Abstract
Three-dimensional architectural motifs are increasingly recognized as determinants of RNA functionality. We submit that such motifs can encode spatial information. RNAs are targeted to subcellular localities in many eukaryotic cell types, and especially in neuronal and glial cells, RNAs can be transported over long distances to their final destination sites. Such RNAs contain cis-acting long-range targeting elements, and recent evidence suggests that kink-turn motifs within such elements may act as spatial codes to direct transport. Kink-turns are complex RNA motifs that feature double- and single-stranded components and introduce a signature three-dimensional structure into helical stems. We propose that the overall architectural design as well as the individual character—as specified by nucleotide identity and arrangement—of kink-turn motifs can serve as RNA targeting determinants.
Keywords: RNA codes, RNA structure, architectural motifs, RNA transport, targeting elements
INTRODUCTION
Kink-turn (K-turn, KT) motifs are recurrent architectural RNA elements that are found in Archaea, Prokarya, and Eukarya.1-3 K-turn (KT) motifs are functionally diverse and have been identified in rRNAs, mRNAs, and small untranslated RNAs (utRNAs, also called non-protein-coding RNAs).4 KT motifs serve as protein recognition sites and are able to engage in multiple intermolecular interactions simultaneously. We have previously reported the presence of KT motifs in neuronal RNAs that are targeted to dendrites, and we have proposed that KT motifs carry codes that specify long-range distal RNA targeting in neurons.5,62 We maintain that RNAs carry various types of codes—of which open reading frames represent only one6—that specify RNA functionality. Here we present an analysis of KT spatial coding motifs in selected neural (neuronal and glial) RNAs. We limit our investigation to such RNAs in which targeting elements have been sufficiently well characterized. This communication intends to alert RNA researchers and neuroscientists to the potential relevance of KT motifs in spatial coding, and to stimulate efforts aimed at dissecting mechanisms of KT-encoded RNA transport.
THE K-TURN (KT) MOTIF
K-turns are three-dimensional (3D) RNA motifs that are characterized by a core of non-canonical — i.e., non-Watson-Crick (non-WC) — base pairs.1,2,7-9 KT motifs, now recognized as key architectural elements in diverse RNAs,3,10-12 adopt a signature 3D conformation in which an internal loop forms a sharp kink in the helical axis of the RNA duplex.1,2,13-15
KT motifs, characterized by double- and single-stranded elements, are organized around two non-canonical base pairs2 (Fig. 1). These base pairs are typically of the sheared G•A type (hence the GA core) and form what is referred to as the non-canonical stem (NC-stem) of the motif (Fig. 1).2 [Sheared base pairs are those involving the shallow groove edge of one base, using trans-Hoogsteen/Sugar-Edge (trans-H/SE) interactions.]3,16,17 K-turns have synonymously also been called GA motifs.9
Figure 1.
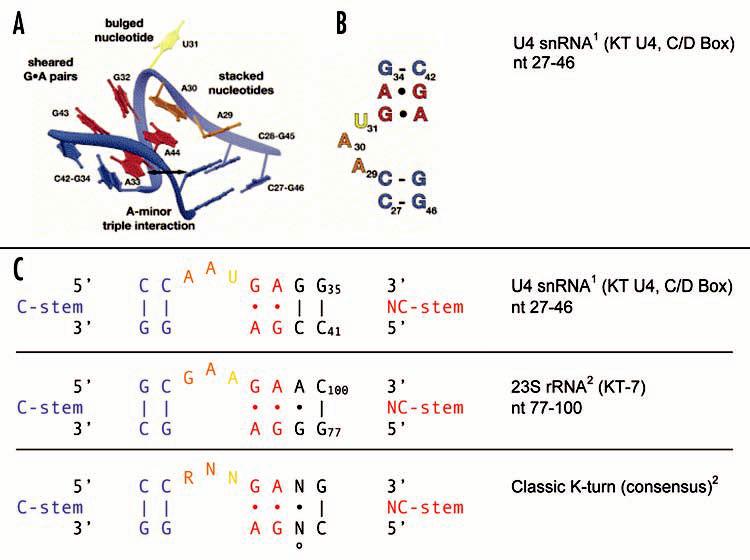
The classic KT motif. K-turns were first described in human U4 snRNA1 and in Haloarcula marismortui rRNAs.2 In this illustration, the KT motif is exemplified by KT U4 in the C/D Box of U4 snRNA (A–C) and by KT-7 in 23S rRNA. The deduced classic consensus2 is shown in (C). (A) Tertiary structure with the C-stem seen towards the front right, the NC-stem towards the rear left. Salient intramolecular interactions are indicated, with A-minor interactions symbolized by a horizontal bidirectional arrow (see also below). (B) Secondary structure, C-stem facing towards the bottom. Illustration reprinted from Hardin and Batey11, with permission from Elsevier. The following intramolecular interactions are universal to classic K-turns. (U4 numbering is used.) Of the tandem sheared G•A pairs, G32•A44 is non-planar. A44 also engages in stacking interactions with both A33 and A30, as a result forming cross-strand triple adenosine stacks.1,2 Thus, A30 stacks onto and thereby caps the NC-stem, and A29 stacks onto and caps the C-stem. These stacking interactions are critical for the 3D architecture of the motif. Also contributing to motif conformation is a type I A-minor interaction.18 In this tri-nucleotide motif, the O2′ atom and the “soft” N1-C2-N3 edge of an adenosine (which itself is typically part of a sheared G•A pair) fit into the minor groove of a receptor G-C WC base pair. In the case of U4 sRNA, this interaction is realized by A33 fitting into the minor groove of the G45-C28 pair. The A-minor motif is supported by trans-SE/SE interactions between A33 and G45. Trans-SE/SE interactions are also maintained between A29 and A44, or their equivalents in other classic K-turns.3 The specificity of these multiple intramolecular interactions form the basis for the unique 3D architecture. U31 bulges out, i.e. is extruded and does not interact with the other nucleotides of the motif. It is located at the apex of the 120° kink in the helical backbone. (C) KT motifs are aligned with their C-stems (G-C pairs) to the left, NC-stems (G•A pairs) to the right of the asymmetric internal loop. In the classic K-turn of the consensus type, nucleotides other than the ones shown are rarely tolerated in the positions indicated, except that distal G-C base pairs (i.e. those at the flanks of the motif) may occur in either the G-C or the C-G orientation. º denotes the transitional position between the NC-stem and the resumption of A-form helical structure by G-C WC base pairs; this position may be occupied by an intervening base pair, by an unpaired residue, or not at all. R, purine; N, any nucleotide. Blue, canonical G-C pairs of the C-stem; red, non-canonical G•A pairs of the NC-stem; orange, nominally unpaired internal loop nucleotides engaged in stacking interactions; yellow, extruded nucleotide.
In the “classic” KT motif, the NC-stem is connected via a purine-rich internal loop to the C-stem, a canonical stem with at least two standard WC base pairs which are usually of the G-C type (Fig. 1). The internal loop is short (typically 1–4 nt) and often asymmetric. On the other side of the NC-stem, the motif reverts to canonical base pairing typical of standard A-form RNA helices, beginning with a G-C pair and often separated from the NC-stem by a transitional base or base pair (Fig. 1).2,3 The KT signature 3D structure is specified by various WC and non-WC interactions between motif nucleotides.1-3 Trans-H/SE interactions, trans-SE/SE interactions—in one case giving rise to a type I A-minor motif18—and cross-strand stacking interactions all contribute to KT motif architecture (Fig. 1). Together, these requisite interactions impose stringent constraints on the types of nucleotides allowed in each motif position, and their relative orientation with respect to each other. On the basis of this information, a KT consensus has been established for the classic KT motif structure (Fig. 1).1,2,11 Identification of this consensus in an RNA is predictive of the presence of the 3D KT motif structure.2
These intramolecular interactions result in the characteristic 3D structure of the classic KT motif (Fig. 1). The helix is severely overwound (i.e. twisted by 81° around its axis), and the helical stem is bent by about 120°, thus causing a sharp axial kink.1,2 At the apex of this kink resides an unpaired residue (yellow in Fig. 1) that is part of the internal loop but does not interact with other nucleotides of the motif. This nucleotide is extruded (i.e., bulges out) and may engage RNA binding pockets in proteins.1,2,11 Other elements of the KT motif, as described above and in the legend to Fig. 1, may also be exploited as recognition motifs by proteins. Previously described KT-interacting proteins range from ribosomal proteins to putative nuclear RNA export factors.2,10 However, while the presence of the consensus KT sequence is diagnostic of the physical presence of a KT motif in an RNA, it does not predict number or types of proteins that it interacts with.2
KT MOTIFS IN RNA TARGETING ELEMENTS
Dendritic mRNAs
We have previously identified targeting elements in dendritic RNAs.5,19,20,62 The mRNA encoding protein kinase Mζ (PKMζ) contains two dendritic targeting elements (DTEs) of which Mζ DTE2 specifies distal targeting.19 Mζ DTE2 is a 44-nt stem-loop structure with a prominent GA internal loop motif that conforms to the classic K-turn consensus (100% match with conserved nucleotides and nucleotide positions, including NC-stem, C-stem, and asymmetric internal loop; Fig. 2). We submit that in Mζ DTE2, a KT motif serves as a code to specify long-range distal targeting.5
Figure 2.

Classic KT motifs in targeted neural mRNAs. See Figure 1 for color coding and motif alignment. Note that the internal loop nucleotide adjacent to the C-stem is consistently A, the long strand intervening nucleotide consistently G. These nucleotides may be specific to targeting KT motifs and are therefore referred to as targeting character nucleotides. For neural mRNAs, GenBank/EMBL/DDBJ accession numbers and nucleotide numbers are given. KT motifs are typically located in the 3′ UTRs except for KT GlyRα which is located at the 3′ end of the open reading frame. A modified version of DNAMAN software (Lynnon Corporation, Vaudreuil-Dorion, Quebec, Canada) was used to query RNAs for KT motifs. Query parameters were written such that searches were limited to classic KT motifs of the consensus type.
Dendritic RNAs are differentially delivered to proximal or distal destinations along the dendritic extent.21 Are KT-code DTEs thus specific for distally delivered dendritic RNAs, but absent from proximal or soma-restricted RNAs? This question is more easily raised than answered because the functional description of DTEs is still in its infancy for most dendritic RNAs.5 Distal dendritic targeting may be encoded by more than one type of architectural RNA motif; in some cases, one might envision, long-range transport may be specified by an interplay of several targeting (or even targeting-repressing) elements. Furthermore, because KT motifs participate in various cellular functions that are unrelated to RNA targeting, the presence of a KT motif in an RNA can by itself not be a predictor of distal dendritic targeting competence.
These caveats in mind, we used a search algorithm (see Fig. 2) to interrogate representative dendritic mRNAs for classic KT motifs. Focusing on transcripts for which DTEs have been identified or sufficiently narrowed down,5 we identified candidate KT motifs (100% congruent with classic consensus) in several neuronal mRNAs that are delivered to distal dendritic domains (Fig. 2). In CaMKIIα mRNA,21,22 a KT motif is predicted in a DTE segment of the 3′ untranslated region (UTR) that has previously been shown to specify targeting.23,24 In Arc mRNA, two KT motifs are predicted in a DTE section of the 3′ UTR that has recently been shown to be required for distal targeting.25
In contrast, no KT motif was detected in a second DTE that is contained in the Arc 3′ UTR, one that mediates proximal dendritic targeting.25 Similarly, we failed to detect KT motifs in the DTE of MAP2 transcripts.26 MAP2 mRNA, although delivered to dendrites, remains restricted to proximal regions.21,27 Also, no KT motif was found in the 3′ UTR of α-tubulin mRNA, a soma-restricted neuronal transcript.28,29
Finally, we compared mRNAs encoding glycine receptor α and β subunits (GlyRα and GlyRβ). While DTEs have not yet been localized within these mRNAs, GlyRα transcripts (which are delivered to distal postsynaptic domains)30 contain a classic KT motif whereas GlyRβ transcripts (which remain restricted to somata) do not (Fig. 2). Interestingly, KT GlyRα is of the same type as KT Mζ (Fig. 2).
In summary, it appears that KT motifs are associated with long-range distal targeting competence in dendritic mRNAs. We hypothesize that KT motifs in these RNAs carry codes to instruct distal targeting.
Myelin basic protein mRNA
A KT search identified a classic KT motif in the 3′ UTR of myelin basic protein (MBP) mRNA (100% match with consensus). MBP mRNA is transported along oligodendrocyte processes, and a relevant targeting element has been identified in the 3′ UTR of the mRNA.31-33 This 21-nt element, initially known as RNA transport signal (RTS) but now recognized as an hnRNP A2 response element (A2RE), contains an 11-nt targeting-competent subdomain (A2RE11).32,33 It turns out that A2RE11 represents the long (3′) strand of the predicted MBP KT motif (Figs. 2 and 3).
Figure 3.
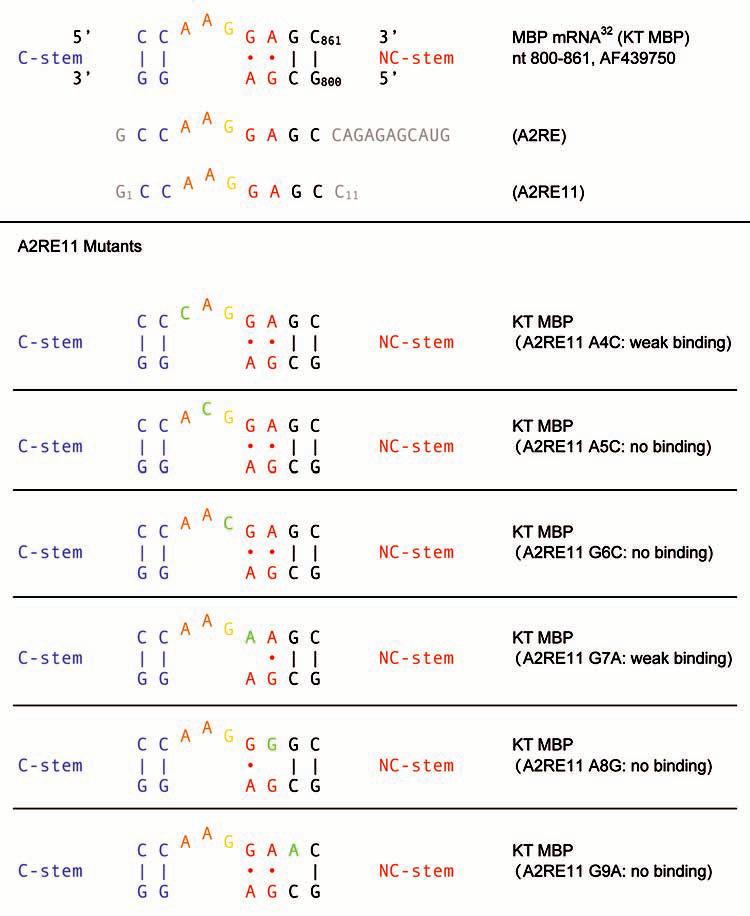
Classic K-turn motif in the 3′ UTR of MBP mRNA (KT MBP). The relationship between KT MBP, A2RE and A2RE11 is shown on top. Mutational analysis of A2RE1132 revealed differential effects of nucleotide replacement on hnRNP A2 binding. The exchange of nucleotides in the KT long strand that are essential for integrity and/or identity of the motif (shown in green) results in drastically diminished or entirely abolished binding to hnRNP A2. In contrast, C2U and C3G mutations result in only moderately diminished binding.32 For A2RE and A2RE11, nucleotides outside the KT motif long strand are shown in gray. A2RE11 numbering is used for mutants.
Mutational analysis has been used to probe the relevance of each nucleotide in A2RE11 for hnRNP A2 binding and RNA transport competence.32,33 Binding of hnRNP A2 was severely compromised by replacement of any of the nucleotides that would occupy strategic positions in the MBP KT long strand: nominally unpaired A4 and A5, engaging in stacking interactions, extruded G6, and G7 and A8 which engage in multiple interactions in the GA core (Fig. 3). G9, part of an intervening base pair that marks the interface between the NC stem and the resumption of standard A-form helix, was also found indispensable. In contrast, replacement of C-residues in the C-stem resulted only moderately diminished binding (Fig. 3). Analogous results were obtained for the targeting competence of these mutants although not all of them were thus examined.32,33 The combined data suggest that binding- and targeting-relevant nucleotides in A2RE are precisely those that are determinants of K-turn identity and functionality.
K-turns in targeted neural mRNAs
A comparison of six targeting KT motifs in neural mRNAs—two in Arc mRNA and one each in PKMζ, CaMKIIα, GlyRα, and MBP mRNAs (Fig. 2) —reveals a remarkable degree of similarity even in those positions where variability is observed in non-targeting KT motifs. Thus, the NC-stem is always of the classic G•A/A•G type, the 5′ unpaired stacking nucleotide in the asymmetric internal loop is A, and the intervening nucleotide in the long strand (at the transitional position between the NC-stem and the resumption of A-form helical structure) is G. It is of course possible that other, yet to be identified targeting KT motifs may show variations in these positions, but the combined available data indicate that the identities of these nucleotides are determinants of the targeting code. These nucleotides are therefore referred to as character nucleotides as they seem to define the character of targeting KT motifs as such. Targeting KT character nucleotides are indispensable for the spatial coding functionality of the motif, thus identifying it as a targeting-encoding subtype of the general classic KT motif. In contrast, motif-essential nucleotides (e.g. those of the GA core) are invariant in most KT motifs—regardless whether they belong to targeting or non-targeting subtypes—and can not be exchanged without destruction of the motif itself.
A second type of character nucleotide in KT motifs is RNA-specific. One example is provided by the nucleotide that pairs with the intervening G in the transitional position of the KT long strand. This short-strand nucleotide may be C, i.e., forming a standard WC base pair with intervening G, it may be another nucleotide that forms a non-canonical pair with intervening G, or it may be missing, leaving intervening G unpaired (Fig. 2). Similarly, in the internal loop, the extruded nucleotide and the 3′ unpaired stacking nucleotide are variable in targeting KT motifs. Such differences may give each of those KT motifs individual character as any of these nucleotides may specify protein recognition.1,2 In fact, motif-essential and both types of character nucleotides may engage in protein interactions, and it is therefore not surprising that exchanges of any of the three types of nucleotides in KT MBP severely impact hnRNP A2 recognition and RNA transport (Fig. 3).32
Untranslated dendritic RNAs
BC1 RNA and BC200 RNA—collectively called BC RNAs (small brain-specific cytoplasmic RNAs)—are untranslated neuronal RNAs that are delivered to distal dendritic domains.20,34-36 Both RNAs have been implicated in local translational control mechanisms.37-40 Dendritic targeting of BC1 RNA is specified by the 5′ BC1 domain which contains two cis-acting targeting elements: DTE1 which is required for somatic export and DTE2 which specifies long-range distal delivery.20,62 The secondary structure of the 5′ BC1 domain has been established by chemical and enzymatic probing,41 and a prominent GA-type KT motif in the apical region of the 5′ BC1 domain was found indispensable for distal targeting.62 KT BC1 (Fig. 4) belongs to the KT-58 subgroup of the classic KT motif, initially identified in 23S rRNA.2,3
Figure 4.
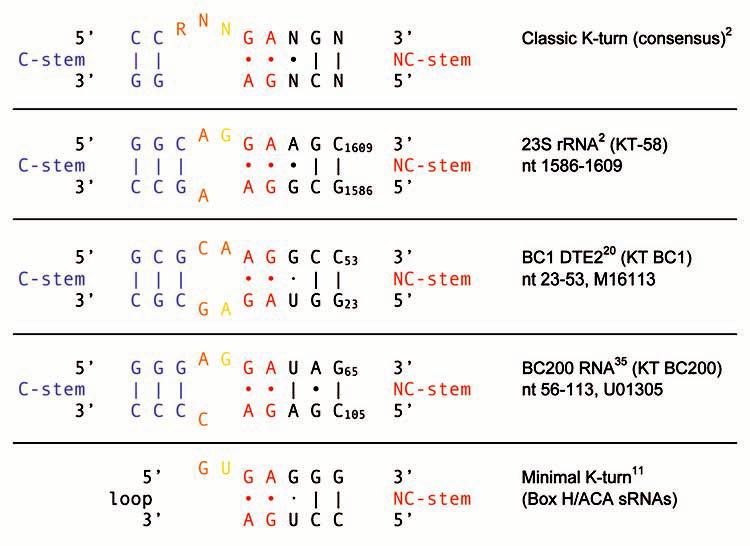
KT BC1 and KT BC200 belong to the KT-58 subgroup of the classic K-turn family, featuring a more symmetric internal loop structure.2,3 In KT motifs of this subgroup, the C-stem G-C pair that is adjacent to the internal loop can also be represented as orange internal loop nucleotides as they are likely to participate in stacking.3 Analogously, the internal loop G/C nucleotides that are adjacent to the C-stem in KT BC1 can also be represented as a blue standard WC pair as they are likely to engage in canonical base pairing. The G·U pair in KT BC1 may be either of the wobble WC type or of the sheared trans-H/SE type.3,12 Note that the geometry of the GA core is inverted in KT BC1, probably an indication that the motif assumes the reverse K-turn conformation.60 The minimal K-turn motif11 is shown for comparison.
Inter-species nucleotide substitutions have been observed in different rodent 5′ BC1 domains.41,42 Such exchanges have been conservative with respect to KT BC1, i.e., the motif structure has been maintained in all cases. The 5′ domain of BC200 RNA, the primate counterpart (analog) of rodent BC1 RNA,35 is also predicted to contain a classic KT motif of the KT-58 type (Fig. 4). DTEs have not yet been established in BC200 RNA, but microinjected BC200 RNA is delivered to distal dendritic segments of sympathetic neurons in culture (Muslimov IA, Tiedge H, unpublished). The KT BC200 motif structure is maintained in all primate species investigated.43 The only nucleotide exchanges observed are in Gorilla and the new world monkey Aotus where C105 has been replaced with a U, resulting in the conversion of a flanking G-C standard WC pair into a flanking G·U wobble WC pair. Such high degree of phylogenetic conservation is further indication of motif functionality.
KT motifs have previously been detected in other small RNAs.7,10,44 In a subset of such RNAs, i.e., some small nucleolar (sno) RNAs, these motifs belong to the class of minimal K-turns.10-12 Minimal K-turns differ from classic K-turns in that they do not contain a C-stem. The NC-stem is typically positioned at the base of a loop which contains, adjacent to the tandem G•A pair, the key extruded nucleotide (Fig. 4). While the KT motifs in BC RNAs conform to the classic type, it is certainly conceivable that targeting elements in other small untranslated RNAs, whether expressed in neurons or other cell types, may use minimal KT motifs as transport codes.
BC1 DTE2 and two other targeting elements, the ones contained in PKMζ and MBP mRNAs, have been narrowed down to fewer than 50 nt. For BC1 RNA, the secondary structure of the targeting competent 5′ domain has been determined experimentally41 and was found to be in agreement with the prediction of the Mfold RNA folding algorithm.45 For Mζ DTE2, the same algorithm predicts a stem-loop structure of 44 nt with a ΔG of −27.9 kcal/mol.19 (Note that Mfold and similar algorithms consider free energy contributions only from WC pairs, not from non-canonical interactions which may nevertheless significantly contribute to ΔG.) Analogously, KT MBP is predicted to be embedded in a stem-loop structure with a ΔG of −21.8 kcal/mol (Fig. 5). In this case, the structure features additional internal loops of the C-loop type, another class of recurrent RNA motifs.3 While the close spatial association of C-loop motifs with targeting-encoding KT motifs is intriguing, its biological significance remains to be explored.
Figure 5.
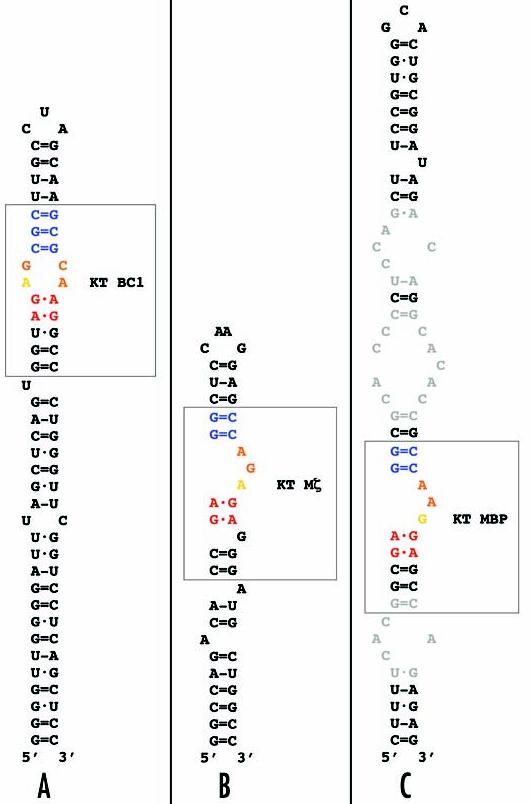
Embedding of KT motifs in stem-loop structures. (A) KT BC1, (B) KT Mζ, (C) KT MBP. In this illustration, base pairings are represented as follows: = (G-C WC); - (A-U WC); · (wobble WC); • (non-canonical). K-turns are boxed and are identified using the same color coding as above. In KT BC1, the internal loop G-C nucleotides that are adjacent to the C-stem can also be represented as a standard WC pair in blue; see Figure 4. In addition to KT motifs, the stem-loop structures feature other non-helical elements that may be functionally relevant, among them unpaired nucleotides61 and internal loops (gray nucleotides in the MBP stem loop, C) that display features of C-loop motifs.3
We can obviously not claim that KT motifs are physically present in all neural RNAs that we have presented above. Our algorithm identifies putative KT motifs, and a predicted KT consensus in an RNA is considered diagnostic of the presence of the 3D KT motif.2 Nonetheless, there can be no certainty that these are the conformations that actually exist in the intracellular milieu in all cases and at all times. Alternative conformations may exist, at least temporarily, e.g., when the motif is engaged with certain proteins (see also below). Thus, with this communication, we solicit efforts to subject the KT model to experimental scrutiny.
KT DECODING
With increasing progress towards an understanding of the functional-molecular architectures of dendritic targeting elements, we expect that common themes (i.e., RNA motifs) will emerge, and common concepts will crystallize. Nevertheless, the world of RNA targeting elements is bound to be complex. While this complexity may reflect a “no-shoe-fits-it-all” biological necessity in RNA targeting, we believe it is also a consequence of evolutionary principles. In evolutionary terms, RNA is much older than proteins, and RNA motifs such as K-turns have been described to subserve diverse functions in the three kingdoms of Archaea, Eubacteria, and Eukarya. Some K-turns may have acquired spatial coding competence in early eukaryotes. In yeast, for example, small nucleolar U14 RNA contains a box C/D element that was shown to be required for intranuclear targeting.46 Subsequently, C/D box elements were identified as KT motifs.1,2 During evolution, such motifs have been modified, adapted, and recruited/exapted into new functions, and they have been redistributed and introduced into other RNAs, for instance by intragenomic horizontal transfer.47-50
Significantly, KT motifs have “learned” to interact with different and novel proteins. In the context of RNA transport, such proteins are trans-acting factors (TAFs)5,51 that decode the information contained in the 3D architecture of the targeting-encoding RNA motif. Decoding is realized through the assembly of ribonucleoprotein (RNP) complexes: RNA motifs such as K-turns engage TAFs and thus act as nucleation sites for RNP assembly. The identity of a KT targeting RNP will be determined by the constellation of specific nucleotides engaged in TAF interactions as both motif-essential and character nucleotides may be recognized by such proteins.
Which TAFs read KT targeting codes? A2RE11 in MBP mRNA interacts with hnRNP A2, a targeting TAF that operates in neurons and glial cells.32,33,51,52 A2RE11 represents the long (3′) strand of KT MBP (Figs. 2 and 3). We reasoned that hnRNP A2 may therefore also interact with some of the other transported RNAs that use KT spatial codes. Recent data62 indeed show that hnRNP A2 also recognizes KT BC1. These data raise the possibility that proteins of the A2 family — several isoforms have been described to date51,53 — act as KT decoders, i.e., recognize KT targeting codes and mediate RNA transport.
Important questions remain. What types of KT targeting codes are recognized by which A2 proteins? Or, from the point of view of the KT motif, which motif-essential and/or character nucleotides are indispensable in engaging A2 proteins? In KT BC1, both strands of the motif are required for A2 recognition. In other cases, recognition of single-stranded (i.e., unpaired) elements in KT motifs may be sufficient for A2 interactions. (A2RE11 by itself binds to hnRNP A2.)32 A2 proteins contain two RNA recognition motifs (RRMs),54 but structural data of RNA engaged by A2 proteins are not available. It is known, however, that KT motifs exist in a dynamic equilibrium between an open structure and the highly ordered kink-turn conformation.55 Different TAFs may thus engage different conformations, and they may also prompt transition between conformational states through induced fit mechanisms.14,56,57
K-turn motifs thus employ highly versatile protein recognition strategies, and it is to be expected that even related RNA motifs—i.e., similar K-turns—may interact with different proteins, and vice versa.2,10,58 We anticipate that the next step towards a functional dissection of RNA transport in dendrites will have to be a comprehensive physico-functional delineation of those RNA motifs that encode dendritic targeting. This analysis will be followed by the identification of factors that recognize and decode such motifs, and ultimately by the establishment of 3D structures that describe motif-TAF complexes.
KT motifs have been implicated in the localization of several box C/D snoRNAs to nucleoli in such diverse species as yeast and Xenopus.46,59 It thus appears likely that KT-encoded targeting is not restricted to neural RNAs, the main focus of this article, but plays a universal—and possibly ancient—role in spatial RNA coding in general.
ACKNOWLEDGEMENTS
I thank Dr. Yue Huang, Lynnon Corporation, for help developing and using search algorithms for the identification of KT-motifs, and Dr. Ross Smith, University of Queensland, for discussions. Work in the author's lab was supported in part by National Institutes of Health grants HD043428 and NS046769.
References
- 1.Vidovic I, Nottrott S, Hartmuth K, Lührmann R, Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell. 2000;6:1331–42. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 2.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–21. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosius J, Tiedge H. RNomenclature. RNA Biol. 2004;1:81–83. doi: 10.4161/rna.1.2.1228. [DOI] [PubMed] [Google Scholar]
- 5.Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–45. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbieri M. The Organic Codes: An Introduction to Semantic Biology. Cambridge University Press; Cambridge: 2003. [Google Scholar]
- 7.Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Lührmann R. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–66. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 8.Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Lührmann R. Functional interaction of a novel 15.5kD [U4/U6·U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 1999;18:6119–33. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winkler WC, Grundy FJ, Murphy BA, Henkin TM. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001;7:1165–72. doi: 10.1017/s1355838201002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Hüttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–77. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardin JW, Batey RT. Curse of the hairpin loop. Structure. 2004;12:731–2. doi: 10.1016/j.str.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Hamma T, Ferré-D'Amaré AR. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 A resolution. Structure. 2004;12:893–903. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Matsumura S, Ikawa Y, Inoue T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 2003;31:5544–51. doi: 10.1093/nar/gkg760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goody TA, Melcher SE, Norman DG, Lilley DM. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 2004;10:254–64. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steitz TA, Moore PB. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem Sci. 2003;28:411–8. doi: 10.1016/S0968-0004(03)00169-5. [DOI] [PubMed] [Google Scholar]
- 16.Westhof E, Fritsch V. RNA folding: beyond Watson-Crick pairs. Structure. 2000;8:R55–65. doi: 10.1016/s0969-2126(00)00112-x. [DOI] [PubMed] [Google Scholar]
- 17.Leontis NB, Westhof E. Analysis of RNA motifs. Curr Opin Struct Biol. 2003;13:300–8. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 18.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci USA. 2001;98:4899–903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muslimov IA, Nimmrich V, Hernandez AI, Tcherepanov A, Sacktor TC, Tiedge H. Dendritic transport and localization of protein kinase Mζ mRNA: implications for molecular memory consolidation. J Biol Chem. 2004;279:52613–22. doi: 10.1074/jbc.M409240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muslimov IA, Santi E, Homel P, Perini S, Higgins D, Tiedge H. RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J Neurosci. 1997;17:4722–33. doi: 10.1523/JNEUROSCI.17-12-04722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paradies MA, Steward O. Multiple subcellular mRNA distribution patterns in neurons: a nonisotopic in situ hybridization analysis. J Neurobiol. 1997;33:473–93. doi: 10.1002/(sici)1097-4695(199710)33:4<473::aid-neu10>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 22.Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, Kelly PT. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J Neurosci. 1990;10:1788–98. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blichenberg A, Rehbein M, Müller R, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. Eur J Neurosci. 2001;13:1881–88. doi: 10.1046/j.0953-816x.2001.01565.x. [DOI] [PubMed] [Google Scholar]
- 24.Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIα impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–19. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi H, Yamamoto S, Maruo T, Murakami F. Identification of a cis-acting element required for dendritic targeting of activity-regulated cytoskeleton-associated protein mRNA. Eur J Neurosci. 2005;22:2977–84. doi: 10.1111/j.1460-9568.2005.04508.x. [DOI] [PubMed] [Google Scholar]
- 26.Blichenberg A, Schwanke B, Rehbein M, Garner C, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J Neurosci. 1999;19:8818–29. doi: 10.1523/JNEUROSCI.19-20-08818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garner CC, Tucker RP, Matus A. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988;336:674–77. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 28.Bruckenstein DA, Lein PJ, Higgins D, Fremeau RT., Jr Distinct spatial localization of specific mRNAs in cultured sympathetic neurons. Neuron. 1990;5:809–19. doi: 10.1016/0896-6273(90)90340-l. [DOI] [PubMed] [Google Scholar]
- 29.Mohr E, Prakash N, Vieluf K, Fuhrmann C, Buck F, Richter D. Vasopressin mRNA localization in nerve cells: characterization of cis-acting elements and trans-acting factors. Proc Natl Acad Sci USA. 2001;98:7072–9. doi: 10.1073/pnas.111146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Racca C, Gardiol A, Triller A. Dendritic and postsynaptic localizations of glycine receptor subunit mRNAs. J Neurosci. 1997;17:1691–700. doi: 10.1523/JNEUROSCI.17-05-01691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ainger K, Avossa D, Diana AS, Barry C, Barbarese E, Carson JC. Transport and localization elements in myelin basic protein mRNA. J Cell Biol. 1997;138:1077–87. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munro TP, Magee RJ, Kidd GJ, Carson JH, Barbarese E, Smith LM, Smith R. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J Biol Chem. 1999;274:34389–95. doi: 10.1074/jbc.274.48.34389. [DOI] [PubMed] [Google Scholar]
- 33.Shan J, Munro TP, Barbarese E, Carson JH, Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J Neurosci. 2003;23:8859–66. doi: 10.1523/JNEUROSCI.23-26-08859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiedge H, Fremeau RT, Jr., Weinstock PH, Arancio O, Brosius J. Dendritic location of neural BC1 RNA. Proc Natl Acad Sci USA. 1991;88:2093–97. doi: 10.1073/pnas.88.6.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tiedge H, Chen W, Brosius J. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci. 1993;13:2382–90. doi: 10.1523/JNEUROSCI.13-06-02382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chicurel ME, Terrian DM, Potter H. mRNA at the synapse: analysis of a preparation enriched in hippocampal dendritic spines. J Neurosci. 1993;13:4054–63. doi: 10.1523/JNEUROSCI.13-09-04054.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sonenberg N, Lomakin IB, Tiedge H. Dendritic BC1 RNA: functional role in regulation of translation initiation. J Neurosci. 2002;22:10232–41. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Iacoangeli A, Lin D, Williams K, Denman RB, Hellen CUT, Tiedge H. Dendritic BC1 RNA in translational control mechanisms. J Cell Biol. 2005;171:811–21. doi: 10.1083/jcb.200506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondrashov AV, Kiefmann M, Ebnet K, Khanam T, Muddashetty RS, Brosius J. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP) J Mol Biol. 2005;353:88–103. doi: 10.1016/j.jmb.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 40.Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006 doi: 10.1146/annurev.neuro.29.051605.112839. in press. [DOI] [PubMed] [Google Scholar]
- 41.Rozhdestvensky T, Kopylov A, Brosius J, Hüttenhofer A. Neuronal BC1 RNA structure: evolutionary conversion of a tRNAAla domain into an extended stem-loop structure. RNA. 2001;7:1–9. doi: 10.1017/s1355838201002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martignetti JA, Brosius J. Neural BC1 RNA as an evolutionary marker: guinea pig remains a rodent. Proc Natl Acad Sci USA. 1993;90:9698–702. doi: 10.1073/pnas.90.20.9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skryabin BV, Kremerskothen J, Vassilacopoulou D, Disotell TR, Kapitonov VV, Jurka J, Brosius J. The BC200 RNA gene and its neural expression are conserved in Anthropoidea (Primates) J Mol Evol. 1998;47:677–85. doi: 10.1007/pl00006426. [DOI] [PubMed] [Google Scholar]
- 44.Watkins NJ, Dickmanns A, Lührmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol Cell Biol. 2002;22:8342–52. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuker M. Calculating nucleic acid secondary structure. Curr Opin Struct Biol. 2000;10:303–10. doi: 10.1016/s0959-440x(00)00088-9. [DOI] [PubMed] [Google Scholar]
- 46.Samarsky DA, Fournier MJ, Singer RH, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–57. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cristofanilli M, Thanas S, Brosius J, Kindler S, Tiedge H. Neuronal MAP2 mRNA: species-dependent differential dendritic targeting competence. J Mol Biol. 2004;341:927–34. doi: 10.1016/j.jmb.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 48.Herbert A. The four Rs of RNA-directed evolution. Nat Genet. 2004;36:19–25. doi: 10.1038/ng1275. [DOI] [PubMed] [Google Scholar]
- 49.Gould SJ, Vrba ES. Exaptation - a missing term in the science of form. Paleobiology. 1982;8:4–15. [Google Scholar]
- 50.Brosius J, Gould SJ. On “genomenclature”, a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc Natl Acad Sci USA. 1992;89:10706–10. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith R. Moving molecules: mRNA trafficking in mammalian oligodendrocytes and neurons. Neuroscientist. 2004;10:495–500. doi: 10.1177/1073858404266759. [DOI] [PubMed] [Google Scholar]
- 52.Carson JH, Barbarese E. Systems analysis of RNA trafficking in neural cells. Biol Cell. 2005;97:51–62. doi: 10.1042/BC20040083. [DOI] [PubMed] [Google Scholar]
- 53.Hatfield JT, Rothnagel JA, Smith R. Characterization of the mouse hnRNP A2/B1/B0 gene and identification of processed pseudogenes. Gene. 2002;295:33–42. doi: 10.1016/s0378-1119(02)00800-4. [DOI] [PubMed] [Google Scholar]
- 54.Shan J, Moran-Jones K, Munro TP, Kidd GJ, Winzor DJ, Hoek KS, Smith R. Binding of an RNA trafficking response element to heterogeneous nuclear ribonucleoproteins A1 and A2. J Biol Chem. 2000;275:38286–95. doi: 10.1074/jbc.M007642200. [DOI] [PubMed] [Google Scholar]
- 55.Dennis PP, Omer A. Small non-coding RNAs in Archaea. Curr Opin Microbiol. 2005;8:685–94. doi: 10.1016/j.mib.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 56.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–7. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 57.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–31. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 58.White SA, Hoeger M, Schweppe JJ, Shillingford A, Shipilov V, Zarutskie J. Internal loop mutations in the ribosomal protein L30 binding site of the yeast L30 RNA transcript. RNA. 2004;10:369–77. doi: 10.1261/rna.2159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Narayanan A, Speckmann W, Terns R, Terns MP. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell. 1999;10:2131–47. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strobel SA, Adams PL, Stahley MR, Wang J. RNA kink turns to the left and to the right. RNA. 2004;10:1852–4. doi: 10.1261/rna.7141504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hermann T, Patel DJ. RNA bulges as architectural and recognition motifs. Structure. 2000;8:R47–54. doi: 10.1016/s0969-2126(00)00110-6. [DOI] [PubMed] [Google Scholar]
- 62.Muslimov IA, Iacoangeli A, Brosius J, Tiedge H. Spatial codes in dendritic BC1 RNA. J Cell Biol. 2006;175:427, 39. doi: 10.1083/jcb.200607008. [DOI] [PMC free article] [PubMed] [Google Scholar]


