Summary
To establish viral infection, SV40 must expose nuclear localization signals (NLSs) that are internal in the virion architecture in order to enter the nucleus via interaction with the host’s nuclear import machinery, which includes importin α and importin β. The time course for SV40 association with the importins in infected cells was examined. The viral DNA associated with importin α by 1.5 hours post infection, before associating with the importin β nuclear import receptor, by 3 hours post infection. Only a small fraction of cell-internalized SV40 that contained viral DNA was bound by the two importins. This fraction, termed “nuclear entry-competent SV40,” was slightly smaller than the virion but, importantly, was larger than the viral chromatin and contained both Vp1 and Vp3. Furthermore, the internalized viral DNA in either anti-importin or anti-Vp3 immune complexes was sensitive to DNase I, whereas the viral DNA in mature virions was resistant. All these results suggest that once SV40 enters the cytoplasm, it undergoes an architectural modification that exposes the virion’s NLSs for nuclear entry.
Many animal viruses replicate their genome in the nucleus. Thus, nuclear import of their genomes is essential for establishing infection (Kasamatsu and Nakanishi, 1998). For adenovirus, simian virus 40 (SV40), herpes simplex virus type 1, hepatitis B virus, canine parvoviruses, and influenza A virus, association of the virion or viral protein(s) with the host’s nuclear import machinery, the α- and β-importins (Gorlich and Kutay, 1999) guides nuclear entry (Clever et al., 1991; Cros et al., 2005; Greber et al., 1997; Ojala et al., 2000; Rabe et al., 2003; Vihinen-Ranta et al., 2002). The nuclear localization signals (NLSs) of virion proteins that recognize host import machinery are not necessarily exposed on the surface of the particles (Cros et al., 2005; Nakanishi et al., 2002; Rabe et al., 2003; Trotman et al., 2001; Vihinen-Ranta et al., 2002). For SV40, Vp3-NLSs function as the virion’s NLSs, but they are not exposed in the intact structure (Nakanishi et al., 2002). Thus, an alteration of the SV40 architecture to expose these signals during infection is essential for the nuclear delivery of its genome.
SV40, family Polyomaviridae, is a small non-enveloped virus that is composed of major capsid protein Vp1, minor capsid proteins, Vp2 and Vp3, a circular double stranded viral DNA genome and host encoded histones. The capsid is comprised of 72 Vp1 pentamers, each containing one minor capsid protein, either Vp2 or Vp3 (Chen et al., 1998; Liddington et al., 1991). The SV40 virion enters the nucleus through the nuclear pore complex (NPC) (Clever et al., 1991), using the NLSs residing within the common amino acids of the minor capsid proteins of Vp2 and Vp3, herein designated as Vp3-NLS. The minor capsid proteins must also associate with SV40 DNA for nuclear entry (Nakanishi et al., 1996). Only a small fraction of virions that enter the cell during polyomavirus infection deliver their genomes into the nucleus (Mannova and Forstova, 2003; Norkin et al., 2002). Because of this small fraction, it has been difficult to identify nuclear entry-competant SV40 in the cytoplasm. This small fraction of SV40 particles are those that expose an NLS and are therefore capable of recognizing the α– and β– importins (Nakanishi et al., 2002). However, importantly, a complete dissociation of the virion within the cell cytoplasm prevents the nuclear entry of the viral DNA (Li et al., 2003).
The size of an intact SV40, 50 nm in diameter, exceeds the functional size of the nuclear pore complex (NPC) (Dworetzky et al., 1988; Pante and Kann, 2002). It is possible that the infecting SV40 can somehow alter its shape to pass through the NPC. To study the alteration, if any, that SV40 might undergo, we examined the time course by which the importins associate with components of the nuclear entry-competent SV40.
TC-7 cells were infected with SV40 for 1.5, 3 and 6 hours, and cytoplasmic lysates containing 20–100 ng/ml of viral DNA were prepared as described (Nakanishi et al., 2002). The lysates from 2.5 x 105 cells were reacted with antibodies against importin α, importin β, or rabbit anti-mouse IgG, a control antibody, and then with protein A-conjugated beads, thereby forming immune complexes (IPs). After the addition of the control DNA (which is slightly shorter in length than the viral genome and thus can serve as product quantity control for PCR), co-precipitated viral DNAs were purified and subjected to semi-quantitative PCR (Nakanishi et al. 2002). In the lysate prepared at 1.5 hours post infection (hpi), no viral DNA was found associated with importin β IP (Fig. 1A; lane 4), whereas a small amount was found in the importin α IP (Fig. 1A, lane 3). Hence, only a few Vp3-NLSs became accessible to and interacted with importins at this time point. By 3 hpi and 6 hpi, substantial amounts of viral DNA were detected in importin α IPs (Fig. 1B and 1C, lane 3). In importin β IPs, the viral DNA was barely detected above the background (lane 2) in the 3-hpi lysate, and its amount increased as infection progressed from 3 hpi (Fig. 1B, lane 4) to 6 hpi (Fig. 1C, lane 4). However, even at 6 hpi only a fraction of input-viral DNA in the lysates (lane1) was found in importin IPs (lanes 3 and 4). The control antibody, anti-mouse IgG, did not co-precipitate viral DNA (lane 2). Thus, the importin association to internalized SV40 occurred in a step-wise manner; first associating to importin α adaptor and then to the nuclear import receptor, importin β. Importin α predominantly localizes in the cytoplasm (Imamoto et al., 1995), and importin β is found mostly at the nuclear rim (Chi et al., 1995). Thus, our results are consistent with the observation that virion particles localize to the perinuclear region at 6hpi but not at the preceding time-points (Mannova and Forstova, 2003; Norkin et al., 2002; Pelkmans et al., 2001; Richards et al., 2002). The simplest interpretation of this result would be that the stepwise association to the respective importins (importin α then to importin β) reflects the intracellular concentration of importins for interaction, and the time for the nuclear entry-competent particles to traverse to the proximity of the nucleus.
Fig. 1. Time course of importins association with cell-internalized SV40.
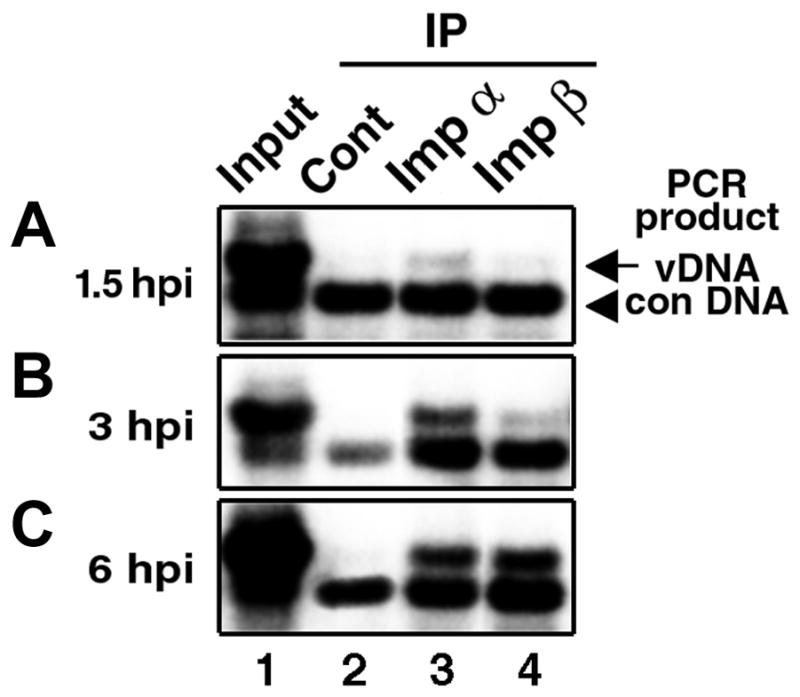
Cells were infected with SV40 at 100 particles per cell, and cytoplasmic lysates prepared after 1.5 (A), 3 (B) and 6 (C) hours of infection (hpi) were reacted with either anti-mouse IgG (lane 2, “Cont”), anti-importin α (lane 3, “Imp α”), or anti-importin β antibodies (lane 4, “Imp β”). The inclusion of an irrelevant antibody such as rabbit anti-mouse IgG in parallel serves as an indicator for background level of detection. The co-immunoprecipitated viral DNA was probed for the presence of viral DNA via semi-quantitative PCR with a fixed amount of exogenously added control DNA, NO-pSV40ΔNcoI (Nakanishi et al. 2002). The expected amplification product of the wild-type SV40 genome is 2.1 kbp (arrow, “vDNA”), and that of the control DNA, 1.7 kbp (arrowhead, “con DNA”). For "Input" (lane 1), 1/5 as much cytoplasmic lysate as the amount used for each immunoprecipitation was directly loaded on the gel (Nakanishi et al., 2002).
We then wished to know if the NLS-exposed virions that associate with importins sediment in sucrose gradients differently than importin-free SV40 virions. Cytoplasmic lysates of SV40 infected cells prepared at either 3 hpi or 6 hpi were sedimented through sucrose gradients and fractionated into 14 samples from the bottom of the tube. The locations in fractions of virion (SV40) or viral minichromosome are marked as described (Nakanishi et al., 2002). The distribution in the gradient of DNA and Vp1 in fractions from 6hpi sample showed that the majority of viral DNA was found in fractions where virions sediment; however, there were smaller or less dense species sedimenting slightly slower than virions. Similarly, Vp1 was mostly associated with intact virions but also was found in slower sedimenting fractions (Fig. 2C). These results suggest that architectural alterations of some of the cell-internalized SV40 have occurred.
Fig. 2. Sedimentation analysis of infected cell lysates.
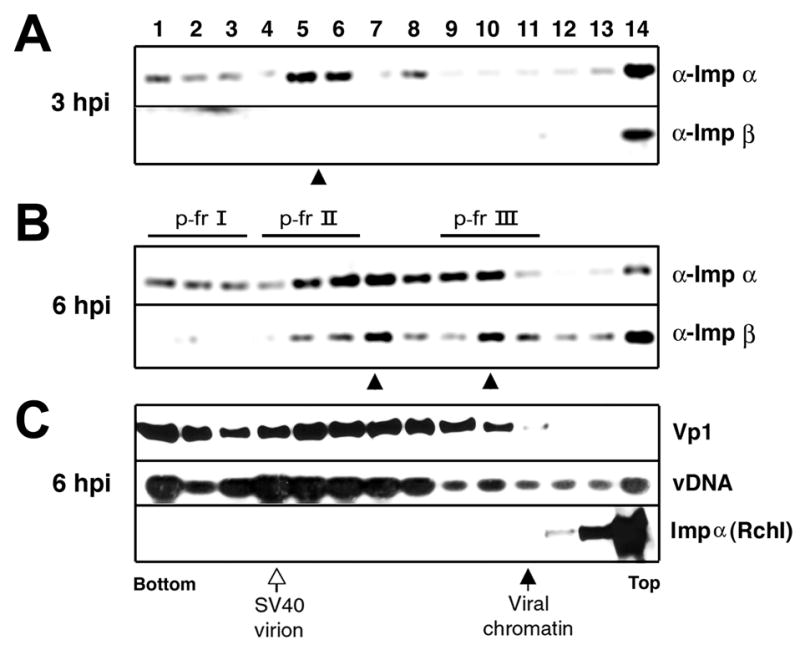
Approximately 500 ul of cytoplasmic lysates prepared at 3 hpi (A) or at 6 hpi [(B) and (C)] as above were sedimented through a 5–32% sucrose gradient in 10 mM HEPES (pH 7.5) using Beckman SW41 rotor at 37,000 rpm for 80 min at 4ºC. Fourteen fractions from the bottom were collected and were immunoreacted with either anti-importin α (α-imp α) or anti-importin β (α-imp β) antibody. A and B: The presence of co-precipitated viral DNA in immunoprecipitates was probed by semi-quantitative PCR. The peak fractions were marked as filled arrowhead at the bottom of each panel. C: The 6-hpi cytoplamic lysate was also probed for the presence in each fraction of either viral DNA (“vDNA”) or Vp1 (“Vp1”) or importin α (“Imp α”). Vp1 or importin was probed by Western blot using rabbit anti-Vp1 serum or mouse anti-importin antibody as the primary antibody followed by detection of the signal by ECL, respectively (Li et al., 2001). The viral DNA was extracted from each fraction and detected by Southern blot using 32P labeled SV40 DNA as a probe. 3H-thymidine labeled SV40 infected cell lysate was sedimented in a parallel tube, and the radioactivities in respective fractions were determined with a scintillation counter. The peak sedimentation positions for “SV40 virion” and “viral chromatin,” correlated with high 3H-counts, were designated at the bottom of the panels (Nakanishi et al., 2002).
The respective fractions of lysates prepared at 3 hpi and 6 hpi were subjected to IP using anti-importin α or anti-importin β, followed by semi-quantitative PCR to determine the amount of co-precipitated viral DNA in each IP (Fig. 2A and 2B). In importin α IPs of the 3-hpi sample, the viral DNAs were found to sediment in two locations, one species that sediments faster-than-virion (fractions 1 through 3) or the other, slightly-slower-than-virion (fractions 5 through 8) (Fig. 2A, arrowhead). In contrast, no viral DNA sedimented as importin β IPs at this time point.
When the 6-hpi lysate was analyzed, both antibodies co-precipitated viral DNAs (Fig. 2B). The DNA profile in importin α IPs was similar to that obtained from the 3-hpi sample; however, much more DNA was found in the 6-hpi sample. The profile also paralleled the distribution of the Vp1 mass (Fig. 2C, Vp1), implying that species that interact with importin α are heterogeneous in size and include the virion. The viral DNA in importin β IPs was found both in virion DNA fractions and those that sedimented more slowly (compare the viral DNA profile of “α-Imp β” in Fig. 2B with that in Fig. 2C). These sedimentation profiles differed from the distribution profiles for viral DNA and Vp1 found in the respective fractions (Fig. 2C). This is consistent with results shown in Fig. 1 that distinct complexes interact with importins and constitute only a small fraction of internalized SV40. The observation that importin α IPs are clearly observed at 3 hpi, whereas importin-β IPs are clearly observed at 6 hpi, taken together with the profile of sedimenting DNA, again argues that a subpopulation of the internalized virions associate first with importin α and then with importin β. That complexes containing both importins, viral proteins and DNA sedimented slower than virions implies that these complexes have a grossly changed architecture from that of the intact virion by 6 hpi.
To see if complexes found in the 6-hpi lysates that interacted with either or both importins (Fig. 2B) contained either Vp1 or Vp3, we examined the presence in the IPs of Vp1 and Vp3. The fractions from gradients that had been prepared in parallel to those shown in Fig. 2B were pooled: Fractions 1 to 3 (referred to as p-fr I), 4 to 6 (referred to as p-fr II), or 9 to 11 (referred to as p-fr III). Portions of each pool were immunoprecipitated with anti-importin α or anti-importin β antibody, and the resulting IPs were probed for Vp1 and Vp3 on separate SDS-PAGE gels and separate Western blots, with a few times more of the IPs being allotted for Vp3 detection than for Vp1 detection (Fig. 3A). Vp1 appeared to be present in the importin α IPs of p-fr I, II, and III (Fig. 3A, Anti-Vp1, lanes 1, 3, and 5) and the importin β IP of p-fr I (lane 2), and Vp1 was hardly detectable in importin β IPs of p-fr II and III (Fig. 3A, “Anti-Vp1,” lanes 4 and 6). Yet because the immunoglobulin (Ig) heavy chain migrated close to the position of the Vp1 band and was reacting strongly with the Western blot reagents in Fig. 2B, obscuring the presence and relative amount of Vp1 in each lane. All importin IPs contained Vp3 to varying extents (Fig. 3A, “Anti-Vp3,” lanes 1 through 6). In an attempt to better detect the importins-associated capsid proteins, 6 hpi infected lysated were prepared by increasing the number of virion particles from 100 to 3000 particles per cell as well as increasing the total number of cells used for infection. In this experiment, fractions 1 through 11 were combined following sedimentation and were used instead of separate pooled fractions I, II, and III. We also used a commercially available combination of anti-mouse resin (in place of protein A beads) for the IP reactions and secondary antibody lacking affinity for denatured Ig chains (in place of regular anti-rabbit secondary antibody) for the Western blots, in order to eliminate interfering detection of Ig chains. Vp1 and Vp3 were found in anti-Imp α and β IPs at greater levels than those found in control anti-β gal IP (Fig. 3B, lanes 3 and 4 of upper and lower panels, compared with lane 2). Under these blotting conditions, we can detect about 0.4 ng of Vp1 or 0.1 ng of Vp3 (Fig. 3B, lanes 3 and 4, compared to the SV40 standard in lane 5). The ratio of Vp1 to Vp3 found in the Imp IPs was close to that detected for purified SV40 (Fig. 3B, lane 5), compatible with the idea that “particle-like” complexes are associated with the importins and enter the nucleus. Despite an increase in the multiplicity of infection (MOI), the signals for Vp1 and Vp3 remained low, suggesting the possibility that there may be a small but unique level of importins destined for specific cargos and the import systems have been fully saturated. Mannova and Forstova have reported that regardless of MOI, only a small amount of polyomavirus DNA reaches the nucleus during polyomavirus infection (Mannova and Forstova, 2003).
Fig. 3. Analysis of importins-bound fraction of infected lysate A and B: Presence of capsid proteins in importin immunoprecipitates.
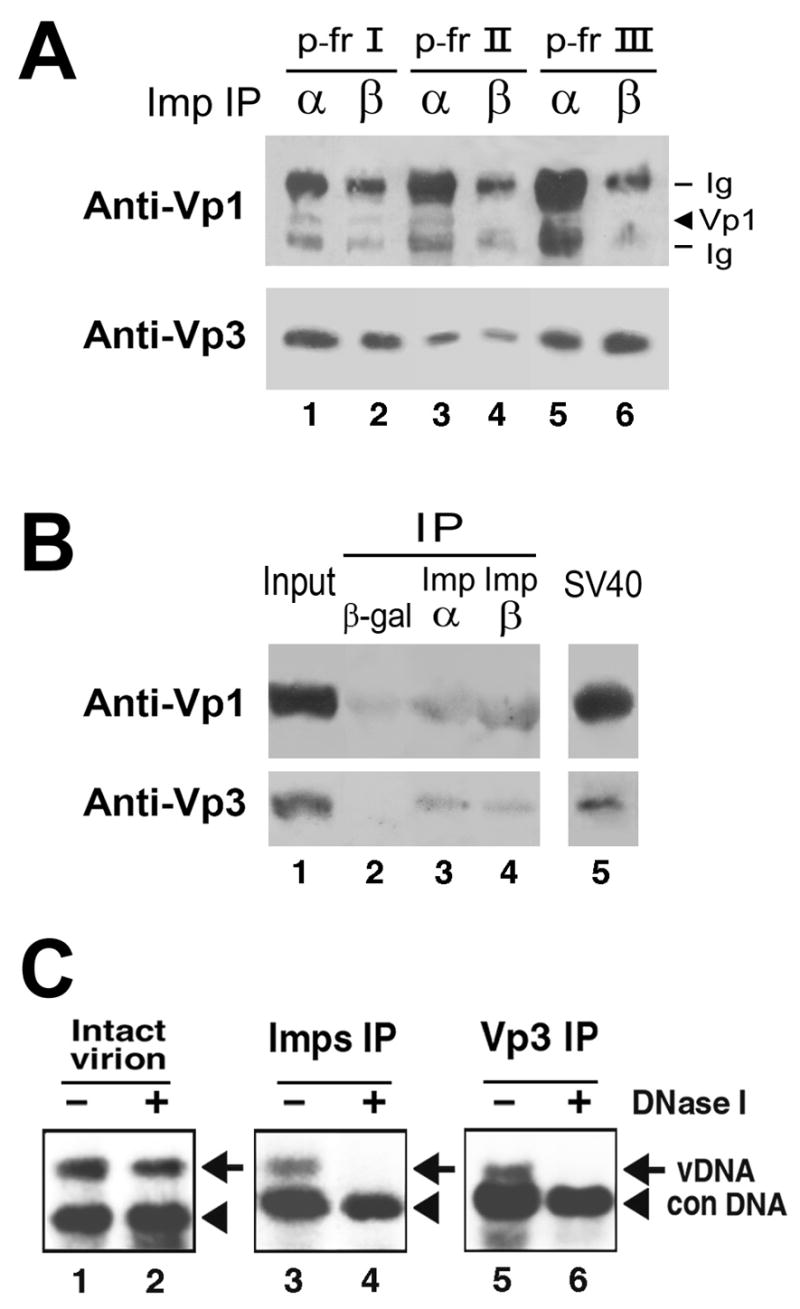
(A) Cytoplasmic lysate prepared at 6 hpi from 2 x 107 cells was sedimented through a 5–32% sucrose gradient as in Fig. 2. Of the 14 fractions collected, fractions 1 through 3 (p-fr I), 4 through 6 (p-fr II), or 9 through 11 (p-fr III) were pooled, and half of each pool was reacted with either anti-importin α (lanes labeled with α) or anti-importin β (lanes with β) antibody, followed by reaction with protein A-sepharose beads. One-fourth and ¾ of each immunoprecipitate were resolved on separate SDS-PAGE gels and Western blotted with anti-Vp1 serum (“Anti-Vp1” panel) and affinity-purified anti-Vp3 antibody (“Anti-Vp3” panel), respectively, followed by ECL detection (Li et al., 2001). Bands originating from the heavy chain of anti-importin immunoglobulin G are also visible (“Ig”). (B). Cytoplasmic lysate was prepared at 6 hpi from 4 x 107 cells that were infected with SV40 at 3000 particles per cell. The lysate was sedimented through sucrose gradient and fractionated as in Fig. 2A. Fractions 1 through 11 were pooled and divided into three equal parts. Each part was reacted with either anti-β-galactosidase (“β– gal”), anti-importin α (“Imp α”, or anti-importin β (“Imp β") antibody. The immune complexes (“IP”) were collected via reaction with TrueBlot anti-mouse Ig beads (eBioscience, San Diego, Calif.) and resolved on one SDS-PAGE gel. The portions of the gel containing the expected Vp1 or Vp3 band were then processed for separate Western blots as above (“anti-Vp1” and “anti-Vp3” panels), except TrueBlot horseradish peroxidase-conjugated anti-rabbit IgG (eBioscience) was used as the secondary antibody for both blots. The fractions-1-through-11 pool, at 1/150 the amount used for each immunoprecipitation in lanes 2, 3, or 4, was included as the “Input” sample in lane 1. Purified SV40 virions containing 2 ng of Vp1 and 0.2 ng of Vp3 were analyzed in lane 5 as an “SV40” Western blot control. C: Viral DNA in importin-bound complex was sensitive to DNase I digestion. Cytoplasmic lysate of 6-hpi cells was immunoprecipitated with either a mixture of anti-importin α and β antibodies (“Imps IP”) or anti-Vp3 antibody (“Vp3 IP”) as described (Nakanishi et al., 2002). The immune-complexes (lanes 3 through 6, right panels) as well as SV40 virion (lanes 1 and 2, left panel, Intact virion) were incubated in 10 mM Tris-HCl, 10 mM NaCl, and 5 mM MgCl2 either in the absence (lanes 1, 3, and 5) or in the presence (lanes 2, 4, and 6) of 0.1 unit/ml of DNase I at 37ºC for 20 min. The samples were probed for viral DNA as in Fig. 1. Arrow and arrowhead indicate the amplified products for viral DNA and control DNA, respectively.
That viral DNA was present in the same sucrose fractions that contained the importins (Fig. 2C) is consistent with the interpretation that viral DNAs are in association with capsid proteins following internalization at 6 hpi, and consistent with our previous observations (Nakanishi et al., 2002). The results again imply that during infection, the SV40 architecture changes in the cytoplasm, but still maintains some of the capsid proteins along with the viral DNA, as the viral complex becomes competent for nuclear entry (Yamada and Kasamatsu, 1993).
The DNA in intact SV40 was resistant to DNase I treatment (Fig. 3C, lanes 1 and 2). In confirming the results shown above, the viral DNAs in IPs were also examined for the nuclease sensitivity. After the nuclease digestion, the viral DNAs in importin IP and Vp3 IP became DNase I sensitive (Fig. 3C, lanes 4 and 6, compared with lanes 3 and 5). When virions were incubated with importins α and β either in the absence or the presence of recombinant Ran, the viral DNA remained resistant to the nuclease (Nakanishi, unpublished observation). Thus, the change in nuclease sensitivity which we observed in the IPs reflects the physiological change caused by the cell entry of virion, prior to the virion’s interaction with the nuclear import machinery.
Finally, recent results suggest a role of Vp1 amino acid, Glu330, which coordinates site-1 calcium binding, in exposing signals that are necessary for the cell attachment and cell internalization of infecting SV40 (Li et al., 2003). Cavaldesi et al. have also postulated an alteration of the structure of murine polyomavirus capsid upon sialic acid binding (Cavaldesi et al., 2004). Our results described here further support the idea that the architecture of the internalized SV40 undergoes an alteration in the cytoplasm in order to expose Vp3-NLSs and become nuclear entry-competent. Further studies are necessary to substantiate if the observed structures found in association with the importins represents genuine intermediates that infecting virions go through prior to the nuclear entry. Understanding of how and where such alteration takes place is important for understanding the nuclear entry of SV40 during infection.
Acknowledgments
We thank Laurie Bankston for editing. This work is supported by a grant from the National Cancer Institute (CA50574).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cavaldesi M, Caruso M, Sthandier O, Amati P, Garcia MI. Conformational changes of murine polyomavirus capsid proteins induced by sialic acid binding. J Biol Chem. 2004;279(40):41573–9. doi: 10.1074/jbc.M405995200. [DOI] [PubMed] [Google Scholar]
- Chen XS, Stehle T, Harrison SC. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 1998;17(12):3233–40. doi: 10.1093/emboj/17.12.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130(2):265–74. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever J, Yamada M, Kasamatsu H. Import of simian virus 40 virions through nuclear pore complexes. Proc Natl Acad Sci U S A. 1991;88(16):7333–7. doi: 10.1073/pnas.88.16.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cros JF, Garcia-Sastre A, Palese P. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic. 2005;6(3):205–13. doi: 10.1111/j.1600-0854.2005.00263.x. [DOI] [PubMed] [Google Scholar]
- Dworetzky SI, Lanford RE, Feldherr CM. The effects of variations in the number and sequence of targeting signals on nuclear uptake. J Cell Biol. 1988;107(4):1279–87. doi: 10.1083/jcb.107.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–60. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Greber UF, Suomalainen M, Stidwill RP, Boucke K, Ebersold MW, Helenius A. The role of the nuclear pore complex in adenovirus DNA entry. Embo J. 1997;16(19):5998–6007. doi: 10.1093/emboj/16.19.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. Embo J. 1995;14(15):3617–26. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamatsu H, Nakanishi A. How do animal DNA viruses get to the nucleus? Annu Rev Microbiol. 1998;52:627–86. doi: 10.1146/annurev.micro.52.1.627. [DOI] [PubMed] [Google Scholar]
- Li PP, Nakanishi A, Shum D, Sun PC, Salazar AM, Fernandez CF, Chan SW, Kasamatsu H. Simian virus 40 Vp1 DNA-binding domain is functionally separable from the overlapping nuclear localization signal and is required for effective virion formation and full viability. J Virol. 2001;75(16):7321–9. doi: 10.1128/JVI.75.16.7321-7329.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PP, Naknanishi A, Tran MA, Ishizu K, Kawano M, Phillips M, Handa H, Liddington RC, Kasamatsu H. Importance of Vp1 calcium-binding residues in assembly, cell entry, and nuclear entry of simian virus 40. J Virol. 2003;77(13):7527–38. doi: 10.1128/JVI.77.13.7527-7538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991;354(6351):278–84. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- Mannova P, Forstova J. Mouse polyomavirus utilizes recycling endosomes for a traffic pathway independent of COPI vesicle transport. J Virol. 2003;77(3):1672–81. doi: 10.1128/JVI.77.3.1672-1681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi A, Clever J, Yamada M, Li PP, Kasamatsu H. Association with capsid proteins promotes nuclear targeting of simian virus 40 DNA. Proc Natl Acad Sci U S A. 1996;93(1):96–100. doi: 10.1073/pnas.93.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi A, Shum D, Morioka H, Otsuka E, Kasamatsu H. Interaction of the Vp3 nuclear localization signal with the importin alpha 2/beta heterodimer directs nuclear entry of infecting simian virus 40. J Virol. 2002;76(18):9368–77. doi: 10.1128/JVI.76.18.9368-9377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norkin LC, Anderson HA, Wolfrom SA, Oppenheim A. Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J Virol. 2002;76(10):5156–66. doi: 10.1128/JVI.76.10.5156-5166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol Cell Biol. 2000;20(13):4922–31. doi: 10.1128/mcb.20.13.4922-4931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13(2):425–34. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L, Kartenbeck J, Helenius A. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat Cell Biol. 2001;3(5):473–83. doi: 10.1038/35074539. [DOI] [PubMed] [Google Scholar]
- Rabe B, Vlachou A, Pante N, Helenius A, Kann M. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc Natl Acad Sci U S A. 2003;100(17):9849–54. doi: 10.1073/pnas.1730940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AA, Stang E, Pepperkok R, Parton RG. Inhibitors of COP-mediated transport and cholera toxin action inhibit simian virus 40 infection. Mol Biol Cell. 2002;13(5):1750–64. doi: 10.1091/mbc.01-12-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Mosberger N, Fornerod M, Stidwill RP, Greber UF. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat Cell Biol. 2001;3(12):1092–100. doi: 10.1038/ncb1201-1092. [DOI] [PubMed] [Google Scholar]
- Vihinen-Ranta M, Wang D, Weichert WS, Parrish CR. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J Virol. 2002;76(4):1884–91. doi: 10.1128/JVI.76.4.1884-1891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Kasamatsu H. Role of nuclear pore complex in simian virus 40 nuclear targeting. J Virol. 1993;67(1):119–30. doi: 10.1128/jvi.67.1.119-130.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]


