Abstract
We present a technique for identifying the amino acids responsible for a loss of immunoreactivity in response to treating an antigen with a chemical modifier. This is of particular interest for the chemical formaldehyde, the cross-linking agent in formalin. Formalin is a commonly used fixative to preserve the cellular architecture of cells and tissues and to prevent degradation from proteases and nucleases. Formalin is also routinely used in the preparation of vaccines, to inactivate both toxins and microbes. Formalin fixation attenuates infectivity and pathogenicity by cross-linking while often preserving antigenicity. However, some epitopes are irreversibly modified by formalin while others are not. An understanding of how formalin affects epitope immunoreactivity may be useful in vaccine development or in the development of diagnostic antibody reagents for formalin-fixed tissues. In this report, we describe a method for systematically identifying formalin-sensitive and formalin-insensitive epitopes in a high throughput fashion, for any particular antibody. The data from this effort underscore the importance of certain amino acids, notably lysine, in affecting antibody immunoreactivity after formalin fixation. The method can be generally applicable in exploring the sensitivity of protein epitopes to an agent or condition of interest.
Keywords: Formalin, formaldehyde, fixation, epitope, antibody, combinatorial library
1. INTRODUCTION
For decades, aldehyde fixatives, such as formaldehyde and glutaraldehyde, have been used in biomedical and pharmaceutical disciplines. Formaldehyde has been used to fix clinical biopsy tissues, preserving the integrity of tissue architecture from the destruction of proteases and nucleases (Fox et al., 1985; Riederer, 1989; Shi et al., 2000). Formaldehyde has also been used to inactivate toxins and viruses, such as for the production of vaccines, including diphtheria, tetanus, hepatitis A, anthrax and polio vaccine (Murdin et al., 1996; Ellis, 1999; Leppla et al., 2002). Formaldehyde, a highly reactive dipolar compound, forms protein-protein and protein-nucleic acid crosslinks (Shi et al., 2000; Srinivasan et al., 2002).
Despite its widespread use, formalin fixation is associated with some serious drawbacks. Most importantly, formalin fixation may cause protein alterations that result in a loss of immunoreactivity to tissue or cellular antigens important for clinical diagnosis by immunohistochemistry (Bertheau et al., 1998; van den Boreck and van de Vijver, 2000; Werner et al., 2000; Wester et al., 2000). Formalin cross-linking of proteins to DNA also introduces new challenges in nucleic acid analysis, such as polymerase chain reaction (PCR) or in situ hybridization (Foss et al., 1994; Ikeda et al., 1998; Masuda et al., 1999; Williams et al., 1999.; Srinivasan et al., 2002). Recently there are reports alluding to the difficulties associated with proteomic or mass-spectrometric (MS) analysis of proteins from formalin-fixed tissues or cells (Crockett et al., 2005). This limits the utility of analyzing proteins from archived pathological tissue samples. Formaldehyde-induced cross-linking causes proteins to become relatively insoluble, complicating biochemical extraction and analysis, and thereby rendering them less amenable to mass spectrometry-based proteomic studies (Ikeda et al., 1998; Crockett et al., 2005).
Similarly, formalin cross-linking of vaccine antigens can result in a diminution or loss of antigenicity. There are numerous instances where formalin-treated vaccine preparations confer minimal or no immunity to the antigen (Murdin et al., 1996; Ellis, 1999; Leppla et al., 2002). The loss of vaccine efficacy is often ascribed to epitope masking by formalin cross-linking. For tissue sections, the deleterious effects of formalin on proteins or nucleic acids can be reversed by treatment with certain enzymes or heating protocols, which facilitate the detection or amplification of proteins or nucleic acids respectively (Shi et al., 1991; Shi et al., 2000). A heat-induced antigen retrieval method, involving heating formalin-fixed tissue sections in a buffer, restores the immunoreactivity of tissue antigens. Similar antigen retrieval methods have been adopted for the retrieval of detectable signal for nucleic acids and proteins, for molecular genetic or proteomic analysis of archived tissue samples (Ikeda et al., 1998; Shi et al., 2002). Although these antigen retrieval methods aid in reversing formalin cross-links, they are not suitable for all proteins or antibodies.
Since formalin plays such an integral role in clinical research and diagnosis, it is important to understand the mechanism of formalin fixation and its reversal with heat. In this regard, investigators have used various methods and model systems to elucidate the underlying chemistry associated with formalin fixation (Metz et al., 2004; Rait et al., 2004a; Rait et al., 2004b). In a series of publications from the 1940’s, Fraenkel-Conrat and associates described formaldehyde reactions with proteins involving various amino acid side chains (Fraenkel-Conrat et al., 1947; Fraenkel-Conrat and Olcott, 1948a; Fraenkel-Conrat and Olcott, 1948b). More recently, Metz and co-workers used a set of model peptides to further describe some of the formaldehyde-induced chemical modifications occurring on each natural amino acid residue (Metz et al., 2004). Formalin fixation and antigen retrieval were also analyzed with biophysical techniques using a pancreatic ribonuclease model antigen system (Rait et al., 2004a; Rait et al., 2004b). In an effort to characterize formalin-sensitive epitopes recognized by commonly used monoclonal antibodies in clinical practice, we recently identified formalin-reactive amino acid residues associated with the recovery of immunoreactivity after antigen retrieval (Sompuram et al., 2004; Sompuram et al., 2006a; Sompuram et al., 2006b). Although the literature reports that most protein epitopes are conformational (Kuby, 1994; Huebner, 2004), we found that epitopes which remain immunoreactive after formalin-fixation and antigen retrieval are comprised of linear amino acid sequences (Sompuram et al., 2006b).
In an effort to understand the underlying immunochemistry of formalin fixation, we developed a new screening method for distinguishing formalin-sensitive and formalin-resistant epitopes. Our method approaches the immunochemistry of formalin fixation in an entirely new fashion. Rather than screening many antibodies for a desired phenotype (e.g., binding to a formalin-resistant epitope), we turn the conventional paradigm around. We start with a single antibody and interrogate the universe of possible epitopes that bind to the antibody. This method involves biopanning from a peptide combinatorial library with an antibody. The peptides that bind to the antibody were tested for their sensitivity to formalin fixation. Some peptides remain immunoreactive whereas others do not. After distinguishing formalin-resistant from formalin-sensitive epitopes, we then sequenced them to see which amino acids, or combinations of amino acids, are associated with resistance or susceptibility to formalin. The technique is broadly applicable to sensitivity analysis of epitopes in response to any chemicals or physical conditions.
2. MATERIALS & METHODS
2.1 Antibodies used in the study
Estrogen receptor (ER)-specific monoclonal antibody (mAb) clone ER1D5 was obtained from DakoCytomation, Carpinteria, CA. Progesterone receptor (PR)-specific mAb clone PR1A6 and Her-2-specific mAb clone CB11 were obtained from Vision Biosystems, Norwell, MA.
2.2 Phage-display and biopanning
Phage libraries contained rationally-designed combinatorial libraries of peptide sequences inserted into the amino terminus of the cpIII minor coat protein of the M13 bacteriophage. The libraries were prepared and supplied by Dyax Corporation (Cambridge, MA). The libraries (termed “TN6” and “TN10”) contain two conserved cysteine residues separated by either four (for the TN6 library) or eight (for the TN10 library) amino acids. The cysteines formed a disulfide bridge, creating a conformationally constrained ring (McLafferty et al., 1993). The amino acids within the ring and the three amino acids on either side of the ring were diversified to allow all amino-acid types (except cysteine) with equal frequency using TRIM technology (Virneckas et al., 1994). TRIM technology involves controlled polymerization of preformed trinucleotides.
The libraries were screened by biopanning using standard methods (Parmley and Smith, 1988; Sparks et al., 1996; Smith and Petrenko, 1997) with a few modifications. Briefly, aliquots of paramagnetic beads were coated with anti-mouse IgG (Dynabeads, Dynal, A.S. Oslo, Norway) by mixing either with the positively-selecting mAb (ER1D5, PR1A6, or CB11) or the negatively-selecting antibody (polyclonal mouse IgG) and incubating overnight at 4°C on a rotator. Antibody-adsorbed Dynabeads were washed 4–5 times with phosphate buffered saline (PBS)-Tween 20 (0.05%) and twice with PBS before use in biopanning of phage libraries. A TN6 or TN10 phage library of 1011–12 plaque forming units (PFU) was depleted of immunoglobulin framework binders by incubation with Dynabeads (100 μl) coated with polyclonal mouse IgG, for one hour at room temperature (RT) on a rotator. With this negative depletion step, phage that express peptides which bind to common, shared regions of mouse IgG are depleted from the library by attaching to the mouse immunoglobulin attached to the paramagnetic beads. The unbound phage in the supernatant were then positively selected on the monoclonal antibody-adsorbed paramagnetic beads. The library was incubated with the mAb-coated beads for 2–3 hours on a rotator. The beads were then washed ten times with (PBS)-Tween 20 (0.05%) and three times with PBS to remove non-specifically bound phage. Phage bound to the mAb-coated beads were then eluted with a 0.1 M glycine-HCl, pH 2.2 buffer containing 1 mg/ml bovine serum albumin (BSA). After elution, the acidic buffer was neutralized with 1M Tris-HCl, pH 9.0. To ensure that high affinity phage were completely eluted, the beads were eluted twice, serially, and the eluates were pooled. The eluted phage were then amplified and used in a second round of biopanning. After two rounds of positive selection, E. Coli were infected by the cultured phage and grown on agar plates. Individual phage colonies were picked and grown for further analysis.
2.3 Phage library screening
Random cyclic peptide libraries, approximately 15–18 amino acids long, are prepared and provided by Dyax Corp. We selected for binders to the ER 1D5, PR 636, p53 DO7, Ki-67 MIB-1, and Her-2 CB11 mAbs. All of these mAbs are suitable for work on paraffin-embedded, formalin-fixed tissues. After the third round of selection and amplification, individual phage clones are isolated by infecting log phase E. coli, expanded and assayed against the original mAb in a phage ELISA. In the past, using this screening strategy (Sompuram et al., 2002), we generally obtained 80–95% positive clones after only two or three rounds of selection.
2.4 Screening for formalin sensitive peptide phage clones
After testing the individual clones in a phage ELISA, XL-1 Blue bacteria in log phase growth are infected with various dilutions of phage and grown on agar plates. About 1,000 well-dispersed individual phage clones from agar plates are transferred to replicate nitrocellulose membranes by a plaque-lift technique, as outlined in Figure 2. A nitrocellulose membrane (82mm diameter HATF filter, Millipore Corp.) is overlaid onto the agar and allowed to sit at RT for 10–15 minutes. In this way, the plaques are partially transferred to the membrane. Without disrupting the agar and plaques, the nitrocellulose membrane is carefully lifted and referred to as Lift #1. A second nitrocellulose membrane is then overlaid onto the same agar plate and plaques are transferred (“Lift #2”). Both filters are air-dried. Plaque Lift # 2 was immersed in 5% non-fat dried milk in Tris-buffered saline pH 7.4 and then stored at 4° C. Lift #1 was fixed in 10% formalin for 24 hrs.
Figure 2.

Flow chart for immunoblot selection of formalin-sensitive and formalin-resistant phage clones. The phage clones were enriched with two rounds of positive selection using a mAb coupled to paramagnetic beads (not shown in the figure). The individual clones were then plated onto LB agar, as shown in the upper left. Replicate blots, formalin-treated (left side) or untreated (right side), distinguish formalin-sensitive from formalin-resistant epitopes expressed by individual phage colonies. The arrows on the left side blot identify phage clones that are no longer immunoreactive with the mAb after formalin fixation. These phage clones are shown as open circles, and express peptide inserts with formalin-sensitive epitopes.
It is often observed that during repeated (serial) plaque-lifts, the first lift acquires more colonies than subsequent lifts. Therefore, we chose plaque lift #1 for formalin fixation. In this way, the absence of subsequent immunostaining that might be due to formalin fixation on lift #1 on various colonies is not likely attributable to fewer bacteria in the colony. This protocol provides for better discrimination of diminished/lost immunoblot signal due to formalin fixation. Plaque-Lift #2 serves as a positive control for antibody- binding phage clones. After formalin fixation of plaque Lift #1, the membrane is rinsed and protein binding sites are blocked with 5% non-fat dry milk in TBS. Both membranes are then immunostained with the mAb used for phage panning. The immunostaining procedure includes an initial incubation with the selecting mAb (e.g., ER 1D5) for 1–2 hrs. The membranes are then rinsed, followed by the application of detecting reagents. The detecting reagents include horseradish peroxidase (HRP)-conjugated goat-anti-mouse IgG followed by color development using 3, 3-diaminobenzidine (DAB) and 0.01% H2O2.
2.5 Sandwich phage ELISA
A sandwich-type ELISA was used to assay for the immunoreactivity of the enriched recombinant phage with the selecting mAb used for phage panning. As a first step, the selecting mAb (or other isotype-matched control mAbs) were adhered to the plastic. The various antibodies used for selecting phage out of the library were unpurified, circumventing the need to purify each mAb. Therefore, we first coated the microtitre plates with a mouse IgG-reactive polyclonal antibody. Briefly, three 96-well microtiter plates (Immulon 1, Dynex, CA) were coated with goat anti-mouse IgG (2 μg/ml, Sigma Chemical Co., St Louis, MO), 100 μl/well in carbonate-bicarbonate buffer, pH 9.5 and incubated overnight at 4° C. The wells were rinsed in PBS and free protein binding sites were blocked with 200 μl of 2% bovine serum albumin (BSA) in PBS for 1 hour at RT. The wells were then washed five times in PBS-Tween 20 (0.05%). These microtiter plates, coated with an anti-mouse IgG antibody, were then used to capture various mouse mAbs.
The ER, PR, or Her-2 mAb, or irrelevant, isotype-matched mAbs, were diluted to 1 μg/ml in PBS-Tween 20 (0.05%), added to the anti-IgG coated 96-well microtitre plates (100 μl/well), and incubated at RT for three hours. As an additional negative control (besides an isotype-matched negative control), microtitre plate wells were coated with 2% BSA in PBS (200 μl/well) and incubated at RT for 2 hrs. Antibody and BSA coated wells were then washed 8 times with PBS-Tween 20 (0.05%).
The immunoreactivity of selected phage clones with the ER, PR, or Her-2 mAb, or control mAbs was assessed by measuring binding to the respective mAbs. These mAbs were already immobilized onto the microtitre wells as described above. Individual phage clones (~108–9 PFU/well), diluted in PBS-Tween 20 (0.1%) with 0.1% BSA, were added to the microtitre wells (100 μl/well) and incubated at RT for 2 hrs. Wells were washed 10 times with PBS-Tween 20 (0.05%) to remove non-specifically adherent phage. The presence and relative amount of plate-adherent phage was determined by adding horseradish peroxidase (HRP)-conjugated sheep anti-M13 antibody (Pharmacia Biotech, Piscataway, NJ, 100 μl/well) and incubated for 1 hour at RT. The microtitre wells were then washed eight times with PBS-Tween 20 (0.05%) to remove non-specifically adherent sheep anti-M13 –HRP antibody conjugate. Finally, an HRP colorimetric enzyme substrate, 2,2-azino-di-[3-ethyl-benzthiaoline sulfonate] (ABTS), 0.5 mg/ml, was added to the microtitre wells. Color development in the microtitre wells was measured with a BioRad microplate reader, model 2550 (BioRad Corporation, Hercules, CA), at a 405 nm wavelength. Data are expressed as optical density (O.D.). Phage clones that gave ELISA signals at least three times the background value were considered positive.
2.6 DNA Insert Sequencing
Of the immunoreactive phage clones, 10–50 each of formalin-sensitive and formalin-insensitive phage clones were picked for subsequent analysis. The DNA inserts were amplified by polymerase chain reaction (PCR) and their peptide-coding inserts sequenced at the DNA Core Facility of Tufts University Medical School, Boston, MA. We sequenced the nucleotide inserts coding for the combinatorial peptides in phage clones that gave high O.D. readings (considered to be specific binders) and several poorly binding clones (for comparison). The sequencing template was prepared by PCR amplification from an overnight phage culture. The primers used for insert amplification were CGGCGCAACTATCGGTATCAAGCTG and CATGTACCGTAACACTGAGTTTCGTC. Thirty rounds of PCR were performed on a MJ Research Tetrad thermocycler (MJ Research, Inc., Waltham, MA). The PCR product was diluted 1:20 with distilled water. Sequencing was performed in both the forward and reverse directions using the following primers: GATAAACCGATACAATTAAAGGCTCC and GTTTTGTCGTCTTTCCAGACGTTAG.
3. RESULTS
3.1. Identification of immunoreactive epitopes
ER, PR, and Her-2-specific mAbs were used for biopanning against random peptide combinatorial phage display libraries. These mAbs were developed and commercialized because they bind to their antigens even in formalin-fixed, paraffin-embedded tissue, after antigen retrieval (Al Saati et al., 1993; Bevitt et al., 1997; Anonymous, 2002b; Anonymous, 2002a). Phage expressing peptides that bind to the antigen-combining region of those mAbs were enriched by serial rounds of selection and amplification. First, we depleted the phage that bind to common, shared (framework) regions of polyclonal mouse IgG (see Materials and Methods). We then positively selected for phage that bind to the mAb specific for ER, PR or Her-2 (see Materials and Methods).
After two rounds of selection (“biopanning”), the input-to-output phage ratio reached a plateau (data not shown), suggesting that the majority of the phage were specific to the target monoclonal antibody used for biopanning. The specificity of the enriched phage clones was confirmed with a phage ELISA, by randomly picking the individual phage clones and testing their ability to bind to mAb-coated microtitre wells. Depending on the target antibody, we typically observe approximately 80–90% of the clones to be specific binders for the selecting mAb. Representative phage ELISA data for PR mAb 1A6-binding phage are shown in Figure 1. Mouse polyclonal IgG was used as a negative control, to assess the assay background (open, unshaded vertical bars). Additional controls are shown on the right side of Figure 1. In the column labeled “M13”, we tested the immunoreactivity of the PR1A6 mAb with wild type M13 phage that did not contain a peptide insert. As expected, the immunoreactivity was low. As a positive control, we also tested the bulk phage preparations that were enriched for immunoreactivity with the 1A6 mAb, after one or two rounds of positive selection, without cloning. These are shown to the right in Figure 1, where “1” and “2” on the x axis refer to the number of positive selection rounds of enrichment against the 1A6 mAb. The data demonstrate progressively increased binding with each round of enrichment. The data from an additional negative control are also shown to the right in Figure 1. Instead of enriching against the PR1A6 mAb, we also enriched the same phage library against the PR636 mAb, which binds to a different epitope on the same protein. As expected, immunoreactivity of the (irrelevant) PR636 mAb-enriched phage clones to the PR1A6 mAb is relatively low.
Figure 1.
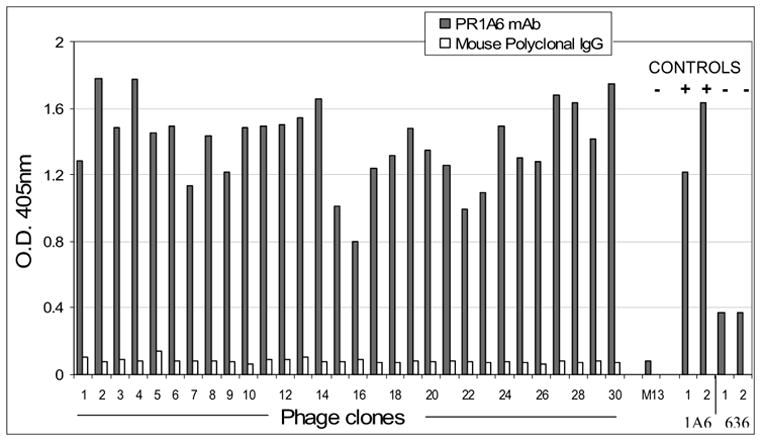
A representative phage ELISA, showing immunoreactivity of each phage clone to the PR 1A6 mAb (shaded bars). Polyclonal mouse antibody was used as negative antibody control (open bars). Wild type M13 phage (“M13”) is an additional negative control. Bulk uncloned round 1 and round 2 enriched phage preparations were also run for comparison. As positive controls, the round 1 and round 2 uncloned phage preparations enriched with the relevant mAb PR1A6 are shown towards the right portion of the figure. The fact that these would be expected to be immunoreactive with the PR1A6 mAb is indicated by the “+”. As an additional negative control, bulk uncloned phage preparations that were enriched with an irrelevant mAb PR 636 were also tested. The fact that these are not expected to be immunoreactive is denoted by the “−” symbol.
3.2. Identification of formalin-sensitive and formalin-insensitive peptides
Since we could enrich phage libraries for peptides that are immunoreactive with a desired mAb, we tested the role of formalin fixation in further sub-selecting those phage clones. We used the second round, highly enriched phage display peptide library to identify the epitopes and amino acid residues that are important in conferring formalin sensitivity and resistance. This approach is uniquely unbiased in that only the mAb, with or without formalin fixation, performs the selection. By virtue of the diversity of immunoreactive peptide sequences found in the second-round enriched phage display peptide library, we can screen for the peptides that are sensitive or resistant to formalin.
We devised a replica plaque assay, as summarized in Figure 2, to identify peptides that are formalin-sensitive and formalin-insensitive. After two rounds of panning, the selected mAb-specific phage were grown as plaques on a bacterial lawn, on agar. Each phage clone expresses a different peptide, almost all of which are immunoreactive with the mAb. Most peptides share an internal consensus motif that is responsible for binding to the mAb. The consensus motif resembles the actual epitope, as found in the native protein to which the mAb binds. Replicate lifts of nitrocellulose blots were created. One of the nitrocellulose blots was incubated in 10% neutral buffered formalin whereas the other in buffer alone. By aligning the replicate immunoblots, we can determine the phage clone’s immunoreactivity with and without formalin fixation. When both blots, formalin treated and untreated, were immunostained with the mAb and appropriately aligned for comparison, we observed that formalin fixation caused 20 to 80% of the phage clones to lose immunoreactivity. We then sequenced the clones in both categories, analyzing their sequence for clues to explain their sensitivity or resistance to formalin.
A representative plaque-lift assay for the PR mAb 1A6, with or without formalin treatment, is shown in Figure 3. Most of the immunoreactive phage clones in the unfixed replicate blot (left) are no longer immunoreactive after formalin fixation (right). However, as highlighted by the arrows, there are some phage clones that bind to the PR 1A6 mAb regardless of formalin fixation. The immunoreactive peptides expressed by these clones are formalin-insensitive. The open circles at the edge of each blot denote the location of marks that we made on the nitrocellulose membrane, for the purpose of aligning the replicate membranes in the same orientation. Phage clones in both groups (formalin-sensitive as well as formalin-insensitive) were grown and further analyzed.
Figure 3.
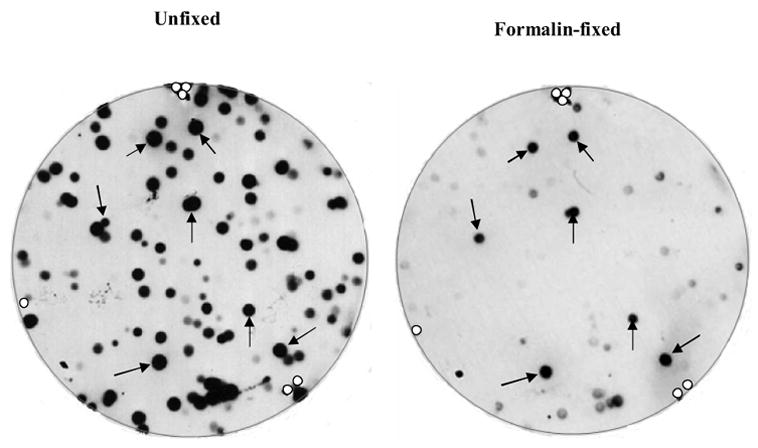
Autoradiograms of duplicate plaque lifts that were untreated (left blot) or treated with formalin (right blot) before immunostaining. The arrows identify the phage clones containing peptide inserts that are immunoreactive in both blots and therefore are not sensitive to formalin. The open circles at the blot edges show where the alignment marks were placed in the membrane, for the purpose of orienting the blot.
We evaluated the phage clones from both groups to identify if there is any difference in binding to the selecting mAb (PR 1A6). Formalin-sensitive or formalin-insensitive phage clones were picked, amplified, and tested for immunoreactivity to mAb 1A6. We tested each phage clone for binding to the PR 1A6 mAb in a phage ELISA (see Materials and Methods). Figure 4 illustrates the mean and standard deviation of the individual phage clones’ immunoreactivity with mAb 1A6. There was no statistically significant difference in the immunoreactivity of the phage clones from the two groups (formalin-insensitive and formalin-sensitive). Both were highly immunoreactive to the target mAb 1A6. This indicates that the strength of antibody binding to the epitopes is comparable between the two groups. Therefore, the strength of antibody binding does not account for sensitivity or insensitivity to formalin fixation.
Figure 4.
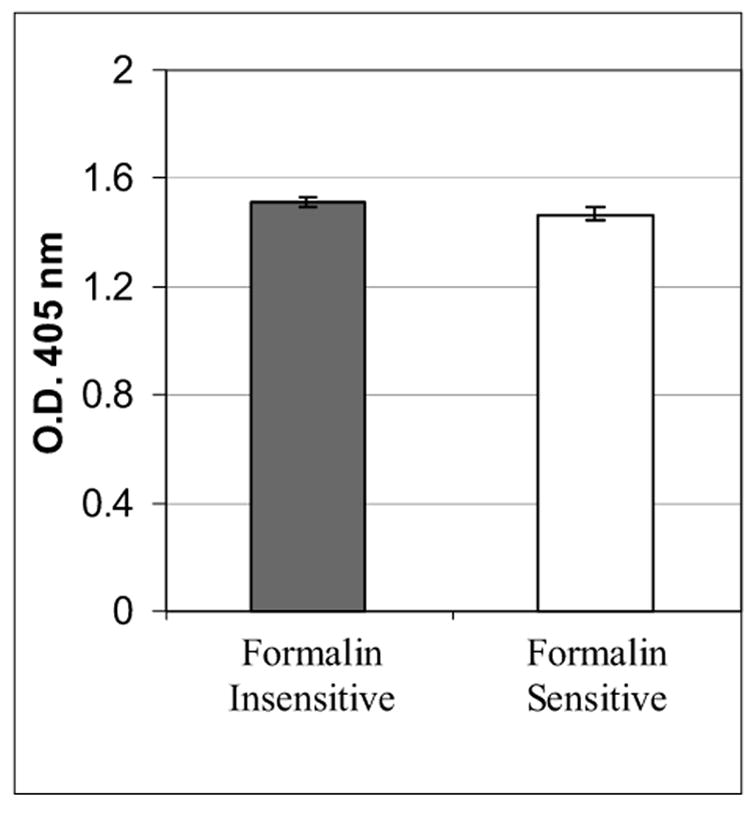
Immunoreactivity of phage clones expressing peptides that are formalin-sensitive and formalin-insensitive, for binding to the PR 1A6 mAb. The vertical bars represent the mean optical density in a phage ELISA ± the standard deviation. For each group, n = 12. There is no statistically significant difference between the two groups.
3.3. Amino acid composition of peptides
To investigate the protein chemistry responsible for formalin-sensitivity, we sequenced the DNA of 10–30 PR1A6 mAb binding phage clones from each group. The amino acid sequence for each phage clone’s peptide insert was deduced from the DNA sequence. Representative data are shown in Figure 5. When the amino acid sequence of the phage clones were aligned, as shown in Figure 5, a consensus amino acid sequence of SRSGX(C)XSYY emerged. This closely matches the epitope in the native sequence, SARSPRSY. In our cyclic peptide phage library, two cysteines (separated by ten intervening residues) are conserved in order to create a cyclic peptide formed by a disulfide bridge between their side chains. Since the cysteines are invariant, no conclusions can be drawn from their presence. The most striking finding from these sequences is the presence of lysines (K) in the formalin-sensitive peptides. Formalin-insensitive peptides had significantly fewer, or no, lysines. Besides PR 1A6, additional analyses were performed using mAbs to the estrogen receptor (mAb ER 1D5), PR 636, and Her-2 CB11 mAb.
Figure 5.

Representative PR mAb 1A6 binding peptides from both formalin-sensitive and formalin-insensitive phage clones. The top panel represents the amino acid sequence of peptide inserts from phage clones that are formalin-insensitive. The bottom panel represents the amino acid sequence of peptide inserts from phage clones that are formalin-sensitive. Each amino acid is represented by its single letter code. The underlined residues represent amino acids that are either identical or conserved substitutions to the native epitope. Lysines (K) are bolded, to facilitate their identification.
Quantitative analysis of the frequency for each amino acid in the peptides of PR 1A6, PR 636, ER 1D5, and Her-2 CB11 mAb binding phage clones, is shown in Figure 6. The open symbol in the graph shows the formalin-insensitive group with a solid black line. The formalin-sensitive group is represented with a dotted line (filled-in symbols). In general, lysine is the only amino acid that is consistently over-represented in the formalin-sensitive group. The difference in the frequency of lysines between the formalin-sensitive and formalin-resistant group is statistically significant (p= .025, one-way ANOVA). This reinforces and reaffirms the importance of lysine residues in formalin reactivity and therefore cross-linking of proteins.
Figure 6.
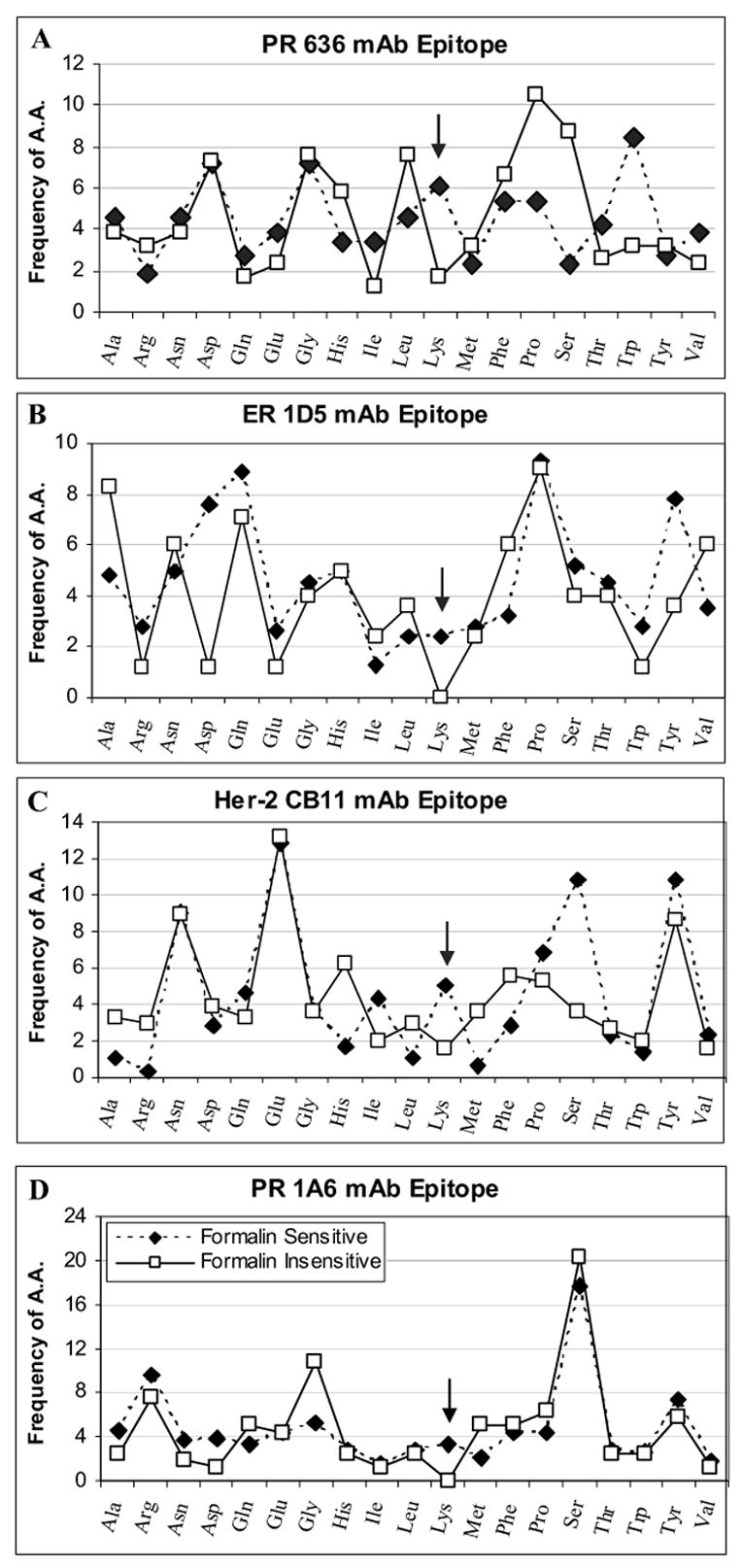
Frequency of each amino acid among formalin-sensitive and formalin-insensitive phage clones. Each box represents the amino acid frequency for PR 636 (A), ER 1D5 (B), Her-2 CB11 (C), and PR 1A6 (D) mAb epitopes. Amino acid frequency is calculated from many peptide sequences for each epitope and the average percentage is presented. The arrows highlight the point in the graphs relevant to lysine frequency, showing their increased frequency in formalin-sensitive epitopes.
Apart from lysines, certain amino acids are over- or under-represented in some, but not all, antibody epitopes. In the PR 1A6 phage clones, glycines (G) are found more often in the formalin-insensitive group. In the PR 636 phage clones, trytophans (W) are over-represented in the formalin-sensitive group, while prolines (P), and serines (S) are over-represented in the formalin-insensitive group. For the ER 1D5 phage clones, aspartic acid (D) and tyrosine (Y) are over represented in formalin-sensitive group. In the Her-2 CB11 phage clones, serine is (moderately) over-represented in the formalin-sensitive group.
4. DISCUSSION
We describe a combinatorial library technique for analyzing antibody epitopes. Phage display of peptide libraries provides us with thousands of immunoreactive peptide sequences after several rounds of selection. By screening this universe of peptides (that bind to an antibody) for sensitivity to a chemical agent, it is now possible to distinguish in a high throughput fashion those peptides that are retain or lose immunoreactivity. Different mAbs will select for different peptides, thereby creating a diverse range of potential chemical reactions. Sequence analysis reveals particular amino acids that are correlated with the phenotype in question. Because of its importance to immunodiagnostics and in vaccine development, we tested the technique using formalin. Once the peptide phage that are sensitive to formalin are identified by plaque lift immunoblots, they can be further amplified and analyzed. By screening so many independent phage clones, this peptide diversity offers a high throughput analysis of the effects of formaldehyde reactions. Peptide phage screening with immunoblot selection method is simple but robust enough to analyze many sequences in a short period of time.
Formalin has a well-established role as a cell and tissue fixative or as an attenuating agent in vaccine preparations. Therefore, there is interest in understanding formaldehyde reactions in the practical contexts for which it is used, as it relates to immunoreactivity. A better understanding of formaldehyde chemistry may help better exploit formaldehyde’s use in pharmaceutical, diagnostic, or biochemical samples, without the adverse effects that are presently encountered. For example, formaldehyde is the most widely used fixative prior to tissue embedding for histopathologic analysis. Unfortunately, many antibodies lose immunoreactivity with their tissue antigens after formalin fixation and paraffin embedding. This study helps delineates the effect of formalin fixation on tissue antigens, at least implicating the role of lysines in the loss of immunoreactivity. Our experimental protocol does not address the possible effect(s) of paraffin embedding postfixation on tissue immunoreactivity.
Of the four mAbs whose epitopes we analyzed, the increased frequency of lysines consistently distinguished the formalin-sensitive from formalin-resistant groups. As previously reported (Fraenkel-Conrat et al., 1947; Fraenkel-Conrat and Olcott, 1948a; Fraenkel-Conrat and Olcott, 1948b; Shi et al., 2000; Metz et al., 2004), lysine residues are reactive with formaldehyde, resulting in various reactions. Formaldehyde can cross-link via a Schiff base, or it can cross-link the epsilon amines of two lysines, or it can cross-link a lysine to hydroxyl group of a serine, or a lysine to a peptide bond. This suggests that lysine is an important residue in conferring sensitivity to formalin fixation, supporting earlier suggestions (Fraenkel-Conrat and Olcott, 1948a; Shi et al., 2000; Metz et al., 2004).
We were surprised that no other amino acid consistently correlated with formalin sensitivity. Several other amino acids’ side chains are capable of reacting with formaldehyde: Asn (N), Gln (Q), Tyr (Y), Arg (R), His (H), and Ser (S). In fact, we previously published data for several mAbs used for clinical immunohistochemistry, which correlated sensitivity to formaldehyde with the presence of tyrosine and arginine (Sompuram et al., 2004). An important distinction between the two studies is that the previous one used mAbs characterized by a reversible loss of immunoreactivity. Namely, immunoreactivity was recoverable after antigen retrieval, a process whereby tissue sections are heated to boiling. By contrast, reversibility of immunoreactivity, such as after antigen retrieval, was not a selecting criterion in this study. Thus, it is possible that, although lysine was a preponderant amino acid in this study, it might not have been had we required reversibility.
Apart from lysines, there were isolated instances where other formaldehyde-reactive amino acids were increased in the formalin-sensitive group. For example, the ER 1D5 mAb formalin-sensitive epitopes showed an increased proportion of tyrosines and, to a smaller degree, arginines (Figure 6, panel B). This was the same mAb whose epitope we previously characterized as being associated with the Mannich reaction, involving tyrosines and arginines, after formalin fixation. Also, the Her-2 mAb CB11 had an increased proportion of serines in the formalin-sensitive group. Therefore, it seems reasonable that other formaldehyde reactions may likely occur in fixed tissues besides those involving lysines, depending upon the amino acid composition of the epitope.
In a broader context, this screening strategy is potentially useful for identifying immunoreactive epitopes with defined chemical sensitivities. With regards to formalin, this might be useful in characterizing vaccine sensitivity, or in the creation of novel mAbs that are immunoreactive after formalin treatment. For either goal, a random combinatorial peptide phage library can be enriched with immune sera and tested for sensitivity to the chemical in question. Sequencing of the peptide inserts will lead to identification of the epitope conferring the desired property. For antibody development, that peptide, or even the phage clone itself (van Houten et al., 2006) can then be used as an immunogen.
Acknowledgments
This work was supported by grants R44CA81950 and R43CA94557 from the National Cancer Institute to S.A. Bogen.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al Saati T, Clamens S, Cohen-Knafo E, Faye J, Prats H, Coindre J, Wafflart J, Caveriviere P, Bayard F, Delsol G. Production of monoclonal antibodies to human estrogen receptor protein (ER) using recombinant ER (RER) Int J Cancer. 1993:651–654. doi: 10.1002/ijc.2910550423. [DOI] [PubMed] [Google Scholar]
- Anonymous. (2002a) 6F11 [package insert].
- Anonymous. (2002b) CB11 [package insert].
- Bertheau P, Cazals-Hatem D, Meignin V, de Roquancourt A, Verola O, Lesourd A, Sene C, Brocheriou C, Janin A. Variability lf immunohistochemical reactivity on stored paraffin slides. J Clin Pathol. 1998;51:370–374. doi: 10.1136/jcp.51.5.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevitt D, Milton I, Pigger N, et al. New monoclonal antibodies to oestrogen and progesterone receptors effective for paraffin section immunohistochemistry. J Pathol. 1997;183:228–232. doi: 10.1002/(SICI)1096-9896(199710)183:2<228::AID-PATH895>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Crockett D, Lin Z, Vaughn C, Lim M, Elenitoba-Johnson K. Identification of proteins from formalin-fixed paraffin-embeded cells by LC-MS/MS. Lab Invest. 2005;85:1405–1415. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- Ellis R. New techniques for making vaccines. Vaccine. 1999;17:1596–1604. doi: 10.1016/s0264-410x(98)00416-2. [DOI] [PubMed] [Google Scholar]
- Foss R, Guha-Thakurta N, Conran R, Gutman P. Effect of ficative and fixation time on the extraction and polymerase chain reaction amplification of RNA form paraffin-embedded tissue. Comparision of two housekeeping gene mRNA controls. Diagn Mol Pathol. 1994;3:148–155. doi: 10.1097/00019606-199409000-00003. [DOI] [PubMed] [Google Scholar]
- Fox C, Jhonson F, Whiting J, Roller P. Formaldehyde fixation. J Histochem Cytochem. 1985;33:845–853. doi: 10.1177/33.8.3894502. [DOI] [PubMed] [Google Scholar]
- Fraenkel-Conrat H, Brandon B, Olcott H. The reaction of formaldehyde with proteins. IV Participation of idole groups Gramicidin. J Biol Chem. 1947;168:99–118. [PubMed] [Google Scholar]
- Fraenkel-Conrat H, Olcott H. The reactions of formaldehyde with proteins. V Cross-linking between amino and primary amide or guanidyl groups. J Am Chem Soc. 1948a;70:2673–2684. doi: 10.1021/ja01188a018. [DOI] [PubMed] [Google Scholar]
- Fraenkel-Conrat H, Olcott H. Reactions of formaldehyde with proteins. VI Cross-linking of amino groups with phenol, imidazole, or indole groups. J Biol Chem. 1948b;174:827–843. [PubMed] [Google Scholar]
- Huebner J. Antibody-antigen interactions and measurements of immunologic reactions, Chap 9. In: Pier GB, Lyczak JB, Wetzler LM, editors. Immunology, Infection, and Immunity. 2004. [Google Scholar]
- Ikeda K, Monden T, Kanoh T, Tsujie M, Izawa H, Haba A, Ohnishi T, Sekimoto M, Tomita N, Shiozaki H, Monden M. Extraction and analysis of diagnostically useful proteins form foramlin-fixed, paraffin-embeded tissue sections. J Histochem Cytochem. 1998;46:397–403. doi: 10.1177/002215549804600314. [DOI] [PubMed] [Google Scholar]
- Kuby J. Immunology. WH Freeman; New York, N.Y: 1994. [Google Scholar]
- Leppla S, Robbins J, Schneerson R, Shiloach J. Development of improved vaccine for anthrax. J Clin Invest. 2002;110:141–144. doi: 10.1172/JCI16204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA form formalin-fixed samples and optimization of molecular biology applicatons for such samples. Nucleic Acids Res. 1999;27:4436–4443. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLafferty M, Kent R, Ladner R, Markland W. M13 bacteriophage displaying disulfide-constrained microproteins. Gene. 1993;128:29–36. doi: 10.1016/0378-1119(93)90149-w. [DOI] [PubMed] [Google Scholar]
- Metz B, Kerstein F, Hoogerhout P, Brugghe H, Timmermans H, de Jong A, Meiring H, ten Hove J, Hennink W, Crommelin D, Jiskoot W. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J Biol Chem. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- Murdin A, Barreto L, Plotkin S. Inactivated poliovirus vaccine: past and present experience. Vaccine. 1996;14:735–746. doi: 10.1016/0264-410x(95)00211-i. [DOI] [PubMed] [Google Scholar]
- Parmley S, Smith G. Antibody-selected filamentous fd phage vectors: affinity purification of target genes. Gene. 1988;73:305–318. doi: 10.1016/0378-1119(88)90495-7. [DOI] [PubMed] [Google Scholar]
- Rait V, O'Leary T, Mason J. Modelling formalin fixation and antigen retrieval with bovine pancreatic ribonuclease A: I-Structural and functional alterations. Lab Invest. 2004a;84:292–299. doi: 10.1038/labinvest.3700045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rait V, Xu L, O'Leary T, Mason J. Modelling formalin fixation and antigen retreival with bovine pancreatic RNase A: II- Interrelationship of cross-linking, immunoreactivity, and heat treatment. Lab Invest. 2004b;84:300–306. doi: 10.1038/labinvest.3700041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer B. Antigen preservation tests for immunocytochemical detection of cytoskeletal proteins: influence of aldehyde fixatives. J Histochem Cytochem. 1989;37:675–681. doi: 10.1177/37.5.2495321. [DOI] [PubMed] [Google Scholar]
- Shi SR, Cote R, Wu L, Liu C, Datar R, Shi Y, Liu D, Lim H, Taylor C. DNA extraction from archival formalin-fixedm paraffin-embedded tissue sections based on the antigen retrieval principle: Heating under the influence of pH. J Histochem Cytochem. 2002;50:1005–1011. doi: 10.1177/002215540205000802. [DOI] [PubMed] [Google Scholar]
- Shi S-R, Gu J, Turrens J, Cote R, Taylor C. Development of the antigen retrieval techniques: Philosophical and theoretical bases. In: Shi S, Gu J, Taylor C, editors. Antigen Retrieval Techniques: Immunohistochemistry and Molecular Morphology. Eaton Publishing.; Natick, MA.: 2000. pp. 17–40. [Google Scholar]
- Shi SR, Key M, Kalra K. Antigen retreival in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochrmical staining based on microwave oven heating of tissue sections. J Histochem Cytochem. 1991;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- Smith G, Petrenko V. Phage display. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- Sompuram S, Kodela V, Ramanathan H, Wescott C, Radcliffe G, Bogen S. Synthetic peptides identified from phage-displayed combinatorial libraries as immunodiagnostic assay surrogate quality control targets. Clin Chem. 2002;48:410–420. [PubMed] [Google Scholar]
- Sompuram S, Vani K, Bogen S. A molecular model of antigen retrieval using a peptide array. Amer J Clin Pathol. 2006a;125:91–98. [PubMed] [Google Scholar]
- Sompuram S, Vani K, Hafer L, Bogen S. Antibodies immunoreactive with formalin-fixed tissue antigens recognize linear protein epitopes. Amer J Clin Pathol. 2006b:82–89. [PubMed] [Google Scholar]
- Sompuram S, Vani K, Messana E, Bogen S. A molecular mechanism of formalin fixation and antigen retrieval. Amer J Clin Pathol. 2004;121:190–199. doi: 10.1309/BRN7-CTX1-E84N-WWPL. [DOI] [PubMed] [Google Scholar]
- Sparks A, Adey N, Cwirla S, Kay B. Screening phage-displayed random peptide libraries. In: Kay B, Winter J, McCafferty J, editors. Phage display of peptides and proteins. Academic Press; New York, NY: 1996. [Google Scholar]
- Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boreck L, van de Vijver M. Assessment of problems in diagnostic and research immunohistochemistry associated with epitope instability in stored paraffin sections. Appl Immunohistochem Mol Morphol. 2000;8:316–321. [PubMed] [Google Scholar]
- van Houten N, Zwick M, Menendez A, Scott J. Filamentous phage as an immunogenic carrier to elicit focused antibody responses against a synthetic peptide. Vaccine. 2006;24:4188–4200. doi: 10.1016/j.vaccine.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virneckas B, Pluckthun A, Scneider K, Wellnhofer G, Moroney S. Trinucleotide phosphoramidites: ideal reagents for the synthesis of mixed oliginucleotides for random mutagenesis. Nucleic Acid Res. 1994;22:5600–7. doi: 10.1093/nar/22.25.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M, Chott A, Fabiano A, Battifora H. Effects of formalin tissue fixation and processing on immunohistochemistry. Am J Surg Pathol. 2000;24:1016–1019. doi: 10.1097/00000478-200007000-00014. [DOI] [PubMed] [Google Scholar]
- Wester K, Wahlund E, Sundstrom C, Ranefall P, Bengtsson E, Russell P, Ow K, Malmstrom P, Busch C. Paraffin section storage and immunohistochemistry: effects of time, temperature, fixation and retrieval protocol with emphasis on p53 protein and MIB1 antigen. Appl Immunohistochem Mol Morphol. 2000;8:61–70. [PubMed] [Google Scholar]
- Williams C, Ponten F, Moberg C, Söderkvist P, Uhlén M, Pontén J, Sitbon G, Lundeberg J. Am J Pathol. Vol. 155. 1999. A high frequency of sequence alterations is due to formalin fixation of archival specimens; pp. 1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]


