formerly known as the Read Codes), but it was also engineered using a different technology: a DL-based authoring system developed by Apelon (http://www.apelon.com/). Other large biomedical terminologies such as the NCI Thesaurus have recently adopted OWL for their representation [52]. With OWL DL becoming a de facto standard ontology language, many attempts to convert existing terminologies and ontologies into OWL DL have taken place recently (e.g. MeSH [53]). However, in most cases, converting to OWL DL is not simply a matter of syntactic translation: information implicit in the formalism of origin may need to be made explicit in OWL DL in order to fully take advantage of the possibilities offered by the language, which often requires enriching the original representation [54, 55].
Semantic Web technologies
In addition to contributing to specialized domains such as healthcare and life sciences, the World Wide Web Consortium (W3C) creates the very infrastructure of the Semantic Web. The W3C originally developed the specifications of HTML, the markup language used to represent documents in the World Wide Web. Similarly, the W3C produced the specifications of other formalisms for representing documents, resources and ontologies, including XML, RDF/S, OWL. Collectively know as Semantic Web technologies, these specifications define the building blocks of the Semantic Web. Building upon them, additional formalisms are defined to represent, for example, rules. Some of these technologies will be briefly reviewed, with emphasis on their relations to biomedical applications. The interested reader is referred to the corresponding chapters in [44] for further information.
The Resource Description Framework (RDF) extends the capabilities of the extensible markup language XML as it enables many-to-many relationships between resources and data. The resulting structure is a graph in which the nodes are resources (identified by a Uniform Resource Identifier or URI) or data (e.g. strings, numerals) and the edges are relationships (called properties). RDF integrates limited inference rules, enabling, for example, to define subclasses and subproperties. Some extensive resources such as UniProt have already been converted to RDF (http://expasy3.isb-sib.ch/~ejain/rdf/) The BioRDF (http://esw.w3.org/topic/HCLSIG_BioRDF_Subgroup) task force of the W3C Semantic Web Health Care and Life Sciences Interest Group currently investigates methods whereby existing resources can be converted to RDF.
OWL plays a central role in bio-ontologies and has been mentioned multiple times already. OWL DL, the description logic flavor of OWL, is particularly well suited for representing bio-ontologies. In addition to many bio-ontologies, BioPAX (http://www.biopax.org/) a data exchange format for biological pathway data, uses OWL for its representation.
The inference supported by RDF and OWL is limited compared to rule-based languages. For example, clinical decision support systems typically require complex knowledge better expressed with rules. The role of ontologies in this context is to provide the vocabulary used in the rules. The Arden Syntax is one example of formalisms developed for representing rules supporting medical practice (e.g. drug interactions). Recent efforts related to Semantic Web technologies include SWRL (http://www.w3.org/Submission/SWRL/) and the rule markup language RuleML (http://www.ruleml.org/).
Formalisms and tools specific to bio-ontologies
Some formalisms and tools have been developed specifically by the bio-ontology community, where they enjoy great popularity. OBO-Edit (http://www.geneontology.org/GO.sourceforge.links.shtml#obo) is ‘an open source, platform-independent application for viewing and editing OBO ontologies’. Formerly known as DAG-Edit, OBO-Edit is a tool for visualizing and editing the graph structure of an ontology. The OBO format is used to represent the majority of the ontologies seen in Figure 4. It is a large subset of that expressivity allowed in OWL (see the section titled Limitations). It allows the creation of types, subtype relationships and other kinds of relationship. It can express disjunction of types and features of relationships such as transitivity, symmetry, etc. It does not express, for example, quantification in relationships nor does it allow expressions to be built using types. Conversely, the OBO format has several built-in features for supporting terminology, as opposed to ontology, that OWL does not. It has built-in support for thesaurus constructs and semantic-free identifiers. It also has mechanisms for supporting view-like mechanisms over a terminology.
As illustrated in Figure 8, the OBO format is informally expressed, but its extensive documentation (http://www.geneontology.org/GO.format.shtml#oboflat) can be used to derive the language semantics which means it can be converted into OWL (that is, the semantics of the language are the same). Indeed, the GO has provided an OWL translation of its ontologies for many years. The directed acyclic graph used by the GO is a subset of the OBO format.
Figure 8.
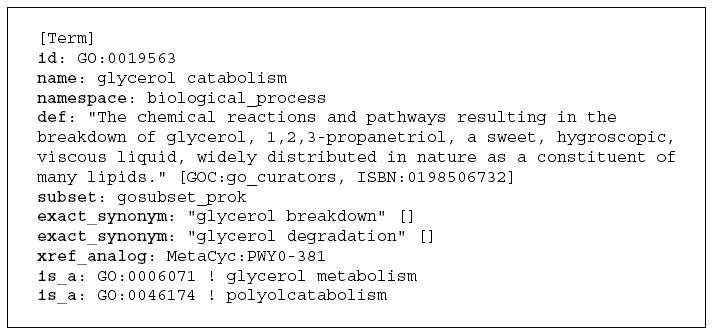
Representation of the Gene Ontology term ‘‘glycerol catabolism’’ in the OBO format.
Seen in the context of how GO and OBO have developed (see the section titled ‘The Gene Onthology Phenomenon’), the development of the language and its tools have been central to the success of biologists’ uptake of ontology. It should be remembered that representations such as OWL are more recent additions to the catalog of representations and their use is still being explored. In addition, the OBO community has paid more attention to the needs of a biologist type of user than the knowledge representation specialist in, for instance, the OWL tools.
Apart from DAG-Edit, the Gene Ontology Consortium and the wider community have built a wide range of tools and resources, such as AmiGO (see Figures 2 and Figure 3), that allow display and querying of the GO and annotations stored in a specialist GO database. Further tools allow searching GO, annotating data using GO, and microarray analysis. A catalog of these tools can be found at the Gene Ontology Web site (http://www.geneontology.org).
COBrA is another ontology editor developed within the bioinformatics community, this time by a group interested in developmental anatomy [56]. COBrA has the standard editing features and can export to both OBO format and Semantic Web languages. It is distinguished by giving prominence to the formation of links between ontologies, for instance, joining a tissue type to a cell type. As various ontologies, especially those in OBO, become cross-linked, such features as the support of modularization in ontologies will become of increasing importance.
CONTRIBUTION OF FORMAL ONTOLOGY TO BIO-ONTOLOGIES
Formal ontology stems from philosophy and provides a rigorous framework for understanding and representing differences between entities. Counter intuitively, formal ontology is not the same as the formal languages used to represent ontologies. Namely, an ontology expressed in a formal language such as OWL does not necessarily adhere to the principles of formal ontology, although the formality of the language can help in making ontological distinctions. This section briefly reviews some important formal ontological distinctions and properties and their applications. The notions of top-level ontology and reference ontology are presented next. We then emphasize the importance of relations in bio-ontologies before illustrating some of the current limitations of formal languages used in bio-ontologies.
Formal-ontological distinctions and properties
Important formal ontological distinctions include the difference between continuants, which continue to exist through time and occurrents (or processes), which unfold through time in successive phases. Continuants are themselves divided into dependent and independent continuants, based on whether or not they require the existence of any other entity in order to exist. Occurrents always depend on some independent continuant. For example, the process oxygen transport and the dependent continuant oxygen transporter both depend on the independent continuant oxygen. These distinctions, along with metaproperties such as identity, rigidity, unity and dependency form the basis for OntoClean, a methodology for analyzing and validating ontologies [57].
In search of a top-level ontology
The top-level distinctions presented in the preceding text can be used as the basis for creating top-level (or upper-level) ontologies, i.e. ontologies in which high-level categories are defined. All entities and processes constitutive of a particular domain can then be defined in reference to (e.g. as subclasses of) these top-level categories. As mundane as it might seem to biologists, upper-level ontologies end up being discussed in mainstream biology journals (e.g. [58]). To date, it is probably fair to say that there has not been an agreement yet on what constitutes a good top-level ontology. Candidates include the Basic Formal Ontology (BFO), the Descriptive Ontology for Linguistic and Cognitive Engineering (DOLCE) and the Suggested Upper Merged Ontology (SUMO). The UMLS Semantic Network (http://semanticnetwork.nlm.nih.gov/) is sometimes regarded as an upper-level ontology for the biomedical domain [59].
Domain reference ontologies
Ontologies defined independently of specific objectives are often referred to as reference ontologies. By definition, top-level ontologies should be reference ontologies as they constitute the top-level structure of many domain ontologies. However, the notion of reference ontology can be extended to domain ontologies [41]. For example, the Foundational Model of Anatomy (FMA), a reference ontology of structural anatomy has been proposed as a reference for describing physiology and pathology [60]. More generally, cell types and chemical entities are often referred to in other entities such as cytotoxicTcell differentiation and 6-alpha-maltosylglucose catabolism. Ontologies of cell types (e.g. the OBO cell ontology [61]) and chemical entities (e.g. ChEBI—the Chemical Entities of Biological Interest) could be used as a reference and guide the development of the ontology of biological processes in the GO. This strategy is being implemented progressively by the Gene Ontology Consortium, in part through the Obol language [14].
OBO relations
The semantics of the relations used in most biomedical terminologies are weak. For example, in the Medical Subject Headings (MeSH), the semantics of A narrower than B simply means that users interested in Bs might also be interested in As. The MeSH terms found under Accidents include kinds of accidents—as expected (e.g. Traffic accidents), but also Accident prevention. In contrast, A isa B implies that all As are also Bs, i.e. that A necessarily inherits all the properties of B. The publication of the OBO relations [62] therefore represents an important contribution to bio-ontologies. This article defines 10 relations: isa, part_of, located_in, contained_in, adjacent_to, transformation_of, derives_from, preceded_by, has_participant and has_agent. Interestingly, these relations were defined and agreed upon by a multidisciplinary group including philosophers, physicians, biologists and computer scientists. Logical definitions are provided for each relation and relations are defined at both class and instance levels whenever appropriate. This core set of relations has been proposed for use in the OBO family of ontologies. Moreover, some relations such as has_participant and has_agent are defined in reference to formal ontological distinctions between continuants (e.g. the lungs) and processes (e.g. breathing), the processes having continuants as their agents or participants.
Limitations
Formality, both in the ontological and representation language sense, is a stern friend. A formal language has a well-defined interpretation of the world and a well-defined language with which to say things about that world [63]. The OBO relations, described in the preceding text, take a standard logical view of binary relationships [63] and describe a world with binary relationships between individuals (instances of a class). Expressed in the OWL [64], each and every instance of a class must hold such a relationship (or none at all hold the relationship). In this sense, OWL talks about universals. These instances form sets or classes. Subclass relationships can hold and, by implication, every instance in a subclass must also be an instance of its superclass.
In OWL, we can place some kind of quantification on what goes at the other end of a relationship (its successor). It is possible to say there is at least one successor (existential quantification) or that an instance of a class of objects is the only kind of instance that may appear as a successor (universal quantification). In OWL, a modeler can use these constructions to describe restrictions on what instances may be members of a class. These conditions can be of two types:
Necessary conditions are those an instance must hold to be a member of a particular class. It is, however, possible for a member to hold that condition and not be a member of that class.
Necessary and sufficient conditions: are such that any object or individual holding those conditions can be recognized to be a member of a particular class and not other classes.
For example, the OWL class expression in Figure 9 shows a complete definition for a ReceptorProteinTyrosinePhosphatase. These OWL statements state that a ReceptorProteinTyrosinePhosphatase is any protein that, among other things, has at least one TyrosinePhosphataseCatalyticDomain and at least one TransmembraneDomain. Any protein having these features can be recognized to be a member of this class of protein phosphatase. Note the phrase ‘among other things’—OWL has an assumption of an open world. Just because our description does not mention other sequence features or domains that are possible, or functions, substrates, processes, etc. it does not mean there are none—we simply have not mentioned them. OWL explicitly states what is known, whether to the positive or negative. Unless explicitly stated, the model simply does not know. As there is much we do not know about biology, OWL’s open-world assumption can make a lot of sense.
Figure 9.
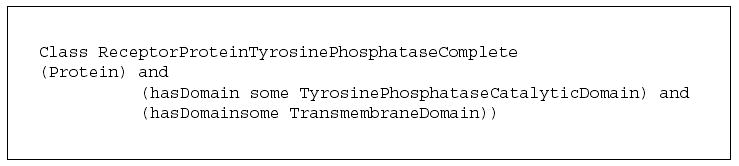
A complete definition of ReceptorProteinTyrosinePhosphatase.
This is only a subset of OWL’s expressivity. An ontologist can use statements in OWL to create ontologies that have precise meanings; precise enough such that a machine can reason over those statements and make inferences [65–67]. As such, the formality (strictness) of the language is good—this precision means that automatic reasoning with the symbols of the ontology can take place. This very strictness is, however, potentially restrictive. OWL has many limitations in what it offers an ontologist [63]:
Only binary relationships are possible between instances and in the natural world relationships of higher degree are possible.
OWL takes a static view of the world and is restricted in how it models temporal aspects.
In OWL’s view of the world there is a lack of fuzziness (or modality). Biologists like words such as ‘mostly’, ‘usually’, etc. and OWL’s two-valued logic, where relationships are universally held is illmatched with this fuzzy view. One argument is, however, that biologists might well model what is true, rather than to model what is not.
OWL allows no exceptions. Again, biologists often wish to model the ‘typical’ case, which is allowed in knowledge representation languages such as Frames [68].
For a more detailed analysis of OWL limitations, the interested reader is referred to [63].
Within its limitations, OWL has patterns that can provide ways to model, for instance, n-ary relationship, lists and exceptions [63]. There are, however, large islands of biology that can be modeled with great success using OWL. The formality of the language not only means that machines are able to use the ontology to make inferences [66, 67, 69, 70], but the formality also makes an ontologist ask hard ontological questions about what he or she is modeling; in this sense ontology and language formality are linked.
POSSIBLE DIRECTIONS FOR THE FUTURE
The successes and, more generally, the developments observed in the field of bio-ontologies over the past 5 years certainly make sense in today’s context, but would not necessarily have been easily predictable. In the rest of this article, we take the risk of outlining some directions, which, in our opinion, may shape biomedical ontology in the years to come.
‘Guruization’ of bio-ontology: the end of an era
Like many new domains, biomedical ontology is still as much an art as it is a science. Methods are just emerging, and beliefs and doctrine make up for the lack of objective metrics for evaluating quality. To the casual observer, being assertive and charismatic seems to be all it takes at this early stage to become a guru in the field of bio-ontology. Would-be ontologists are eager to embrace bio-ontologies and become disciples. We are, however, near the end of this era. More than individuals, multidisciplinary fora, such as the National Center for Biomedical Ontology, will now shape the discipline. Biologists interested in ontologies for their usefulness also increasingly recognize the importance of rigor in building these ontologies. As scientific techniques become available for building ontologies [71], and as objective metrics are developed for measuring their quality [31, 72, 73], today’s gurus who have contributed to promoting such techniques will be remembered as visionaries in the history of bio-ontologies.
Ontology validation and certification
Not all ontologies are equal. As mentioned earlier, some ontologies called reference ontologies, representing a limited domain rigorously and consistently, can serve as a reference for developing domain and application ontologies. A new organization, the OBO Foundry (http://obofoundry.org/) promotes guidelines for ontology development in relation to the OBO family of ontologies. It also selects ‘high-quality’ ontologies and promotes their use as reference ontologies within the OBO family. This certification process relies on objective metrics for evaluating bio-ontologies, most of which are still to be defined.
Ontological needs for tomorrow
By and large, there has been a wide use of ontology to generate vocabularies with which to describe biomedical data. With the increasing volumes of data—it is hoped, described data—there is an increasing need to automate analyses of these data. The precise capture of biological knowledge in a computational form means it is possible to compute with knowledge as we compute with continuous mathematics and strings. To accommodate this severe need, there needs to be an increase in formality and richness in biomedical ontologies.
We can see in Figure 4 the expansion of topics covered in bio-ontologies. At present these are orthogonal, but implicit relationships are implicit between, for example, GO’s biological processes and ChEBI’s chemicals. Plans exist to formalize this cross-linking, and this trend will increase. This will help querying of data, analyzing data and also help in the building and maintenance of the ontologies themselves.
We also expect to see medical research and biological research join ontologies at the level of anatomy, drugs, etc. The two communities might need different granularities or even different views of the same ontology, but they have interests in common.
Collaborative development and curation of bio-ontologies
With the success of collaborative resources such as Wikipedia (http://en.wikipedia.org/wiki/Main_Page), the computer science community is increasingly interested in the social organization supported by the Web and Semantic Web technologies (see, for instance, the 2006 World Wide Web conference (http://www2006.org/)). Extended to ontologies, the notion of Semantic Wikipedia has arisen [74]. Harnessing the knowledge resource of the community, to an even greater extent than has been seen with the GO, has the potential to shift knowledge gathering and defining from a small community of experts to a larger number of ‘eyeballs’, i.e. to knowledgeable scientists who would not be otherwise involved with ontology development for geographical or other reasons. Experiments of collaborative development of biomedical ontologies have been reported already (see, e.g. [75]). While it is still unclear as to what the correct framework for collaborative development is or whether it will even work, this phenomenon should certainly not be ignored without investigation.
Analogously, a collaborative approach could be used also for the curation of bio-ontologies. Every assertion in an ontology could be commented upon by users, and the result of such critical evaluation can be recorded on the form of annotations added to the ontology. Early implementations of this emerging trend are becoming available [76]. There are obvious dangers to such an approach, in building, curating and annotation, but this approach has potential, especially where funding is scarce. In such a situation, where a community decides if an ontology is necessary, a decision has to be made about whether something is better than nothing.
Key Points
Use of ontology within biomedicine is now a mainstream activity.
There is a recognized need to be able to compute with the knowledge component that is vital to biology and medical research.
The widespread uptake of the technique has now led to the institutionalization of the activity in national centers.
There is a growing formality within the resources being developed, both ontologically and in their representation languages.
In biology in particular, ontologies have largely been used to deliver vocabularies for describing data. The future will see greater analysis of data due to increasing formality of these ontologies.
This formality will also see the growth of reference ontologies in biomedicine.
Acknowledgments
R. S. would like to acknowledge funding by the Sealife project (IST-2006-027269). This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM).
Appendix 1
See Appendix Table
Contributor Information
Olivier Bodenreider, Staff Scientist in the Cognitive Science Branch of the Lister Hill National Center for Biomedical Communications at the U.S. National Library of Medicine. His research interests include terminology, knowledge representation and ontology in the biomedical domain, both from a theoretical perspective and in their application to natural language understanding, reasoning, information visualization and interoperability.
Robert Stevens, Senior Lecturer in bioinformatics in the School of Computer Science, University of Manchester, U.K. He has degrees in biochemistry, biological computation and computer science. He was a member of the ground breaking TAMBIS project, which was the first in bioinformatics to use description logic ontology to form a homogenizing query layer over bioinformatics resources. Interest in the use of formal ontology has continued in the development of semantic similarity metrics over ontologically annotated corpora. His other work includes the development of methodologies to migrate ontologies from the informal to formal and use reasoning to increase structural validity. His current work includes the use of protein family ontologies to catalog proteins in genomes and the use of ontologies to describe in silico experiments. He has co-chaired the annual bio-ontologies meeting at ISMB for many years and is a co-developer of a highly successful OWL training course..
References
- 1.Stevens R, Goble CA, Bechhofer S. Ontology-based knowledge representation for bioinformatics. Brief Bioinformatics. 2000;1:398–416. doi: 10.1093/bib/1.4.398. [DOI] [PubMed] [Google Scholar]
- 2.Davidson SB, Overton C, Buneman P. Challenges in integrating biological data sources. Comput Biol. 1995;2:557–72. doi: 10.1089/cmb.1995.2.557. [DOI] [PubMed] [Google Scholar]
- 3.Karp P. A strategy for database interoperation. Comput Biol. 1995;2:573–86. doi: 10.1089/cmb.1995.2.573. [DOI] [PubMed] [Google Scholar]
- 4.Gruber TR. The role of common ontology in achieving sharable, reusable knowledge bases. In: Allen JF, Fikes R, Sandewall E, editors. Proceedings of KR’ 1991: Principles of Knowledge Representation and Reasoning. San Mateo, California: Morgan Kaufmann; 1991. pp. 601–2. [Google Scholar]
- 5.McCray AT. Conceptualizing the world: lessons from history. J Biomed Inform. 2006;39:267–73. doi: 10.1016/j.jbi.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Guarino N. Formal ontology in information systems. Proc FOIS’98. 1998:3–15. [Google Scholar]
- 7.Karp PD, Riley M, Saier M, et al. The EcoCyc database. Nucl Acids Res. 2002;30:56–58. doi: 10.1093/nar/30.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altman R, Bada M, Chai XJ, Carillo MW, Chen RO, Abernethy NF. RiboWeb: an ontology-based system for collaborative molecular biology. IEEE Intell Syst. 1999;14:68–76. [Google Scholar]
- 9.Klein TE, Chang JT, Cho MK, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenomics. 2001;1:167–70. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 10.Goble CA, Stevens R, Ng G, et al. Transparent Access to Multiple Bioinformatics Information Sources. IBM Syst Special Issue on Deep Computing for the Life. Sciences. 2001;40:532–52. [Google Scholar]
- 11.Schulze-Kremer S. Adding semantics to genome databases: towards an ontology for molecular biology; Proceedings of the Fifth International Conference for Intelligent Systems for Molecular Biology Conference (ISMB); 1997. pp. 272–5. [PubMed] [Google Scholar]
- 12.Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucl Acids Res . 2006;34:D322–6. doi: 10.1093/nar/gkj021. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bada M, Stevens R, Goble C, et al. A short study on the success of the Gene Ontology. J Web Semantics. 2004;1:235–40. [Google Scholar]
- 14.Mungall CJ. Obol: integrating language and meaning in bio-ontologies. Comp Funct Genomics. 2004;5:509–20. doi: 10.1002/cfg.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eilbeck K, Lewis SE, Mungall CJ, et al. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whetzel PL, Parkinson H, Causton HC, et al. The MGED Ontology: a resource for semantics-based description of microarray experiments. Bioinformatics. 2006;22:866–73. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]
- 17.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–70. doi: 10.1093/nar/gkh061. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donnelly M, Bittner T, Rosse C. A formal theory for spatial representation and reasoning in biomedical ontologies. Artif Intell Med. 2006;36:1–27. doi: 10.1016/j.artmed.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Goble C, Stevens R, Bechhofer S. The Semantic Web and Knowledge Grids BioSilico. Drug Disco Today. 2005 doi: 10.1016/j.ddtec.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Kumar A, Yip YL, Smith B, Grenon P. Bridging the gap between medical and bioinformatics: an ontological case study in colon carcinoma. Comput Biol Med. 2006;36:694–711. doi: 10.1016/j.compbiomed.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Charlet J, Bachimont B, Jaulent MC. Building medical ontologies by terminology extraction from texts: an experiment for the intensive care units. Comput Biol Med. 2006;36:857–70. doi: 10.1016/j.compbiomed.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Steichen O, Le Bozec CD, Thieu M, Zapletal E, Jaulent MC. Computation of semantic similarity within an ontology of breast pathology to assist inter-observer consensus. Comput Biol Med. 2006;36:768–88. doi: 10.1016/j.compbiomed.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Henegar C, Bousquet C, Lillo-Le Louet A, Degoulet P, Jaulent MC. Building an ontology of adverse drug reactions for automated signal generation in pharmacovigilance. Comput Biol Med. 2006;36:748–67. doi: 10.1016/j.compbiomed.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Pinciroli F, Pisanelli DM. The unexpected high practical value of medical ontologies. Comput Biol Med. 2006;36:669–73. doi: 10.1016/j.compbiomed.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Surjan G, Szilagyi E, Kovats T. A pilot ontological model of public health indicators. Comput Biol Med. 2006;36:802–16. doi: 10.1016/j.compbiomed.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Eccher C, Purin B, Pisanelli DM, Battaglia M, Apolloni I, Forti S. Ontologies supporting continuity of care: the case of heart failure. Comput Biol Med. 2006;36:789–801. doi: 10.1016/j.compbiomed.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Fox J, Alabassi A, Patkar V, Rose T, Black E. An ontological approach to modelling tasks and goals. Comput Biol Med. 2006;36:837–56. doi: 10.1016/j.compbiomed.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Masseroli M, Pinciroli F. Using Gene Ontology and genomic controlled vocabularies to analyze high-throughout gene lists: Three tool comparison. Comput Biol Med. 2006;36:731–47. doi: 10.1016/j.compbiomed.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Dieng-Kuntz R, Minier D, Ruzicka M, Corby F, Corby O, Alamarguy L. Building and using a medical ontology for knowledge management and cooperative work in a health care network. Comput Biol Med. 2006;36:871–92. doi: 10.1016/j.compbiomed.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 30.Lee Y, Supekar K, Geller J. Ontology integration: experience with medical terminologies. Comput Biol Med. 2006;36:893–919. doi: 10.1016/j.compbiomed.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 31.Zhang S, Bodenreider O. Law and order: assessing and enforcing compliance with ontological modeling principles in the Foundational Model of Anatomy. Comput Biol Med. 2006;36:674–93. doi: 10.1016/j.compbiomed.2005.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Rey D, Maojo V, Garcia-Remesal M, et al. ONTOFUSION: Ontology-based integration of genomic and clinical databases. Comput Biol Med. 2006;36:712–30. doi: 10.1016/j.compbiomed.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Orgun B. Vu J, HL7 ontology and mobile agents for interoperability in heterogeneous medical information systems. Comput Biol Med. 2006;36:817–36. doi: 10.1016/j.compbiomed.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 34.Cimino JJ, Smith B. Introduction: International Medical Informatics Association Working Group 6 and the 2005 Rome Conference. J Biomed Inform. 2006;39:249–51. [PMID: 16580886] [Google Scholar]
- 35.Blake JA, Bult CJ. Beyond the data deluge: data integration and bio-ontologies. J Biomed Inform. 2006;39:314–20. doi: 10.1016/j.jbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Rector A, Rogers J, Bittner T. Granularity, scale and collectivity: when size does and does not matter. J Biomed Inform. 2006;39:333–49. doi: 10.1016/j.jbi.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Schulz S, Kumar A, Bittner T. Biomedical ontologies: what part-of is and isn’t. J Biomed Inform. 2006;39:350–61. doi: 10.1016/j.jbi.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Cimino JJ. In defense of the Desiderata. J Biomed Inform. 2006;39:299–06. doi: 10.1016/j.jbi.2005.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fellbaum C, Hahn U, Smith B. Towards new information resources for public health—from WordNet to MedicalWordNet. J Biomed Inform. 2006;39:321–32. doi: 10.1016/j.jbi.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 40.Smith B. From concepts to clinical reality: an essay on the benchmarking of biomedical terminologies. J Biomed Inform. 2006;39:288–98. doi: 10.1016/j.jbi.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Burgun A. Desiderata for domain reference ontologies in biomedicine. J Biomed Inform. 2006;39:307–13. doi: 10.1016/j.jbi.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Ceusters W, Smith B. Strategies for referent tracking in electronic health records. J Biomed Inform. 2006;39:362–78. doi: 10.1016/j.jbi.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Johansson I. Bioinformatics and biological reality. J Biomed Inform. 2006;39:274–87. doi: 10.1016/j.jbi.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Baclawski K, Niu T. Computational molecular biology. Cambridge, MA: MIT Press; 2005. Ontologies for bioinformatics. [Google Scholar]
- 45.Staab S, Studer R. International Handbooks on Information Systems. Berlin: Springer; 2004. Handbook on Ontologies; p. 660. [Google Scholar]
- 46.Hahn U, Schulz S, Staab S, Studer R, editors. Building a very large ontology from medical thesauri, in Handbook on ontologies. Berlin; New York: Springer; 2004. pp. 133–50. [Google Scholar]
- 47.Stevens R, Wroe C, Lord P, Goble C. In: Ontologies in bioinformatics. Handbook on ontologies. Staab S, Studer R, editors. Berlin; New York: Springer; 2004. pp. 635–57. [Google Scholar]
- 48.Pisanelli DM. Studies in Health Technology and Informatics. Vol. 102. Amsterdam, Burke, VA: IOS Press; 2004. Ontologies in Medicine; p. 165. [Google Scholar]
- 49.Baader F, Horrocks I, Sattler U. In: Description logics. Handbook on ontologies. Staab S, Studer R, editors. Berlin; New York: Springer; 2004. pp. 3–28. [Google Scholar]
- 50.Antoniou G, van Harmelen F. Web Ontology Language: OWL. In: Staab S, Studer R, editors. Handbook on ontologies. Berlin; New York: Springer; 2004. pp. 67–92. [Google Scholar]
- 51.Rector AL, Bechhofer S, Goble CA, Horrocks I, Nowlan WA, Soloman WD. The GRAIL concept modelling language for medical terminology. Artif Intell Med. 1997;9:139–71. doi: 10.1016/s0933-3657(96)00369-7. [DOI] [PubMed] [Google Scholar]
- 52.Golbeck J, Fragoso G, Hartel F, Hendler J, Oberthaler J, Parsia B. The National Cancer Institute’s thesaurus and ontology. Journal of web Semantics. 2003;1:75–80. [Google Scholar]
- 53.Soualmia L, Golbreich C, Darmoni S. Representing the MeSH in OWL: Towards a semi-automatic migration. Proceedings of the KR 2004 Workshop on Formal Biomedical Knowledge Representation. 2004:81–87. http://CEUR-WS.org/Vol-102/soualmia.pdf.
- 54.Wroe CJ, Stevens R, Goble CA, Ashburner M. A methodology to migrate the Gene Ontology to a description logic environment using DAML+OIL. Pac Symp Biocomput. 2003:624–35. doi: 10.1142/9789812776303_0058. [DOI] [PubMed] [Google Scholar]
- 55.Zhang S, Bodenreider O, Golbreich C. Experience in reasoning with the Foundational Model of Anatomy in OWL DL. In: Altman RB, et al., editors. Pacific Symposium on Biocomputing 2006. World Scientific; 2006. pp. 200–11. [PMC free article] [PubMed] [Google Scholar]
- 56.Aitken S, Korf R, Webber B, Bard J. COBrA: a bio-ontology editor. Bioinformatics. 2005;21:825–6. doi: 10.1093/bioinformatics/bti097. [DOI] [PubMed] [Google Scholar]
- 57.Guarino N, Welty C. In: An overview of OntoClean. Handbook on ontologies. Staab S, Studer R, editors. Berlin; New York: Springer; 2004. pp. 151–72. [Google Scholar]
- 58.Soldatova LN, King RD. Are the current ontologies in biology good ontologies? Nat Biotechnol. 2005;23:1095–08. doi: 10.1038/nbt0905-1095. [DOI] [PubMed] [Google Scholar]
- 59.McCray AT. An upper-level ontology for the biomedical domain. Comp Funct Genomics. 2003;4:80–84. doi: 10.1002/cfg.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 61.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith B, Ceusters W, Klagges B, et al. Relations in biomedical ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stevens R, Aranguren ME, Wolstencroft K, et al. Using OWL to Model biological Knowledge. Int Human Comput Stud. 2006 (in press). [Google Scholar]
- 64.Dean M, Connolly D, Harmelen FV, et al. [last accessed: 29 July, 2006.];OWL Web Ontology Language 1.0 Reference. [Web Page] [cited 2006 10 June] http://www.w3.org/TR/owl-ref/
- 65.Wolstencroft K, Brass A, Horrocks I, et al. International Semantic Web conference. Galway; Ireland: 2005. A little Semantic Web goes a long way in biology. [Google Scholar]
- 66.Stevens R, Goble C, Horrocks I, Bechhofer S. OILing the way to Machine Understandable Bioinformatics Resources. IEEE Inform Technol in Biomed (Special Issue on Bioinformatics) 2002;6:135–41. doi: 10.1109/titb.2002.1006300. [DOI] [PubMed] [Google Scholar]
- 67.Stevens R, Goble C, Horrocks I, et al. Building a bioinformatics ontology using OIL. IEEE Trans Inf Technol Biomed. 2002;6:135–41. doi: 10.1109/titb.2002.1006301. [DOI] [PubMed] [Google Scholar]
- 68.Ringland GA, Duce DA. Knowledge-Based and Expert Systems Series. John Wiley; Chichester: 1998. Approaches to knowledge representation: An introduction. [Google Scholar]
- 69.Wolstencroft K, Lord P, Tabernero L, Brass A, Stevens R. Classifying Proteins Using Ontological Classification, in Intelligent Systems for Molecular biology (ISMB) 2006. 2006. Fort e Lezza. [DOI] [PubMed] [Google Scholar]
- 70.Baker PG, Goble CA, Bechhofer S, Paton NW, Stevens R, Brass A. An ontology for bioinformatics applications. Bioinformatics. 1999;15:510–20. doi: 10.1093/bioinformatics/15.6.510. [DOI] [PubMed] [Google Scholar]
- 71.Yu AC. Methods in biomedical ontology. J Biomed Inform. 2006;39:252–66. doi: 10.1016/j.jbi.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 72.Kohler J, Munn K, Ruegg A, Skusa A, Smith B. Quality control for terms and definitions in ontologies and taxonomies. BMC Bioinformatics. 2006;7:212. doi: 10.1186/1471-2105-7-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rogers JE. Quality assurance of medical ontologies. Methods Inf Med. 2006;45:267–74. [PubMed] [Google Scholar]
- 74.Völkel M, Krötzsch M, Vrandecic D, Haller H, Studer R. Semantic Wikipedia; Proceedings of the 15th international conference on World Wide Web, WWW 2006; Edinburgh, Scotland. 23–26 May, 2006. [Google Scholar]
- 75.Good BM, Tranfield EM, Tan PC, et al. Fast, cheap and out of control: A zero curation model for ontology development. In: Altman RB, et al., editors. Pacific Symposium on Biocomputing 2006. World Scientific; 2006. pp. 128–39. [PubMed] [Google Scholar]
- 76.Supekar K. Ontology metadata to support the building of a library of biomedical ontologies. AMIA Annu Symp Proc. 2005:1127. [PMID: 16779414] [PMC free article] [PubMed] [Google Scholar]


