Abstract
YFR041C/ERJ5 was identified in Saccharomyces cerevisiae as a gene regulated by the unfolded protein response pathway (UPR). The open reading frame of the gene has a J domain characteristic of the DnaJ chaperone family of proteins that regulate the activity of Hsp70 chaperones. We determined the expression and topology of Erj5p, a type I membrane protein with a J domain in the lumen of the endoplasmic reticulum (ER) that colocalizes with Kar2p, the major Hsp70 in the yeast ER. We identified synthetic interactions of Δerj5 with mutations in genes involved in protein folding in the ER (kar2-159, Δscj1Δjem1) and in the induction of the unfolded protein response (Δire1). Loss of Erj5p in yeast cells with impaired ER protein folding capacity increased sensitivity to agents that cause ER stress. We identified the ERJ5 mRNA and confirmed that agents that promote accumulation of misfolded proteins in the ER regulate its abundance. We found that loss of the non-essential ERJ5 gene leads to a constitutively induced UPR, indicating that ERJ5 is required for maintenance of an optimal folding environment in the yeast ER.
Keywords: DnaJ, chaperones, protein folding, Saccharomyces cerevisiae, endoplasmic reticulum, unfolded protein response
1. Introduction
Molecular chaperones play a variety of functional roles in cellular processes leading to the acquisition of native conformation of cellular proteins. Members of the family of DnaK/Hsp70 and DnaJ/Hsp40 are conserved in organisms of all kingdoms and are found ubiquitously in different cellular compartments of the living cell. The Hsp70 chaperone machines perform different roles, including folding of nascent polypeptides, refolding of denatured proteins, protein translocation across membranes, and targeting of misfolded proteins to degradation. Hsp70s bind unfolded or partially folded polypeptides in an ATP-regulated cycle [1]. DnaJ proteins, defined by the presence of a roughly 70 amino acid region termed J domain, interact with Hsp70s partners, and act as co-factors that stimulate the ATPase activity at the ATPase domain of Hsp70. In some cases, DnaJs may deliver specific substrates to Hsp70s and/or recruit Hsp70s at specific subcellular locations [2, 3].
The endoplasmic reticulum (ER) is the organelle where the biosynthesis and maturation of secretory proteins takes place. Reducing agents and glycosylation inhibitors interfere with protein folding in the ER, and induce the unfolded protein response (UPR) pathway leading to a general transcriptional program that allows the cell to cope with the accumulation of misfolded proteins [4, 5]. Thus, the UPR regulates the expression of ER resident chaperones and enzymes required for folding, assembly, and modification of secretory and membrane proteins. Proteins accumulated in the ER that cannot be folded despite the increased folding capacity elicited by the UPR, are degraded in a pathway termed ERAD (ER-associated degradation). In that process, many functions involved in protein folding are also required for retrotranslocation of the misfolded proteins back to the cytosol where they are degraded by the proteasome [6].
Two Hsp70s, Kar2p and Lhs1p (Cer1p/Ssi1p), whose expression is regulated by the UPR, perform diverse functions in the yeast ER. The essential Kar2p, the yeast homologue of mammalian Bip [7], is a resident lumenal protein of the ER involved in protein translocation across the ER membrane, protein folding in the ER lumen, and ERAD [8–10]. This diversity of functions of Kar2p is believed to rely on the intrinsic properties of the Hsp70s, and on its interactions with different DnaJs partners. Three DnaJs of the yeast ER have been described: Sec63p, an essential membrane protein that interacts with Kar2p to drive translocation of proteins into the ER lumen [2], and two non essential UPR regulated lumenal proteins Scj1p and Jem1p, that are involved in Kar2p functions required for protein folding and ERAD [11, 12]. The KAR2 gene has a direct role in nuclear membrane fusion during karyogamy, which is independent of its role in translocation and folding [13, 14]. JEM1 has also been found to function in karyogamy in conjunction with KAR2 [15, 16]. Loss of the nonessential Lhs1p causes a translocation defect of a subset of proteins into the ER and reduced ability to fold proteins in the ER lumen, suggesting a partial overlap of functions between the two Hsp70s [17–19]. Recent studies demonstrated that Kar2p and Lhs1p interact with each other to couple their respective activities, and that the two Hsp70s do not share a common complement of co-chaperones [20].
In S. cerevisiae, the transcriptional scope of the UPR has been recently determined at a genomic scale. Under the stringent criteria employed in that work, 381 ORFs were identified as targets of the UPR including unknown and previously characterized genes that were classified in several functional categories [21]. One of the uncharacterized genes reported as targets of the UPR was YFR041C, encoding an ORF with homology to DnaJ and a putative signal sequence for translocation across the ER membrane. Sequence analysis of the S. cerevisiae genome identified 22 ORFs encoding members of the J protein family [22]. In that work, the YFR041C locus was named ERJ5 based upon global protein localization data and the fact that besides the three J proteins Sec63p, Scj1p and Jem1p, [15, 23, 24] there is a fourth membrane anchored protein in the organelle (Hlj1p) with a J domain facing the cytosol [25]. To date, several members of the J protein family have been functionally characterized in experimental studies, whereas many others for which a function and cellular localization is inferred from sequence analysis and genome-wide protein localization data await confirmation and detailed analysis from a direct experimental approach. Here, we have performed a biochemical and genetic analysis to get insight into the function of the YFR041C/ERJ5 gene. Our results indicate that the non-essential ERJ5 gene is required to preserve the folding capacity of the yeast ER.
2. Materials and methods
2.1. Yeast strains, growth conditions, and transformation procedure
Saccharomyces cerevisiae strains and plasmids used in this study are listed in Table 1. Standard yeast genetic techniques were used to construct strains by crossing. Heterozygous diploids were sporulated, tetrads were dissected, and the genotype of each resulting haploid was determined by genetic markers and/or phenotype.
Table 1.
Strains and plasmids used in this study
| Strain | Genotype | Source |
|---|---|---|
| YPH274 | Mata/a ura3-52 ura3-52 leu2Δ1leu2Δ1 lys2-801lys2-801 ade2-101 ade2-101 trp1-Δ1 trp1-Δ1 his3Δ200 his3Δ200 | Sikorski 1989 [31] |
| MS1380 | Matα ura3-52 kar2-159 | Rose lab. |
| DNY421 | Mata ire1::TRP1 ura3-1 can1-100 ade2-1 ade3 leu2-3-112 his3-11::HIS3-UPRE LacZ | Ng 2000 [4] |
| BJ5464 | Matα ura3-52 trp1 leu2-Δ1 his3-Δ200 pep4::HIS3 prb1-D1.6R can1 GAL | ATCC |
| W303α | Matα trp1-1 ura3-1 can1-100 ade2-1 leu2-3-112 trp1-Δ1 his3-11 | Ng lab. |
| RGY131 | Mata haploid derived from YPH274 | Silberstein 1998 [11] |
| RGY132 | Matα haploid derived from YPH274 | Silberstein 1998 [11] |
| RGY145 | as in strain RGY132 but Δscj1::HIS3 | Silberstein 1998 [11] |
| RGY147 | as in strain RGY132 but Δjem1::HIS3 | Silberstein 1998 [11] |
| RGY148 | as in strain RGY132 but Δscj1::TRP1 Δjem1::HIS3 | Silberstein 1998 [11] |
| SSY201 | as in strain RGY131 but Δerj5::kanMX4 | This study |
| SSY211 | as in strain RGY132 but ERJ5-3HA::HIS3MX6 | This study |
| SSY381 | as in strain MS1380 but ERJ5-3HA::kanMX6 | This study |
| SSY301 | as in strain W303α but Δerj5::kanMX4 | This study |
| Plasmid | ||
| pRS314-UPRE-GFP | pRS314 derivative (CEN/ARS, TRP1) | Xu 2005 [35] |
| pRS426-Erj5-Flag | pRS426 derivative (2μ, URA3) | This study |
Yeast were cultured in liquid or solid YPD (1% yeast extract, 2% peptone, and 2% dextrose) supplemented with adenine and/or tryptophane for strains ade2 or trp1. For selective plates, synthetic minimal media (0.67% yeast nitrogen base, 2% dextrose) was supplemented with appropriate amino acids or uracil. Yeast transformations were performed using the lithium acetate method [26].
2.2. Deletion of the YFR041C/ERJ5 locus
A one step PCR-based gene disruption of ERJ5 was performed in two different genetics backgrounds: RGY131 (a YPH274 derived a-haploid), and W303α to obtain SSY201 and SSY301, respectively. A 1.5 kb deletion cassette containing the KanMX4 marker was generated by PCR using pFA6-KanMX4 (kindly provided by P. Philippsen, University of Basel, Switzerland) as a template and ERJ5-S1 and ERJ5-S2 as the primers [27]. The KanMX4-containing PCR fragment was flanked at each end by 45 bp (nucleotides 15 to 60) and 46 bp (nucleotides 1214 to 1260) of the ERJ5 open reading frame, resulting in replacement of the ERJ5 gene with the selection marker. Yeast transformants were selected on YPD plates containing 200 mg/l G-418 (Invitrogen). Correct gene replacement was confirmed by PCR on genomic DNA using a combination of primers that annealed within the transformation cassette and outside of the integration region.
2.3. Carboxy-terminal epitope tagging of Erj5p in yeast strains
A PCR-based strategy to introduce epitope tags to chromosomal loci [28] was used to generate Erj5p-3HA in strains RGY132 and MS1380, obtaining strains SSY211 and SSY381, respectively. A DNA fragment containing three copies of the HA epitope and a selection marker flanked by 5′ (nucleotides 846 to 886) and 3′ (nucleotides 890 to 932) regions of the ERJ5 gene was generated by PCR using as template pYM1 or pYM2 (carrying the selection markers HIS3MX6 and KanMX6, respectively, that were kindly provided by Elmar Schiebel, CRC Beatson Laboratories, U.K.). Correct insertion of the module was confirmed by PCR on genomic DNA of transformed yeast strains using a combination of primers that annealed within the transformation cassette and outside of the integration region.
2.4. Yeast cell homogenate and microsomal membrane isolation
Yeast strains were grown in liquid YPD to mid-log phase; aliquots were taken and added to an equal volume of 20 mM sodium azide and processed immediately or stored at −80°C. Pelleted cells were resuspended in Buffer A (20 mM Tris-Cl pH: 7.4, 5 mM Mg Cl2, 1mM EDTA, 1mM EGTA, 1mM DTT, 0.1mM PMSF, 16% sucrose, and protease inhibitor mixture (0.1 μg/ml each of pepstatin A, chymostatin, and antipain; 1 μg/ml aprotinin and 5 μg/ml leupeptin). Cell lysis was performed at 4°C by agitation with glass beads. The supernatant obtained after removing unbroken cells and debris by centrifugation for 5 min at 2000g was used as the cell homogenate. Cell membranes were obtained after centrifugation of the cell homogenate at 120,000g for 60 min. The membrane pellets were resuspended in Buffer B (20 mM Tris-Cl pH: 7.4, 1 mM DTT, 10 % glycerol and protease inhibitor mixture) and recentrifuged as above. Microsomal membranes obtained after the second centrifugation were resuspended in Buffer B and stored at −80°C.
Rapid lysis of cells with glass beads [29] were performed after pelleting cells from liquid cultures added to an equal volume of cold 20 mM sodium azide and kept on ice.
2.5. Alkali extraction, endoglycosidase H digestion, protease digestions and protein immunoblots
Cell homogenates adjusted to 0.1 M sodium carbonate were incubated on ice for 30 min before being centrifuged at 120,000g for 60 min. The supernatant and pellet fractions were collected and resuspended in SDS loading buffer for SDS/PAGE analysis and immunoblot. Endoglycosidase H was purchased from New England Biolabs: digestions were performed following the manufacturer’s recommendations. Yeast microsomal membranes were digested with trypsin (100 μg/ml) either in the presence or absence of 0.5% Triton X-100 following a procedure described previously [30]. Proteins resolved by SDS-PAGE were transferred to PVDF membranes (New England Biolabs) by semi-dry electroblotting. The membrane blots were blocked with 5% nonfat dried milk and incubated overnight with the monoclonal anti-HA antibody 1:1000 (HA-probeF-7, Santa Cruz Biotechnology) or with antiserum that recognize Kar2p (1:10000) or Wbp1p (1:1000). Immunoreactive bands were visualized with alkaline phosphatase-labeled second antibodies (anti-mouse or anti-rabbit IgGs).
2.6. Construction of the Carboxy-terminal Flag tagged Erj5p in a plasmid
A PCR fragment containing nucleotides −242 to 1160 of the ERJ5 sequence, flanked by SmaI and HindIII restriction sites was cloned in the corresponding sites of the pRS426 polylinker [31] to obtain pRS426-ERJ5. Correct sequence of the insert was confirmed. The DDDDK epitope was added to pRS426-ERJ5 using two complementary synthetic oligonucleotides (Forward: 5′ GTAACGATTATAAAGATGATGATGATAAAG; Reverse: 5′ GTTACCTTTATCATCATCATCTTTATAATC) encoding the DDDDK flanked by BstEII restriction sites. Ligation was preformed with an excess of annealed oligonucleotides added to the BstEII digested pRS426-ERJ5. Correct in-frame insertion of the epitope was confirmed by sequencing. The resulting plasmid (pRS426-ERJ5-Flag) was introduced in strain BJ5464 by transformation and selection in media lacking uracil.
2.7. Immunofluorescence microscopy
Yeast cells were fixed and stained following a modified standard protocol [32]. Cells were washed after formaldehyde treatment with 40 mM potassium phosphate (pH 7.5) and 0.5 mM MgCl2, and then resuspended in the same buffer plus 1.2 M sorbitol and 30 mM DTT. Cells were spotted onto polylysine microscope slides and allowed to settle for 10 minutes. After removing excess liquid, the slides were dunked in cold methanol for 6 minutes then quickly dunked into cold acetone for 30 seconds. After blocking in PBS with 3% BSA, slides were incubated in primary antibody, rabbit anti-Kar2p and goat anti-DDDDK (Abcam Inc.), at a dilution of 1:500 overnight, then washed and incubated with secondary antibody, goat anti-rabbit Alexa 633 and donkey anti-goat Alexa 546 (Molecular Probes) at a dilution of 1:1000 for 1.5 hours and then washed again. Slow Fade lite (Molecular Probes) was added to the slides before sealing. Cells were imaged on an LSM 510 NLO system (Zeiss) using a 40X Plan-Neofluor objective. Alexa Fluor 546 was excited by a 543 laser (HeNe) and the emission captured through a BP565-615 filter. Alexa Fluor 633 was excited with a 633 laser (HeNe) and captured through a LP650 filter.
2.8. RNA extraction and Northern blot analysis
Yeast cells were grown at 24°C until mid-log phase. As indicated, tunicamycin (10 μg/ml) or DTT (10 mM) were added 120 min and 60 min, respectively before cells were collected. Total RNA was prepared using the hot phenol procedure [33]. RNA samples (20 μg) were resolved by 1.2 % agarose/formaldehyde gel electrophoresis and transferred to Hybond-N+ membranes (Amersham Corp.). Prehybridization and hybridizations were carried out by standard techniques [34] with probes specific for ERJ5 (nucleotides 338 to 870), KAR2 (nucleotides 1088 to 2376) and ACT1 (nucleotides 2 to 844) generated by PCR. Hybridization probes were 32P-labeled using the Random primers DNA labeling System (Invitrogen). Radioactive bands were quantified with a PhosphorImager (Molecular Dynamics Inc.) and visualized by autoradiography.
2.9. Fluorescence measurement of GFP
GFP fluorescence was measured from cultures of the strains bearing pRS314-UPRE-GFP as previously described [35] using an F-4500 Fluorescence Spectrophotometer (Hitachi). Briefly, cells were grown overnight to a density below 1 OD before treatment with or without DTT. At the end of the treatment cells were collected, the media removed, and resuspended in PBS for GFP fluorescence measurement.
3. Results
3.1. YFR041C/ERJ5 encodes a novel type I membrane protein with a J domain inside the lumen of the endoplasmic reticulum
The YFR041C locus encodes an open reading frame of 295 amino acids that by sequence comparison analysis was found to contain a J domain. Based upon the presence of a J domain and the subcellular localization data compiled in the SGD, this gene was named ERJ5 [22]. We will refer to this gene as ERJ5 hereafter. Hydropathy analysis revealed two hydrophobic protein segments in the Erj5p sequence; one amino terminal preceding the J domain and a second one located roughly in the middle of the protein sequence. From this distribution of hydrophobic sequences the J domain is not expected to be exposed to the cytosol.
To experimentally address the expression and topology of the protein encoded by ERJ5 we generated strains in which three copies of the HA epitope were fused to the carboxy-terminus of the open reading frame by genomic integration of a PCR-amplified module. An immunoreactive band of a mass relative of roughly 37 kD was detected by Western blot of crude extracts with a monoclonal anti-HA antibody only in strains carrying Erj5p-3HA (not shown and Fig. 1). The kar2-159 haploid strain carrying the carboxy-terminal epitope-tagged protein in the genome did not show the synthetic growth defect of the kar2-159Δerj5 strain (see below) suggesting that the Erj5p-3HA fusion protein was functional in the cell.
Figure 1.
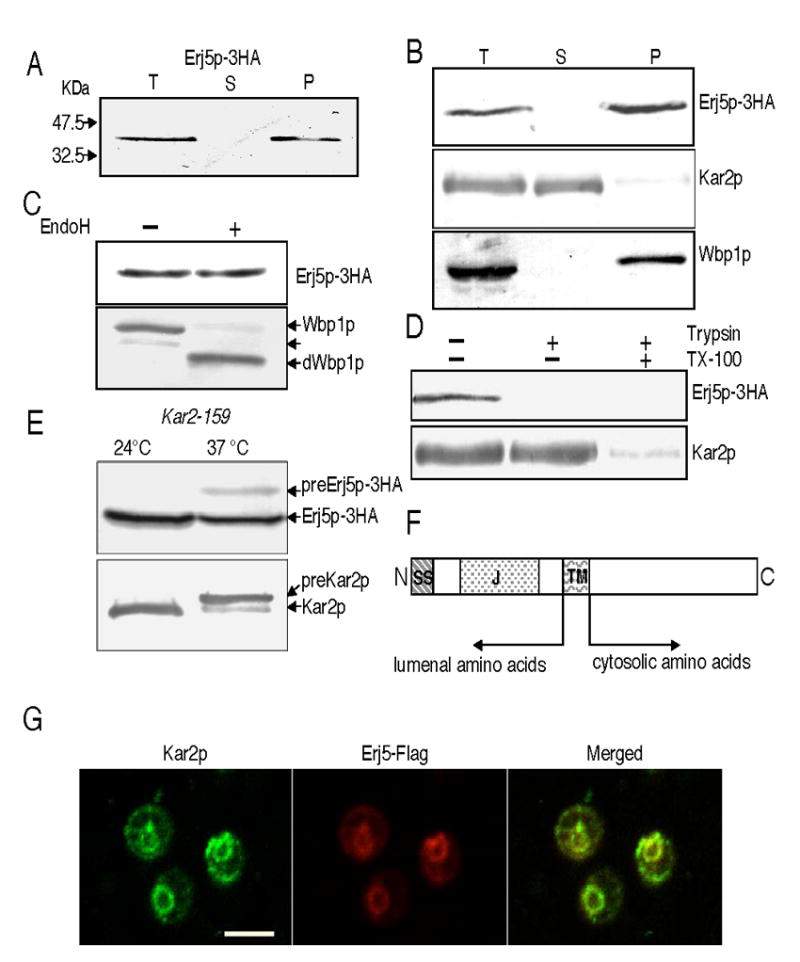
ERJ5 encodes a novel type I ER membrane protein. The Erj5p-3HA protein was detected with anti HA antibodies, the soluble ER lumenal Kar2p and the integral ER membrane Wbp1p were detected with specific antisera. Experiments were performed as described in Materials and Methods. A) Erj5p-3HA is detected in yeast microsomes. Cell homogenates from SSY211 cells that express the Erj5p-3HA fusion protein were centrifuged to obtain microsomal membranes. Supernatant (S) and pellet (P) fractions derived from the equivalent amount of total cell homogenate (T) were resolved by SDS/PAGE and immunobloted for detection of the HA-epitope. B) Erj5p remains in the membrane fraction after treatment of the cell homogenates with 0.1M sodium carbonate pH: 11.5. Supernatant (S) and pellet (P) derived from the equivalent amount of total cell homogenate (T) were immunobloted for detection of Erj5p-3HA. Soluble Kar2p and the integral membrane Wbp1p were immunodetected as controls. C) Erj5p is not N-glycosylated. Microsomes were incubated in presence or absence of Endoglycosidase H before SDS/PAGE and immunodetections of Erj5p-3HA and the Wbp1p glycoprotein as a control. Wbp1p glycoforms containing one or two N-oligosaccharydes or fully deglycosylated (dWbp1p) are indicated with arrows. D) The carboxy-terminus of Erj5p is accessible to proteases in intact microsomes. Three aliquots of yeast microsomes were incubated with or without trypsin in the presence or absence of Triton X-100. Erj5p-3HA and the lumenal soluble Kar2p were immunodetected. E) preErj5p is detected in yeast strains defective in ER protein translocation. A kar2-159 strain expressing the Erj5p-3HA fusion protein (SSY301) was grown to mid-log phase at 24ºC in liquid YPD. Cultures were divided, half was kept at 24ºC, and half was shifted at 37ºC as indicated. After 3 h of incubation, aliquots were removed, cells were lysed with glass beads, and proteins resolved in SDS/PAGE for immunoblotting. Labeled arrows indicate the migration position of preErj5p-3HA and Erj5p-3HA, preKar2p and Kar2p. F) Schematic representation of the overall structure of Erj5p. SS: signal sequence for translocation across the ER; J: J domain, TM: transmembrane region. G) Colocalization of Erj5p and Kar2p in the ER by immunofluorescence. BJ5464 cells expressing Erj5-Flag protein were analyzed by double label immunofluorescence microscopy using anti-Kar2p and anti-Flag antibodies. Left: fluorescent image of Kar2p; middle: fluorescent image of Erj5-Flag; right: merged image. Bar indicates 5 μm.
Biochemical analysis of Erj5p-3HA shows it behaves as an integral membrane protein, consistent with predicted topology. Cell fractionation done on strains carrying the Erj5p-3HA showed that the protein is associated with the ER membrane pellets (Fig. 1A). Sodium carbonate (pH 11.5) treatment, used to extract soluble and peripherally associated membrane proteins from microsomes, showed that it cofractionated with a known ER membrane protein, Wbp1p, in the membrane pellet while the lumenal soluble Kar2p remained in the supernatant (Fig. 1B). Two consensus N-glycosylation sites are present in the sequence of Erj5p; however none of the sites are glycosylated in vivo. Endoglycosidase H treatment of cell extracts did not change the electrophoretic mobility of the immunoreactive band of Erj5p-3HA, although faster migrating bands corresponding to different glycoforms, and predominantly to the deglycosylated form of the membrane glycoprotein Wbp1p were detected in the Endoglycosidase H digested cell extract (Fig. 1C). The amino terminal portion including the J domain appears to localize to the lumenal side. Protease digestion experiments (Fig. 1D) showed that the carboxy-terminus of Erj5p is exposed to the cytosol. Trypsinization of intact yeast microsomes eliminated the immunoreactive band of Erj5p-3HA without producing any polypeptide of greater mobility, consistent with digestion of the HA epitope. The lumenal protein Kar2p became accessible to the protease upon addition of detergent, but was inaccessible in intact membranes.
Although both hydrophobic segments in the Erj5 protein sequence are of sufficient length and hydrophobicity to be membrane-spanning segments, the amino-terminal hydrophobic segment could function as a cleavable signal sequence for translocation across the ER. To experimentally test this possibility, a conditional kar2 ER translocation mutant was tagged in the ERJ5 locus with the HA epitope as described above. ER proteins are translocated at an approximately normal rate in a kar2-159 mutant incubated at the permissive temperature, but a severe translocation defect is evidenced after a shift to the restrictive temperature [8]. Accumulation of precursor forms of secretory proteins with cleavable signal sequences are evidenced as slower-migrating bands in protein immunoblots. As it can be seen in Fig. 1E, besides the immunoreactive band corresponding to Erj5p-3HA, a second band of reduced mobility becomes apparent in crude extracts of kar2-159 mutant cells that were incubated at 37ºC and probed with the anti-HA antibody. As expected, precursor forms which contains a cleavable signal sequence uncleaved, and mature forms of Kar2p were readily apparent in crude extracts of kar2-159 incubated at the restrictive temperature. These findings are consistent with the prediction that the amino-terminal hydrophobic sequence of Erj5p functions as a cleavable signal sequence for translocation across the ER. A schematic representation of the overall structure of Erj5p is summarized in Fig. 1F.
Our results show that Erj5p is a type I membrane protein that enters the secretory pathway. GFP tagging of a genomic encoded Erj5p in a global analysis of protein localization in yeast localized Erj5p to the ER [36]. However, Erj5p lacks an obvious ER retention/retrieval sequence. We confirmed the subcellular localization of Erj5p by expressing the protein from a plasmid containing Erj5p tagged in the carboxy-terminus with the Flag epitope in BJ5464 yeast cells. Immunostaining of the yeast cells was done with the anti-Flag and anti-Kar2p antibodies (Fig. 1G). The similar staining patterns, typical of resident ER proteins, observed with both antibodies, and the merged images allow us to conclude that Erj5p colocalizes with Kar2p in the yeast ER.
Topology and subcellular localization identify Erj5p as a fourth DnaJ homologue with a J domain in the ER lumen. Fig. 2 shows a multiple sequence alignment of the conserved J domain of E.coli DnaJ and the J domains located in the lumen of the ER. The J domain of Erj5p is 30%, 26% and 23% identical with the J domains of Scj1p, Jem1p and Sec63p, respectively. Close examination of the Erj5p J domain sequence showed that it possesses all the structural features required for being a functional J domain that would interact with Hsp70s[37, 38].
Figure 2.
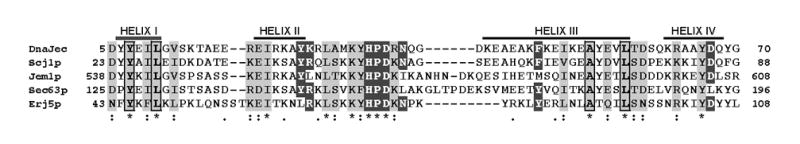
Sequence alignment of the conserved J domains of E. coli DnaJ and the S. cerevisiae DnaJ proteins with J domains located in the lumen of the ER. The multiple alignments were performed with ClustalX [51]. Numbers in the left and the right indicate first and last amino acids of each aligned sequence. The four α-helical regions of the J domain of E. coli DnaJ and its human homologue HDJ1 that were determined by NMR structure [37] are indicated. The highly conserved HPD tripeptide, a hallmark of the J domains that mediate interactions with Hsp70s, and essential amino acids identified in E.coli J domain are in black boxes.
3.2. Synthetic growth defects are identified when ΔERJ5 is combined with mutations in chaperones of the endoplasmic reticulum
We generated disruptions of the ERJ5 gene in different yeast backgrounds and found that Δerj5 haploid yeast cells were viable as it was reported by the yeast systematic deletion project (compiled at SGD: http://www.yeastgenome.org) and did not show any noticeable growth defect phenotype in a variety of media and growth temperatures tested (data not shown and Fig. 3). Given the localization of Erj5p to the ER and the lumenal orientation of its J domain, we investigated whether ERJ5 would show genetic interactions with genes encoding proteins that perform chaperone functions in the ER. This approach has been widely used to demonstrate cooperativity among components of the chaperone machinery of the yeast ER [11, 19, 39].
Figure 3.
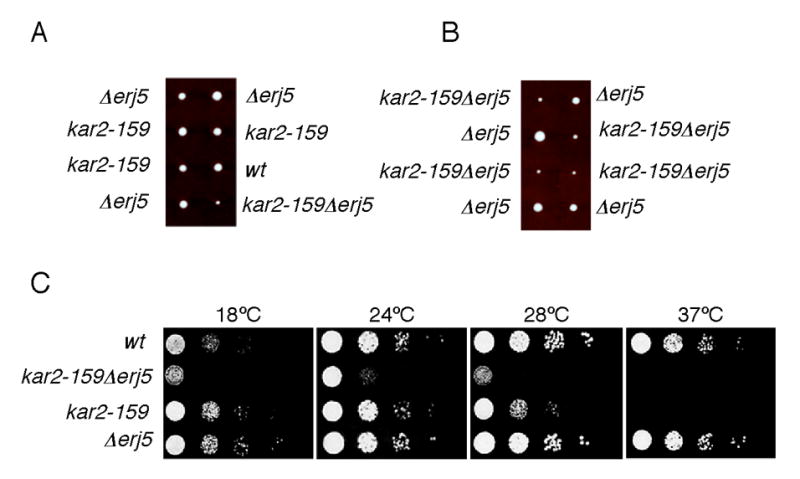
Growth of strains combining the kar2-159 conditional allele and the erj5 deletion. A) Diploids that were heterozygous for kar2-159 and for Δerj5 were obtained by crossing strains MS1380 and SSY201. To minimize effects of different strain backgrounds, a kar2-159 haploid isolated from the initial cross was backcrossed to SSY201 three times. A diploid from the last cross was sporulated and tetrads were dissected. B) Tetrads dissected from a diploid obtained by crossing haploids kar2-159 Δerj5 and Δerj5 obtained from the last backcross described in A. Plates were incubated at 24ºC for 3 d. C) Strains obtained from a tetratype tetrad as of Panel A were grown in liquid YPD at 24ºC and diluted to a density of 106 cells/ml. 5-μl aliquots of 10-fold serial dilutions of the cultures were plated on YPD-agar and incubated at the indicated temperatures for 3 d.
We combined a Δerj5 deletion with a conditional mutation of the essential KAR2 gene that encodes the most extensively studied Hsp70 of the yeast ER. We crossed a Δerj5 mutant strain with the kar2-159 temperature-sensitive strain [8]. After sporulation and tetrads dissection, plates were incubated at the kar2-159 permissive temperature for growth. Although four viable spores were obtained in all the tetrads analyzed, discarding a possible synthetic lethality by combination of the two mutations, colonies bearing both Δerj5 and kar2-159 mutations were significantly smaller in size compared to those developed from wild-type spores or spores carrying only one of the mutations. Fig. 3A shows colonies obtained from two tetrads dissected from diploids which were heterozygous for the Δerj5 and for the kar2-159 alleles, yielding four normal size colonies (parental ditype) or three normal size plus one smaller colony (tetratype). Fig. 3B shows colonies obtained from diploids that were homozygous for the Δerj5 mutations and heterozygous for the kar2-159 allele, yielding two normal size and two smaller size colonies. Smaller size colonies developed at 24°C did not grow at 37°C, indicating that there were all kar2-159. The growth of haploid strains derived from a tetratype tetrad was compared at different temperatures on YPD plates (Fig. 3C). Wild-type and Δerj5 strains grew at all the temperatures tested, and kar2-159 showed its characteristic temperature-sensitive phenotype at 37°C. The kar2-159Δerj5 double mutant exhibited a severely impaired growth at 18°C, 24°C and 28°C, temperatures that did not affected significantly the growth of the single mutants Δerj5 and kar2-159 strains.
The synthetic phenotype generated when Δerj5 and kar2-159 mutations are combined suggests that both proteins might be involved in a common functional pathway. Kar2p has two functions that can be genetically differentiated by analyzing different mutant alleles. The class I kar2-159 allele [8] carries a single point mutation in the ATPase domain of Kar2p [40]. At the non-permissive temperature of 37°C, the kar2-159 allele blocks translocation of proteins at an early step, and yeast cells rapidly die. kar2-159 mutants are translocation-proficient at temperatures below 37°C ([8]; see also Fig.1E), but exhibit defects in protein folding [9] and ERAD [12] at permissive temperatures. Since we did not detect translocation defects in Δerj5 mutants (data not shown), the genetic negative interaction between kar2-159 and Δerj5 suggests that loss of Erj5p in a kar2-159 strain could be causing an aggravation of the kar2-159 post-translocational functions that are already affected at permissive temperatures in this mutant.
LHS1/CER1/SSI1 encodes a non-essential Hsp70 of the ER. Δlhs1 cells are viable at all temperatures but exhibit a partial translocation block of some ER proteins and defects in protein folding [17–19, 39]. We generated a double disrupted Δlhs1Δerj5 strain and found that the strain was viable and, different than the kar2-159Δerj5 mutant, did not show any detectable growth defect at any temperature tested or in presence of agents that disturb the folding capacity of the ER. We did not find an aggravation of the translocation defect in the Δlhs1Δerj5 strain compared to the Δlhs1 single mutant (data not shown).
As observed for Δerj5 strains, single deletions of any of the two characterized DnaJs that have a role in protein folding in the ER, Scj1p and Jem1p, do not cause a growth phenotype. However, Δscj1Δjem1 double mutants that grow at wild-type rates at 24°C arrest growth after a few cell divisions at 37°C, evidence for a partially overlapping function of these two proteins [11]. Compared to the single deletion mutants, double null combinations Δerj5Δscj1 and Δerj5Δjem1 strains did not show any noticeable difference in growth rates on YPD plates at any temperature tested (data not shown). However, the growth rate of a Δscj1Δjem1 strain was reduced in absence of the ERJ5 gene at 18°C, 24°C, and 37°C (Fig. 4). At 24°C, the triple Δscj1Δjem1Δerj5 mutant exhibited colonies of smaller size than the double Δscj1Δjem1. At this temperature, a significantly reduced growth of the Δscj1Δjem1Δerj5 strain was observed on plates containing the reducing agent β-mercaptoethanol, suggesting that loss of ERJ5 makes Δscj1Δjem1 yeast cells more sensitive to the stress caused by accumulation of unfolded proteins in the ER.
Figure 4.
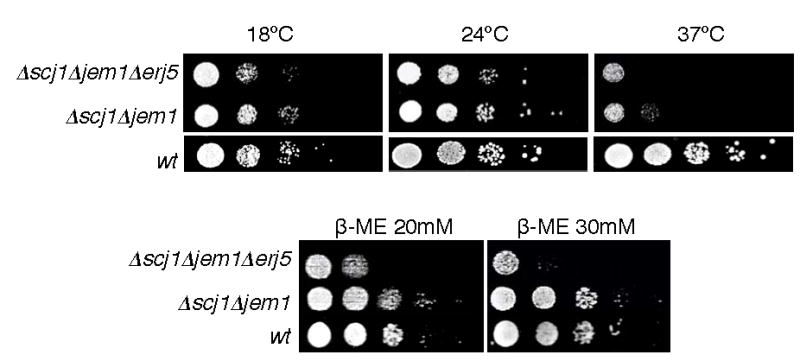
Loss of Erj5p aggravates the growth defect and sensitivity to agents that produce stress in the ER of a Δscj1Δjem1 strain. Isogenic Δscj1Δjem1Δerj5, Δscj1Δjem1, and wild-type strains were grown in liquid YPD at 24ºC and diluted to a density of 106 cells/ml. 5-ul aliquots of 10-fold serial dilutions of the cultures were plated on YPD-agar or on plates with the same medium containing β-mercaptoethanol at the indicated concentrations. YPD plates were incubated at the indicated temperatures for 3 d. Plates containing β-mercaptoethanol were incubated at 24ºC for 4 d.
Scj1p overexpression suppresses the temperature sensitive phenotype of Δscj1Δjem1 strains [11, 15]. We asked if ERJ5 could function as a high copy suppressor of the temperature-sensitive phenotype of the Δscj1Δjem1 strain. Expression of Erj5p from a high-copy plasmid did not rescue the temperature-sensitive phenotype of the Δscj1Δjem1 strain (data not shown) indicating that the sole overexpression of Erj5p is not able to compensate for loss of any of this other two DnaJs of the ER.
3.3. Loss of ERJ5 in a strain unable to induce the unfolded protein response results in a growth defect and aggravated sensitivity to ER stress
In response to the accumulation of misfolded proteins in the ER, cells of all eukaryotic organisms activate the UPR. Strains defective in signaling components of this mechanism are sensitive to situations that promote accumulation of misfolded proteins in the ER, therefore synthetic interactions with this type of mutations is a very sensitive assay of ER stress.
We asked if loss of ERJ5 would affect growth and sensitivity to ER stress agents in a Δire1 strain. Ire1p encodes an ER membrane protein that acts as a sensor of the folding capacity of the ER lumen and transduces the signal to the nucleus, which results in increased levels of chaperones and enzymes required to cope with misfolded proteins accumulated in the ER. Δire1 strains grow normally in rich media because the UPR pathway is dispensable for cell viability under normal cell conditions but are sensitive to agents that promote accumulation of misfolded proteins in the ER [41]. We generated a Δerj5 mutant in the same strain background of a Δire1 mutant and a double Δire1Δerj5 strain was obtained by crossing and tetrads dissection. Δire1Δerj5 cells showed a reduced growth rate compared to Δire1 cells on YPD plates incubated at 18°C and 24°C but not at 37°C (Fig. 5). At the growth temperature of 28°C, at which a very modest growth defect was observed on YPD plates for the Δire1Δerj5 strain, a more pronounced sensitivity to β-mercatoethanol was observed relative to the Δire1 strain. As previously described, Δire1 cells showed a slight sensitivity to reducing agents [41], whereas Δerj5 cells grew at wild-type rates when tested on plates containing β-mercaptoethanol (Fig. 5). The diminished growth rate and the aggravation of the β-mercaptoethanol sensitivity of the Δire1 strain that lacks the ERJ5 gene suggest that loss of Erj5p results in an impaired folding capacity of the ER.
Figure 5.
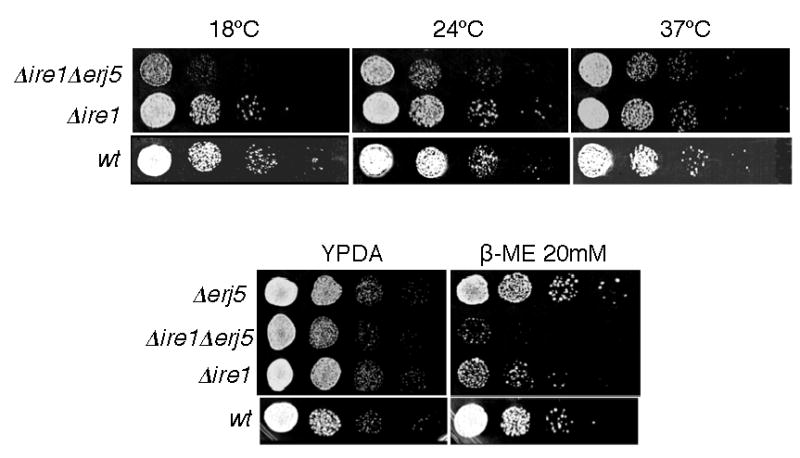
Loss of Erj5p in a Δire1 strain results in a growth defect and hypersensitivity to ER stress. Isogenic Δire1Δerj5, Δire1, Δerj5, and wild-type strains were grown in liquid YPD at 24ºC and diluted to a density of 106 cells/ml. 5-ul aliquots of 10-fold serial dilutions of the cultures were plated on YPD-agar or on plates with the same medium containing β-mercaptoethanol at the indicated concentrations. YPD plates were incubated at the indicated temperatures for 3 d. Plates containing β-mercaptoethanol were incubated at 28ºC for 4 d.
3.4. The ERJ5 mRNA is induced under conditions that lead to the accumulation of misfolded proteins in the ER, and loss of ERJ5 induces the UPR
We identified the ERJ5 mRNA and examined its levels in cells that were exposed to agents that promote stress in the ER. Northern blot analysis showed that ERJ5 mRNA accumulated in wild-type yeast cells incubated in presence of the N-glycosylation inhibitor tunicamycin or the reducing agent DTT (Fig. 6A). Both treatments increased roughly 2.4-fold the ERJ5 mRNA expression. This result is in agreement with a previous classification of this gene as a transcriptional target of the UPR in a genome-wide analysis [21].
Figure 6.
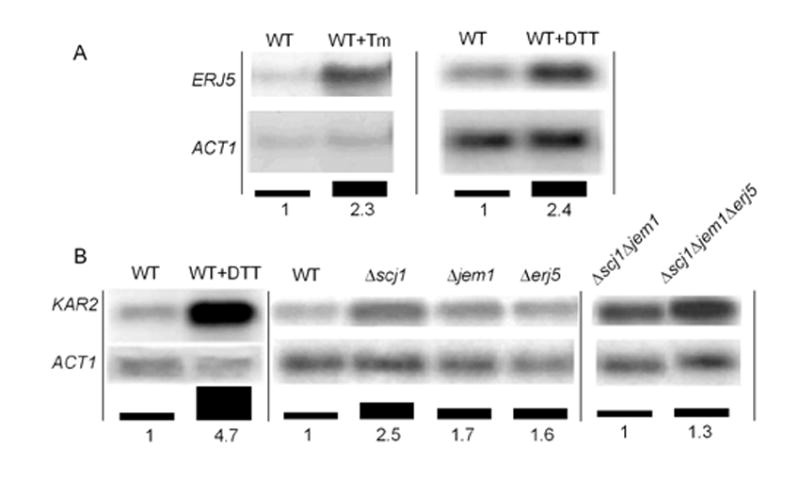
Northern blot analysis. Yeast cells RNA preparation and Northern blot analysis were performed as described in Materials and Methods. A) Induction of the ERJ5 mRNA by agents that promote the accumulation of unfolded proteins in the ER. A wild type strain (RGY132) was grown at 24ºC in the absence or presence of 10 μg/ml of tunicamycin (Tm) or 10 mM DTT. ERJ5 mRNA, normalized to ACT1 mRNA probed on the same blot, is expressed as bars corresponding to the fold-induction of ERJ5 mRNA relative to the expression in the wild-type in absence of the treatments. B) Induction of the unfolded protein response. KAR2 mRNA normalized to ACT1 mRNA probed on the same blots, is expressed as bars corresponding to the fold-induction of KAR2 mRNA relative to the expression in the wild-type in absence of treatments (left and center panels), or as fold-induction relative to the Δscj1Δjem1 strain (right panel). Representative Northern blots of several independent repetitions are shown.
The transcription of the KAR2 gene is indicative of an increased content of unfolded proteins in the yeast ER [42]. Incubation of wild-type yeast cells with tunicamycin [11] or with DTT (Fig. 6B) resulted in approximately 4.7-fold increase in KAR2 mRNA expression. This transcriptional increase of the UPR-induced KAR2 gene represents the maximal induction of the UPR in these strains. The levels of KAR2 mRNA were determined in wild-type, Δscj1, Δjem1 and Δerj5 strains incubated at 24°C (Fig. 6B center panel). Compared to the wild-type, Δscj1 and Δjem1 mutant strains showed a 2.5-fold and 1.7-fold increase in KAR2 mRNA expression respectively. This result is in agreement with previous reports of the constitutive induction of the UPR in these strains [11]. KAR2 mRNA was also induced in the Δerj5 mutant, and this induction of roughly 1.6-fold was comparable to the exerted by loss of the JEM1 gene. From this result we conclude that Δerj5 strains have a constitutive induction of the UPR.
Since loss of ERJ5 aggravates the growth phenotype and enhanced sensitivity to reducing agents of the Δscj1Δjem1 strain (Fig. 4), we examined if loss of ERJ5 would lead to an increased KAR2 mRNA expression in the Δscj1Δjem1 mutant. As it can be seen in Fig. 6B right panel, the Δscj1Δjem1Δerj5 mutant showed an increase of 1.3-fold in KAR2 mRNA levels respect to the Δscj1Δjem1 strain. This increase of the UPR by loss of Erj5p is likely reflecting a decreased ability to deal with protein folding in the Δscj1Δjem1Δerj5 strain.
The effect of loss of Erj5p on the UPR was also tested directly using a sensitive fluorescent sensor of UPR induction [35]. A centromeric plasmid containing GFP driven by four repeats of the unfolded protein response element (UPRE: [41, 43]) [21] was transformed into the Δerj5 and control W303 cells. Under normal growth conditions, GFP fluorescence levels in the Δerj5 strain were significantly higher than in the wild-type (Fig. 7). A constitutive induction of the UPR of roughly 2-fold was obtained for the Δerj5 mutant respect to the wild-type strain. This result is consistent with the obtained analyzing KAR2 mRNA levels by Northern blot (Fig. 6B center panel). Treatment with DTT to induce the UPR in the cells increased fluorescence in both wild-type and Δerj5 strains, but higher levels of GFP fluorescence were obtained in the Δerj5 strain. These results suggest that the UPR induced levels measured in Δerj5 cells after DTT treatment reflects a combination of ER stresses: the accumulation of misfolded proteins promoted by the reducing agent, and the ER perturbation caused by loss of the ERJ5 gene.
Figure 7.

UPR induction in Δerj5 cells measured by the GFP-fluorescence of a UPR reporter. W303 yeast cells bearing pRS314-UPRE-GFP were grown in minimal medium without tryptophane to mid-log phase. GFP-fluorescence was quantified as described in Materials and Methods from cells incubated for 90 min in the absence or presence of 5 mM DTT. The mean fluorescence of wild-type (WT) and Δerj5 (erj5) cells were normalized to the fluorescence values of the WT in absence of DTT treatment. The mean fold induction was calculated from five experiments and the standard deviation is represented as error bars.
4. Discussion
Our results show that ERJ5 encodes a protein with an N-terminal signal sequence that is cleaved after translocation across the endoplasmic reticulum. Erj5p contains a lumenally exposed J domain, a single membrane spanning sequence, and a cytoplasmic carboxy-terminal tail. Protein localization analysis of Erj5p, showed that it colocalizes with Kar2p in the ER, confirming the assigned subcellular localization in a global analysis of yeast strains expressing GFP fusion proteins [36]. These data support the identification and classification of Erj5p as the fourth DnaJ homologue with a J domain in the S. cerevisiae ER lumen.
Sequence comparison using available public databases identify Erj5p orthologues in all the fungal genomes sequenced to date, suggesting that Erj5 plays a role important enough to be preserved through evolution in yeast. All the sequences maintain the overall structure of Erj5p summarized in Fig. 1F. The aligned sequences (http://db.yeastgenome.org/fungi/YFR041C.html) show the highest level of conservation throughout the lumenal portion of the proteins but exhibit some variability at the carboxy-terminus in terms of amino acid identities and length.
The DnaJ homologues, possessing the highly conserved J domain, a signature of this family of proteins, have been classified into three groups based upon the domains shared with E. coli DnaJ, the paradigmatic member of the group. Type I DnaJ proteins possess all three domains: the highly conserved J domain, the glycine-phenylalanine rich region, and a zinc finger-like domain. Type II J proteins lack the zinc finger-like domain, and Type III only possess the J domain [44]. In the yeast ER, Scj1p is the only member of the Type I group. Jem1p, Sec63p and Erj5p are type III DnaJ homologues. Erj5p as well as Sec63p are integral membrane proteins whereas Scj1p and Jem1p are soluble proteins. These four DnaJ homologues have a J domain located in the lumen of the ER. The HPD motif, a hallmark in all J domains predicted to mediate interaction with Hsp70s, is present in Erj5p as well as in the other three J domains of the yeast ER (Fig. 2).
J-domain proteins are co-factors that regulate the ATP hydrolysis of their Hsp70 partners. Several lines of biochemical, functional and genetic evidences support the notion that Sec63p, Scj1p, and Jem1p act as co-factors of Kar2p, the main Hsp70 of the yeast ER [42]. Having determined that Erj5p has a J-domain in the ER we tested for a role in Kar2p associated activities.
The essential Sec63p is an integral component of the yeast translocation complex [45] that interacts with Kar2p to promote nascent chain transport into the ER. Erj5p would not appear to play a role similar to Sec63p, since we did not detect protein translocation defects by loss of Erj5p in wild-type strains, and the partial translocation block of a Δlhs1 mutant was not aggravated in a ΔlhsΔerj5 double mutant (data not shown). Therefore, Erj5p could function after the nascent chain has moved beyond the translocation channel.
A role for Erj5p in post-translocational Kar2p functions is supported by our results. Loss of the nonessential Scj1p and Jem1p DnaJ proteins that are required for Kar2p functions in the ER lumen, also yield translocation-proficient strains. Our genetic interaction data suggest that Kar2p may interact with Erj5p, in addition to interacting with Scj1p and Jem1p, in the ER lumen. Whereas Δscj1 and the kar2-159 mutation are synthetically lethal [24], we found that although kar2-159Δerj5 strains are still viable, there is a synthetic negative interaction between Δerj5 and the kar2-159 mutation. Furthermore, the aggravation of the growth phenotype and sensitivity to ER stress that is generated by the loss of Erj5p in a Δscj1Δjem1 yeast strain would suggest a partial overlap of functions for Scj1p, Jem1p and Erj5p.
We next tested Erj5p in folding assays that disclosed a role for Scj1p and Jem1p in protein folding. Simultaneous loss of Scj1p and Jem1p causes a dramatic reduction of the transport rate of an unglycosylated mutant of carboxypeptidase Y (CPY) [11], a soluble protein widely used to monitor protein folding in the yeast ER [9]. We found that unglycosylated CPY is transported at similar rates in Δscj1Δjem1 and Δscj1Δjem1Δerj5 mutant strains (data not shown). As for the group of genes with a role in protein maturation in the yeast ER, ERJ5 mRNA expression has been found elevated in a strain expressing a single misfolded secretory protein [6]. However, a Δerj5 strain showed no changes in the secreted levels of a heterologous single chain antibody (scFv) compared to the wild-type strain (Xu, P., personal communication). It remains possible that the substrates tested may not be the optimal to reveal an ER folding delay caused by the loss of Erj5p. It is conceivable that whereas soluble lumenal Scj1p and Jem1p may participate with Kar2p in processes that take place in the ER lumen [11, 12], the topological restriction of the J domain of Erj5p to the ER membrane proximity might limit its function as a Kar2p cofactor to a more restricted environment or to a different and specific subset of untested folding proteins.
The co-existence in the ER of several members of the chaperone families with partially overlapping functions together with the ability of the UPR to compensate for the loss of components of the folding machinery by inducing a wide number of ER functions, make defining the specific role of each individual protein difficult. The wild-type growth rate of the Δscj1 and Δjem1 mutants can be explained by the induction of the UPR [11] that enhances expression of Kar2p and the others DnaJs of the ER. This explanation can be extended to understand the non-essential nature of Erj5p, an ER protein that, like Scj1p and Jem1p, is up-regulated by the UPR.
Without the UPR, key components of the ER folding machinery become limiting and the contribution of a gene to the ER homeostasis is evidenced. In a strain unable to induce the UPR (Δire1), loss of Erj5p causes a growth defect under normal conditions and is poorly tolerated under conditions that promote accumulation of misfolded proteins in the ER. This observation suggests that the Ire1p-dependent UPR is required to induce one or more factors that compensate for loss of Erj5p.
Our results show that ERJ5 mRNA is induced under conditions that promote stress in the ER, supporting a previous identification of the ERJ5 gene as an UPR target in a genomic analysis [21]. Regulation of gene transcription by the UPR does not necessarily imply that loss of the gene will up-regulate the UPR in the cell. We have measured the UPR in the Δerj5 mutant by two independent approaches (KAR2 mRNA levels and activation of a GFP reporter sensor), and found that loss of Erj5p function leads to a small but significant chronic UPR induction. Constitutive induction of the UPR has been observed in yeast strains with mutations in genes whose functions are required for protein maturation in the ER: N-glycosylation [46], chaperones [11, 17, 19, 39], and in genes required for ERAD that do not cause a detectable growth phenotype [21]. The most likely explanation for a constitutive induction of the UPR is that a perturbation of the folding capacity of the ER is caused by loss of functions required for efficient performance of the ER biosynthetic machinery. Since the UPR is triggered by accumulation of misfolded proteins, constitutive induction of the UPR in these mutants is indicative of an altered folding capacity in the ER in absence of ER stress.
We extended our genetic analysis to get insight into the function of this new non-essential DnaJ of the yeast ER, combining the erj5 deletion with mutations in several ER genes whose functions are required for protein maturation, in which we had previously observed that loss of Scj1p caused growth defects at different temperatures whereas loss of Jem1p had very mild or no effect ([11]and data not shown). We did not detect any growth phenotype associated to the loss of Erj5p in a N-glycosylation mutant lacking a non-essential subunit of the oligosaccharyltransferase (Δost3), in a glucosidase1-deficient (Δgls1) strain, or in a strain lacking calnexin (Δcne1) (data not shown). These results are consistent with evidence that point to Scj1p as the main DnaJ homologue involved in ER protein folding. Jem1p, and also Erj5p, would be Kar2p regulatory co-factors whose absence results in a less pronounced perturbation of the ER folding capacity as indicated by a lower constitutive induction of KAR2 mRNA in the null strains, compared to Δscj1.
A critical function of the UPR is to reduce the lumenal concentration of misfolded proteins by either directly refolding proteins or removing them from the ER. Consistent with the dynamic requirements of the living cell, chaperones have been often found functionally involved in both processes. We have not tested for a possible role of Erj5p in ERAD. However, a number of groups have performed ERAD assays on strains in which the ERJ5 gene was deleted using a variety of ERAD substrates. The soluble substrate CPY* [47], an integral membrane with the CPY* ERAD motif [48], and the soluble mutant A1PiZ [49]. No significant changes on the degradation rate of these proteins were detected by loss of Erj5p.
Although a global effect of Erj5p on ER protein maturation acting as a cofactor for Kar2p would be consistent with the genetic interactions detected, with the increased sensitivity to agents that promote accumulation of misfolded proteins in the ER, and with the effect on the UPR observed by loss of this J domain protein, it remains possible that Erj5p could perform a particular substrate-specific chaperone activity. In yeast, several genes involved in the biogenesis of specific secretory proteins have been identified, although the level at which they act is not fully understood in many cases [50]. They may be required for folding and/or secretion, or they may act as specific quality control factors, since their deletion often result in the accumulation of specific substrate molecules in the ER. Although further studies will be required to define the exact function of Erj5p in the ER, the results of this work clearly establish that this DnaJ homologue is required for optimal performance of the yeast ER folding machinery.
Acknowledgments
We thank Dr. T.W. Ng for providing yeast strains, Dr. Markus Aebi and P. Walter for antibodies, Dr. P. Philippsen and Dr. E. Schiebel for plasmids for yeast gene disruption and chromosomal tagging, respectively. We also thank Dr. R. Gilmore and Dr. A. J. Parodi for advice during the course of the experiments, and Dr. A. Colman-Lerner and Dr. M. L. Cantore for critical reading of the manuscript.
This work was supported by grants from Fundación Antorchas and Agencia Nacional de Promoción Científica y Tecnológica (to S.S.) and NIH R01 GM065507 (to D.R. and A.S.R.). M.C.F., N.Z. and D.R.L. are graduate students from Universidad de Buenos Aires. S.S. is an Investigator of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET-Argentina).
Abbreviations
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- ERAD
ER-associated degradation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 2.Corsi AK, Schekman R. The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J Cell Biol. 1997;137:1483–1493. doi: 10.1083/jcb.137.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng DT, Spear ED, Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Casagrande R, Stern P, Diehn M, Shamu C, Osario M, Zuniga M, Brown PO, Ploegh H. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol Cell. 2000;5:729–735. doi: 10.1016/s1097-2765(00)80251-8. [DOI] [PubMed] [Google Scholar]
- 7.Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 8.Vogel JP, Misra LM, Rose MD. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.te Heesen S, Aebi M. The genetic interaction of kar2 and wbp1 mutations. Distinct functions of binding protein BiP and N-linked glycosylation in the processing pathway of secreted proteins in Saccharomyces cerevisiae. Eur J Biochem. 1994;222:631–637. doi: 10.1111/j.1432-1033.1994.tb18906.x. [DOI] [PubMed] [Google Scholar]
- 10.Simons JF, Ferro-Novick S, Rose MD, Helenius A. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J Cell Biol. 1995;130:41–49. doi: 10.1083/jcb.130.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silberstein S, Schlenstedt G, Silver PA, Gilmore R. A role for the DnaJ homologue Scj1p in protein folding in the yeast endoplasmic reticulum. J Cell Biol. 1998;143:921–933. doi: 10.1083/jcb.143.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latterich M, Schekman R. The karyogamy gene KAR2 and novel proteins are required for ER-membrane fusion. Cell. 1994;78:87–98. doi: 10.1016/0092-8674(94)90575-4. [DOI] [PubMed] [Google Scholar]
- 14.Holkeri H, Paunola E, Jamsa E, Makarow M. Dissection of the translocation and chaperoning functions of yeast BiP/Kar2p in vivo. J Cell Sci. 1998;111(Pt 6):749–757. doi: 10.1242/jcs.111.6.749. [DOI] [PubMed] [Google Scholar]
- 15.Nishikawa S, Endo T. The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J Biol Chem. 1997;272:12889–12892. doi: 10.1074/jbc.272.20.12889. [DOI] [PubMed] [Google Scholar]
- 16.Brizzio V, Khalfan W, Huddler D, Beh CT, Andersen SS, Latterich M, Rose MD. Genetic interactions between KAR7/SEC71, KAR8/JEM1, KAR5, and KAR2 during nuclear fusion in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:609–626. doi: 10.1091/mbc.10.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 1996;15:2640–2650. [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton TG, Norris TB, Tsuruda PR, Flynn GC. Cer1p functions as a molecular chaperone in the endoplasmic reticulum of Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5298–5307. doi: 10.1128/mcb.19.8.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baxter BK, James P, Evans T, Craig EA. SSI1 encodes a novel Hsp70 of the Saccharomyces cerevisiae endoplasmic reticulum. Mol Cell Biol. 1996;16:6444–6456. doi: 10.1128/mcb.16.11.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- 21.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 22.Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–71. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlenstedt G, Harris S, Risse B, Lill R, Silver PA. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with Hsp70s. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J Biol Chem. 2003;278:8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- 26.Kuo CL, Campbell JL. Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8-1 mutation. Mol Cell Biol. 1983;3:1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 28.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 29.Hann BC, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- 30.Silberstein S, Collins PG, Kelleher DJ, Gilmore R. The essential OST2 gene encodes the 16-kD subunit of the yeast oligosaccharyltransferase, a highly conserved protein expressed in diverse eukaryotic organisms. J Cell Biol. 1995;131:371–383. doi: 10.1083/jcb.131.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 33.Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 35.Xu P, Raden D, Doyle FJ, 3rd, Robinson AS. Analysis of unfolded protein response during single-chain antibody expression in Saccaromyces cerevisiae reveals different roles for BiP and PDI in folding. Metab Eng. 2005;7:269–279. doi: 10.1016/j.ymben.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 37.Szyperski T, Pellecchia M, Wall D, Georgopoulos C, Wuthrich K. NMR structure determination of the Escherichia coli DnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues 2-108) containing the highly conserved J domain. Proc Natl Acad Sci U S A. 1994;91:11343–11347. doi: 10.1073/pnas.91.24.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genevaux P, Schwager F, Georgopoulos C, Kelley WL. Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ (Hsp40) J-domain. Genetics. 2002;162:1045–1053. doi: 10.1093/genetics/162.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamilton TG, Flynn GC. Cer1p, a novel Hsp70-related protein required for posttranslational endoplasmic reticulum translocation in yeast. J Biol Chem. 1996;271:30610–30613. doi: 10.1074/jbc.271.48.30610. [DOI] [PubMed] [Google Scholar]
- 40.Kimata Y, Kimata YI, Shimizu Y, Abe H, Farcasanu IC, Takeuchi M, Rose MD, Kohno K. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol Biol Cell. 2003;14:2559–2569. doi: 10.1091/mbc.E02-11-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 42.Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- 43.Mori K, Sant A, Kohno K, Normington K, Gething MJ, Sambrook JF. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- 46.Silberstein S, Gilmore R. Biochemistry, molecular biology, and genetics of the oligosaccharyltransferase. FASEB J. 1996;10:849–858. [PubMed] [Google Scholar]
- 47.Caldwell SR, Hill KJ, Cooper AA. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J Biol Chem. 2001;276:23296–23303. doi: 10.1074/jbc.M102962200. [DOI] [PubMed] [Google Scholar]
- 48.Taxis C, Hitt R, Park SH, Deak PM, Kostova Z, Wolf DH. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J Biol Chem. 2003;278:35903–35913. doi: 10.1074/jbc.M301080200. [DOI] [PubMed] [Google Scholar]
- 49.Palmer EA, Kruse KB, Fewell SW, Buchanan SM, Brodsky JL, McCracken AA. Differential requirements of novel A1PiZ degradation deficient (ADD) genes in ER-associated protein degradation. J Cell Sci. 2003;116:2361–2373. doi: 10.1242/jcs.00439. [DOI] [PubMed] [Google Scholar]
- 50.Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 51.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]


