Abstract
We previously used a yeast-based enhancer trap to identify a strong, retinoic acid response element (RARE). We have now characterized testis and eye transcripts that are adjacent to this regulatory element. Bioinformatics analysis of expressed sequence tag (EST) clones and RNase protection, reverse transcription-PCR, and northern blot assays indicate that these two RNAs are transcribed from the same locus on opposite template strands. This positions the RARE upstream of the testis transcript and downstream of the eye transcript. Additionally, these two RNAs are embedded within the third intron of the 329 kbp gene that encodes the Zinc Finger and BTB domain containing 7C protein (Zbtb7C). We present the evidence indicating that the testis transcript is expressed primarily in spermatocytes and/or early round spermatids. Furthermore, our analyses of transcript levels in eyes and testes isolated from vitamin A deficient mice or from mice with defects in retinoid storage or signaling indicate that retinoids are required for expression in vivo.
Keywords: retinoids, spermatogenesis, gene regulation, eye, RAREs
Introduction
Retinoids (vitamin A and related compounds) are key molecules necessary for growth, vision, reproduction, immunity and embryonic patterning and organogenesis. Testis, heart, limbs, brain, eyes, central nervous system, and craniofacial structures are major targets of retinoid action. Many basic cellular processes such as proliferation, migration, differentiation and apoptosis are either positively or negatively regulated by retinoids (Chung and Wolgemuth, 2004; Clagett-Dame and DeLuca, 2002; Kastner et al., 1995; Ross et al., 2000; Zile, 1998).
Retinoids exert these effects by altering gene expression via nuclear receptors, retinoic acid receptors (RARs) and retinoid X receptors (RXRs), which act as retinoid-dependent transcription factors. Genes that are “directly” activated by retinoid receptors initiate critical signaling cascades controlling numerous developmental processes. Canonical retinoid signaling involves binding of the ligand (RA) to a heterodimer complex, containing one RAR and one RXR. The receptor dimers recognize and bind to RA response elements (RAREs) in gene regulatory regions. Ligand binding relieves repression by coregulators and recruits coactivator complexes to activate gene transcription. Combinatorial control by different receptors, receptor cofactors, and RAREs contribute to the diverse effects of retinoids on different physiological processes in different cell types (Balmer and Blomhoff, 2002; Colbert, 2002; Lonard and O'Malley, 2005; Mangelsdorf et al., 1994; McKenna and O'Malley, 2002). Elucidating the genes that RA directly controls is essential to understanding the effect of retinoids on embryogenesis and physiology.
Many studies have found that retinoids regulate a plethora of genes (Arima et al., 2005; Flentke et al., 2004; Truckenmiller et al., 2001; Williams et al., 2004; Zheng et al., 2005). Experimental analyses must focus on a limited number of cell types. Consequently, the number of genes regulated by retinoids in one cell type or another is likely to be considerably larger than those currently identified to date. Distinguishing the genes that are directly controlled by retinoids from those that retinoids regulate indirectly is challenging. We developed an enhancer trap assay in yeast, wherein RAREs in the mouse genome were identified directly by virtue of their ability to induce reporter gene expression in yeast that were co-transfected with mammalian retinoid receptors (Glozak et al., 2003). Reporter gene analyses in yeast and mammalian cells and electrophoretic mobility shift assays (EMSA) showed that the trapped sequences were bona fide RAREs. Our RARE trap study (Glozak et al., 2003) and an extensive literature survey (Balmer and Blomhoff, 2002) indicate that a relatively small fraction of RA-regulated genes are directly regulated.
We previously showed that a motif in a trapped clone was a strong, RA-responsive enhancer (Glozak et al., 2003). Clone C13 activated the yeast β-gal reporter gene by 10-fold in response to RA treatment of yeast expressing RARβ and RXRγ. In F9 embryonal carcinoma cells, this sequence strongly activated two different promoters (SV40 in pGL3pro and thymidine kinase in pLucTK2) to induce the mammalian luciferase reporter gene by up to 9-fold in the presence of RA. Sequencing and site-directed mutagenesis found that the C13 clone contained a motif resembling a classical DR5 RARE (two PuG(G/T)TCA motifs separated by five nt.). In vitro synthesized RARβ and RXRγ heterodimers bound wild type oligonucleotides with the DR5 element, but not to oligonucleotides with a mutated DR5 motif. These data proved that the C13 clone contained a strong RARE that could activate heterologous gene expression in response to RA.
We now show that the C13 RARE is closely associated with previously uncharacterized testis and eye transcripts embedded within an intron of a 329 kbp gene. We present the first characterization of this complex locus and evidence indicating that these transcripts require retinoids for expression in vivo.
Results
The C13 RARE is within a complex locus
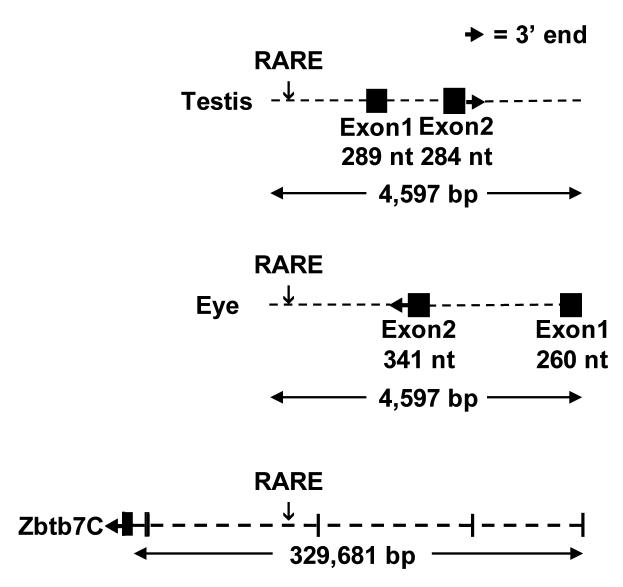
The genomic sequence flanking the C13 RARE was searched for transcripts submitted to GenBank using Blast and UniGene (http://www.ncbi.nlm.nih.gov/dbEST, http://www.ncbi.nlm.nih.gov/MapViewer and www.ensembl.org). This genomic analysis indicates that the C13 RARE lies in a complexly transcribed region (Fig. 1). Six expressed sequence tag (EST) clones generated from testis RNA map near the C13 RARE on chromosome 184. The longest EST (AK005654) predicts two exons of 289 bp, and 284 bp. The C13 RARE lies 1,040 bp upstream of the testis mRNA. The testis RNA contains an open reading frame with an initiator methionine, but without a consensus Kozak sequence, that could encode a peptide of 38 amino acids. This putative peptide (Ensemble #, ENSMUSESTP00000002566) has no obvious similarity to known proteins.
Fig. 1.
The C13 RARE lies within a complex genomic region. A diagram illustrating the relative locations of the RARE and RNAs transcribed in testes, neonatal eyes, and the Zinc finger and BTB domain containing 7C (Zbtb7C) protein-coding gene. The RARE is indicated relative to each transcription unit and has been lined up between individual transcript diagrams. The testis and eye RNAs lie entirely within intron 3 of Zbtb7C.
Fourteen ESTs5 generated by the RIKEN Consortium from neonatal (day 0) eyeball RNA map to the opposite strand relative to the testis ESTs. The longest of these (BB542970 and BB606706) would predict a transcript with exons of at least 318 bp and 268 bp. The C13 RARE would be 1,825 bp downstream of the 3′ end of this eye transcript. The short open reading frames within these ESTs did not align to any known proteins or protein motifs.
Interestingly, the testis and eye RNAs lie within the third intron of an unusually large gene coding for the Zinc Finger and BTB domain containing 7C protein (also known as Zbtb36, NM_145356.3). The 4,591 nt. mRNA that encodes Zbtb7C is distributed over 329 kbp. According to an analysis of the 97 cDNA sequences submitted to GenBank, Zbtb7C is widely expressed in brain and many other tissues. The 5′ end of the Zbtb7C transcript would be 198 kbp upstream of the C13 RARE.
Expression analysis
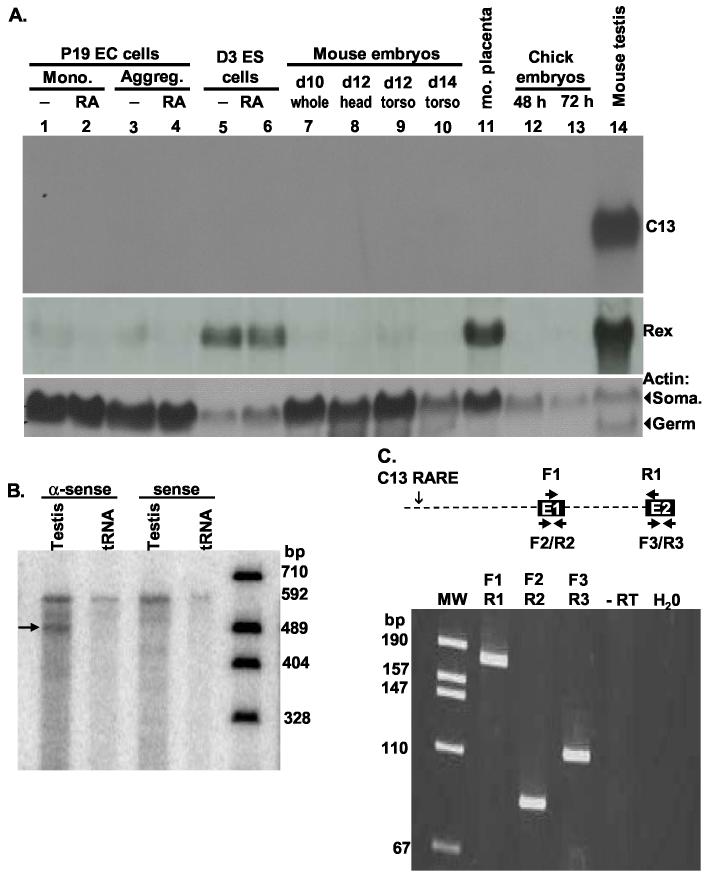
Because only six ESTs support the testis transcript, we performed northern blot analysis, reverse transcription (RT)-PCR and RNase protection assays (RPAs) to confirm the predicted expression patterns. Probes specific to the mouse testis transcript were hybridized to northern blots of RNA from several different tissues and cell lines. These probes hybridized intensely to a transcript in mouse testis RNA, but not to RNA from mouse or chick embryos at various developmental stages containing diverse cell types (Fig. 2A). Nor did these probes hybridize to murine embryonal carcinoma or embryonic stem cells grown in the presence or absence of RA or to many adult mouse organs, including whole brain and dissected parts, uterus, liver, kidney, and ovary (Fig. 2A and data not shown). We also used northern blot analysis (Fig. 4B) and the more sensitive RPA and RT-PCR techniques (not shown) to test if this transcript was present in unstaged mouse ovaries or ovaries from proestrus, metaestrus, diestrus, or estrus stages. The absence of detectable transcript in any ovary RNA suggests that the transcript is specific to male reproductive cells.
Fig. 2.
The C13 RARE lies upstream of an RNA expressed only in testis. A. Northern blot of RNA isolated from P19 embryonal carcinoma (EC) grown as monolayers (lanes 1, 2) or aggregates (lanes 3, 4) in the presence (lanes 2, 4) or absence (lanes 1, 3) of 0.5 μM RA, D3 embryonic stem (ES) cells grown as aggregates in the presence (lane 6) or absence (lane 5) of 0.5 μM RA, whole day 10 mouse embryos (lane 7), day 12 mouse embryo heads (lane 8) and torso (lane 9), day 14 mouse embryo torso (lane 10), day 14 placenta (lane 11), 48 hr or 72 hour chick embryos (lanes 12, 13), and adult strain 129 testis (lane 14). The same blot was probed with Rex1 (Zfp-42, expressed in stem cells, placenta, and spermatocytes (Rogers et al., 1991) and the constitutively expressed Actin (Act). Actin has both somatic (Soma.) and germ cell mRNA isoforms as illustrated by the two bands in testis RNA. B. A probe that is anti-sense to nt. 1 - 527 of testis EST BU936305, but not sense, protected an RNA of this size in RNA isolated from testis. The same probes failed to protect any RNAs from unstaged ovaries or proestrus, metaestrus, diestrus, or estrus ovaries (not shown). C. An RT-PCR using the primer pairs shown in the diagram amplified the products predicted by the alignment of the ESTs to the genome. Primers spanning the intron between exon 1 (E1) and 2 (E2) (F1 and R1) yielded a fragment of 181 nt., but not the fragment of 1064 bp that would have been generated from genomic DNA or unspliced RNA. Primers F2 and R2 in exon 1 and primers F3 and R3 in exon 2 yielded fragments of 85 nt and 99 nt. No amplification occurred in reactions lacking reverse transcriptase (-RT) or RNA (H20) or in reactions with ovary RNA (not shown).
Fig. 4.
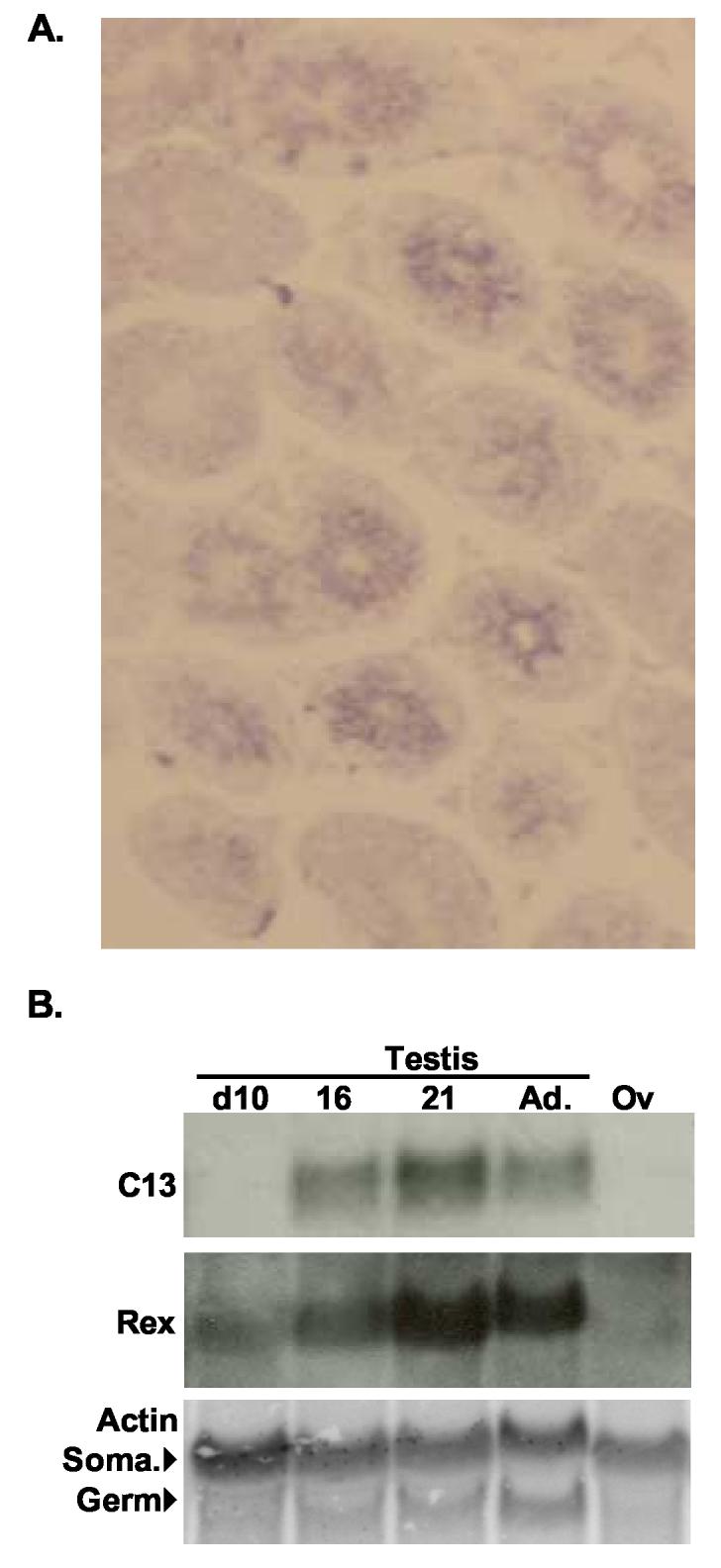
Testicular expression of the RNA associated with the C13 RARE. A. Cryosections of mature mouse testis hybridized to anti-sense testis transcripts labeled with digoxygenin-UTP and detected with alkaline phosphatase-conjugated antibodies. (100X) B. Northern blot of RNA isolated from day 10, 16, 21, and adult (Ad.) testes and adult ovaries (Ov) was probed with cDNA probes for the C13 testis transcript, Rex1 (Zfp-42), and Actin. Rex1 is a spermatocyte marker (Rogers et al., 1991). In the actin panel, the upper bands are the somatic (Soma.) mRNAs expressed in Sertoli and Leydig cells and ovaries and the lower bands are the germ cell mRNAs expressed in developing spermatids. The onset of C13 testis RNA expression at day 16 after the expression of Rex1, but before the expression of the germ cell actin isoform, suggests that this RNA is produced in secondary spermatocytes or early round spermatids.
The migration of the testes transcript relative to the ribosomal, actin, and Rex1 (Zfp42, NM_009556) RNAs is consistent with a transcript of 573 nt. as predicted by the longest EST (Figs. 2A, 4B). Antisense, but not sense, RPA probes spanning the first 527 nt. of the predicted RNA were fully protected by testis RNA (Fig. 2B), but not RNA isolated from ovaries at any stage of estrus (not shown). Finally, RT-PCR using primer pairs within exon 1 or 2 or spanning these exons yield fragments of the sizes predicted by the mapped ESTs only from testis RNA (Fig. 2C). Together these data indicate that the C13 RARE is located upstream of the first exon of a short testis transcript. Independent rapid analysis of cDNA ends (RACE) confirmed the 5′ and 3′ ends of this transcript6.
The 14 RIKEN eye ESTs from this region were unusual in that each EST was only 90 to 94% identical to the mouse genomic sequence. The sequence mismatches occurred in specific nucleotides suggesting RNA editing; however, the changes do not adhere to known editing rules (Keegan et al., 2001). To confirm that RNA was indeed transcribed from the strand opposite to the testes transcript, we used an RPA probe derived exclusively from the testes intron (Fig. 3A). A 261 nt. fragment of this probe was protected in day 10 neonatal mouse eye RNA, but not in mouse testis or ovary RNA (Fig. 3B) as would be predicted by an mRNA with a 3′ end at the position shown in Fig. 1. Probes from this region that were antisense to the testes mRNA were completely unprotected by RNA from any tissue, including testes. RT-PCR using primer pairs specific to the eye transcript, but not the testis transcript, within exon 1 or 2 or spanning these exons yield fragments of the sizes predicted by the mapped ESTs only from eye RNA (Fig. 3C). The sequence variation observed in the 14 RIKEN ESTs could have been due to RNA editing or to sequence error. Because we were unable to obtain the EST clones from RIKEN, we sequenced six independent clones derived from an RT-PCR using the F4 to R4 primers shown in Fig. 3C. The sequences of these clones from both ends were 100% identical to the genomic sequence and to each other (DQ367877). Thus, our empirical data indicate that this region is transcribed in eye.
Fig. 3.
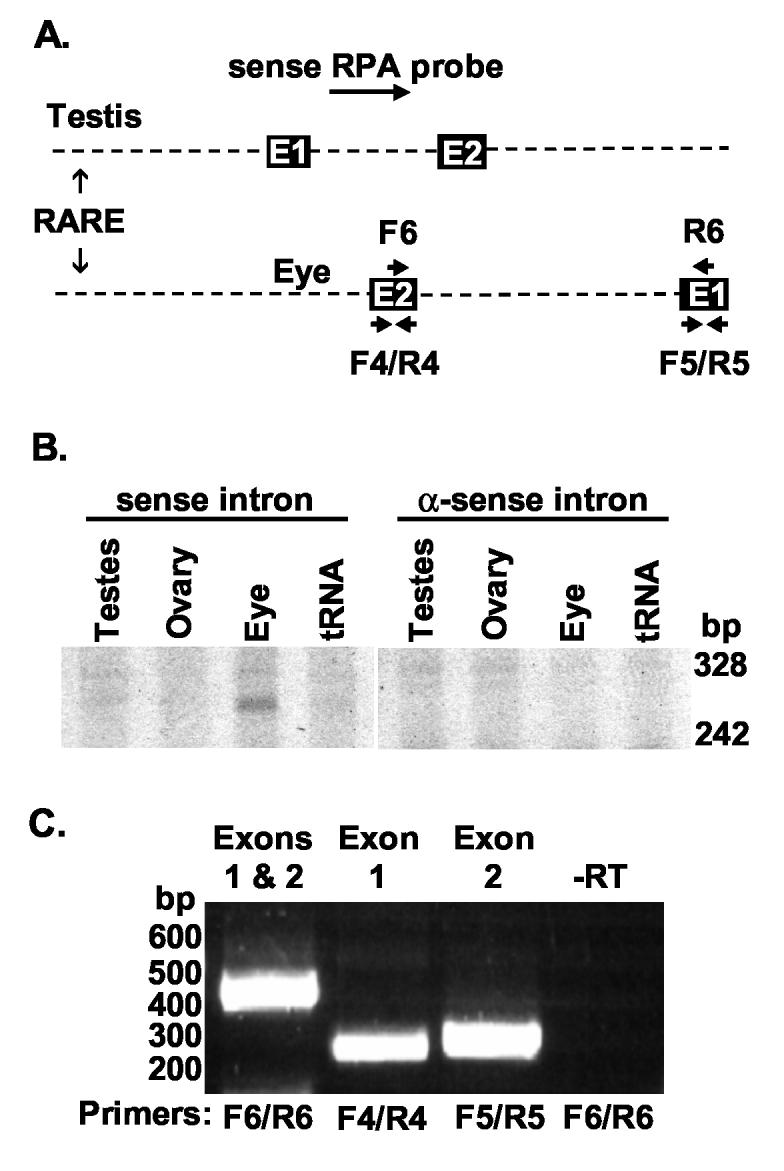
The C13 RARE is downstream of an RNA expressed in neonatal eye. A. A diagram showing the relative locations of the RPA probe and RT-PCR primers used in B and C. E1 and E2 refer to exons 1 and 2 of each transcript. B. RPAs using probes derived from the intron of the testes RNA were performed on RNAs from day 10 eyes and mature testes and ovary. These RNAs overlapped the last 261 nt. of the eye transcript. Sense probes relative to the testis transcript, but not anti-sense probes, protected an ∼261 nt. RNA in eye, but not ovary or testis, RNA. C. An RT-PCR using the primer pairs shown in the diagram amplified the products predicted by the alignment of the ESTs to the genome. Primers spanning the intron between exon 1 (E1) and 2 (E2) (F6 and R6) yielded the expected fragment of 415 nt., but not the greater than two kb fragment that would have been generated from genomic DNA or unspliced RNA. Primers F4 and R4 in exon 2 of the eye transcript yielded the expected fragments of 260 nt. Primers F5 and R5 in exon 1 of the eye transcript yielded the expected fragments of 271 nt. This reaction is representative of several reactions. No amplification ever occurred in reactions containing RNA but lacking reverse transcriptase (-RT) or in those lacking RNA (not shown).
In summary, a 573 nt. transcript located downstream of the C13 RARE is transcribed exclusively in testis. In eye, but not in testis, another short transcript is transcribed from the opposite strand. The eye RNA is transcribed from the same template strand as the Zbtb7C mRNA. After splicing, none of these RNAs would overlap or be anti-sense to each other. Because we have a long-standing interest in the role of retinoids in spermatogenesis, we chose to analyze the testicular transcript at this time.
The testis transcript is expressed in secondary spermatocytes
The adult testis is composed of many different cell types, including somatic cells and germ cells at various stages of differentiation. Spermatogenesis occurs in the seminiferous tubules in which the Sertoli cell and germ cells are located, from the outer edge towards the central lumen of the tubules. Mitotically renewing spermatogonial stem cells are located in the outermost layer of the tubule. The primary and secondary spermatocytes undergo meiosis I and II in more interior layers. Finally, the post-meiotic spermatids undergo spermiogenesis in the most central layers prior to extrusion into the lumen as spermatozoa. These germ cells are intimately associated with somatic cells, called the Sertoli cells, throughout their development. The somatic Leydig cells are located in the interstitial space outside of the tubules. To localize the C13 RARE associated RNA in the testis, in situ hybridization was performed on adult testicular sections using a C13 testis-specific probe. Fig. 4A shows a representative cross-section of testis hybridized to probes specific to the testis transcript. The highest level of hybridization was localized over the central layers occupied by spermatocytes and round spermatids.
The germ cells within the seminiferous epithelium form defined cellular associations, the complete set of which is termed a spermatogenic cycle. In mice, the spermatogenic cycle is composed of 12 cellular associations or stages (Leblond and Clermont, 1952; Oakberg, 1956). For example, primary spermatocytes in the last stages of meiosis I (diplotene and diakinesis), secondary spermatocytes, and early round spermatids are limited to few specific stages of the spermatogenic cycle (Oakberg, 1956). An RNA expressed specifically in these cells would be present in some but not all cross-sections. As seen in Fig 4A, we observed that some cross-sections of tubules hybridized strongly while others did not. This suggests that the RNA is synthesized by germ cells that are restricted to particular stages of the spermatogenic cycle. The staining pattern also suggests that the RNA is not located in Sertoli cells, which are present in all stages of the spermatogenic cycle.
To clarify this issue, we isolated RNA from testes obtained from mice at various days following birth. This method has been used to determine gene expression in specific developing germ cells as the mouse approaches sexual maturity (Bellvé et al., 1977). The data shown in Fig. 4B indicate that the transcript was not detected at day 10, but was observed at day 16 and day 21. In contrast, the mRNA of Rex1 (Zfp42, NM_009556), a transcription factor expressed in primary spermatocytes (Hosler et al., 1993; Rogers et al., 1991), was present in the testis mRNA samples from day 10 mice, 6 days before C13 transcript expression. Additionally, the germ cell-specific actin transcript was not detected until day 21, when round spermatids are present in the seminiferous tubules. These data together with the in situ hybridization signal in the central layers of the tubules, but not in all cross-sections suggest that the RARE-associated, testis transcript is expressed primarily in late spermatocytes and/or round spermatids7.
The eye and mouse RNAs are regulated by RA in vivo
To test the hypothesis that the C13 RARE functioned to transmit a retinoid signal in vivo, we compared the expression of the eye RNA in lecithin:retinol acyltransferase (LRAT) knockout mice (Liu and Gudas, 2005) fed a vitamin A sufficient diet relative to mice that were fed a vitamin A-deficient (VAD) diet. Unlike normal mice that resist vitamin A depletion, LRAT−/− mice are much more susceptible to vitamin A deficiency and thus, are a more convenient VAD model (Liu and Gudas, 2005; O'Byrne et al., 2005). As shown in Fig. 5A, we were unable to detect the eye transcript in RNA extracted from the eyes of VAD LRAT−/− mice using RT-PCR (lane 1). In contrast, an amplified product was readily detected in eye RNA from both LRAT−/− and LRAT+/+ mice fed a vitamin A-sufficient diet (lanes 4, 7). LRAT−/− mice fed a vitamin A-sufficient diet appear normal, but have been reported to have significantly reduced retinoid stores compared to well-fed LRAT +/+ mice (Liu and Gudas, 2005; O'Byrne et al., 2005). Consistent with a requirement for retinoid signaling in inducing the C13 RARE-associated eye transcript, LRAT+/+ mice express more of the eye transcript relative to well-fed LRAT−/− mice (compare lanes 4 and 7). This is in contrast to the observed levels of the constitutive actin RNA (lanes 3, 6, 9). Together, these results strongly suggest that the decreased retinoid levels in the LRAT−/− mice reduced the expression of the C13 RNA.
Fig. 5.
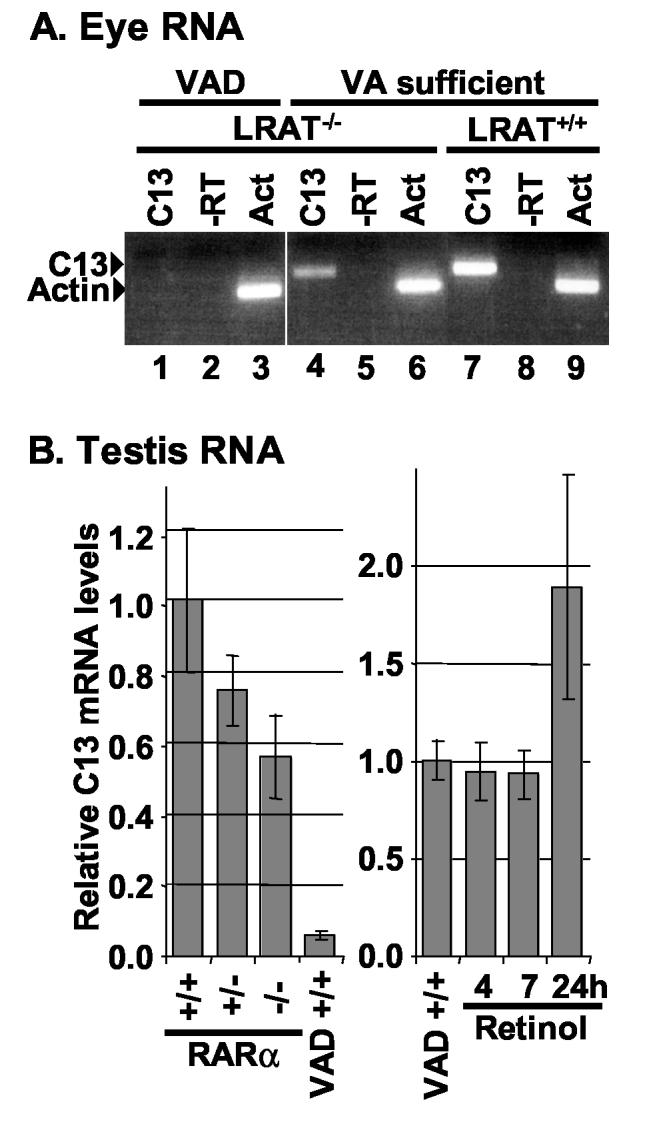
Expression of the eye and testis transcripts depends on normal retinoid signaling. A. RT-PCR reactions using the primer pairs F4 and R4 (C13) shown in Fig. 3 amplified products of the expected size in eye RNA isolated from LRAT−/− or LRAT+/+ mice fed a vitamin A (VA)-sufficient diet (lanes 4, 7) but not in vitamin A-deficient (VAD) LRAT−/− mice (lane 1). No amplification occurred in reactions containing RNA and primers but lacking reverse transcriptase (-RT, lanes 2, 5, 8). Similar levels of amplified products were obtained from each RNA sample using actin-specific primers (Act, lanes 3, 6, 9). B. The relative level of C13 RNA isolated from the testes of RARα wild type (+/+), heterozygous (+/−), and knockout (−/−) RARα mice and of vitamin A-deficient (VAD) and retinol-replenished mice was measured using real-time PCR (means ± S.D.). The data are representative of two primer sets and at least two mice. The real-time PCR was conducted on cDNA in triplicate. The heterozygous, knockout, and vitamin A-deficient values differed significantly from wild type (p ≤ 0.05). The 24 hour retinol repletion values differed significantly from VAD (p ≤ 0.05).
Similarly, real time RT-PCR analyses showed that the testis transcript was significantly reduced in RNA isolated from the testis of VAD (LRAT+/+) mice generated by standard methods (Fig. 5B). Vitamin A deficiency blocks spermatogenesis at an early stage of meiosis (Griswold et al., 1989; Ismail et al., 1990; Morales and Griswold, 1987; Wolbach and Howe, 1925). Because VAD testes have mainly Sertoli cells, spermatogonia, and early spermatocytes, this result is consistent with the hypothesis that the C13 testis transcript is expressed in spermatocytes or round spermatids. The levels of C13 mRNA increased by 89% in the testes from retinol replenished VAD mice after 24 hrs compared with those observed in the testes from VAD mice. Because late spermatocytes and round spermatids require over a week to differentiate (Bellvé et al., 1977), these data are consistent with the hypothesis that retinoids induce the testis transcript in the few remaining spermatocytes.
Mice with a disrupted RARα gene have been reported to have a testicular phenotype resembling that of VAD animals (Lufkin et al., 1993), although in our hands the degeneration of the germinal epithelium in RARα knockout mice varies from moderate to severe (unpublished data). Thus, although other retinoid receptors (Kim and Griswold, 1990) can partially compensate for the loss of RARα, these mice provide a genetic alternative to our nutritional test of the C13 RARE's functionality. After normalizing the C13 mRNA level to an internal control in each sample (the S2 ribosomal protein mRNA), we found that testicular C13 mRNA levels in RARα heterozygous (+/−) and knockout (−/−) mice were significantly decreased by 26% and 45%, respectively (p ≤ 0.05), compared with those from the normal (+/+) mice. The dissociation curves showing the melting temperature for the C13 and S2 real-time PCR products indicated that each primer set generated a single product (data not shown). Together, the absence of the eye and testis transcripts in VAD mice and the down-regulation of the testis transcript in RARα knockout mice suggests that retinoids are required to induce these two transcripts in vivo.
Discussion
Retinoids bind RARs and RXRs to directly activate genes. Separating the genes induced directly by retinoid bound receptors from those induced subsequently by other regulatory factors is laborious. The loose consensus defining known RAREs further complicates this effort. We developed a yeast-based system to trap functional RAREs in the mouse genome (Glozak et al., 2003). Out of 11 RAREs chosen for expression analysis, we have so far mapped five near RA-regulated transcripts. Four of these genes encoded known proteins: the nonclassical MHC class I gene, T20d; the spermatid-specific RING zinc-finger protein coding gene (sperizin/Zinc and ring finger 4, Znrf4) ; the highly conserved trinucleotide repeat containing 5 gene (Tnrc5); and Pax3, a transcription factor in which mutations cause Waardenburg syndrome (Pruitt, 1992).
A fifth RARE, C13, is adjacent to polyadenylated RNAs expressed in eye and testis. Most importantly, we have presented data derived from VAD mice and mice that are defective in retinoid signaling consistent with the hypothesis that the C13 RARE regulates these transcripts. Vitamin A and its metabolites are essential for both eye development and function, and for spermatogenesis (Chung and Wolgemuth, 2004; Ross et al., 2000). We have now trapped RAREs that appear to control two genes expressed in male germ cells (sperizin (Glozak et al., 2003) and the C13-associated testis RNA). Thus, the trap approach can identify genes regulated by RA in complex organs, like testis and eye, which would be challenging to identify by other means.
The longest open reading frame in the testis RNA would encode a novel peptide of 38 residues. The sequence leading up to the methionine in this peptide does not conform to the Kozak consensus sequence nor were we able to detect a peptide after in vitro transcription and translation (not shown). Therefore, it is also possible that this mRNA is non-coding. We confirmed the gene structure of this RNA and that of the small RNA transcribed from the opposite strand in eye using RPAs, RT-PCRs, and computational analyses of EST clones. The mature testis and eye RNAs predicted by our analysis do not overlap, although their primary transcripts do. Interestingly, the testis and eye RNAs lie entirely within the intron of an unusually large gene (329 kb) coding for the Zinc Finger and BTB domain containing 7C protein (Zbtb7C). The absence of mRNA complementarity would rule out regulation of one RNA by another via an anti-sense or RNA interference-mediated cytoplasmic process such as translational repression. The significance of small transcripts within the introns of large transcripts remains to be determined. However, analyses of the genomes from diverse organisms have begun to reveal many examples of such complex transcription patterns (Mattick, 2005; Willingham and Gingeras, 2006).
Indeed, genomic analyses indicate that much more of the genome is transcribed than had been predicted. Furthermore, diverse classes of transcripts have been identified. These include mRNAs that encode short, but functional, open reading frames (Basrai et al., 1997; Brent, 2005), and non-coding RNAs including essential regulators such as micro RNAs and riboswitches (Katayama et al., 2005; Reichow and Varani, 2006; Shendure and Church, 2002). Non-coding RNAs are highly tissue specific. For example, an abundant class of small RNAs that bind protein regulators of stem cell maintenance and meiosis are uniquely expressed in germ line cells (Girard et al., 2006). Indeed, half of the transcripts synthesized by cells may not code for proteins (Mattick, 2005; Willingham and Gingeras, 2006). These developmentally regulated non-coding RNAs are likely to play specific signaling roles (Willingham et al., 2005). Given that RA initiates signaling cascades, it is not surprising that RA would regulate non-coding RNAs. Indeed, one genomic study found that RA regulated nearly a quarter of non-coding RNAs (Cawley et al., 2004). Our trap may have identified the first RARE to control non-coding RNAs.
Experimental Procedures
Animals
To generate vitamin A-deficient (VAD) mice for testis RNA analysis, adult female B6-129 mice (LRAT+/+) were placed on a VAD diet (Harland Teklad, Madison, WI) for 3 wk before they were mated with males that had been maintained on a normal diet. The females were continued on a VAD diet through gestation and until pups were weaned. After weaning, male offspring were fed the VAD diet for an additional 12 wk to obtain VAD male mice. Histology of testicular sections from the VAD and the retinol-replenished mice showed typical germ cell degeneration (data not shown). Retinol-replenished VAD male mice were generated by treating the VAD male mice once with 17 mg/kg retinol (Sigma) in 50% ethanol by i.p. injection. Mice heterozygous for RARα were acquired from Dr. Pierre Chambon (CNRS INSERM Universite Louis Pasteur, Strasbourg, France) (Lufkin et al., 1993) and mated to produce the RARα wild type, heterozygous, and knockout mice. RNA from VAD eyes was obtained from Drs. L. Liu and L.J. Gudas. Briefly, lecithin:retinol acyltransferase (LRAT−/−) knockout mice were fed a normal vitamin A-sufficient diet (Harlan Teklad, Madison, WI) or a vitamin A-deficient diet (Harlan Teklad, Madison, WI) as described in (Liu and Gudas, 2005). Serum samples were analyzed to confirm that retinol and retinyl esters were depleted at 6 weeks. Animal experimentation was approved by the Institutional Animal Care and Use Committees of UMDNJ-NJ Medical School, Washington State University, and Weill Medical College of Cornell University, and conducted in accordance with the highest standards of humane animal care as outlined in the National Institutes of Health guide for the Care and Use of Laboratory Animals.
Plasmid constructs and genomic PCR cloning
Various murine-specific primers (see below for sequences) were designed from exons 1 and 2 of the testis or eye transcripts for PCR. 100 μl PCR reactions contained 1 unit Taq recombinant DNA polymerase (Sigma), 100 ng genomic DNA, 50 pmols of each primer, 0.25 mM dNTPs, 1.5 mM MgCl2, and buffer conditions as recommended by the manufacturer (Sigma). Reactions were incubated as follows in a Mastercycler gradient PCR machine (Eppendorf): denaturation 94°C, 2 min; annealing 59°C, 90 secs; extension, 72°C, 60 secs for 25 cycles, followed by a final extension at 72°C for 5 min. PCR products were separated by 6% polyacrylamide gel electrophoresis (37.5:1 acrylamide: bis-acrylamide), visualized by ethidium bromide staining and confirmed by restriction enzyme digestion and Southern blotting. The PCR products were cloned into the pCRII TA cloning vector (Invitrogen) according to the manufacturer's instructions and further sequenced (Molecular Resource Facility, UMDNJ). An EST containing the testis transcript (BU936305; pDNRlib, Clontech) was digested with EcoRI and ligated into the EcoRI site of pGEM4 (Promega) for in vitro transcription of probes for RNase Protection Assays (RPAs) and in situ hybridization.
Northern Blotting
Total RNA was isolated from prepuberty and mature mouse testis using the standard guanidium isothiocyanate gradient method (Ausubel et al., 1997) and hybridized to mouse cDNA clones as described previously (Fritz et al., 2004; Rogers et al., 1991; Rogers et al., 1992). The hybridized bands were quantified using a Molecular Dynamics PhosphorImager and ImageQuant software.
Reverse transcriptase-polymerase chain reactions (RT-PCR)
Oligo d(T) primed or random hexamer primed testis cDNA was used as a template for amplification. Forward and reverse primer pairs specific to the testis transcript (diagrammed in fig. 2C) were designed from exon 1 (F2: 5′ GCAGAAAGGTAGAGGGGAGCA 3′; R2: 5′ CTGGACCTGAAGAGGGTAGCG 3′), exon 2 (F3: 5′ ACAAAGGGCTGACAGTGCAGA 3′; R3: 5′ TGGCAAACCGTTGAGGTATTG 3′) and exon 1 and 2 spanning the intronic region (F1: 5′ AAACATTGCCTTGCCTCTTGG 3′; R1: 5′ CTCCCCTGTGGTAGGAAGCAC 3′). Forward and reverse primer pairs specific to the eye transcript (diagrammed in fig. 3A) were designed from exon 1 (F5: 5′GTGGAGTACAGCAGAGGGCT3′ and R5: AGAGCCCACCTTCCGATTTC); exon 2 (F4: 5′CAAGTCTTCAAGGGAGTGC 3′ and R4: 5′TTGGCTCACTCTGCCTACCT3′); and exon 1 and 2 spanning the intronic region (F6: 5′CTCTTGGGGCTGGAGCAGGCCAT3′ and R6: 5′ACTTCACCGCATTTTGGTTC3′). Mouse β-actin primers or water as template were used for positive or negative controls, respectively. The primers for actin were (F 5′ GGGAAATCGTGCGTGACATCAAAGAG 3′ and R 5′ CATGGTGCTAGGAGCCAGAGCAGTAA 3′). Minus RT reactions also were performed to check for genomic DNA contamination in the RNA preparations. 20 μl RT reactions contained 4 μl Gibco buffer, 0.5 mM dNTP mixture, 0.05 μg/μl random hexamers or oligo d(T), 0.01 M DTT, 0.5 μl RNasin (Promega), 0.5 μl MMLV RT, and 1 μg total RNA. 100 μl PCR reactions contained 10 μl buffer (Applied Biosystems, 500 ng of each primer, 0.2 mM dNTPs, 1.5 mM MgCl2, 1 unit Taq recombinant DNA polymerase (Sigma), and 5 μl RT template. Reactions were incubated as follows: denaturation 94°C, 1.5 min; annealing 59°C, 2 min; extension, 72°C, 3 min for 25 cycles, followed by a final extension at 72°C for 10 min. The PCR products were electrophoresed and analyzed as described above.
Ribonuclease Protection Assay (RPA)
RPAs were done as described in (Strijker et al., 1989) with slight modifications. Linearized plasmids were transcribed with SP6 or T7 RNA polymerase (New England Biolabs) in the presence of α32P-UTP for antisense and sense transcripts. Briefly, gel-purified radiolabeled RNA probe, total RNA, and yeast tRNA were co-precipitated with ethanol, denatured at 80°C for 10 min, and hybridized overnight at 45°C. After 20 min RNase A/T1 (Ambion) digestion at 37°C, reactions were inactivated with SDS and proteinase K, followed by phenol-chloroform extraction. Subsequently, RNAs were ethanol precipitated, dissolved in 15 μl of gel loading buffer, and electrophoresed on denaturing 5% polyacrylamide gels (8 M urea; 37.5:1, acrylamide:bis-acrylamide). Protected RNAs were visualized using autoradiography.
In situ hybridization
Plasmids were transcribed as described above except digoxygenin d-UTP was used to label the synthetic RNAs. In situ hybridization was done as described in (Shen, 2001).
Realtime PCR
RNA from testes of vitamin A-deficient (VAD) and RARα knockout mice was collected using an RNaqueous kit (Ambion RNA Company). Real-time PCR primers were designed using Primer Express software version 2.0 (Applied Biosystems). The primers used to amplify exon 1 of the testis RNA (F2-R2) were 5′ GCAGAAAGGTAGAGGGGAGCA 3′ and 5′ CTGGACCTGAAGAGGGTAGCG 3′. The primers used to amplify exon 2 of the testis RNA (F3-R3) were 5′ CCAAGGAAGTCACAAAGGGCT 3′ and 5′ TGGCAAACCGTTGAGGTATTG 3′. The forward and reverse primers for the housekeeping gene encoding the ribosomal S2 protein were 5′ CTGACTCCCGACCTCTGGAA 3′ and 5′ GAGCCTGGGTCC TCTGAACA 3′, respectively. Complementary DNA was synthesized from 500 ng RNA samples using iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's protocol. Subsequently, 20 ng cDNA was used as template for real-time PCR assays of mRNAs isolated from wild type, heterozygote, RARα knockout, VAD, and retinol replenished testis with a Gene Amp 7000 thermocycler (Applied Biosystems). Threshold (Ct) values for C13 and S2 were determined using Prism SDS software version 1.2 (Applied Biosystems), and the level of C13 real-time PCR product was evaluated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Specifically, the Ct value for C13 real-time PCR product was normalized to that for S2 real-time PCR product in each sample, and then the fold change for C13 real-time PCR product was calculated relative to the C13 real-time PCR product level in RARα wild-type mice or to the C13 real-time PCR product level in VAD mice. The real-time PCR was conducted on cDNA in triplicate. Statistical analysis of C13 real-time PCR product levels consisted of one-way ANOVA followed by pair wise comparison of the means by the Tukey method (JMP 5.1, SAS Institute Inc., Cary, NC).
Acknowledgements
We thank Drs. Limin Liu and Lorraine J. Gudas for the kind gift of RNA from vitamin A sufficient and deficient mouse eyes and David Fritz for assistance and consultation. This work was supported in part by the Molecular Resource Facility at the UMDNJ – NJ Medical School, and by grants from the National Institute of Child Health and Human Development (#HD31117) and the March of Dimes (#FY02-31).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Eye EST accession #: BB542970, BB606706, BB542698, BB606627, BB542898, BB606659, BB542849, BB606644, BB606926, BB542900, BB6066655, BB543815, BB544354, BB544384
H. Lim, personal communication
This RNA was independently identified in a screen for peroxisome proliferators activated receptor (PPAR) response elements (H. Lim, personal communication). After obtaining in situ hybridization results consistent with our analyses, the sequence was entered in GenBank under the name Secondary Spermatocyte-Specific Marker (Ssm, DQ284430).
References
- Arima K, Shiotsugu J, Niu R, Khandpur R, Martinez M, Shin Y, Koide T, Cho KW, Kitayama A, Ueno N, Chandraratna RA, Blumberg B. Global analysis of RAR-responsive genes in the Xenopus neurula using cDNA microarrays. Dev Dyn. 2005;232:414–31. doi: 10.1002/dvdy.20231. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Wiley and Sons; NY: 1997. [Google Scholar]
- Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43:1773–808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- Basrai MA, Hieter P, Boeke JD. Small open reading frames: beautiful needles in the haystack. Genome Res. 1997;7:768–71. doi: 10.1101/gr.7.8.768. [DOI] [PubMed] [Google Scholar]
- Bellvé AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse: Isolation and morphological characterization. J. Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent MR. Genome annotation past, present, and future: How to define an ORF at each locus. Genome Res. 2005;15:1777–1786. doi: 10.1101/gr.3866105. [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, Wheeler R, Wong B, Drenkow J, Yamanaka M, Patel S, Brubaker S, Tammana H, Helt G, Struhl K, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Chung SS, Wolgemuth DJ. Role of retinoid signaling in the regulation of spermatogenesis. Cytogenet Genome Res. 2004;105:189–202. doi: 10.1159/000078189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clagett-Dame M, DeLuca HF. The role of vitamin A in mammalian reproduction and embryonic development. Annu Rev Nutr. 2002;22:347–81. doi: 10.1146/annurev.nutr.22.010402.102745E. [DOI] [PubMed] [Google Scholar]
- Colbert MC. Retinoids and cardiovascular developmental defects. Cardiovasc Toxicol. 2002;2:25–39. doi: 10.1385/ct:2:1:25. [DOI] [PubMed] [Google Scholar]
- Flentke GR, Baker MW, Docterman KE, Power S, Lough J, Smith SM. Microarray analysis of retinoid-dependent gene activity during rat embryogenesis: increased collagen fibril production in a model of retinoid insufficiency. Dev Dyn. 2004;229:886–98. doi: 10.1002/dvdy.10489. [DOI] [PubMed] [Google Scholar]
- Fritz DT, Liu D, Xu J, Jiang S, Rogers MB. Conservation of Bmp2 post-transcriptional regulatory mechanisms. J Biol Chem. 2004;279:48950–8. doi: 10.1074/jbc.M409620200. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Li Y, Reuille R, Kim KH, Vo MN, Rogers MB. Trapping and characterization of novel retinoid response elements. Mol Endocrinol. 2003;17:27–41. doi: 10.1210/me.2002-0192. [DOI] [PubMed] [Google Scholar]
- Griswold MD, Bishop PD, Kim K-H, Ping R, Siiteri JE, Morales C. Function of vitamin A in normal and synchronized semeniferous tubules. Ann. NY Acad. Sci. 1989;564:154–172. doi: 10.1111/j.1749-6632.1989.tb25895.x. [DOI] [PubMed] [Google Scholar]
- Hosler B, Rogers M, Kozak C, Gudas L. An octamer motif contributes to the expression of the retinoic acid-regulated zinc finger gene Rex-1 (Zfp-42) in F9 teratocarcinoma cells. Mol. Cell. Biol. 1993;13:2919–2928. doi: 10.1128/mcb.13.5.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N, Morales C, Clermont Y. Role of spermatogonia in the stage-synchronization of the seminiferous epithelium in vitamin-A-deficient rats. Am J Anat. 1990;188:57–63. doi: 10.1002/aja.1001880107. [DOI] [PubMed] [Google Scholar]
- Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Keegan LP, Gallo A, O'Connell MA. The many roles of an RNA editor. Nat Rev Genet. 2001;2:869–78. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- Kim KH, Griswold MD. The Regulation of Retinoic Acid Receptor mRNA Levels during Spermatogenesis. Molec. Endocrin. 1990;4:1679–1688. doi: 10.1210/mend-4-11-1679. [DOI] [PubMed] [Google Scholar]
- Leblond CP, Clermont Y. Spermiogenesis of rat, mouse, hamster and guinea pig as revealed by the periodic acid-fuchsin sulfurous acid technique. Am J Anat. 1952;90:167–215. doi: 10.1002/aja.1000900202. [DOI] [PubMed] [Google Scholar]
- Liu L, Gudas LJ. Disruption of the lecithin:retinol acyltransferase gene makes mice more susceptible to vitamin A deficiency. J Biol Chem. 2005;280:40226–34. doi: 10.1074/jbc.M509643200. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW. Expanding functional diversity of the coactivators. Trends Biochem Sci. 2005;30:126–32. doi: 10.1016/j.tibs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Lufkin T, Lohnes D, Mark M, Dierich A, Gorry P, Gaub MP, LeMeur M, Chambon P. High postnatal lethality and testis degeneration in retinoic acid receptor alpha mutant mice. Proc Natl Acad Sci U S A. 1993;90:7225–9. doi: 10.1073/pnas.90.15.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Umesano K, Evans RM. The Retinoid Receptors. In: Sporn M, et al., editors. The Retinoids: Biology, Chemistry, and Medicine. 2d ed. Raven Press, Ltd.; New York: 1994. pp. 319–349. [Google Scholar]
- Mattick JS. The functional genomics of noncoding RNA. Science. 2005;309:1527–8. doi: 10.1126/science.1117806. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–74. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Morales C, Griswold MD. Retinol-induced stage synchronization in seminiferous tubules of the rat. Endocrinology. 1987;121:432–4. doi: 10.1210/endo-121-1-432. [DOI] [PubMed] [Google Scholar]
- Oakberg EF. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am J Anat. 1956;99:391–413. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- O'Byrne SM, Wongsiriroj N, Libien J, Vogel S, Goldberg IJ, Baehr W, Palczewski K, Blaner WS. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT) J Biol Chem. 2005;280:35647–57. doi: 10.1074/jbc.M507924200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt SC. Expression of Pax-3- and neuroectoderm-inducing activities during differentiation of P19 embryonal carcinoma cells. Development. 1992;116:573–83. doi: 10.1242/dev.116.3.573. [DOI] [PubMed] [Google Scholar]
- Reichow S, Varani G. Structural biology: RNA switches function. Nature. 2006;441:1054–5. doi: 10.1038/4411054a. [DOI] [PubMed] [Google Scholar]
- Rogers MB, Hosler BA, Gudas LJ. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast, and spermatocytes. Devl. 1991;113:815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- Rogers MB, Rosen V, Wozney JM, Gudas LJ. Bone Morphogenetic Proteins-2 and 4 are Involved in the Retinoic Acid-induced Differentiation of Embryonal Carcinoma Cells. Molec Biol Cell. 1992;3:189–196. doi: 10.1091/mbc.3.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SA, McCaffery PJ, Drager UC, De Luca LM. Retinoids in embryonal development. Physiol Rev. 2000;80:1021–54. doi: 10.1152/physrev.2000.80.3.1021. [DOI] [PubMed] [Google Scholar]
- Shen MM. Identification of differentially expressed genes in mouse development using differential display and in situ hybridization. Methods. 2001;24:15–27. doi: 10.1006/meth.2001.1152. [DOI] [PubMed] [Google Scholar]
- Shendure J, Church GM. Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0044. RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijker R, Fritz DT, Levinson AD. Adenovirus VAI-RNA regulates gene expression by controlling stability of ribosome-bound RNAs. Embo J. 1989;8:2669–75. doi: 10.1002/j.1460-2075.1989.tb08407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truckenmiller ME, Vawter MP, Cheadle C, Coggiano M, Donovan DM, Freed WJ, Becker KG. Gene expression profile in early stage of retinoic acid-induced differentiation of human SH-SY5Y neuroblastoma cells. Restor Neurol Neurosci. 2001;18:67–80. [PubMed] [Google Scholar]
- Williams SS, Mear JP, Liang HC, Potter SS, Aronow BJ, Colbert MC. Large-scale reprogramming of cranial neural crest gene expression by retinoic acid exposure. Physiol Genomics. 2004;19:184–97. doi: 10.1152/physiolgenomics.00136.2004. [DOI] [PubMed] [Google Scholar]
- Willingham AT, Gingeras TR. TUF love for “junk” DNA. Cell. 2006;125:1215–20. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–3. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Wolbach S, Howe P. Tissue changes following deprivation of fat soluble A vitamin. J. Exp. Med. 1925;42:753–777. doi: 10.1084/jem.42.6.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng PZ, Wang KK, Zhang QY, Huang QH, Du YZ, Zhang QH, Xiao DK, Shen SH, Imbeaud S, Eveno E, Zhao CJ, Chen YL, Fan HY, Waxman S, Auffray C, Jin G, Chen SJ, Chen Z, Zhang J. Systems analysis of transcriptome and proteome in retinoic acid/arsenic trioxide-induced cell differentiation/apoptosis of promyelocytic leukemia. Proc Natl Acad Sci U S A. 2005;102:7653–8. doi: 10.1073/pnas.0502825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zile M. Vitamin A and Embryonic Development: An Overview. J Nutr. 1998;128:455S–458S. doi: 10.1093/jn/128.2.455S. [DOI] [PubMed] [Google Scholar]