Abstract
Target-assisted iterative screening (TAIS) has been applied to a random phage-displayed peptide library in a search for novel ligands of the third baculovirus IAP (‘inhibitors of apoptosis’) repeat (BIR) domain of cIAP1. The peptides selected in the screen fall into two distinct specificity groups, one that conforms to a known IAP-binding motif (IBM) and another one that reveals a novel BIR domain interactingmotif, NH2-SR(V/P)W. The biochemical profiling of selected sequences with synthetic peptides, which included alanine scanning and N- and C-terminal truncations as well as competition with the Smac peptide, suggests a major energetic contribution of tryptophan at the +4 position of peptide ligands to binding and identifies the latter together with the respective pocket on the BIR domain surface as a ‘hot spot’ of the interaction. A peptide featuring the novel motif selectively binds the full-length cIAP1 protein in cell lysates. A ‘two-pocket’ model of BIR domain recognition mechanism is proposed as the basis of differential BIR domain interactions with different IBMs.
Keywords: BIR domain, IAP-binding motif, cIAP1, TAIS, peptide libraries
INTRODUCTION
Baculovirus IAP (‘inhibitors of apoptosis’) repeat (BIR) domains are small, approximately 70 amino acids, zinc-containing protein interaction modules. As the name suggests, they were first identified as repetitive elements within baculovirus IAP proteins (Crook et al., 1993; Birnbaum et al., 1994). Later the repeats were found in various species in a number of different gene products (Duckett et al., 1996; Salvesen and Duckett, 2002). The BIR domain has become a defining feature of the IAP protein family, though not all members of the family are believed to function as bona fide inhibitors of apoptosis. Prototypical inhibitors such as XIAP, cIAP1, and cIAP2, each carrying three consecutive BIR domains at their N-termini and a RING domain at their C-termini, are thought to inhibit apoptosis through direct physical association with activated caspases, an interaction which is mediated by the BIR domains of IAPs and/or the linker region connecting BIR1 and BIR2 domains (Chai et al., 2001; Huang et al., 2001; Riedl et al., 2001). The mammalian pro-apoptotic proteins Smac/DIABLO, Omi/HtrA2, and GSPT1/eRF3 promote apoptosis by competing off activated caspases from their complexes with IAPs (Du et al., 2000; Suzuki et al., 2001; Hegde et al., 2003). Accumulating evidence suggests that XIAP, cIAP1, and cIAP2 may regulate their own protein levels through auto-ubiquitination and proteosomal degradation (Vaux and Silke, 2005). It has been suggested that the ubiquitination activity of IAPs is mediated by their RING domain and is regulated by BIR domain interactions (Yang et al., 2000; Yang and Du, 2004; Vaux and Silke, 2005). IAPs appear also to target for ubiquitination and down-regulation of the proteins they associate with, such as caspases, Smac, and TRAFs (Li et al., 2002; Vaux and Silke, 2005). Though the primary function of XIAP, cIAP1, and cIAP2 is thought to be the inhibition of apoptosis, they have been implicated in a number of other cellular functions such as cell-cycle regulation, protein degradation, and receptor-mediated signal transduction (Salvesen and Duckett, 2002; Galvan et al., 2004; Vaux and Silke, 2005).
Tremendous research efforts have been focused on the biochemical and structural characterization of protein interactions mediated by BIR domains, as these protein modules appear to play a critical role in the majority, if not all, functions attributed to IAPs. The BIR domains bind selectively and with high affinity to the N-termini of proteins containing the so-called IAP-binding motif (IBM). A number of proteins, such as caspases 3, 7, and 9, Smac/DIABLO, Omi/HtrA2, and GSPT1/eRF3 as examples, have been shown to possess functional IBMs that are normally hidden, but become exposed upon proteolytic processing of these proteins (Salvesen and Duckett, 2002). All of the characterized IBMs appear to interact as short linear peptides with the same peptide-binding groove on the surface of structurally conserved members of the BIR domain family. The competition between the IBMs of different proteins has been suggested as a mechanism by which a balance between pro- and anti-apoptotic activities is regulated. Indeed, the Smac-derived peptides were reported to sensitize cancer cells to apoptosis in both cell culture and animal models (Arnt et al., 2002; Fulda et al., 2002). As the overexpression of IAPs has been observed in a variety of cancers (Vucic et al., 2000; Li et al., 2001; Krajewska et al., 2003), thus suggesting a common mechanism of apoptosis evasion in malignant cells, the interaction between BIR domains and their IBM ligands has become the focus of intense drug design/screening efforts (Wu et al., 2003; Oost et al., 2004; Schimmer et al., 2004). The comparison of the known functional IBMs combined with the data from structural and biochemical studies addressing their interactions with BIR domains led to the establishment of a paradigm defining IBM as a short linear peptide in which the invariable N-terminal alanine (A) residue is followed by the somewhat vague and not all-inclusive consensus sequence—A(K/T/V/I)(P/A/E)(F/E/I/S/Y) (Vaux and Silke, 2005).
In the study presented here we applied a novel screening format, target-assisted iterative screening (TAIS), to a random phage-displayed linear peptide library using the BIR3 domain of cIAP1 as target. We isolated two families of peptides that selectively bound the BIR3-cIAP1. Sequences of one family conform to the known IBM consensus, while sequences of the other suggest a novel BIR3-cIAP1-binding motif, NH2-SR(V/P)W, thus expanding the spectrum of potential interacting partners of IAPs. The biochemical profiling of the isolated sequences with synthetic peptides indicates that the position +4 of the novel IBM, which is occupied by tryptophan in the selected peptides, represents an energetic ‘hot spot’ of the BIR3-cIAP1-peptide interaction interface. New findings pertaining to the recognition specificity of BIR domains are discussed in the context of existing experimental data.
MATERIALS AND METHODS
Target-assisted iterative screening
A detailed description of the TAIS method is presented by Kurakin et al. (2004). The TAIS flowchart and protocols can be found on the Internet (http://www.buckinstitute.org/TAIS). Briefly, 30 μg of a GST–BIR3–cIAP1 domain fusion immobilized on sepharose beads were blocked with 0.5% bovine serum albumin (BSA) in Tris-buffered saline, pH 7.4 + 0.1% Tween 20 (TBS-T) and incubated with a random peptide library aliquot (approximately 108–109 pfu). After 90 min of incubation at room temperature (RT) the beads were thoroughly washed with TBS-T and bound phages were eluted with 200 μl of 1% SDS for 15 min at RT. Following elution, the phages were immediately mixed with a molten 0.6% top agarose-containing host cells and plated onto two pre-warmed 150 mm agar plates. When phage plaques became visible, the plates were cooled down for 30 min at 4°C and overlaid with 132 mm nitrocellulose membranes (Schleicher & Schuell) for 5 min. Following plaque lift, the membranes were blocked in 1% BSA in TBS for 1 h at RT and incubated overnight in 25 ml of TBS-T on a rocker at 4°C with 10 μg of the BIR3-cIAP1 domain that had been cleaved from the GST moiety, biotinylated, and complexed with streptavidin–alkaline phosphatase (STRAP) at a ratio of 4:1. After extensive washing with TBS-T, positive plaques were developed on the membranes with insoluble alkaline phosphatase (AP) substrate BCIP/NBT (Sigma). Individual positive plaques were identified on the plates and phages from these plaques were propagated separately in the appropriate host for production of phage DNA. M13 single-stranded DNA was prepared with QIAprep Spin M13 kit (Qiagen).
Phage ELISA
Streptavidin- or GST fusion-coated microtiter ELISA plates (COSTAR) were prepared by passive immobilization of 1 μg of streptavidin or 2 μg of the indicated GST fusion proteins (Figure 2) per well in 200 μl of 0.1M NaHCO3, pH 8.0, overnight at 4°C. Following protein coating, plates were blocked by adding 150 μl of 1% BSA in TBS for 1 h at RT. After five washes with TBS-T, 0.5 μg of the biotinylated BIR3-cIAP1 domain in 200 μl of TBS-T were added to streptavidin-coated wells and incubated additionally for 1 h at RT. Following incubation, the wells with immobilized target proteins were washed five times with TBS-T. One hundred microliters of freshly prepared individual phage lysate was added to the ELISA plate wells and incubated for 1 h at RT. Following incubation, unbound phages were washed away with TBS-Tand the amount of retained phages was determined with phage-specific antibodies. Monoclonal anti-M13 antibodies conjugated to horseradish peroxidase (HRP) (Amersham Pharmacia) were used for detection of M13 phages.
Figure 2.
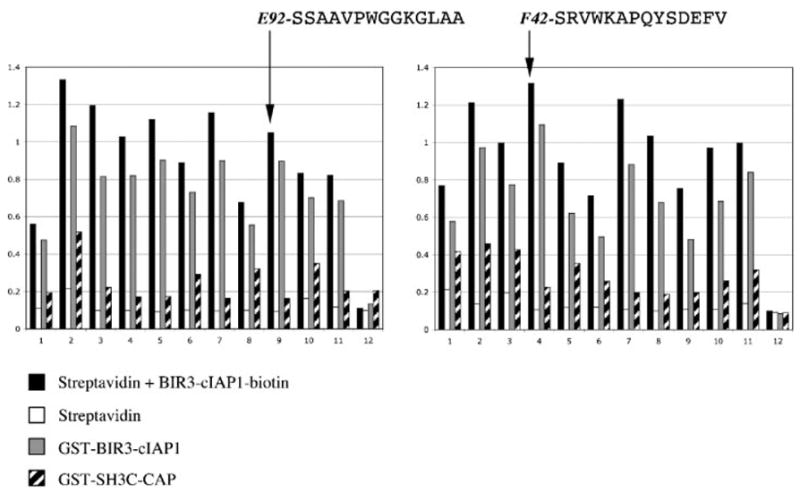
Example of individual phage ELISA. Phages from individual positive plaques from a TAIS experiment were tested for binding to the indicated proteins immobilized on microtiter plates. A representative example of 24 phages out of approximately 96 is shown. The two sequences indicated, E92 and F42, were selected for detailed biochemical profiling (see text and other figures). The y axis on the histograms indicates A values at 405 nm.
Peptide ELISA
To study the interaction of the BIR3-cIAP1 domain with the peptides selected in TAIS experiments outside the phage context, all selected peptides as well their variants were synthesized as C-terminally biotinylated peptides. Wells of microtiter plates were coated passively with 2 μg of GST–BIR3–cIAP1 fusion as indicated above for phage ELISA. Individual biotinylated peptides (30 ng) were pre-incubated with 1 μg of streptavidin-HRP conjugate (Sigma) in 300 μl of TBS-T for 30 min at RT. One hundred microliters of the peptide-streptavidin-HRP conjugate were added to 100 μl of TBS-T left in each coated well after the final wash of the protein immobilization procedure. In the case of competition experiments, the non-biotinylated Smac peptide AVPIAQKS or irrelevant peptide was added to the wells in the amounts indicated in Figure 6 at this moment. Microtiter plates were incubated for 1 h at RT and then washed five times with TBS-T. The amounts of peptides retained were quantified colorimetrically by adding soluble HRP substrate (ABTS/H2O2). ELISA readings were taken on a SpectraMAX190 plate reader (Molecular Devices) at 405 nm. To ensure reproducibility, all peptide ELISA experiments presented were repeated at least three times.
Figure 6.
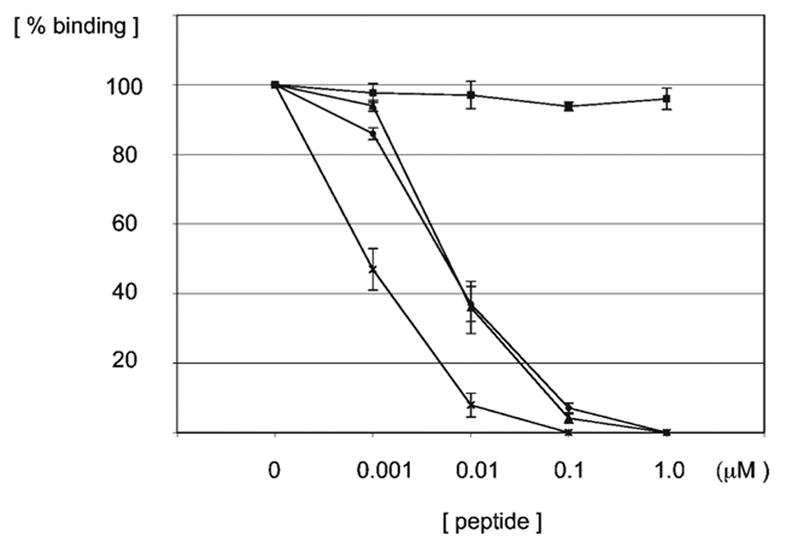
The Smac peptide (AVPIAQKS) competes with the F42 (SRVWKAP-QYSDEFV) peptide for binding to the BIR3-cIAP1 domain. Equivalent amounts of a caspase 9-derived peptide (ATPFQEGL) (triangles), F42 peptide (dots), and Dronc peptide (SRPPFISLNERR) (crosses) were conjugated to streptavidin-HRP and incubated with the BIR3-cIAP1 domain immobilized in microtiter plate wells in the presence of the indicated amounts of non-biotinylated Smac peptide. As a control, the competition between the F42 peptide and an irrelevant non-biotinylated peptide was assayed in the same manner (boxes). The amount of peptides that remained in the wells after the incubation and washing steps was determined colorimetrically by following the kinetics at 405 nm for triplicate samples. Error bars indicate standard error of the mean. Values were normalized to the binding levels measured in the absence of non-biotinylated peptide competitor (not shown).
Peptide Pull Downs
The 293 cell’s lysates were prepared by extraction of cultured cells in lysis buffer (150 mM NaCl, 20 mM Tris-HCl, pH 8.0, 1 mM EDTA, 5 mM EGTA, 10 mM NaF, 0.5% Na deoxycholate, 1% Triton X-100) in the presence of a protease inhibitor cocktail (Roche, mini-tablets). Following extraction on ice, the insoluble fraction was pelleted by centrifugation at 13 000 rpm at 4°C for 5 min. Individual biotinylated peptides (1.2 μg) were pre-immobilized for 30 min at RT on 10 μl of streptavidin-coated beads (UltraLink, PIERCE) in separate 1.5 ml eppendorf tubes, washed three times with 1 ml of TBS-T and mixed with approximately 250 μg of the total lysate protein in 0.5 ml of TBS-T. The mixture was tumbled for 1 h at RT. Ten microliters of beads without peptides were used as negative control. Following incubation, beads were washed five times with 1 ml of TBS-T and the retained proteins were eluted into loading buffer at 70°C for 10 min. Precipitated protein complexes were separated by SDS–PAGE (pre-cast 4–12% Bis-Tris gel/MOPS buffer, Novex), blotted to a PVDF membrane, blocked in 1% BSA, and developed with specific antibodies. Twenty-five micrograms (10%) of total cell lysate protein was used as input. HRP-conjugated secondary antibodies, the ECL detection system (Amersham Pharmacia), and BioMax film (Kodak) were used for protein visualization.
Phage display library, peptides, proteins and antibodies
A 12-mer random peptide library was kindly provided by Dr B. K. Kay (Argonne National Laboratory). The GST–BIR3–cIAP1 fusion protein expression construct was a generous gift by Dr C. Day (University of Otago, New Zealand). Synthetic peptides were purchased from JPT Peptide Technologies GmbH (Berlin, Germany). Mouse monoclonal antibodies for human cIAP1 and XIAP proteins were obtained from BD Pharmingen and Stressgen, correspondingly.
RESULTS
TAIS: identification of unconventional BIR domain ligands
TAIS is a rapid and unbiased screening format for large molecular repertoires (Kurakin and Bredesen, 2002). Detailed description of the method and relevant protocols pertaining to the application of TAIS to phage surface display libraries can be found elsewhere (Kurakin et al., 2004). In short, during the first pre-selection step, a protein target is immobilized on a solid support, such as sepharose beads and incubated with a phage-displayed peptide library in solution. After washing away unbound phages, the retained ones are eluted and plated on a bacterial lawn. During the second step, the plaques of the pre-selected phage subset are transferred from the bacterial lawn and immobilized onto a nitrocellulose membrane. The protein target presented in a different molecular context and conjugated with a reporter, such as AP, is used then as one-step detection reagent to identify the interacting plaques on the membrane. The identities of the displayed polypeptides that interact with the target are deduced by sequencing of the corresponding inserts of the phages from individual positive plaques. Omitting a competition between phages altogether, TAIS eliminates propagation biases inherent to classical phage display panning.
The library of random 12-mer peptides displayed as N-terminal fusions to the pIII coat protein of a filamentous phage was screened to select specific binders for the BIR3-cIAP1 domain. The peptide–pIII fusions are synthesized inside the bacterial host as pro-polypeptides containing a leader sequence that mediates export into the periplasmic space of bacteria where the final phage assembly takes place. Following export the leader is cleaved off by processing proteases, thus revealing the peptide sequences displayed. Due to the idiosyncrasy of the library construct used in this work, all the peptides displayed on mature phages are presumed to have NH2-SS… or NH2-SR… N-terminal sequences (Figure 1A) (Kay et al., 1996). Therefore, none of the peptide ligands selected by the BIR3-cIAP1 domain conforms to the known IBM consensus, as defined by the current BIR domain recognition paradigm, A(K/T/V/I)(P/A/E)(F/E/I/S/Y) (Figure 1B), neither they feature a critical alanine as their first residue. Nevertheless, all peptides selected in the screen specifically bound to the BIR3-cIAP1 domain as confirmed by phage ELISA analysis (Figure 2).
Figure 1.
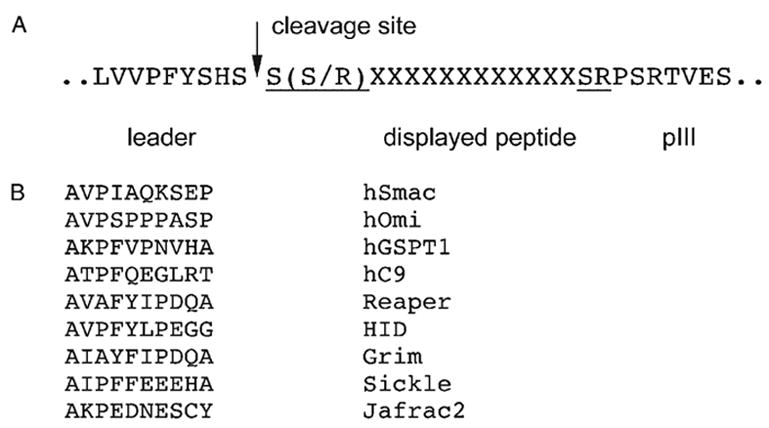
Comparison of the known IBMs and the phage-displayed peptide library used. (A) Due to insertion of a randomized cassette between the leader peptide sequence and the pIII protein of M13 phage, all the peptides displayed in the library used in this work are flanked by S(S/R) and SR amino acid pairs, reflecting the identities of the respective restriction sites used to insert the cassette (Kay et al., 1996). (B) Sequences of the known mammalian and fly IBMs.
Residue frequency patterning delineates two distinct specificity groups among BIR3-cIAP1 peptide ligands
As the deduction of the recognition consensus of the BIR3-cIAP1 domain from the sequences selected did not appear straightforward, we devised a simple statistical procedure, residue frequency patterning (RFP). First, we analyzed what types of amino acid residues are over-represented in the selected peptides, independent of their position within the individual sequences. Second, we looked for repetitive patterns in the relative distribution of the overrepresented residues within individual peptides. It has been noticed that the interaction energy is not uniformly distributed over protein interaction interfaces, but tends to concentrate in a few ‘hot spots’ (Stites, 1997). We rationalized that if each interaction interface is characterized by its unique distribution of ‘hot spots’ it would be possible, given sufficient statistics, to tease out the different patterns corresponding to different interfaces which may potentially be present within the set of peptides selected in the screen. Figure 3A shows the representation of all 20 natural amino acids within the 33 peptides selected by the BIR3-cIAP1 domain relative to their average occurrence in the SWISS-PROT database. As a control, the same type of analysis was performed on the peptide dataset of similar size that we reported recently for the SH3 domains of CIN85/SETA protein (Kurakin et al., 2003). A higher than threefold overrepresentation of proline (P) and arginine (R) in the control dataset is in correspondence with the SH3-CIN85 recognition consensus, Px(P/A)XXR (Kurakin et al., 2003) (Figure 3A). The twofold overrepresentation of cysteine (C) in the control came as a surprise retrospectively, and the reason for this is currently under investigation. In the case of the BIR3-cIAP1 ligands, a remarkable ninefold over-representation of tryptophan (W) residue suggested its critical role in the interaction. In fact, every peptide selected in the screen except one has at least one W in its sequence and more than 30% of the peptides feature two tryptophans. On the other extreme, cysteine was never observed in any of the 33 identified peptides. The meaning of this observation is not clear at present. The small hydrophobic residues A, G, and P occurred at frequencies significantly higher than the average. Analysis of the patterns composed of W, A, G, and P residues within the peptides selected by BIR3-cIAP1 unambiguously revealed at least two specificity families of ligands, which henceforth we refer to as the AVPW group and the SRVW group (Figure 3B). Notably, the AVPW ligands feature a motif that is very similar to the known IBMs, albeit not positioned at the very N-terminus, as required for IBMs.
Figure 3.
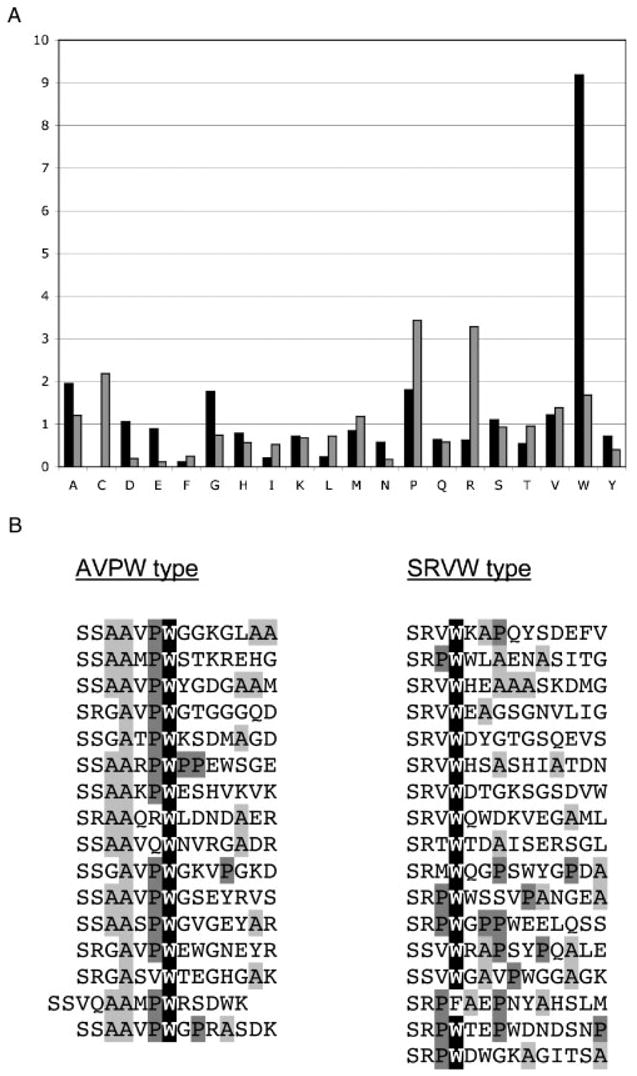
Residue Frequency Patterning: (A) The relative frequency distribution of individual amino acids within the peptides selected by the BIR3-cIAP1 domain (black columns) as compared to the analogous distribution within the peptides selected by the SH3 domains of CIN85/SETA (gray columns) is shown. The y axis indicates the ratio of individual amino acid frequencies in the experimental dataset to the frequencies of the same amino acids in the SWISS-PROT database. (B) The positional patterns of overrepresented residues within the BIR3-cIAP1 selected peptides define two specificity groups, AVPW and SRVW.
AVPW-type peptides require exposure of their IBMs for binding to BIR3-cIAP1 domain
To test BIR domain–ligand interactions outside the context of the phage, we synthesized all the selected sequences as biotinylated peptides and used peptide-streptavidin-HRP conjugates as detection reagents to follow the interaction between synthetic peptides and a GST–BIR3–cIAP1 fusion passively immobilized on ELISA plates. To achieve a meaningful cross-experiment comparison and high reproducibility, we followed the kinetics of peptide–domain interactions, rather than relying on endpoint readout as an estimate of the number of complexes formed. As a positive control, a number of known and putative BIR domain binders were synthesized and tested in peptide ELISA assays (Figure 4A). The strong interaction of the peptide derived from the N-terminus of Chk1 kinase with the BIR3-cIAP1 domain, which is a new finding revealed in the control experiments, is consistent with the recently reported Chk1–XIAP interaction presumably mediated by the BIR domains of the XIAP protein (Galvan et al., 2004).
Figure 4.
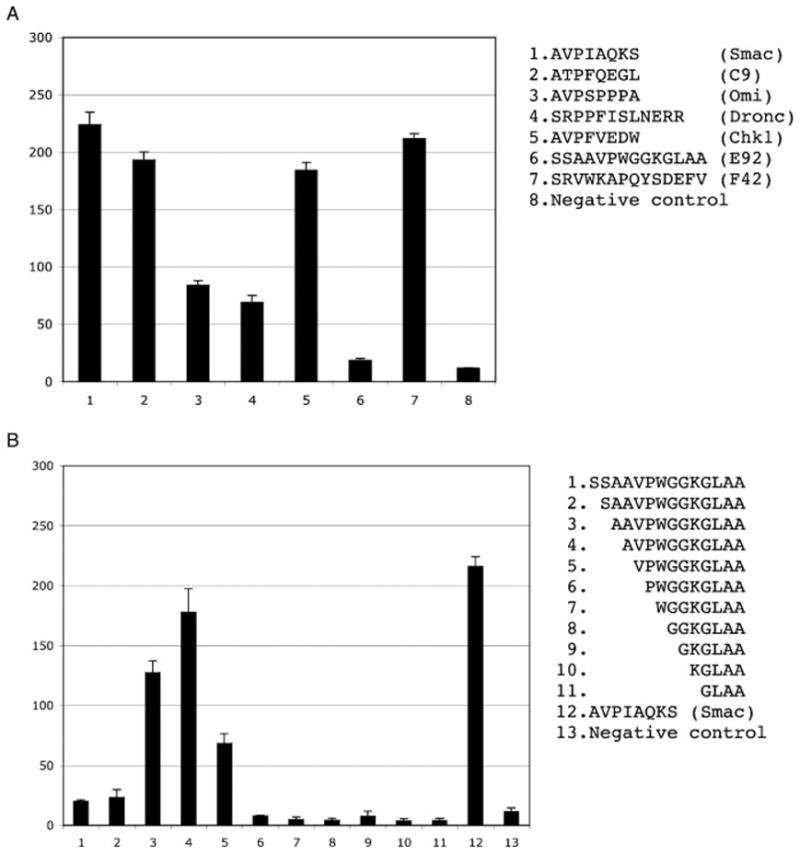
Controls and N-terminal deletion analysis of the E92 (SSAAVPWGGKGLAA) peptide. Interaction histograms of control peptides (A) and N-terminal truncations of the E92 peptide (B) are shown. The sequences indicated were synthesized as biotinylated peptides, conjugated to streptavidin-HRP and tested for their binding to the GST–BIR3–cIAP1 fusion passively immobilized on a microtiter plate. The y axis indicates relative ELISA kinetics slopes expressed in arbitrary units. Error bars correspond to the SD of the mean for triplicate samples.
Quite unexpectedly, none of the synthetic peptides from the AVPW specificity group bound the BIR3-cIAP1 domain, while all of the peptides from the SRVW group retained their ability to bind the target domain outside the phage context (not shown). Therefore, we selected one representative member from each specificity group for a more detailed analysis, E92-SSAAVPWGGKGLAA (AVPW group) and F42-SRVWKAPQYSDEFV (SRVW group) (see Figure 2). Given the similarity between the AVPW motif and the previously reported IBMs, we hypothesized that the peptides from the AVPW specificity group were most likely processed in the context of the phage, thus exposing the IBM motif at their extreme N-termini. Indeed, an N-terminal deletion analysis supports this hypothesis (Figure 4B). Omitting the first three amino acids from the N-terminus of the E92 peptide and exposure of the AVPW motif restored binding of the E92 peptide. It should be noted that deletion of either two amino acids, which results in the AAVPW N-terminus, or of four, leading to the VPW N-terminus, also gave rise to peptides that bound the BIR3-cIAP1 domain with appreciable affinities. Mass spectrometry analysis of the respective peptide preparations revealed that, although the binding of the AAVPWGGKGLAA truncation could be attributed, at least in part, to the presence of the incomplete synthesis product AVPWGGKGLAA in the peptide preparation, the VPWGGKGLAA peptide indeed bound the target domain (not shown). The results of the N-terminal deletion analysis of synthetic peptides strongly suggest that the phage-displayed peptides are processed to expose the AVPW motif required for binding. It is unlikely that the AVPW motif is exposed due to random degradation of the N-termini when the peptides are displayed on the phage, because all of the selected peptides but one feature an IBM in the same position in relation to their N-termini. The latter fact is suggestive of a systematic processing activity, presumably a protease. These results demonstrate that in certain cases a significant discrepancy may exist between what is assumed to be displayed on the phage and what is actually displayed.
SRVW-type ligands define a novel BIR3-cIAP1-binding motif
To evaluate the contribution of individual residues of peptide ligands to binding, the F42 peptide was subjected to alanine-scanning mutagenesis and N- and C-terminal truncation analyses. Alanine substitution of tryptophan at the +4 position of the ligand proved to be the most detrimental for binding (Figure 5A). The ninefold over-representation of tryptophan within the peptide sequences selected and the sensitivity of the interaction to the W → A substitution at ligand position +4 strongly suggest a major energetic contribution of tryptophan to binding, defining the +4 position as a ‘hot spot’ of the peptide–BIR3-cIAP1 domain interaction. This fact may help to rationalize the design of drugs targeting the BIR domain interaction interface. The deletion of the N-terminal serine residue and the substitution of proline at the +7 position attenuate but do not abolish the interaction. The C-terminal deletion analysis appears to suggest that at least nine first residues of the F42 peptide are required for its interaction with the BIR3-cIAP1 domain. It should be pointed out, however, that the detection system employed, which consists of the C-terminally biotinylated peptides conjugated to STRAP complex, is likely to introduce a steric obstacle upon interaction of the conjugated peptide ligands and BIR domain, especially when shorter ligands are assayed, and thus is unsuitable for accurate evaluation of the footprint.
Figure 5.
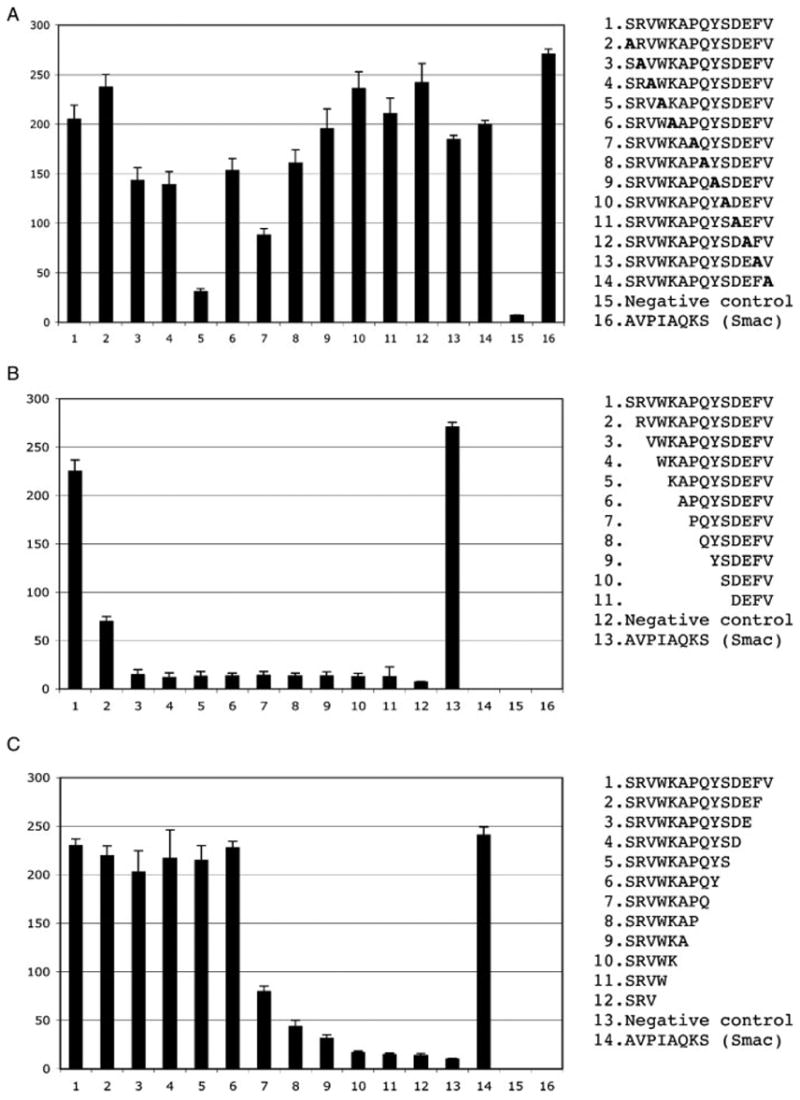
Biochemical profiling of the F42 (SRVWKAPQYSDEFV) peptide ligand. (A) Alanine scanning mutagenesis. (B) N-terminal deletion analysis. (C) C-terminal deletion analysis. The sequences indicated were synthesized as biotinylated peptides, conjugated to streptavidin-HRP and tested for their binding to the GST–BIR3–cIAP1 fusion passively immobilized on a microtiter plate. The y axis indicates relative ELISA kinetics slopes expressed in arbitrary units. Error bars correspond to the SD of the mean for triplicate samples.
The conservation of the SR(V/P)W sequence at the N-termini of all the peptides in the unconventional specificity group suggests a novel BIR3-cIAP1 recognition motif, which has not been reported before, thus expanding a spectrum of potential interacting partners of the cIAP1 protein. It is worth noting that the novel consensus is similar to Dronc-derived peptide that binds to BIR2-DIAP1 domain, SRPPFISLNERR (Chai et al., 2003), and is reminiscent of the neo-termini of processed caspases, SGVD (caspase 3), and SGPI (caspase 7) that have been shown to bind BIR domains of XIAP (Riedl et al., 2001; Scott et al., 2005).
SRVW-type peptides compete with the Smac-derived peptide for binding to the BIR3-cIAP1 domain
To ensure that the peptides selected by TAIS bound the known interaction surface on the BIR3-cIAP1 domain, we performed a series of competition experiments using the non-biotinylated Smac-derived peptide as a competitor. The results presented in Figure 6 suggest that the F42-SRVWKAPQYSDEFV peptide bound the same or a largely overlapping surface on the BIR3-c-IAP1 domain as that bound by the Smac peptide. In fact, the Smac peptide efficiently displaced all synthetic peptides from the SRVW specificity group as well as the control peptides, suggesting a competition for the same binding groove (not shown).
SRVW- and AVPW-type peptide ligands selectively interact with the endogenous full-length cIAP1 protein in cell lysates
To demonstrate that the peptides that bind the isolated BIR3 domain in vitro are able to interact with the full-length cIAP1 protein, we carried out pull-down experiments with the F42 and the truncated E92 (AVPWGGKGLAA) peptides immobilized on streptavidin-coated beads. As shown in Figure 7, both peptides precipitated cIAP1 from cell lysates, albeit with different efficiencies. The truncated E92 peptide, which features a classical IBM, also pulled down the XIAP protein, presumably through interaction with its BIR domain(s). The fact that the AVPW-type ligand precipitated virtually all cIAP1 present in the lysate sample while pulling down only about 10% of the available XIAP suggests a preference of the AVPW-type ligands for the cIAP1 BIR domain(s) combined with a relative promiscuity toward different BIR domains. The SRVW motif failed to pull down XIAP and thus is likely to be specific for the BIR3-cIAP1 domain only. Substitution of the critical tryptophan in the novel IBM abolished the interaction with full-length cIAP1 as expected.
Figure 7.
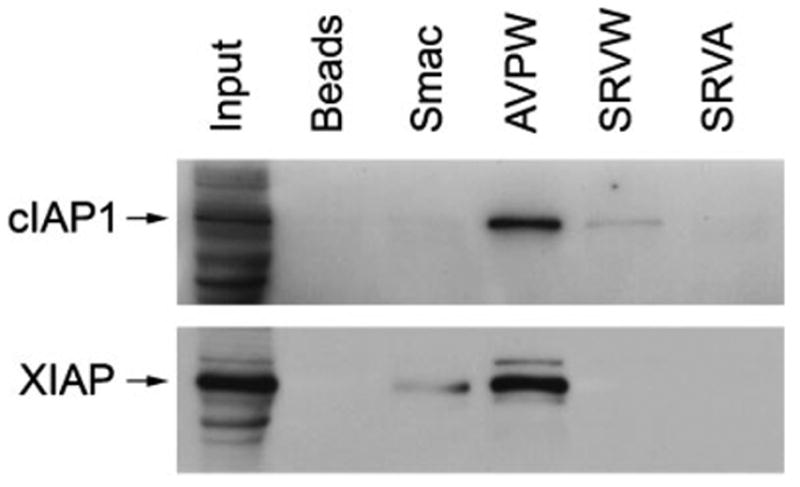
Affinity precipitation of cIAP1 and XIAP from cell lysates with the E92 truncation (AVPWGGKGLAA), the F42 (SRVWKAPQYSDEFV) peptide, and the W → A F42 mutant (SRVAKAPQYSDEFV). Approximately 250 μg of total protein from a 293 cell lysate were incubated with 1.2 μg of the indicated peptides immobilized on streptavidin-coated beads. The bound proteins were resolved by SDS–PAGE and immunoblotted with a cIAP1 monoclonal antibody. The same blot was re-probed with a XIAP monoclonal antibody. The input lane contains 10% of the lysate protein used in the pull-down experiments.
The truncated E92 peptide (AVPWGGKGLAA) appears to interact with the full-length cIAP1 protein significantly stronger than the F42 peptide (Figure 7), despite the fact that both peptides exhibit similar affinities for the isolated BIR3-cIAP1 domain (Figure 4, compare A, pept. #7 and B, pept. #4). This apparent contradiction may be explained by the ‘two-pocket’ recognition mechanism of BIR domain-ligand interaction proposed below. In short, the AVPW motif, which features an N-terminal alanine, a known major contributor to the energetics of BIR domain–IBM interactions, is likely to bind many different BIR domains and thus to act as a relatively non-specific high-avidity binder for proteins-containing multiple BIR domains. On the other hand, the SRVW-binding motif, which lacks the N-terminal alanine may be exclusively specific for only the third BIR domain of cIAP1, which may or may not be readily available for interaction in the context of a full-length folded protein.
DISCUSSION
We screened an N-terminal 12-mer random peptide library displayed on a filamentous phage using the BIR3-cIAP1 domain as a target. Instead of the classical panning procedure, which as a rule leads to propagation biases, a novel screening format, TAIS was employed (Kurakin and Bredesen, 2002; Kurakin et al., 2004). As a result, we identified two distinct families of peptide ligands, one family containing the expected IBM within its sequences and another family featuring a novel-binding motif for the BIR3-cIAP1 domain. Biochemical profiling of the selected sequences with synthetic peptides, which included alanine-scanning mutagenesis and N- and C-terminal deletion analyses as well as competition experiments with a Smac-derived peptide, led us to the following conclusions:
The selected peptides featuring conventional and unconventional IBMs bind specifically to the BIR3-cIAP1 domain in vitro with affinities comparable to those of the Smac N-terminal peptide and of the processed caspase 9–derived peptide (Figures 4 and 5).
Consistent with previous reports, exposure of the conventional IBM at the very N-terminus of ligands is required for the BIR domain–peptide interaction to occur.
As no significant homology between the unconventional ligands selected in the screen was observed beyond their first four amino acids, the novel minimal BIR3-cIAP1-binding motif is defined as SR(V/P)W.
The position +4 within the novel IBM appears to represent a ‘hot spot’ of the BIR3-cIAP1 domain–peptide ligand interaction.
Both conventional and unconventional IBMs interact with the full-length endogeneous cIAP1 in cell lysates. While the conventional IBM preferentially interacts with cIAP1, it also readily cross-reacts with XIAP, which is consistent with the previous reports suggesting a relatively poor selectivity of conventional IBMs toward different BIR domains. On the other hand, the unconventional IBM appears to be specific for the cIAP1 protein only.
Alanine-scanning analysis of the F42 peptide (unconventional ligand) showed that its interaction with the BIR domain is sensitive to mutation of the ligand position +7. One plausible hypothesis explaining this sensitivity is that the F42 peptide, and most likely the other peptides from the unconventional specificity group that feature proline at the +7 position, may bind the BIR3-cIAP1 domain in two opposite pseudo-symmetrical orientations, with the central hook/axis role played by the +4 tryptophan. This hypothesis is supported by the following observations. First, the position +7 in peptides of the SRVW type exhibits a significant preference for proline and is almost exclusively occupied by overrepresented small hydrophobic residues, that is, P, A, or G. At the same time, the corresponding position in AVPW peptides does not show any particular propensity for these residues. Second, the Dronc-derived peptide, SRPPFISLNERR, which is reminiscent of SRVW type peptides, was recently shown in structural studies to bind the BIR2 domain of DIAP1 in reverse orientation as compared to conventional BIR ligands (Chai et al., 2003). Pertinently, the phenylalanine in the Dronc peptide, identified as the most critical residue for binding, occupies a conserved pocket of the BIR2-DIAP1 domain that is distinct from the classical alanine-binding site. Another critical hydrophobic residue, leucine, anchors in the vicinity of the alanine pocket through networks of van der Waals interactions. However plausible, our hypothesis of the two alternative orientations of the SRVW type ligands requires experimental validation through structural analysis.
Positions +1, +4, and +7 of the BIR3-cIAP1 ligands are sensitive to either deletion (+1) or alanine substitution (+4 and +7). It should be noted, however, that the deletions of the N-terminal alanine and serine residues from the IBMs of the E92 truncation (AVPWGGKGLAA) and F42 peptides, respectively, diminish but do not abolish their interaction with the BIR3-cIAP1 domain (see histograms in Figure 4B, column 5 and in Figure 5B, column 2). Similarly, alanine substitution of the +7 proline in the F42 peptide does not abrogate binding completely; only mutation of position +4 appears to disrupt the interaction. In natural IBMs, position +4 is often occupied by aromatic residues such as F and Y, although other types of amino acid residues are also observed (Figure 1B). Analysis of the reported structures of the BIR domains complexed to their ligands indicates that the +4 residues of different IBMs occupy a structurally conserved but chemically variable pocket on the surface of BIR domains, which is distinct from the one occupied by the N-terminal alanine (Figure 8 and not shown). The importance of both pockets for BIR domain-mediated interactions is underscored by the fact that the peptidomimetics developed to disrupt BIR3-XIAP domain interactions target both of these sites simultaneously (Oost et al., 2004). As a practically relevant model, we propose therefore to consider two functionally distinct regions within the peptide-binding groove of BIR domains. One of these regions is the classical ‘alanine pocket’ and the other one is a ‘specificity pocket’ interacting with the +4 residues of IBMs. Within this model, the peptide ligands featuring an N-terminal alanine residue are expected to be relatively non-specific toward different BIR domains, because the contribution of alanine to the overall binding energetics is dominating, and most residue types are tolerated in the IBM positions following the N-terminal alanine (Kipp et al., 2002). On the other hand, peptides featuring a non-alanine residue at their extreme N-termini as well as ligands that may bind BIR domains in the orientation that is opposite to the orientation of classical IBMs, as exemplified by the Dronc-derived peptide (Chai et al., 2003), are expected to rely mainly on the interaction that involves their +4 residues and the ‘specificity’ pocket of BIR domains, and thus to discriminate significantly better between different BIR domains. It is worth noting that the potential communication between these two pockets, which share common residues, is likely to regulate their occupancy in an interdependent fashion (Figure 8).
Figure 8.
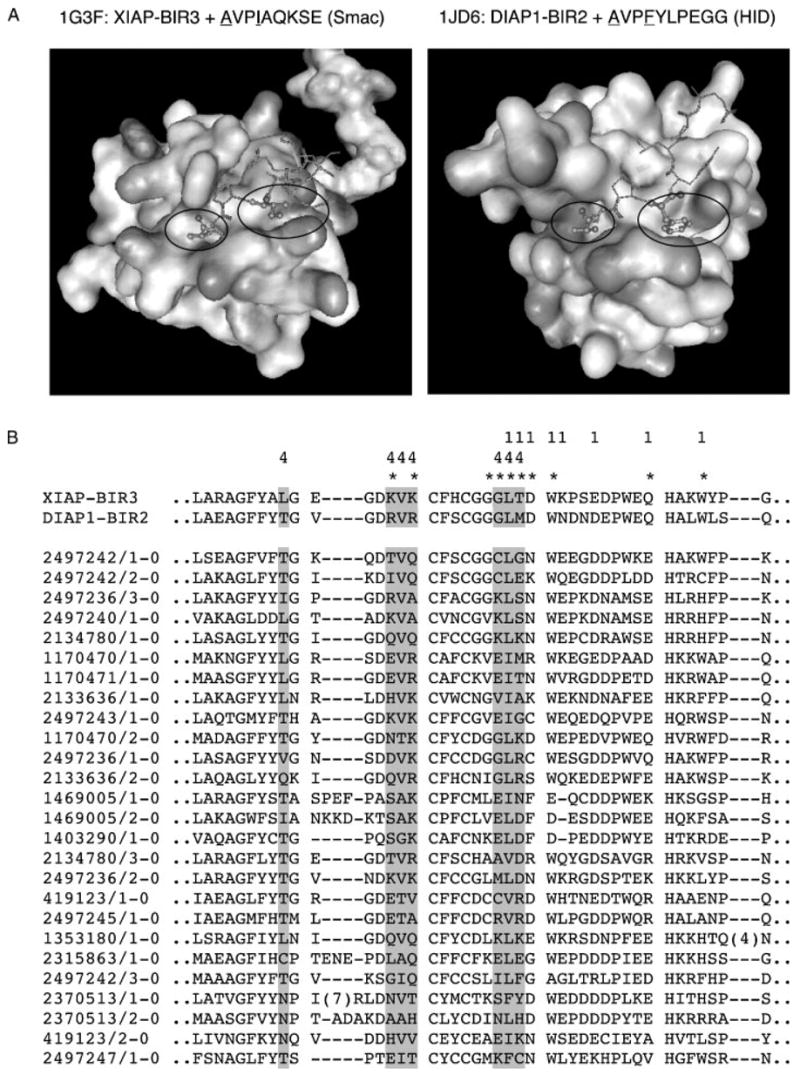
The hypothetical ‘two-pocket’ recognition mechanism employed by BIR domains. (A) Structures of BIR3-XIAP and BIR2-DIAP1 domains complexed with Smac and HID peptides, correspondingly. The smaller oval indicates the alanine pocket and the larger oval indicates a proposed specificity pocket (see Discussion section). The first and the fourth residues of the peptide ligands are shown in ball and stick representation. (B) Partial alignment of the BIR domain family from the SMART database. ‘1’ denotes BIR domain residues that are less than 5 Å away from the N-terminal alanine of the peptide ligands on the structures shown. ‘4’ indicates BIR domain residues that are less than 5 Å away from the residue at the position +4 of the peptide ligands. Residues that have intermolecular NOEs to the Smac peptide are indicated with asterisks (Liu et al., 2000). Highlighted in gray are residues that may constitute the hypothetical specificity pocket. Visualization was done with the WebLab™ ViewerLite 3.2 software.
The numerous biochemical and structural studies focused on the BIR domain peptide recognition mechanism gradually led to the emergence of an IBM paradigm: ‘…The IBM, is defined by an unmodified alanine residue at the N-terminus of the protein (which can be generated by post-translational cleavage or by removal of the initiator methionine) and can be identified by the pattern A(K/T/V/I)(P/A/E)(F/E/I/S/Y)’ (Vaux and Silke, 2005). Though undoubtedly useful for the identification of a certain set of proteins that interact with IAPs in a BIR domain-dependent manner, the IBM paradigm tends to deter researchers from entertaining the possibility of alternative modes of BIR domain–ligand interactions and alternative classes of IBMs. It is our opinion that the IBM paradigm in its current form implies and thus concentrates research efforts more on the similarities rather than on the differences in the regulation/function of the distinct BIR domains and IAPs. However, accumulating experimental evidence suggests a critical importance of the differences: (i) Smac/DIABLO appears to selectively reduce levels of cIAP1 and cIAP2 but not that of XIAP in cultured cells in an IBM-dependent manner (Yang and Du, 2004); (ii) cIAP1 interacts with caspase 7 in an exclusively IBM-dependent, but active site independent, manner (as does its Drosophila homolog DIAP1 but not cIAP2, which fails to interact with the IBM of caspase 7 altogether (Tenev et al., 2005)). At the same time, XIAP appears to inhibit caspase 7 in a two-site interaction mechanism involving both the IBM and the active site of the caspase (Scott et al., 2005); (iii) the overexpression of the IAP antagonists HID and Reaper, which are believed to act through an analogous IBM-dependent mechanism, exert diametrically opposite effects in fly cells carrying a DIAP1 RING mutant protein, reducing and increasing sensitivity to apoptosis, correspondingly (Vaux and Silke, 2005). To explain the daunting complexity and often seemingly contradictory observations pertaining to the biology of BIR domains may require systematic studies focused on differential recognition properties and functions of different BIR domains within the same protein, as well as of different BIR domains within the individual IAP family members.
Acknowledgments
We acknowledge the generosity of Dr B. K. Kay (Argonne National Laboratory) and Dr C. Day (University of Otago, New Zealand) for providing invaluable reagents. We thank Dr B. Shilling and Emily Gaman for mass spectrometry analysis of synthetic peptides. We express our gratitude to Dr V. Galvan for useful comments and help in manuscript preparation, and to Afanasy Kurakin for software tools development. We thank the anonymous reviewer whose thoughtful comments and suggestions helped to improve the manuscript. This study was supported in part by N545093 and NS33376 from the NIH to D.E.B.
References
- Arnt CR, Chiorean MV, Heldebrant MP, Gores GJ, Kaufmann SH. Synthetic Smac/DIABLO peptides enhance the effects of chemotherapeutic agents by binding XIAP and cIAP1 in situ. J Biol Chem. 2002;277:44236–44243. doi: 10.1074/jbc.M207578200. [DOI] [PubMed] [Google Scholar]
- Birnbaum MJ, Clem RJ, Miller LK. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol. 1994;68:2521–2528. doi: 10.1128/jvi.68.4.2521-2528.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J, Shiozaki E, Srinivasula SM, Wu Q, Datta P, Alnemri ES, Shi Y. Structural basis of caspase-7 inhibition by XIAP. Cell. 2001;104:769–780. doi: 10.1016/s0092-8674(01)00272-0. [DOI] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol. 2003;10:892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Duckett CS, Nava VE, Gedrich RW, Clem RJ, Van Dongen JL, Gilfillan MC, Shiels H, Hardwick JM, Thompson CB. A conserved family of cellular genes related to the baculo-virus iap gene and encoding apoptosis inhibitors. EMBO J. 1996;15:2685–2694. [PMC free article] [PubMed] [Google Scholar]
- Fulda S, Wick W, Weller M, Debatin KM. Smac agonists sensitize for Apo2L/TRAIL- or anticancer drug-induced apoptosis and induce regression of malignant glioma in vivo. Nat Med. 2002;8:808–815. doi: 10.1038/nm735. [DOI] [PubMed] [Google Scholar]
- Galvan V, Kurakin AV, Bredesen DE. Interaction of checkpoint kinase 1 and the X-linked inhibitor of apoptosis during mitosis. FEBS Lett. 2004;558:57–62. doi: 10.1016/S0014-5793(03)01488-1. [DOI] [PubMed] [Google Scholar]
- Hegde R, Srinivasula SM, Datta P, Madesh M, Wassell R, Zhang Z, Cheong N, Nejmeh J, Fernandes-Alnemri T, Hoshino S, Alnemri ES. The polypeptide chain-releasing factor GSPT1/eRF3 is proteolytically processed into an IAP-binding protein. J Biol Chem. 2003;278:38699–38706. doi: 10.1074/jbc.M303179200. [DOI] [PubMed] [Google Scholar]
- Huang Y, Park YC, Rich RL, Segal D, Myszka DG, Wu H. Structural basis of caspase inhibition by XIAP: differential roles of the linker versus the BIR domain. Cell. 2001;104:781–790. [PubMed] [Google Scholar]
- Kay BK, Winter J, McCafferty J. Phage Display of Peptides and Proteins. Academic Press, Inc.; San Diego: 1996. [Google Scholar]
- Kipp RA, Case MA, Wist AD, Cresson CM, Carrell M, Griner E, Wiita A, Albiniak PA, Chai J, Shi Y, Semmelhack MF, McLen-don GL. Molecular targeting of inhibitor of apoptosis proteins based on small molecule mimics of natural binding partners. Biochemistry. 2002;41:7344–7349. doi: 10.1021/bi0121454. [DOI] [PubMed] [Google Scholar]
- Krajewska M, Krajewski S, Banares S, Huang X, Turner B, Buben-dorf L, Kallioniemi OP, Shabaik A, Vitiello A, Peehl D, Gao GJ, Reed JC. Elevated expression of inhibitor of apoptosis proteins in prostate cancer. Clin Cancer Res. 2003;9:4914–4925. [PubMed] [Google Scholar]
- Kurakin A, Bredesen D. Target-assisted iterative screening reveals novel interactors for PSD95, Nedd4, Src, Abl and Crk proteins. J Biomol Struct Dyn. 2002;19:1015–1029. doi: 10.1080/07391102.2002.10506805. [DOI] [PubMed] [Google Scholar]
- Kurakin A, Wu S, Bredesen DE. Target-assisted iterative screening of phage surface display cDNA libraries. Methods Mol Biol. 2004;264:47–60. doi: 10.1385/1-59259-759-9:047. [DOI] [PubMed] [Google Scholar]
- Kurakin AV, Wu S, Bredesen DE. Atypical recognition consensus of CIN85/SETA/Ruk SH3 domains revealed by target-assisted iterative screening. J Biol Chem. 2003;278:34102–34109. doi: 10.1074/jbc.M305264200. [DOI] [PubMed] [Google Scholar]
- Li J, Feng Q, Kim JM, Schneiderman D, Liston P, Li M, Vander-hyden B, Faught W, Fung MF, Senterman M, Korneluk RG, Tsang BK. Human ovarian cancer and cisplatin resistance: possible role of inhibitor of apoptosis proteins. Endocrinology. 2001;142:370–380. doi: 10.1210/endo.142.1.7897. [DOI] [PubMed] [Google Scholar]
- Li X, Yang Y, Ashwell JD. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature. 2002;416:345–347. doi: 10.1038/416345a. [DOI] [PubMed] [Google Scholar]
- Liu Z, Sun C, Olejniczak ET, Meadows RP, Betz SF, Oost T, Herrmann J, Wu JC, Fesik SW. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature. 2000;408:1004–1008. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- Oost TK, Sun C, Armstrong RC, Al-Assaad AS, Betz SF, Deck-werth TL, Ding H, Elmore SW, Meadows RP, Olejniczak ET, Oleksijew A, Oltersdorf T, Rosenberg SH, Shoemaker AR, Tomaselli KJ, Zou H, Fesik SW. Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer. J Med Chem. 2004;47:4417–4426. doi: 10.1021/jm040037k. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Liddington RC, Salvesen GS. Structural basis for the inhibition of caspase-3 by XIAP. Cell. 2001;104:791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- Schimmer AD, Welsh K, Pinilla C, Wang Z, Krajewska M, Bonneau MJ, Pedersen IM, Kitada S, Scott FL, Bailly-Maitre B, Glinsky G, Scudiero D, Sausville E, Salvesen G, Nefzi A, Ostresh JM, Houghten RA, Reed JC. Small-molecule antagonists of apoptosis suppressor XIAP exhibit broad antitumor activity. Cancer Cell. 2004;5:25–35. doi: 10.1016/s1535-6108(03)00332-5. [DOI] [PubMed] [Google Scholar]
- Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J. 2005;24:645–655. doi: 10.1038/sj.emboj.7600544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stites WE. Proteinminus signProtein interactions: interface structure, binding thermodynamics, and mutational analysis. Chem Rev. 1997;97:1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol. 2005;7:70–77. doi: 10.1038/ncb1204. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- Vucic D, Stennicke HR, Pisabarro MT, Salvesen GS, Dixit VM. ML-IAP, a novel inhibitor of apoptosis that is preferentially expressed in human melanomas. Curr Biol. 2000;10:1359–1366. doi: 10.1016/s0960-9822(00)00781-8. [DOI] [PubMed] [Google Scholar]
- Wu TY, Wagner KW, Bursulaya B, Schultz PG, Deveraux QL. Development and characterization of nonpeptidic small molecule inhibitors of the XIAP/caspase-3 interaction. Chem Biol. 2003;10:759–767. doi: 10.1016/s1074-5521(03)00157-1. [DOI] [PubMed] [Google Scholar]
- Yang QH, Du C. Smac/DIABLO selectively reduces the levels of c-IAP1 and c-IAP2 but not that of XIAP and livin in HeLa cells. J Biol Chem. 2004;279:16963–16970. doi: 10.1074/jbc.M401253200. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]


