Abstract
Colicins are proteins produced by and toxic for some strains of Escherichia coli. They are produced by strains of E. coli carrying a colicinogenic plasmid that bears the genetic determinants for colicin synthesis, immunity, and release. Insights gained into each fundamental aspect of their biology are presented: their synthesis, which is under SOS regulation; their release into the extracellular medium, which involves the colicin lysis protein; and their uptake mechanisms and modes of action. Colicins are organized into three domains, each one involved in a different step of the process of killing sensitive bacteria. The structures of some colicins are known at the atomic level and are discussed. Colicins exert their lethal action by first binding to specific receptors, which are outer membrane proteins used for the entry of specific nutrients. They are then translocated through the outer membrane and transit through the periplasm by either the Tol or the TonB system. The components of each system are known, and their implication in the functioning of the system is described. Colicins then reach their lethal target and act either by forming a voltage-dependent channel into the inner membrane or by using their endonuclease activity on DNA, rRNA, or tRNA. The mechanisms of inhibition by specific and cognate immunity proteins are presented. Finally, the use of colicins as laboratory or biotechnological tools and their mode of evolution are discussed.
INTRODUCTION
Colicins are proteins produced by some strains of Escherichia coli that are lethal for related strains of E. coli. The first colicin was identified by Gratia in 1925 as a heat-labile product present in cultures of E. coli V and toxic for E. coli φ (235). Further on, numerous colicins produced by different strains of the enteric group of bacteria (E. coli, Shigella, and Citrobacter) were characterized. The name colicin was coined by Gratia and Fredericq in 1946, who demonstrated their protein nature and the specificity of their activity spectra (236). Afterwards, the term bacteriocin was introduced to designate toxic proteins produced by a given strain of bacteria and active against related species but not on the producing cells (296). By analogy with colicins, the new families of bacteriocins carry the name of the producing species of bacteria followed by the suffix -cin. Thus, pyocins from Pseudomonas pyogenes strains, cloacins from Enterobacter cloacae, marcescins from Serratia marcescens, megacins from Bacillus megaterium, etc., have been identified. A nomenclature using the genus name in place of the species name of the producing bacteria has been proposed by Fredericq to avoid redundancies (209). According to the nomenclature, the bacteriocins produced by Pasteurella pestis and by Yersinia pestis would not have both been called pesticins (175, 547) but would have been called pasteurellacins and yersiniacins. Fredericq's advice has not been followed, perhaps in order to retain the word colicin in place of escherichiacin. In our days, the meaning of the word bacteriocin has changed, since it is now used mainly to designate antibiotic peptides produced by gram-positive bacteria and active on a wide range of bacteria. The producers of these toxic peptides, as the strains producing protein bacteriocins, possess a specific immunity mechanism to protect themselves against their own bacteriocin (reviewed in references 128 and 162). Confusions in the nomenclature must be heeded, although they are as old as colicin studies: the first identified colicin, colicin V, is now classified among the microcins but is still called colicin (225, 687). The microcins are a family of low-molecular-weight antibiotics produced by Enterobacteriaceae and are active against phylogenetically related microbial strains (reviewed in references 11 and 294).
The narrow target range of colicins has been shown by Fredericq to be due to the presence of specific receptors at the surface of the sensitive strains on which colicin binds before killing (208). Mutation of the receptor can lead to the loss of sensitivity to the corresponding colicin. Mutants that are resistant to each colicin have been isolated and used as the basis to name each colicin by the alphabet letter used, at the time, to designate the receptor to which it binds. When more than one colicin binds to the same receptor, they are designated by the alphabet letter of the receptor followed by a number, as, for instance, the nine colicins E: E1 to E9. The receptors have been shown to be outer membrane (OM) proteins that allow the entry of specific nutrients such as nucleosides, siderophores, and vitamins (103, 104, 158). BtuB, the receptor of vitamin B12, of the nine colicins E, and of the phage BF23, was the first colicin receptor purified by Sabet and Schnaitman in 1973 (567).
Interest in colicin studies started up in earnest with the work of Jacob et al. in 1952 (297). Using colicin E1 produced by E. coli ML30, those authors demonstrated that (i) the production of colicin by colicinogenic E. coli cells is induced by SOS agents, as is seen with lysogenic phages, and is lethal for producing cells; (ii) the produced colicin is released into the medium late after synthesis (later shown not to be the case for all colicins); (iii) colicin kills sensitive cells according to single-hit kinetics; and (iv) colicin is not active against the producing bacteria due to the presence of a specific antagonist protein called the immunity protein. They compared colicins to bacteriophages, with which they share various properties including specificity of the activity spectra, binding on specific receptors (some of which are common for a given colicin and phage), single-hit mode of action, specific immunity, and lethal production after treatments with mutagenic agents (297, 430). This major contribution triggered numerous studies on the mode of action of colicins.
In 1963, Nomura demonstrated that the various colicins have different modes of action: colicins E1 and K inhibit all macromolecular synthesis without arrest of respiration, colicin E2 causes DNA breakdown, and colicin E3 stops protein synthesis (484). In every case, the colicin lethal action appears to be reversed by treatment with trypsin during a given time period (486). To explain this rescue, a model was proposed in which colicin remains on the bacterial surface at the receptor site and kills the cell from there, in a single-hit process, by sending a signal that is not lethal until it is amplified and reaches its target (484, 485). Rescue from colicin bound to the receptor by trypsin takes place during transmission of the signal, seeing as the time available for rescue is prolonged by energy poisons such as azide and 2,4-dinitrophenol (486). In subsequent work, rescue of cells treated with colicin was obtained with various agents that inactivate free colicin, such as sodium dodecyl sulfate (SDS), salts, and antibodies to colicins (80, 100, 437, 438). It has been proposed that colicin binding is reversible and that rescue might be obtained as long as colicin adsorption does not reach an irreversible state (80, 100, 131a, 437, 583). However, according to the agent used, rescue is obtained with different kinetics, as would be expected to occur if particular agents acted at particular steps of colicin action (100). One of the irreversible events of the colicin lethal action was thought to be the activation of OmpLA, the outer membrane phospholipase A of sensitive cells (100, 101). Thus, one stage of colicin action does not provoke cellular damage as cells are rescued by trypsin treatment, while damage occurs in a second stage (523).
Mutations of the cellular components required for colicin bound on its receptor to transmit its signal have thus been researched. Insensitive mutants that nevertheless possess colicin-specific receptors were isolated in 1967 and were called either tolerant (475, 487) or refractory (266) to distinguish them from the resistant mutants described previously. Different tolerant mutants were afterwards characterized and have been shown to map to either the tol or the tonB gene, defining two machineries used by colicins to enter into the cell, a given colicin always using the same pathway to reach its target. That allowed a classification of colicins into two groups, groups A and B, based on cross-resistance (133, 134). Group A comprises colicins that are translocated by the Tol system, such as colicins A, E1 to E9, K, L, N, S4, U, and Y, while group B comprises colicins that use the TonB system, such as colicins B, D, H, Ia, Ib, M, 5, and 10. It was later shown that the A and B groups are also distinguished by their mechanism of release from the producing cell. In general, group A colicins are encoded by small plasmids and are released into the medium, whereas group B colicins are encoded by large plasmids and are not secreted. However, some colicins might belong to one group and share homologies with colicins of the other group. That is the case of colicins 5 and 10 (515, 516).
It took time to demonstrate that colicin itself is translocated through the cell envelope rather than any putative constituents that play an intermediate role during its action. The demonstration in 1971 that colicin E3 is a specific RNase that makes one cut in the 16S rRNA gene (35, 45, 581) significantly changed the model of colicin action. Further on, the endonuclease activity of numerous colicins was demonstrated, with each one specifically cleaving a particular nucleic acid at a precise site. Colicins E2, E7, E8, and E9 cleave DNA (108, 574, 628), and colicins E3, E4, and E6 and cloacin DF13 hydrolyze rRNA (35, 45, 138), while colicins D and E5 cleave tRNA (491, 629). The killing action of colicins that stop cell metabolism has been more difficult to elucidate. In 1978, Finkelstein's team demonstrated that colicin A, which had been misidentified as colicin K (429), acts by making tiny pores in phospholipid bilayers, thus allowing the leakage of ions across them (575). The inner membrane (IM) was known to be the target of colicins that trigger the arrest of protein synthesis and active transport, since such colicins provoke various membrane perturbations (131a, 202, 203, 231a, 428a, 513a, 552).
The three steps of colicin action have thus been described. The colicin molecule causes killing after binding to a specific receptor on the outer membrane and being translocated through the cell envelope by either the Tol or TonB machinery to its target, which is the inner membrane for ionophoric colicins and the cytoplasm for nuclease colicins. Colicin M is a unique colicin acting on peptidoglycan synthesis through the enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors (176, 252).
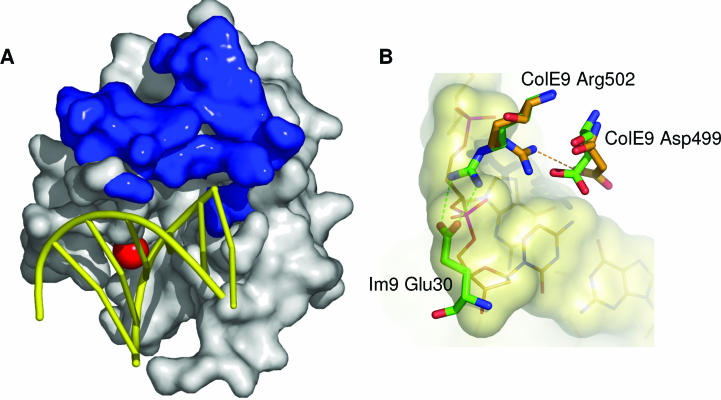
No colicin acts on its own producing bacteria since each bacterium produces a specific inhibitor called the immunity protein. The immunity protein of pore-forming colicins is located in the inner membrane of producing cells (673), blocking colicin when it reaches its target after its entry into sensitive cells. In contrast, the immunity protein of nuclease colicins forms a complex with the cognate colicin in the producing cell, neutralizing its catalytic activity. It is this complex that is released. It dissociates only during colicin action on sensitive bacteria (35, 165a, 304, 590). The specificity of the interaction between the nuclease colicins and their immunity proteins has been extensively studied. The affinity of colicin E9 binding to Im9, its immunity protein, has been found to be in the femtomolar range; i.e., it is one of the strongest associations observed for a complex of two proteins (667).
In 1953, it was suggested that the ability to produce colicin resides in an extrachromosomal genetic element, named the colicinogenic factor, after the demonstration that the determinant for colicin is transmitted in mating experiments (122, 210). In 1965, De Witt and Helinski presented the first evidence that the colicin genetic determinants are located on plasmids (155). Further on, the various plasmids encoding the most studied colicins were isolated (20, 122, 356, 626). Two classes of colicinogenic plasmids, designated pCol, have been identified: small multicopy plasmids that contain the colicin operon and a mobilization factor and large monocopy plasmids that carry numerous genes besides the genes for colicin activity and are able to conjugate (251). Plasmid pColE1 was the first plasmid used as a cloning vehicle at the start of the era of genetic engineering in 1974 (263). Its genetic map was first drawn in 1978 (160), and its 6,646 bp were sequenced in 1985 (110). Since then, the complete sequences of pCloDF13 (9,957 bp) and pColA (6,720 bp) have been published (465, 480).
The organization of the colicin operon carried by the colicinogenic plasmids was first demonstrated in 1978 for colicin E1, an ionophoric colicin (160), and for cloacin DF13, a nuclease colicin (8). Both operons contain the SOS promoter followed by the structural gene for colicin. In the operon of enzyme colicins, the structural gene of the immunity protein is located downstream from the colicin gene and upstream from the gene encoding the lysis protein responsible for colicin release. The operon of pore-forming colicins does not contain the structural gene of the immunity protein: it is located on a specific operon under constitutive regulation present on the opposite DNA strand. The gene encoding the colicin lysis protein is always the last gene of the operon. It is present in the operons of group A colicins but not in those of group B colicins.
Colicins have been purified and found to be proteins of high molecular mass ranging from 40 to 80 kDa (131, 261, 262, 358, 579). Their amino acid sequences have been deduced from the nucleotide sequences of their structural genes; the first to be determined was colicin E1 in 1982 (681). None of the sequences contains disulfide bonds. All colicins are organized into three domains, each corresponding to one step of colicin action, as shown first by de Graaf et al. in 1978 and then by Ohno-Iwashita and Imahori in 1980 (140, 495, 496). The N-terminal domain is involved in translocation through the membrane, and the central domain is involved in binding to the receptor, while the C-terminal domain contains the active part. The pore-forming colicins are monomeric, whereas the enzyme colicins are heterodimers of colicin and the immunity protein.
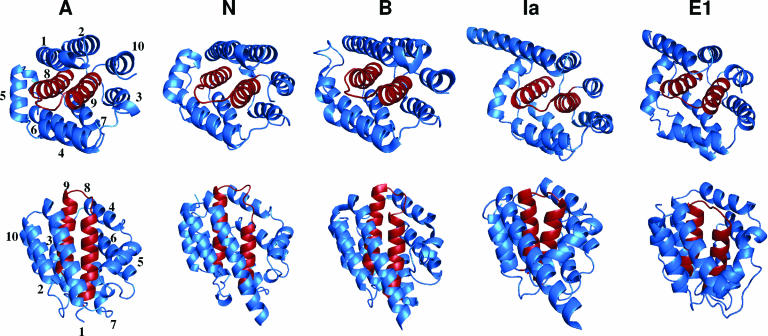
Information on the structures of colicins has slowly emerged. They are elongated proteins (358) and are hard to crystallize. The crystal structure of the C-terminal domain of colicin A was obtained in 1989 (502), long before that of an entire colicin was obtained, which did not appear until 1997, when the structure of colicin Ia was solved (675). This has now been joined by several other colicin structures (177, 267, 607, 650). In most cases, the domain structure deduced from experiment is well explained by the crystal structure.
This report on the landmarks of colicin research during more than 80 years demonstrates that colicins have yielded numerous results pertinent to a variety of fields. The main data obtained on their mechanism of production and release, on the three steps of their mode of killing, on the specificity of the immunity towards them, and on the possible route of their evolution are considered below. Many of them have been reported in previous reviews (258a, 269a, 308, 356a, 552a). It was of interest to collect and compare them and to look forward to where colicins may lead us in the future.
COLICIN SYNTHESIS
Colicinogenic Plasmids
Colicins are produced by strains of Escherichia coli that harbor one colicinogenic plasmid, pCol. Such strains, called colicinogenic strains, are widely distributed in nature and are particularly abundant in the guts of animals. They usually contain many different plasmids, among them only one specific colicinogenic plasmid. There are two classes of pCol: type I and type II (251). The type I plasmids are small plasmids of 6 to 10 kb present in about 20 copies by cell. They can be amplified and are mobilizable in the presence of a conjugative plasmid. They encode mainly colicins of group A and have been abundantly used for genetic engineering and biotechnology. The type II pCol plasmids are large monocopy plasmids of about 40 kb that usually encode colicins of group B. They are conjugative and promote the horizontal transfer of genetic material between donor and recipient cells by physical contact, as do the sexual factors. They can thus transmit the colicin operon and even small mobilizable plasmids present in the same cell to other strains. These large pCol plasmids might contain either one or two colicin operons located side by side. The cells that carry them then produce two different colicins, for instance, colicins B and D, B and M, and Ia and V.
Various plasmids may encode a similar colicin. The best known case is that of colicin E1, which is encoded by type I plasmids as different as pML30 and pJC411. The sequence and organization of both plasmids and the amino acid compositions of their gene products are different, although the encoded colicin has the same characteristics: it is a pore-forming protein using BtuB as a receptor and the Tol system as a transit machinery. Other plasmids producing colicin E1 are known (D. Cavard, unpublished results). A nomenclature should be established to designate them using either the name of the plasmid, as colicin E1-ML30, or a letter, as colicin E1a. This case is not unique. Different plasmids encode colicins A, E2, E3, B, and D (561).
One case of a chromosomally encoded colicin has been reported: colicin-like bacteriocin 28b produced by Serratia marcescens, a colicin very homologous to pore-forming colicins. In this case, the structural gene of colicin is not associated with immunity and release genes in contrast to what was observed with plasmid-encoded colicins (239).
Col plasmid replication has been thought to be coregulated with colicin production since phage induction has often been compared to colicin induction and involves the replication of the phage genome (4, 155). That has been ruled out, since colicin biosynthesis can be induced when plasmid synthesis is inhibited (170, 289, 353).
Colicin Operons: Gene Organization and Regulation
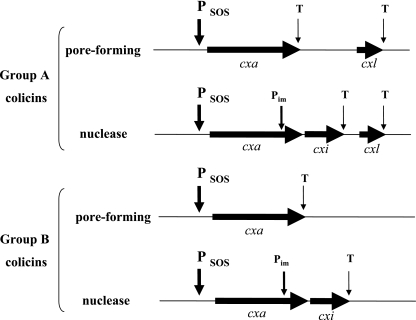
The genetic organization of almost all known colicin operons was reviewed by Riley in 1993 (555, 556) and is summarized in Fig. 1. In all the colicin operons, the first gene is the gene encoding colicin, called cxa, for colicin X activity. It might be the unique gene in the case of operons of pore-forming colicins of group B (347, 433, 578, 649).
FIG. 1.
Organization of the colicin operons. The genes are represented by arrowheads. SOS promoters (PSOS), the immunity promoter (Pim), and transcription terminators (T) are indicated by arrows. Names of the colicin gene (cxa, in which x is specific to the colicin) and its immunity gene (cxi) and lysis protein gene (cxl) follow the nomenclature.
In the operons encoding a nuclease colicin, the gene encoding the immunity protein, designated either cxi, for colicin X immunity, or imX, is located downstream from the structural gene for colicin (2, 108, 123, 129, 244, 303, 383, 564, 634). It is under the regulation of two promoters: the LexA promoter of the colicin operon and its own constitutive operon that allows a constant production of the immunity protein in order to ensure that there is never free colicin, which would kill the producing cell. This separate promoter is located within the structural gene of the nuclease colicin (106, 304, 445, 609a) There is no immunity gene in operons encoding an ionophoric colicin: it is located on the opposite DNA strand of the intergenic space between the colicin and the lysis structural genes and is transcribed from its own promoter under constitutive regulation.
The last gene of colicin operons is the gene encoding the lysis protein, named cxl for colicin X lysis protein, whose product allows the release of colicin into the medium and is responsible of the cell death after induction (colicins A [99], E1 [568, 660], E2 [123], K [515], N [535], U [599], and Y [558] and cloacin [244, 245]). It is present in the operons of group A colicins and in some operons of group B colicins such as those of colicins 5 (515), 10 (516), and D (268, 534, 564).
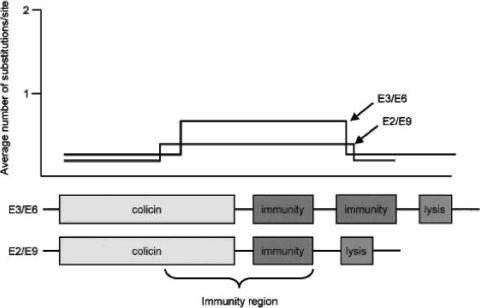
The colicin operons thus contain one to three genes. However, redundancies have been found in the organizations of many colicin operons. For instance, the colicin E3 operon contains two different immunity genes, one to colicin E3 and one to colicin E8 (105, 445), as does that of colicin E6, which contains the immunity genes to E6 and to E8 (2). The colicin E9 operon contains the immunity genes to colicin E9 and to colicin E5 and two lysis genes, that to colicin E9 and that to colicin E5 (107, 129, 383). A common origin of all colicin operons and an evolutional relationship of the colicinogenic plasmids have been suggested. The various colicins seem to have been assembled from a few DNA fragments that encode the functional domains of the proteins (2, 107, 129, 383, 446, 555, 564, 627, 671).
The organization of the colicin Js operon differs from that reported above. The cjl gene encoding the lysis protein is located upstream from the gene for colicin activity, cja (600). Colicin Js is a 95-amino-acid polypeptide of 10.4 kDa with no sequence similarity to other known colicins. It resembles microcins, although the pColJs plasmid that encodes it shows striking similarities with pColE1, and cannot be classified among colicins as previously reported (601).
Transcription of the colicin operons is strongly repressed by the LexA protein, the repressor of the SOS genes (reviewed in reference 409). Except for cloacin DF13, colicin operons contain two LexA boxes in tandem but overlapping by one or two bases (106, 123, 171, 411, 433, 445, 466, 537, 578, 599, 638, 649). The two LexA boxes have been found in every colicin operon studied so far and in the same organization and location. They are located just downstream from the Pribnow box. Each box binds one dimer of LexA. The fixation of two dimers provokes DNA bending, which adds to the blockade of the operon transcription (414). After DNA damage by mutagenic and carcinogenic agents, RecA is activated and stimulates LexA autocleavage and release from the LexA boxes, allowing transcription of the colicin operon. The agents that are able to trigger the SOS response and to induce colicin production are numerous and of different natures: from physical agents such as UV light to chemical drugs and stress conditions (430). The most popular agent used to induce colicin production in research laboratories is the antibiotic mitomycin C (286). Such DNA damage regulation is found for colicins and every class of protein bacteriocins but neither for peptide bacteriocins nor for microcins (128, 262).
Although LexA is the common repressor of colicin transcription, other repressors or activators play a role to modulate the expression of some colicin operons. For instance, transcription of the colicin E1 operon is stimulated by catabolite repression (172). A site of the cyclic AMP receptor protein-cyclic AMP complex has been identified on the promoter of the colicin E1 operon (171, 172), and a potential site for cloacin DF13 (638), colicin Ib (649), and colicin B (578) promoters has been found. Thus, regulators other than SOS agents interfere with colicin transcription. That and DNA bending may explain the pronounced lag observed in colicin expression after SOS induction (262), compared with the expression of other SOS genes (569). The lag is more or less significant according to growth conditions and to the bacterial strain used. Colicin synthesis has been shown to be stimulated in various cases by thymine starvation (589), stringent response (419), catabolite repression (172, 535), ompR mutation (543), the stationary phase of growth (185, 569), anaerobiosis (184), high temperatures (85, 86, 333), or nutrient depletion (368). In contrast, it is significantly reduced by low temperatures and in pldA null mutants (85, 88). Thus, various global regulatory proteins and environmental signals influence colicin synthesis, and some specific gene products are required. The expression of the colicin A operon is activated by both of its gene products: colicin A and Cal, the colicin A lysis protein (86, 87). A role of the E8 lysis protein in the regulation of colicin E8 synthesis has been reported (386). An activator has been found to be required for the transcription of colicin-like bacteriocin 28b (201). Regulation of the colicin operons is complex and may vary from one operon to the other. It looks like that of the virulence genes of various pathogenic bacteria, which depends on growth conditions (455), indicating that colicins, like virulence factors, play a competitive role in the wild environment. However, colicins are primarily under SOS control, and the reason for this is not yet clear.
Transcription from the SOS promoter of the colicin operons of group A results in the formation of two mRNA transcripts due to the presence of two terminators of transcription (Fig. 1). The major mRNA corresponds to the colicin gene for the operons of pore-forming colicins and to the colicin and the immunity genes for the enzyme-colicin operons. In this case, both colicin and immunity genes are coordinately transcribed and translated, and both genes products associate immediately after synthesis to form a dimeric complex devoid of enzymatic activity. In order to block nuclease colicins, an additional promoter is present upstream of the immunity gene (Fig. 1), allowing a higher production of the immunity protein than that of the colicin (105, 445). The minor mRNA is the largest one, as it corresponds to a transcript of the entire operon that is of both the colicin and the lysis genes for the pore-forming colicin operons and of the colicin, the immunity protein, and the lysis protein genes for the nuclease colicin operons (106, 411, 412, 637). Thus, the lysis gene is transcribed at lower levels than the colicin gene.
The translation of the mRNAs of colicins A, E2, and E3 is discontinuous. Discrete elongation intermediates are observed during colicin synthesis. That appears to be due to the tRNA availability for the various codons and to the presence of a high proportion of codons corresponding to rare tRNA in colicin genes (648). The presence of several rare codons in the colicin K mRNA allows ppGpp to regulate colicin K synthesis via a variable cognate tRNA availability (367). An autoregulation of the translational expression of the colicin E7 operon by the immunity protein to colicin E7 has been suggested (283).
Colicin Expression
Colicins are not synthesized under normal conditions since the colicin operon is repressed by LexA, but a small amount of colicin is always present in the culture of colicinogenic cells and is increasing with growth. After treatment of cells with SOS agents, the amount of colicin starts to increase exponentially after a lag period of variable length, which depends upon growth conditions. It reaches a maximum level after 60 to 90 min of induction. At this time, it is about 1,000 times higher than the amount present before induction (262). Colicin is expressed in huge amounts, as its transcription is under the control of a strong promoter and its structural gene is carried by multicopy plasmids in the case of group A colicins. Colicin then becomes the major protein of the cell. Colicin A has been shown to be expressed in various forms, many of them of higher molecular mass than colicin, suggesting the presence of multimers and oligomers of colicin (88).
Whether colicin is produced in small amounts by all the cells of a culture or in large amounts by a fraction of cells during either the spontaneous or the induced production of colicin has long been a subject of controversy. In 1959, Ozeki et al. tried to determine the number of individual cells producing colicin E2 at a given time by measuring the number of lacunae on a lawn of sensitive cells on a petri dish (501). One lacuna is due to the growth inhibition provoked by the amount of colicin produced by a single colicinogenic cell. They concluded that 0.1% of cells produce colicin E2 under normal conditions compared to 50% after UV irradiation (501). Recent work with colicin K labeled with green fluorescent protein (GFP) demonstrated that only 3% of cells produce colicin upon induction by nutrient starvation (471). That does not seem to be the case upon induction of a group A colicin by an SOS agent such as mitomycin C, which is always accompanied by the death of the total population of the colicinogenic culture (80a, 92, 93, 94, 262, 297). The death is not due to the produced colicin, as colicinogenic cells are protected against it by the immunity protein, but rather is due to the production of the lysis protein coexpressed with colicin.
Lethality of Colicin Production
The synthesis of group A colicins is a lethal event for the cell. After induction, the number of viable cells immediately starts to decrease while the amount of colicin is increasing (92, 262, 297, 353, 501, 540, 613). Cell death is due to the colicin lysis protein coexpressed with colicin. Its killing process against its host is unknown. It seems to be responsible for the shutoff of chromosomal protein synthesis reported during colicin induction.
Colicin operons of group B colicins contain a lysis gene when carried by plasmids of type I, as colicins 5 and 10, but do not contain a lysis gene when present on type II plasmids, except that of colicin D (268). Thus, their synthesis is not lethal for the producing cells and is not followed by the release of colicin into the extracellular medium.
COLICIN RELEASE
As first observed by Gratia, colicins are present in the culture medium of producing bacteria (235), and their release into the environment has long since been thought to be a unique form of protein secretion by E. coli. Colicin release involves only one gene product, the colicin lysis protein, also referred to as the killing (kil) protein or bacteriocin release protein (BRP) (245, 303, 500, 541, 568, 641), and takes place following the synthesis of both colicin and the lysis protein. Lysis proteins allow colicins to be released into the medium. The mechanism of colicin secretion differs from that of the five secretion pathways in gram-negative bacteria known to be involved in the release of proteins into the extracellular medium (reviewed in references 481 and 623). It also differs from the mechanism of action of phage lysis proteins that provoke the total lysis of phage-producing bacteria (reviewed in reference 690). It does not involve autolytic enzymes, which hydrolyze the peptidoglycan, as do many phage lysis proteins (279), nor is it related to the secretion system of microcins such as colicin V, which is mediated by an exporter system consisting of two specific cytoplasmic proteins, CvaA and CvaB, and of the host outer membrane protein TolC. CvaB is a member of the ATP binding cassette (ABC) superfamily. The amino-terminal export signal of colicin V, which is a double-glycine leader sequence specific for the CvaA-CvaB-TolC exporter, is processed concomitant with secretion (226, 242). The export of colicin V instead resembles that of various peptide bacteriocins produced by gram-positive bacteria, which also require the activity of a dedicated ABC transporter. The transporter is the maturation protease that cleaves off the typical double-glycine leader sequence of the peptide bacteriocins concomitant with their translocation across the membrane (162, 254, 294).
The gene encoding the lysis protein is the last gene of the operons of group A colicins. Release does not occur in cells missing the gene, such as bacteria producing a group B colicin, nor does it occur in bacteria containing mutations, insertions, or deletions in the lysis gene (99, 244, 245, 303, 541, 568, 627). Transcription of the lysis gene relies upon transcriptional readthrough from the promoter of the colicin structural gene across the entire operon. Thus, the colicin lysis protein is coexpressed with colicin; conversely, colicin might be synthesized without the lysis protein due to a transcription terminator at the end of the gene (Fig. 1). The lysis protein is always produced in lower amounts than colicin (106, 411, 412, 637) but in significant amounts (similar to that of Lpp, the murein lipoprotein). Induction of the lysis gene cloned under various promoters provokes protein release and death of the host, even in noncolicinogenic cells (3, 94, 423, 541).
Sequence, Synthesis, and Localization of the Colicin Lysis Proteins
Colicin lysis proteins are small lipoproteins of 27 to 35 amino acids whose sequences exhibit a high degree of similarity, which is suggestive of a common origin. They are synthesized as precursors of around 4.5 kDa. The first lysis protein to be identified and sequenced was that of colicin E1, CelA, in 1979 (497). The amino acid sequences of the 21 lysis proteins determined thus far by nucleotide sequencing of lysis genes are presented in Fig. 2. The sequences of the two lysis proteins of colicin E1 (497, 660) and of colicin E5 (306, 383) differ by one or two residues, probably due to different plasmid sources.
FIG. 2.
Sequence alignment of colicin lysis proteins. Amino acid sequences of the colicin lysis proteins are shown. The sequences of the colicin lysis proteins encoded by the colicinogenic plasmids indicated at the left are presented. Identical amino acids are in boldface type. The numbers of residues in the signal peptide and in the mature form are indicated. X63621, EMBL/GenBank/DDBJ accession number X63621.
The sequences of all lysis protein precursors are similar. They possess a classical signal sequence that contains 15 to 22 residues, with an N-terminal methionine followed by basic residues and a hydrophobic core containing numerous leucine and isoleucine residues. The site of cleavage is contained in a lipobox, LXYC, in which X is A,V, or S and Y is either A or G, which allows the acylation and processing of the lipoproteins. The lysis protein of colicin E9 does not contain a lipobox and is not functional, as its sequence is truncated by a naturally occurring transposon (383). The mature forms of lysis proteins are similarly homologous to each other. Those of the colicins E, K, Y, 5, and 10 differ by only a few residues, while their signal sequences exhibit greater diversity. All lysis proteins contain a cysteine at their N termini, which becomes N and S acylated, a glutamine at position 2, an arginine-aspartic acid pair at positions 7 and 8 (with the exception one out of the two presented for pColE1 and pColE5), one proline in the middle of the sequence, numerous glycine and serine residues, and one charged residue among the C-terminal residues (that of pColN is especially rich in charged residues). The consensus sequence indicates 10 identical residues and 7 related ones in the mature forms of the 21 lysis proteins presented in Fig. 2.
Lysis proteins are acylated and processed in many steps, as are all bacterial lipoproteins. They are synthesized in the cytoplasm as precursor forms, which are driven by the signal sequence to the inner membrane and translocated by the Sec machinery to the outer leaflet of the inner membrane, where they are modified by Lgt, the lipoprotein glyceryl-transferase, with a diglyceride on the sulfydryl group of the cysteine residue of the lipobox. Next, the modified precursor forms are processed by LspA, the signal peptidase II specific for lipoproteins, into apolipoprotein and signal peptide. The amino group of the acylated cysteine is then acylated by Lnt, the lipoprotein N-acyltransferase (679, 689).
The rate of acylation and processing varies from one lysis protein to another; this is rapid for CelA, the colicin E1 lysis protein, but of the order of several minutes for Cal, the colicin A lysis protein, and BRP, the cloacin DF13 lysis protein. Consequently, three forms of Cal and BRP are present following induction: the precursor, the modified precursor, and the mature forms. The kinetics of Cal maturation do not vary regardless of whether the lysis protein is coexpressed with colicin or alone. However, the levels of the three forms of Cal vary with time and depend on the genotype of the host strain (81, 84). The signal peptides of Cal, CelB (colicin E2 lysis protein), and BRP are stable after cleavage and accumulate in the inner membrane, while that of CelA is immediately degraded (80a, 84, 91, 99, 424, 425, 539). The dependence on the Sec machinery also varies from one lysis protein to another: CelA and BRP are Sec dependent, while Cal is Sec independent (81, 499).
The final location of lipoproteins depends on the residue at position 2 (218, 683). They are anchored to the periplasmic leaflet of either the inner or outer membrane through fatty acyl chains covalently linked to the N-terminal cysteine. The inner membrane retention signal for lipoproteins has Asp at position 2 in combination with certain residues at position 3, which functions as a Lol avoidance signal, since this inhibits the recognition of lipoproteins by the five proteins of the Lol system, which release lipoproteins from the inner membrane (622). All colicin lysis proteins possess a glutamine residue at position 2 and so should be recognized by the Lol system and located in the outer membrane. The mature forms of cloacin DF13 and colicin A, E2, and E3 lysis proteins have been observed in both the inner and outer membranes, while that of colicin N has been found only in the outer membrane (123, 277, 303, 500, 538). The presence of lysis proteins in both membranes is due to their slow assembly in the outer membrane. Maturation and assembly have been measured for Cal by sucrose gradient analysis of radiolabeled membranes of colicin A-producing cells at various times of induction (277). Both Cal and CelA take the same time to acquire the specific electrophoretic behavior of outer membrane proteins; both lysis proteins are soluble in SDS after maturation, but, many minutes later, they have to be heated in SDS to be solubilized at the time of colicin release. They are then partly released into the extracellular medium with colicin. The exported lysis proteins share the electrophoretic property of outer membrane proteins, indicating that they are kept in a structure similar to that before export (80a, 81). No colicin lysis protein has been purified to homogeneity thus far, which has limited their biochemical characterization. In addition, lysis proteins are not readily soluble and are prone to aggregation.
Functions of Colicin Lysis Proteins
The main function of the lysis protein is to promote colicin release, and so they are more correctly termed colicin release proteins, Cxr (for colicin X release). Concomitantly, they provoke quasilysis; modifications of the structure of the cell envelope; activation of OmpLA, the outer membrane phospholipase A; and death of the producing cell. However, the chronology of the various events is not well understood and seems to appear simultaneously. The functioning of lysis proteins does not depend on colicin production or on RecA-LexA regulation. It does, however, require that the lysis protein be produced in reasonable amounts. The various physiological changes provoked by lysis proteins occur late after synthesis, indicating that a critical concentration of the lysis protein in the host cell is needed. There is no specificity associated with lysis functions since colicin lysis proteins are similar (Fig. 2) and therefore interchangeable (541).
Colicin release.
After induction by DNA-damaging agents, colicins are expressed and accumulate in the cytoplasm of the producing cells (96). Group A colicins that are synthesized with a mutated or a deleted lysis protein and colicins of group B (which are not coexpressed with a lysis protein) remain in the cytoplasm during and after induction. Colicins do not possess signal export domains; they have neither cleavable N-terminal signal sequences, which can mediate their transport to the periplasm, nor internal export domains, which could help them to be secreted by the lysis protein (17). Some mutations of colicins A, E1, and E3 and cloacin DF13 that inhibit release have been identified (7, 15, 461, 646, 682), but this could be due to aggregation or to changes in conformation, as these mutated colicins were inactive. On the other hand, mutations of the colicin lysis proteins have been reported to block release, as described below.
After induction, colicins A and E2 and cloacin DF13 are found in both the cytoplasm and periplasm (88, 96, 500, 540). Part of colicin A has been shown to be localized in the outer membrane, associated with porins and other outer membrane proteins (88). Colicins are progressively released into the extracellular medium 60 to 90 min after induction. Some periplasmic and cytoplasmic proteins are found in the medium with colicin (17, 99, 303, 461, 540, 541, 613, 647), but it is not known whether these proteins come from some lysed cells or through the lack of specificity of colicin release. They are found similarly after induction of a cloned lysis gene.
The level and timing of colicin release vary with growth conditions. Release is slowed down in the presence of divalent cations (20 mM), as was seen for colicins A, E1, and E2 and cloacin DF13 (3, 93, 426, 542). It is also significantly retarded in colicin A-producing bacteria induced at low temperatures. In contrast, colicin A release is sped up at elevated temperatures, after a heat shock, in the presence of either EDTA or Triton X-100, or in hosts carrying a mutation in the degP gene (85, 93, 98). Such variations seem to be due to the cellular concentration of the lysis protein, since the amount of Cal is higher in the presence than in the absence of EDTA and is lower in the presence than in the absence of Mg2+ ions (84). The level of the colicin lysis protein is modulated by the same environmental factors as colicin and is directly responsible for the rate of colicin release. The cellular concentration of the lysis protein might also be regulated by proteolysis, as some stress conditions provoke the induction of proteases. In summary, the amount of lysis protein appears to be critical for its function in colicin release from bacteria.
Quasilysis.
The optical density of a culture of bacteria producing a group A colicin increases for about 60 min after induction, as does that of a noninduced culture. It then decreases significantly, reaching an optical density similar to that at the time of induction (297). This drop of absorbance was called quasilysis by Jakes and Model in 1979 (300) and occurs whether or not colicin is produced (94, 423). It does not correspond to complete lysis as seen after the induction of lysogenic bacteria, as no lysed bacteria or membrane fragments have been observed during this period (92).
Quasilysis is a good reporter of the functioning of colicin lysis proteins. Both quasilysis and colicin release take place simultaneously and vary according to growth conditions as described above. Neither occurs in pldA cells (542). In colicin A-producing cells, both are reduced in an rpoH mutant but increased in a degP mutant (85, 98). However, colicin A release occurs without quasilysis in cells incubated in the presence of 20 mM divalent cations and in cells grown at low temperatures. In both cases, the synthesis of colicin A and Cal is significantly retarded, allowing the growth of cells, which may mask the decrease of culture turbidity normally linked to colicin A export (85, 93).
Modifications of cell envelope structure.
After colicin induction, changes in the structure of the cell envelope occur. Analysis on sucrose density gradients of cell envelopes for cells overproducing the colicin A lysis protein showed that both inner and outer membranes cannot be separated during the latter stages of induction (277).
Activation of OmpLA, the outer membrane phospholipase A.
Neither colicin release nor quasilysis occurs in bacteria containing a mutated pldA gene (542). OmpLA, the pldA gene product (60), is an inactive monomeric protein which dimerizes when activated (141, 603). OmpLA is activated after the induction of colicins A and E2 and of cloacin DF13 (91, 426, 542). Its activation is due to the production of the lysis protein, as demonstrated by OmpLA dimerization after the induction of a subcloned BRP, the cloacin DF13 lysis protein (142). OmpLA activation has been postulated to cause colicin release, quasilysis, and killing of host cells. It provokes the formation of lysophospholipids, which are detergents and would permeabilize the outer membrane and, subsequently, the inner membrane of the cells (542). However, both colicin A release and quasilysis occur in the absence of an active OmpLA in a tolQ mutant (277). Functioning of lysis proteins is decreased in the presence of divalent cations, and that of Cal is increased in the presence of EDTA, despite the fact that OmpLA requires Ca2+ ions to be active (60, 141). OmpLA activation seems to be a consequence of the mechanism of lysis protein action and might be the cause of death of producing cells, as various lethal agents are known to provoke OmpLA activation (101).
The presence of an inactive OmpLA in the cell induces the Cpx and σE regulons and consequently switches on many genes including genes encoding proteases such as degP (375). The amount of Cal is significantly reduced in cells with a missense mutation in the pldA gene compared to that of wild-type (WT) cells, suggesting proteolysis. The introduction of a degP mutation in pldA cells does not restore the level of Cal, indicating the presence of a protease(s) other than DegP that might be able to degrade the various Cal forms in cells with an inactive OmpLA (84). The induction or activation of proteases by a pldA mutation might explain why cells containing a null pldA mutation produce only traces of colicin A, in contrast to cells containing an inactive OmpLA (88). The induction of a protease(s) would be less significant in the missense pldA mutant than in the null pldA mutant in which both colicin A and Cal might be degraded by proteases.
Death of the host cell.
The production of lysis protein, induced via the SOS or lac promoter, causes the death of the host cell whether or not colicin is expressed (3, 93, 98, 244, 245, 423, 541). Cell death is thought to be caused by OmpLA activation (542), which is a lethal event in bacteria (101, 141). However, the colony-forming ability of colicin A-producing cells that possess an inactive OmpLA decreases after induction although less significantly than that of wild-type cells. The survival of bacteria is altered by various treatments that affect quasilysis. Heat shock and the addition of EDTA or Triton X-100, for example, increase both the lethality and quasilysis of colicin A-producing cells, while divalent cations and low temperatures decrease them (85, 93). Surviving bacteria seem to be uninduced cells, cells in which colicin induction is retarded, or cells in which the two gene products of the colicin operon have been proteolysed. These cells multiply during induction, and their progeny may mask the death of the induced cells and quasilysis.
Mutants of E. coli that are resistant to both the lethal and lytic action of celA (kil) gene expression have been isolated. Some mutants are unable to produce colicin, while others are leaky to periplasmic proteins. The latter type of mutant releases colicin without OmpLA activation, but none of them die after induction. This suggests that CelA, the colicin E1 lysis protein, acts in various steps, each one involving various cell components (3, 614).
Regulator of expression of the colicin operon.
Lysis proteins appear to play a role in the expression of the related colicin and in their own expression. The presence of a transposon in the lys gene of colicin E8 has been shown to reduce colicin E8 synthesis significantly, suggesting a role for the E8 lysis protein in the regulation of colicin E8 synthesis (386). A similar role for Cal (the colicin A lysis protein) as an activator of the transcription of the colicin A operon has been reported, as colicin A synthesis does not occur in the presence of globomycin in cells with a nonfunctional cal gene (86, 87).
Structure-Function Relationships of Colicin Lysis Proteins
A surprising feature of colicin lysis protein biology is that although the proteins share a high degree of sequence conservation, mutagenesis experiments have shown in fact that only acylation, processing, and length of the lipopeptide are required for function. Acylation of the cysteine residue of the lipobox is essential for function. Changing the lipobox cysteine inactivates the protein. However, some colicin release has been observed when the cysteine of the colicin E2 lysis protein is replaced by a glutamine (539). The colony-forming ability is inhibited when the cysteine of BRP is replaced by glycine (425). Mutated Cal lysis proteins, which contain either proline or threonine in place of the cysteine residue in position 1, are rapidly hydrolyzed (91). The enzyme responsible for the degradation of the mutated precursor forms of Cal has been shown to be DegP, a periplasmic heat shock protein that combines refolding and proteolytic activities (363). In a degP null mutant, the precursor forms of C1P and C1T Cal remain stable and unprocessed in the inner membrane but nonfunctional (90).
Processing is absolutely required for function. Globomycin, an inhibitor of LspA, the signal peptidase of the lipoproteins (288), blocks the processing of the modified precursor form of lysis proteins and inhibits colicin release. In the presence of globomycin, the modified precursor form of CelA is rapidly hydrolyzed, while that of Cal is cleaved into two acylated fragments in wild-type cells. It accumulates uncleaved in a degP mutant, indicating that DegP hydrolyzes the modified precursor form of Cal, pCalm (98), as the unmodified Cal precursor, pCal (90). The two sites of DegP cleavage on Cal seem to be located between Val14-Ser15 and Val23-Ser24, upstream of Met25, in agreement with reported DegP sites (355). They are therefore present in the three forms of Cal, pCal, pCalm, and Cal, all of which might be substrates for DegP. In the absence of globomycin, two truncated Cals are observed by immunoblotting, while they are not seen with radioactive methionine labeling (84, 98). These short Cals might be formed either by DegP cleavage of the Cal mature form or by acylation and processing of pCal and pCalm fragments produced after DegP action.
Despite the presence of DegP in wild-type cells, the three forms of Cal always seem to be present, indicating that the majority of Cal avoids DegP degradation. Various hypotheses might explain such an avoidance mechanism. First, DegP sites on Cal may be masked, for example, due to the formation of protein complexes with chaperones and/or other proteins or to polymerization. Second, the maturation of Cal might take place in locations that do not contain DegP, although it is known to occur at the periplasmic side of the inner membrane, where DegP is located. Third, Cal is produced in large quantities that might overwhelm DegP, which is a relatively minor protein. Synthesis of DegP is induced after heat shock, during stress conditions (610), and upon the overproduction of some outer membrane lipoproteins (459). Membrane anchoring of the lipoprotein NlpE is essential for degP induction, since the nonlipidated derivative of NlpE does not induce it (604). DegP synthesis may be induced during globomycin treatment by the accumulation of pCalm. Finally, DegP combines two activities and may be switched to chaperone activity during colicin induction and to protease in the presence of globomycin. It is known that the chaperone function dominates at low temperatures, while the proteolytic activity is present at elevated temperatures (610). However, the level of the three forms of Cal present in degP cells is higher and Cal functioning is more significant than in degP+ cells, indicating that a certain amount of either form of Cal cannot escape DegP cleavage (84).
DegP sites are not present in all lysis proteins (Fig. 2), explaining why DegP degradation is not observed for CelA, the colicin E1 lysis protein. The modified precursor form of CelA accumulates neither in wild-type nor in degP cells after globomycin treatment, indicating that it is the substrate of a protease(s) other than DegP (81).
The signal peptide of the lysis protein is either unstable, like that of CelA and all lipoproteins, or stable, as seen in Cal and BRP. In these latter cases, it accumulates in the inner membrane in contrast to other leader sequences, which are hydrolyzed by signal peptide peptidases. This stability might be the cause of lysis and death of the host cell. However, quasilysis and cell death occur in colicin A- and colicin E1-producing cells even though the Cal signal peptide is stable and that of CelA is unstable (81). Cloning of the signal peptides of the colicin E2 and cloacin DF13 lysis proteins causes quasilysis, lethality, and some colicin release (322, 642). The replacement of the stable BRP signal peptide by the unstable signal peptide of the murein lipoprotein Lpp inhibits cloacin DF13 release but provokes killing, quasilysis, and leakage of periplasmic proteins. The construction of these hybrid precursors indicates that the BRP signal peptide is responsible for the slow processing and contributes with mature BRP to the transfer of cloacin DF13 across the cell envelope (642, 643). In vitro mutagenesis of the Cal signal peptide demonstrates that the Ile residue at position 13 is important for stability and that both Ile13 and Ala18 contribute to the slow modification and processing of the Cal precursor. However, it does not influence colicin A release, quasilysis, and death of the cell host, which are caused only by the mature form of Cal (280).
Although the composition of the mature form of colicin lysis proteins is particularly well conserved, mutagenesis of Cal has shown that many residues/regions are unimportant for function; changes to the conserved residues of the N-terminal half of Cal are without effect, except when a negative charge is present, indicating that charge plays some role in its function (276). More important is the length of the mature lysis protein. Truncated colicin E2, colicin E3, and cloacin DF13 lysis proteins of 20 amino acids are active, while a shortened Cal of 18 residues and a BRP of 16 residues are not (276, 422, 627). The length of the mature form plays a role in the rate of processing and maturation. Truncated Cal containing the first 16 or 18 residues is neither acylated nor processed, except when overproduced, indicating that its maturation is slower than that of wild-type Cal, explaining its lack of activity (82). In contrast, amino acid extensions to lysis proteins do not seem to modify their functions. A hybrid protein of BRP-lactamase functions as BRP for the export of cloacin DF13 (424).
Colicin lysis proteins are homologous to VirB7, the lipoprotein of the type IV secretion system of Agrobacterium tumefaciens (90, 586). VirB7 is a 41-amino-acid lipoprotein that forms homodimers and heteromultimeric complexes with other putative outer membrane proteins of the VirB system (reviewed in reference 76).
Colicin lysis proteins share some similarities with the lysis proteins of single-stranded phages (384). The main difference between them is that the colicin lysis proteins are lipoproteins, while phage lysis proteins are not. However, a lipoprotein homologous to the colicin lysis protein, called Rz1, is encoded by a reading frame embedded within the Rz lysis gene of phage λ. It is localized in the outer membrane and plays a role in the timing of lysis in lysogenic cells following induction (328, 700). Similar lipopeptides have been shown to be encoded by the lysis genes of other bacteriophages such as P2 and N4, which form heteromeric complexes and are required for host lysis (436).
Mechanism of Action of Colicin Lysis Proteins
How the colicin lysis protein allows colicin release has not been fully elucidated. The majority of secretion systems thus far described for transporting proteins across the membranes of gram-negative bacteria utilize multiprotein machines. Colicin release differs from all these secretion systems not only by the number of proteins required for transport but also by the amounts of both the protein transported and the protein transporter, the timing of the release, and the lack of specificity. Both colicins and their lysis proteins are produced in large amounts in the cell, with colicin in higher concentrations than the lysis protein. Most of the colicin is released into the medium, while only some of the lysis protein is released, and this occurs only late in the induction/release process. The lag before release is greater than a generation time, suggesting that modifications and/or the buildup of structures might occur.
Various models to explain the mode of action of lysis proteins have been proposed. For example, they could form pores through both the inner and outer membranes. The colicins located in the cytoplasm would then cross both membranes through these putative trans-envelope pores. The stable signal peptide present in the inner membrane might participate in the formation of such pores, which would destabilize the cell envelope and provoke OmpLA activation (139, 643).
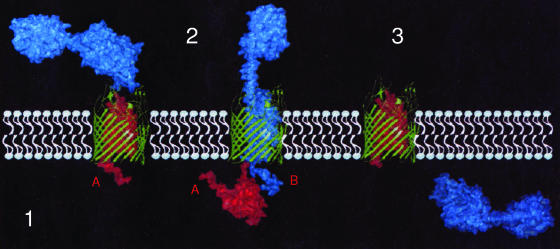
Recent studies described two steps of colicin A release by Cal, the colicin A lysis protein (90). In the first step, colicin A produced in the cytoplasm moves to a location where it can be extracted by washing. This location is presumably the periplasm, in which colicin A and other colicins produced in the presence of a lysis protein have been detected. Cal is required for this step, suggesting that it helps colicin A cross the inner membrane. This step is slowed down in cells with a nonacylated Cal precursor, pCal, suggesting that both pCal and colicin A may be associated and translocated simultaneously to the periplasm. In the second step, colicin A is released into the medium with some Cal. During both steps, colicin A is found in various forms, as described previously (90). In particular, hetero-oligomers of colicin A bound to OmpF and OmpC porins, called colicin Au, are present during each step. Colicin A present in the periplasm would associate by its C-terminal domain with porins and also to other outer membrane proteins already incorporated into the outer membrane, constituting colicin Au, the formation of which is Sec independent. In vitro binding of the C-terminal part of colicin N with porins has been reported (161). It has been proposed that the colicin Au structure might be similar to that of TolC (88), i.e., a β-barrel of porins fused to an α-helical tunnel made of the C-terminal domain of colicin A (359). This structure would allow cellular material to go out and would be dangerous for the producing cells except in the presence of Cal. In the absence of Cal, colicin Au is unstable, and the cells stop synthesizing OmpF, indicating that OmpF is lethal for them. Cal seems to associate with colicin Au, as it is found in purified colicin Au fractions, and to stabilize and detoxify them. It is proposed that this multimeric complex constitutes part of the machinery for colicin export.
Colicin A present in the periplasm might also associate with the Tol proteins, which would become nonfunctional. Such an interaction in cells producing the N-terminal domain of colicins has been described (43). The producing cells exhibit phenotypes similar to those of tol mutants, which are leaky and produce outer membrane vesicles (28). In this instance, vesicles would contain Cal located in the outer membrane and colicin A present in the periplasm. The outer membrane vesicles would be released from bacteria, explaining the presence of both lysis proteins and colicin in the medium (90). Vesicle-mediated export for the release of toxins and for sending signals from cell to cell has been described (447, 657). However, the formation of such vesicles, as that of the colicin export machinery, requires periplasmic colicin and does not explain the mode of action of separately expressed lysis proteins.
Current evidence suggests that colicin release has similarities to colicin entry. As described above, colicins are able to interact with outer membrane proteins such as porins and the Tol machinery in the periplasm. Both the uptake and the release of colicin provoke OmpLA activation (91, 101). Such a model would require the association of the lysis protein precursor with colicin in order to translocate through the inner membrane to the periplasm, despite the fact that no colicin sequence required for interaction with the lysis protein has yet been identified (17). This association might explain the slow rate of acylation and processing of the lysis protein. The mature lysis protein might then become involved in the formation of colicin-porin complexes, which would constitute the exit gate of the colicin. This would draw the inner membrane near the outer membrane in order to allow release. In this way, the small amount of lysis protein relative to that of colicin would succeed in fulfilling its many functions with the help of the molecule it is designed to release. We speculate that the lack of release specificity might allow the lysis protein to associate with any major protein cosynthesized with it, regardless of whether it is a colicin, although such a mechanism remains pure speculation at the present time.
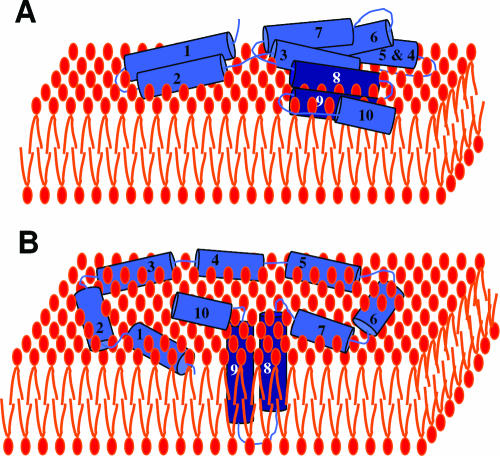
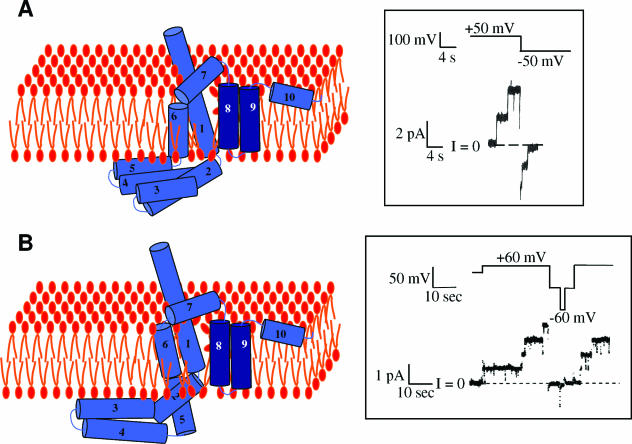
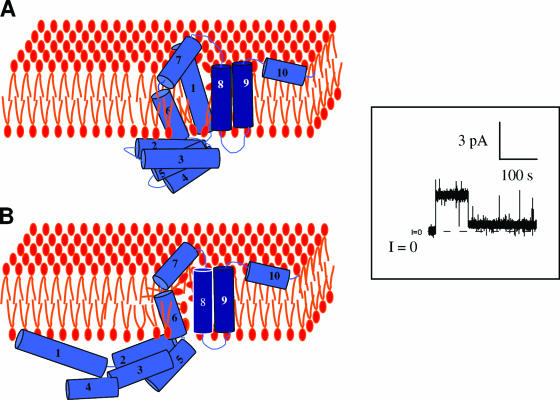
STRUCTURAL ORGANIZATION OF COLICINS
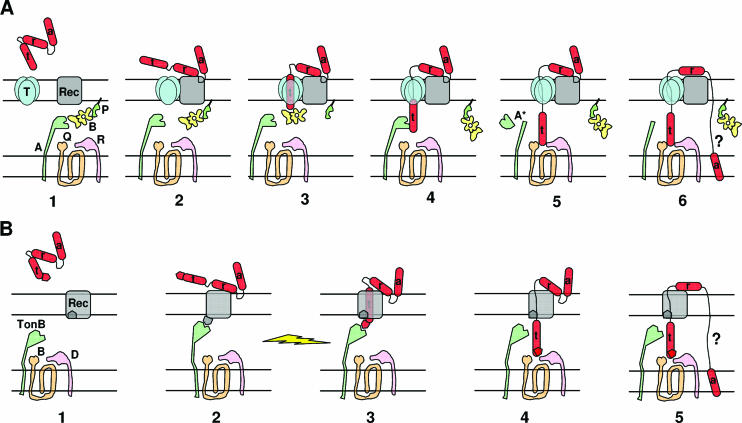
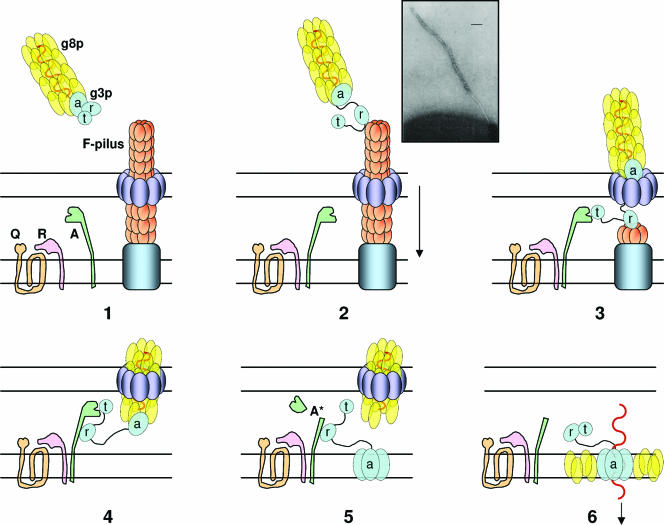
Colicin organization has been elucidated by isolating insensitive mutants. Characterization of the insensitive colicin mutants led to the discovery of the various steps of colicin action and demonstrated that each step of colicin action depends on one domain of the colicin molecule. Binding of colicins to bacterial receptors has been demonstrated by the characterization of mutants that are insensitive to colicins, called resistant mutants. More recently, a new kind of insensitive mutant in which colicin action, but not colicin binding, is prevented has been found (158, 243, 266, 475, 485, 566). These mutants have been called tolerant and allowed a classification of colicins into groups A and B based on the dependence of colicin entry using either one of two different systems, the Tol and Ton systems, respectively (133, 134). These two systems are also parasitized by phages to infect bacteria. To inject single-stranded DNA through the envelope of gram-negative bacteria, filamentous bacteriophages (Ff phages) use the minor coat protein g3p located at the tip of the particle (234). The minor coat protein g3p interacts with the tip of the conjugative pilus and requires the Tol system to further translocate the single-stranded DNA through the cell envelope (180, 479, 565). Like colicins, g3p follows a two-step import process that includes receptor binding and translocation across the outer membrane. On the other hand, the T1 and φ80 phages use the outer membrane ferrichrome receptor FhuA and the Ton system. Nevertheless, the phage proteins interacting with the Ton system are poorly described. While phages interact with bacteria to inject their genome and to multiply, colicins kill the susceptible bacteria during a third step of action either by nuclease activity or by pore formation in the cytoplasmic membrane (see Colicin Activities).
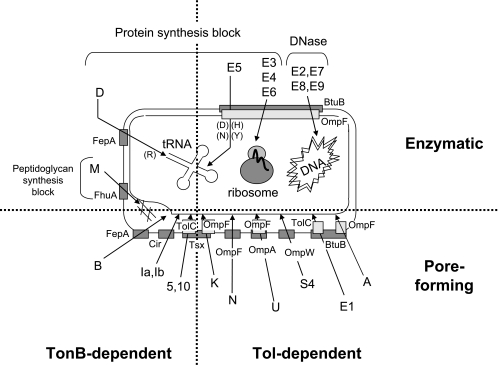
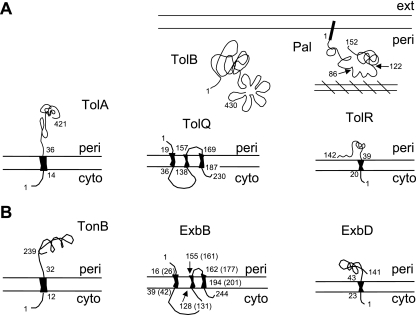
The Ton system is formed by three inner membrane proteins, TonB, ExbB, and ExbD (530), while the Tol system contains proteins of the same topology and localization, TolA, TolQ, and TolR, plus a periplasmic protein, TolB. It also includes Pal, an outer membrane-anchored lipoprotein, which appears not to be required for colicin and phage import (388, 413, 674) (see Transit through the Periplasm). Table 1 and Fig. 3 show the cell envelope proteins involved in colicin import. Colicins of group A require the Tol proteins or a subset of them; only TolA and TolQ are essential for colicin E1, while TolA, TolQ, and TolR are needed for colicin N import. In contrast, TonB, ExbB, and ExbD are required for all group B colicins, associated in most cases with a single outer membrane gated porin receptor. Cloacin DF13, which is naturally produced by Enterobacter cloacae, is active against E. coli strains producing the TonB-dependent OM transporter IutA. Strikingly, using various tol, tonB, and porin mutants, cloacin DF13 has been found to bind to the IutA siderophore receptor and to use the OmpF and TolAQR proteins (624, 677a). The import mechanism of colicins 5 and 10, which use Tsx as a receptor, requires the additional outer membrane protein TolC. Colicin K, which also uses Tsx as a receptor, needs both OmpF and OmpA (536) and the TolABQR proteins. The nine E colicins and colicin A, which use BtuB as a receptor, also need OmpF, except for colicin E1, which requires TolC for its translocation step (475). Some colicins, such as colicins E2 and E3 or colicins B and D, use the same receptor-translocator proteins but exert different lethal activities. In contrast, other colicins, such as colicins 5 and 10 or colicins Ia and Ib, use the same receptor-translocator and act similarly, although their active domains are not identical. They can be distinguished by subtle differences in activity or by the specificities of their immunity proteins. Colicin M is the unique colicin that exerts its lethal activity in the periplasm of sensitive cells (252), where it hydrolyzes the peptidoglycan lipid intermediates I and II (176). The specific requirements for the receptor and translocator proteins and the intrinsic lethal activities demonstrated the modular organization of colicins.
TABLE 1.
Tol- and Ton-dependent imports: cell envelope proteins required for reception and translocation steps of colicins and bacteriophages
| Colicin or bacteriophage | Receptor | Import | Cytotoxicity |
|---|---|---|---|
| Colicinsa | |||
| Group A | |||
| A | BtuB | OmpF, TolABQR | Pore |
| E1 | BtuB | TolC, TolAQ | Pore |
| E2-E7-E8-E9 | BtuB | OmpF, TolABQR | DNase |
| E3-E4-E6 | BtuB | OmpF, TolABQR | 16S RNase |
| E5 | BtuB | OmpF, TolABQR | tRNA-(Y-H-N-D)-specific RNase |
| K | Tsx | OmpF, OmpA, TolABQR | Pore |
| N | OmpF | OmpF, TolAQR | Pore |
| U | OmpA | OmpF, TolABQR | Pore |
| Cloacin DF13 | IutA | OmpF, TolAQR | 16S RNase |
| Group B | |||
| B | FepA | TonB-ExbBD | Pore |
| D | FepA | TonB-ExbBD | tRNA-(R)-specific RNase |
| Ia-Ib | Cir | TonB-ExbBD | Pore |
| M | FhuA | TonB-ExbBD | Degradation of the C55 phosphate-linked peptidoglycan precursors |
| 5-10 | Tsx | TolC, TonB-ExbBD | Pore |
| Bacteriophagesb | |||
| f1-fd-M13/Ike | Tip-pilus F/N | TolAQR | |
| T1-φ80 | FhuA | TonB-(ExbBD-TolQR) |
FIG. 3.
Schematic summary of reception, translocation, and mode of action of most studied colicins. Colicins are distinguished by their general modes of action (upper section, enzymatic; lower section, pore forming) and transit machineries (right section, TonB; left section, Tol) separated by dotted lines. For each colicin (the name is indicated at the arrow base), the outer membrane protein used for the reception step (and sometimes for outer membrane translocation) (ColB, D, Ia, Ib, M, and N) and the OM protein involved in the translocation step (OmpF, colicins A, E2 to E9, K, and U; TolC, colicins 5, 10, and E1) are indicated. For enzymatic colicins, the mode of action at the physiology level (peptidoglycan synthesis block [colicin M], protein synthesis block by cleavage of tRNA [colicins D and E5] or 16S rRNA [colicins E3, E4, and E6], and DNA degradation [colicins E2 and E7 to E9]) is also indicated. For tRNA colicins (colicins D and E5), the specific tRNA targeted is indicated by the one-letter code in parentheses (D, aspartate; H, histidine; N, asparagine; R, arginine; Y, tyrosine). See the text for details.
Domain Organization of Colicins
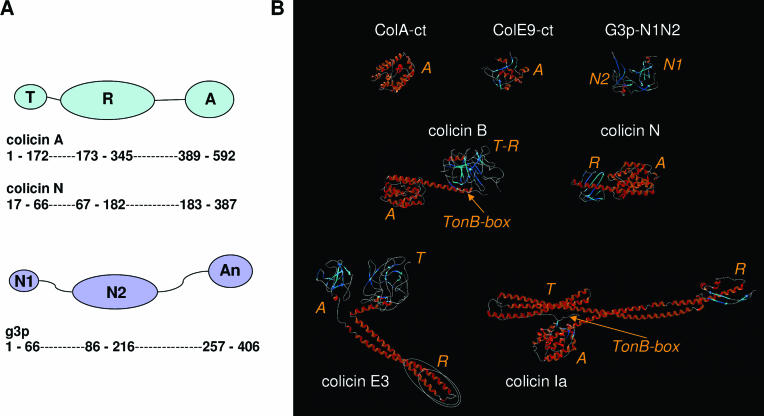
The multistep processes of colicin and Ff phage DNA import point out that both colicins and g3p possess different domains. Both Tol- and Ton-dependent colicins are organized into three domains in relation with the three steps of their action: reception, translocation, and killing. Binding to specific receptors located in the outer membrane, translocation across the cell envelope, and cytotoxic activities are dependent on the central, N-terminal, and C-terminal domains, respectively (14, 132, 140, 187, 441, 495, 496). It is noteworthy that colicins are macromolecules containing an average of 500 to 600 residues, with a maximum of 697 residues for colicin D and a minimum of 271 residues for colicin M (54).
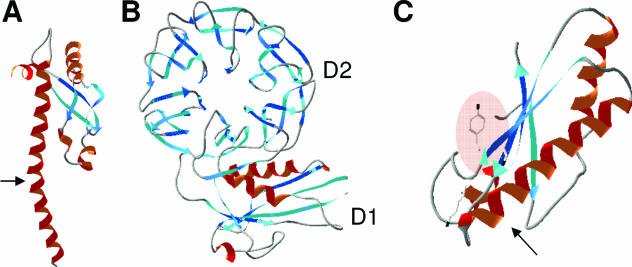
The minor coat protein g3p, also called the adsorption protein of Ff phages (71), contains a central domain and an N-terminal domain devoted to pilus reception and translocation essential for phage infection, respectively (612). Its C-terminal domain is required for membrane anchoring and further capsid assembly (Fig. 4).
FIG. 4.
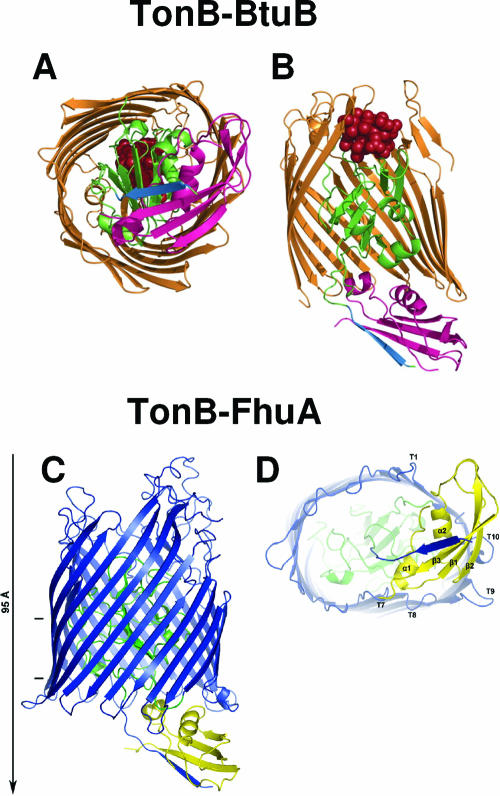
Colicin and g3p domain organization and crystal structures. (A) Schematic representation of the domains involved in reception (R, N2), translocation (T, N1), and activity (A) (for colicins) or anchoring (An) (for g3p). Positions of residues surrounding the various domains are indicated together with hinge regions connecting the domains. Colicin A, N, and g3p domains are presented. (B) Structures of the C-terminal domains of colicin A (ColA-ct) (PDB accession number 1COL; resolution, 2.40 Å) and colicin E9 (ColE9-ct) (PDB accession number 1EMV; resolution, 1.70 Å) and the structures of intact colicin E3 (PDB accession number 1JCH; resolution, 3.02 Å), colicin Ia (PDB accession number 1CII; resolution, 3.00 Å), colicin B (PDB accession number 1RH1; resolution, 2.50 Å), colicin N (PDB accession number 1A87; resolution, 3.10 Å), and g3p (reception-N2 and translocation-N1 domains) (PDB accession number 1G3P; resolution, 1.46 Å). The Im9 and Im3 proteins bound to colicin E9 and colicin E3, respectively, have been removed from the complex structures. The TonB box sequences present in the first β-strand of colicin B and in the first residues of colicin Ia are indicated. The receptor binding domains of colicin E3 and E9 (76 residues from colicin E9) (512) or corresponding to a cyclic peptide of 34 residues from colicin E3 (463) are indicated by circles on the colicin E3 three-dimensional structure.
Specific Functions of Colicin Domains
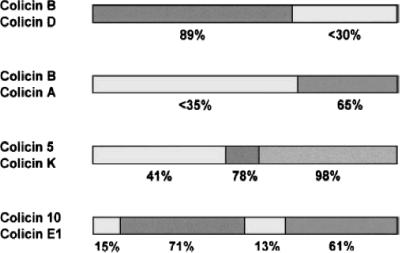
The first step of colicin action consists of binding to a specific receptor. Biochemical analyses of proteolytic fragments of colicins have been used to map the peptide domain involved in binding. The receptor binding domain is located within the central region of all colicins (61, 495). Both the N-terminal and the central domains of colicin B share 90% identity with those of colicin D, since both colicins depend on the same receptor, FepA, and on the same translocator proteins, TonB-ExbBD (see Fig. 23). However, the central receptor binding domains of these two colicins (residues 130 to 291) share 30% identity with those of colicins A, E2, E3, E6, E7, and E9, which require the BtuB gated receptor (156). The minimal receptor binding domain of colicin E9, which confers cell protection against colicin E9, has been identified to contain residues 343 to 418 (512). The crystal structure of a shorter receptor binding sequence has been determined recently by using a synthetic cyclic peptide of 34 residues corresponding to residues 366 to 399 of colicin E3. This peptide has been shown to bind to BtuB with a high affinity in the presence of calcium and to overlap with the binding site of cyanocobalamin, the natural BtuB substrate (463) (see also the BtuB-ColE3 costructure shown in Fig. 5A). However, the receptor binding step appears not to be essential for g3p and colicin E3 imports, since neither Ff phage infection nor colicin activity is abolished in some receptor mutants (565, 625). Moreover, in low-ionic-strength buffers, the activity of a colicin A mutant lacking its receptor binding domain is similar to that of full-length colicin A incubated with bacteria that either possess or lack BtuB (22, 96). For the TonB-dependent phages, adsorption of T1 and φ80 on the FhuA receptor has been shown to be irreversible on wild-type cells while becoming reversible in the absence of TonB (247). All these results demonstrate that both the Tol and Ton systems are able to import colicins that exert either a membrane or a cytoplasmic activity and that the translocation mechanism is the essential step for the passage through the outer membrane.
FIG. 23.
Pairwise comparisons of pore-forming colicin protein sequences. Values below each comparison indicate the percent sequence identity for the region indicated. Colicins are not drawn to scale.
FIG. 5.
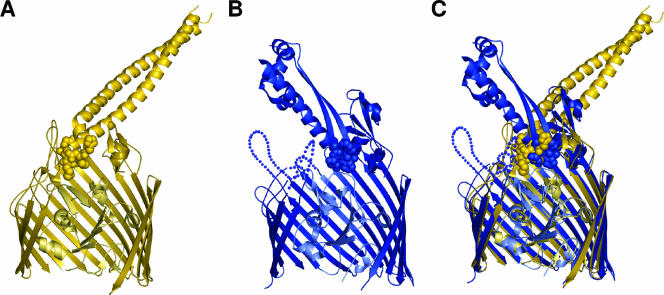
Ribbon diagrams of BtuB-colicin E3 and Cir-colicin Ia. (A) BtuB-colicin E3 complex (371) (PDB accession number 1UJW; 2.75 Å). BtuB and part of the receptor binding domain of colicin E3 are colored dark gold, while the plug domain of BtuB is colored light gold. Several β-strands have been removed to show the plug domain more clearly. Colicin E3 is seen at the top of the figure as a long coiled coil with a loop between helices that interacts with extracellular loops of BtuB. Residues of this loop that participate in receptor recognition are shown in the space-filling representation. (B) Cir-colicin Ia complex (Buchanan et al., unpublished). Cir and colicin Ia are shown in dark blue, with light blue representing the plug domain. Again, several β-strands have been removed to visualize the plug domain. The receptor binding domain of colicin Ia (at the top of the figure) consists of two β-strands wrapped around a long α-helix, and the space-filling representation depicts colicin Ia residues that interact with Cir. Dotted lines indicate residues not seen in the crystal structure due to high mobility (disorder). (C) A superposition of the two complex structures shows that the tips of colicin E3 and colicin Ia extend to about the same depth in the respective transporters. Each colicin binds its transporter at an angle of approximately 45° with respect to the lipid bilayer although from opposite directions. Courtesy of Petra Lukacik, reproduced with permission.
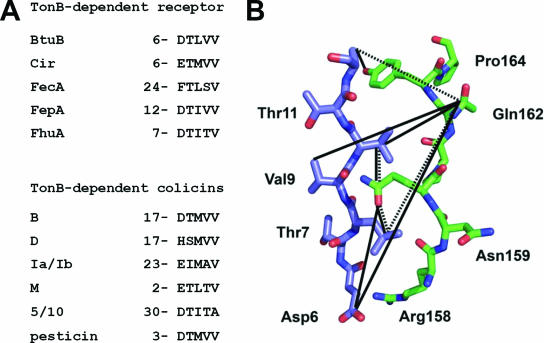
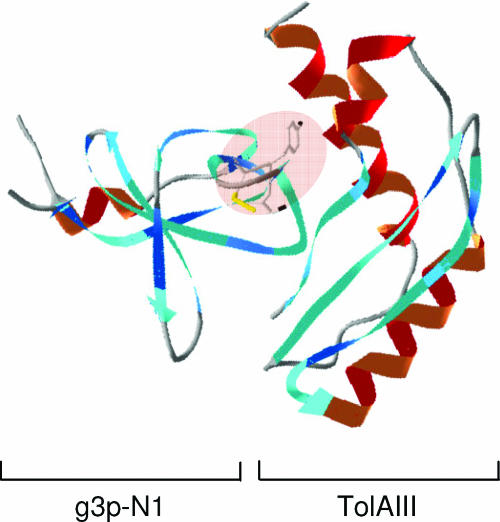
Homologies between the Tol and Ton systems and the domains responsible for translocation have been further delineated using protein deletions and protein fusions. Domains of colicins A and E1 required for the import machinery have been defined by colicin A-colicin E1 fusions, which allowed the mapping of the OmpF, TolB, and TolR binding sequences of colicin A and the TolC binding region of colicin E1 in the N-terminal region (21). Further results demonstrated the direct interaction of the N-terminal domain of colicins with the Tol proteins: colicins A and E1 with TolA (23), colicins A and E3 with TolB (42, 43), and colicin A with TolR (313). Using biochemical methods, colicin N domains have also been defined. Probably due to its OmpF requirement for both receptor and translocation, it possesses the smallest reception and translocation domains (67 and 115 residues, respectively), while its active domain has a length similar to those of other pore-forming colicins (179, 537). A point mutation that is localized in the N-terminal domain of colicin E3 abolishes colicin activity (187, 462) and has contributed to the identification of the TolB box, which is found in various TolB-dependent colicins and is required for the interaction of colicin with TolB (42, 72, 313) (see Fig. 8C). The specific TonB box determinants map to the N-terminal domains of TonB-dependent colicins as well as to the N termini of TonB-dependent receptors (259, 456, 464) (see Transit through the Periplasm and Fig. 11A). The sequences of colicins K, 5, and 10, which require the Tsx receptor, have also been compared. While their receptor binding and C-terminal pore-forming domains present strong sequence identity, their N-terminal domains were found to differ, since they use either the Tol or Ton system (515) (see Fig. 23).
FIG. 8.
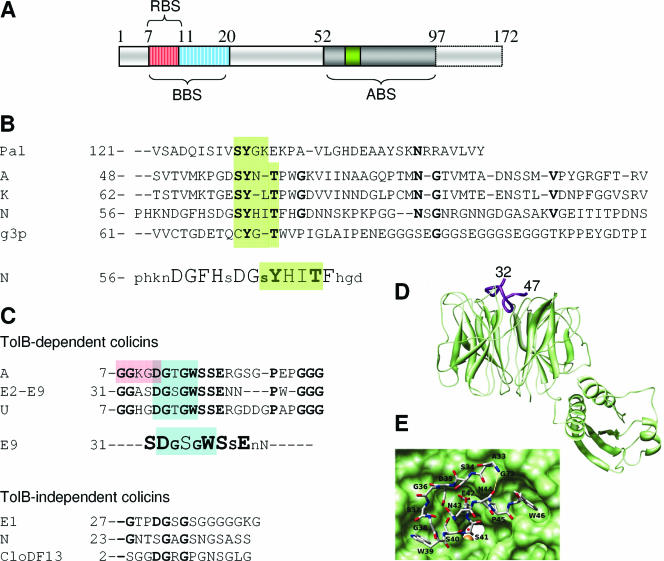
Tol binding sequences. (A) Schematic illustration of the colicin A N-terminal domain and the three binding sequences identified (RBS, TolR-binding sequence, residues 7 to 11; BBS, TolB-binding sequence, residues 7 to 20; ABS, TolA-binding sequence, residues 52 to 97) (42, 43, 313). (B) Sequence alignments of the Pal, colicins A, K, and N, and g3p TolA binding sequences. Conserved residues are in boldface type, and the TolA binding motif with the conserved and critical tyrosine residue is highlighted in green (78, 527). Lower panel, results from alanine-scanning mutagenesis of residues 56 to 75 of colicin N (228). Uppercase letters, essential residues; lowercase letters, nonessential residues. (C) Sequence alignments of the TolB-dependent (upper panel) and -independent (lower panel) colicin TolB binding sequences. Conserved residues are in boldface type. Colicin TolB boxes are highlighted in blue, whereas the colicin A TolR binding sequence is framed. Note that the WSSE motif present in TolB-dependent colicin TolB binding sequences is not found in TolB-independent colicins. Middle panel, results from alanine-scanning mutagenesis of residues 34 to 44 of colicin E9 (248). Large letters, critical residues; small letters, nonessential residues; medium letters, mutations decreasing but not abolishing TolB-colicin E9 N-terminal domain affinity. (D) Crystal structure of the TolB protein (green) with the residues 32 to 47 of colicin E9 (purple) (417) showing localization of the TolB binding sequence at the entrance of the β-propeller. (E) Top view of the same complex, which emphasizes the binding site. The colicin E9 peptide is represented in a backbone. (Panels D and E are reprinted from reference 417 with permission of the publisher. Copyright 2006 National Academy of Sciences U.S.A.)
FIG. 11.
The TonB box. (A) Sequence alignments of TonB boxes. Shown are sequence alignments of the BtuB, Cir, FecA, FepA, and FhuA (upper panel) and of the colicin B, D, Ia/Ib, M, and 5/10 and pesticin TonB boxes (lower panel). Conserved residues are in boldface type. (B) BtuB TonB box interactions with TonB. Shown are in vivo cysteine disulfide cross-linking data for the TonB region around Gln160 (green) and the BtuB TonB box (blue). The antiparallel interaction between the β-strands is shown, as observed in the crystal structure. (Reprinted from reference 588 with permission of AAAS.) Solid lines indicate cross-links observed between TonB and BtuB (67). Dotted lines indicate cross-links observed between TonB and FecA, with the FecA TonB box aligned with the BtuB TonB box (492).
The lethal activities on specific cellular targets correspond to the third step of the colicin mode of action. The C-terminal domains of colicins have been isolated and were shown to carry either a pore-forming or a nuclease activity (64, 441, 496). Some nuclease domains (495, 617) have been shown to possess channel activity (467). A colicin-producing cell synthesizes a specific nuclease inhibitor, called an immunity protein, which associates with the C-terminal toxic domain of a colicin in order to protect the colicinogenic bacteria against the produced colicin. However, the 16S rRNase active domains of both E3 and E6 colicins are well conserved and differ only in a small number of residues responsible for the interaction of colicin with its specific immunity protein (2) (see Fig. 24). Similarly, pore-forming domains of colicin A and colicin B share 60% identity but differ in specific transmembrane (TM) determinants required for the association with their cognate immunity protein (217).
FIG. 24.
Graph indicating the average number of total nucleotide replacements between pairs of nuclease-type colicin gene clusters (colicin pairs E2/E9 and E3/E6). Most of the divergence between colicins occurs in the immunity region of the gene cluster (composed of the immunity gene and the immunity binding region of the colicin gene).
Homologies between the import mechanisms of colicins and g3p have been determined by studying the mode of action of a g3p-colicin E3 hybrid protein (301). The recombinant molecule carrying the reception and translocation domains of g3p followed by the 16S rRNase domain of colicin E3 is active on sensitive cells harboring the F pilus and the TolAQR proteins. This demonstrates that the F pilus replaces the outer membrane protein BtuB and that the g3p-colicin E3 hybrid is able to kill sensitive cells as wild-type colicin E3 does.
Three-Dimensional Structures of Colicins
Structural knowledge on colicins started with analytical ultracentrifugation experiments that demonstrated the elongated shape of colicins. Axial ratios ranging from 7 to 20 were found for colicins E2, E3, Ia, and Ib, with I colicins being more elongated (358). Further hydrodynamic analyses of colicin A have shown that it forms tetramers at acidic pHs and that while full-length colicin A is an asymmetric molecule, the isolated translocation domain is globular (102, 345). The structure of the C-terminal domain of colicin A has been solved by X-ray crystallography, in part due to the emergent technique of site-directed mutagenesis. Cysteine-containing variants were constructed to fix mercurial derivatives. The pore-forming domain of the colicin A structure was solved and refined at 2.5-Å resolution. It represents the first structure of a soluble protein containing a hairpin formed by two hydrophobic α-helices completely buried within a bundle of eight α-helices (502) (Fig. 4 and see Fig. 16). This hairpin inserts into the membrane, a process accompanied by tertiary structural changes in the absence of any secondary structure modification (503). Shortly thereafter, the structure of the colicin E1 C-terminal fragment was determined and found to be similar to that of colicin A (678) (see Fig. 16). The first entire colicin to be crystallized was colicin Ia. Its structure resolution reveals a high α-helical content and an elongated Y shape of the molecule in which three regions can be distinguished (222). The structure was further refined at 3.0 Å, and two 160-Å coiled-coil α-helices have been found to separate the three colicin domains (675) (Fig. 4). Another structure of an entire colicin corresponding to the pore-forming, Tol-dependent, and BtuB-independent colicin N at a resolution of 3.1 Å has been determined (650). However, the N-terminal 90 residues are missing. The N-terminal region containing a β-sheet wrapped around the long α-helix forms the receptor binding domain, while the C-terminal domain corresponds to the 10 helix bundles similarly to other pore-forming colicins (see Fig. 16). Because colicin N is the smallest pore-forming colicin, the absence of the 90 N-terminal residues corresponding to the whole translocation domain is striking, although the lack of resolution in portions of N-terminal domains is a general feature of colicin crystal structures (Fig. 4).
FIG. 16.
Comparison of the structures of the pore-forming domains of colicins A, N, B, Ia, and E1. Each molecule is shown in two orientations: one approximately parallel to the hydrophobic hairpin (upper) and the other perpendicular to it (lower). The hydrophobic helices are red, whereas the other eight helices are blue. Helix numbers are shown explicitly for colicin A. The overall similarity in the folds of these molecules is apparent.
After the determination of the structures of pore-forming colicins, the three-dimensional structures of enzymatic colicins were determined. The cocrystals formed with the soluble DNase domain of colicins E7 and E9 bound to their immunity proteins, Im7 and Im9, respectively (343, 346), have been solved, and the inhibitory actions of the Im proteins have been deduced from the residues in interactions (see Fig. 21 and Colicin Activities). The comparison of the interactions of both complexes further indicates that the immunity proteins interact with their cognate DNase domains differently (369). The first complex of an entire colicin bound to its specific immunity protein to be solved was that of colicin E3 and its 16S rRNase activity inhibitor Im3 (607). As found in colicin Ia, colicin E3 possesses two 100-Å antiparallel coiled-coil α-helices that separate the three domains. According to its high binding affinity for colicin E3, the Im3 protein is found in tight association with the nucleasic C-terminal domain but is also found to interact with the translocation domain. As previously observed, the N-terminal residues (residues 1 to 83) of the colicin E3 translocation domain exhibit no electron density in the crystal, indicating the presence of a disordered region. The most recent colicin structure solved at 2.5 Å corresponds to that of colicin B (267). As found in other colicins, colicin B is an elongated molecule harboring a central α-helix of 74 Å that separates both the receptor and the translocation domains from the C-terminal pore-forming domain. Here, the dumbbell structure of colicin B differs from those of colicins Ia and E3 (which form a Y shape in which the translocation, reception, and active domains are distinct) but resembles colicin N, which contains a single coiled-coil helix (Fig. 4). While the N-terminal residues of colicin B are not visible in the crystal structure, the TonB box (residues 11 to 28) is located in a β-strand, tightly associated with polar residues from a parallel β-strand, leaving the flanking residues (positions 1 to 10 and 29 to 43) highly flexible. In contrast to colicin B, the colicin Ia TonB box is present within an antiparallel α-helix bundle. In relation with structure resolutions, no three-dimensional information on Tol binding sequences of group A colicins is currently available (except the ColE932-47 peptide) (417), as they are localized within disordered glycine-rich regions. Indeed, sequences of the N-terminal translocation domains contain ∼20 to 40% glycine residues for group A colicins and ∼10 to 20% glycine residues for group B colicins.
FIG. 21.
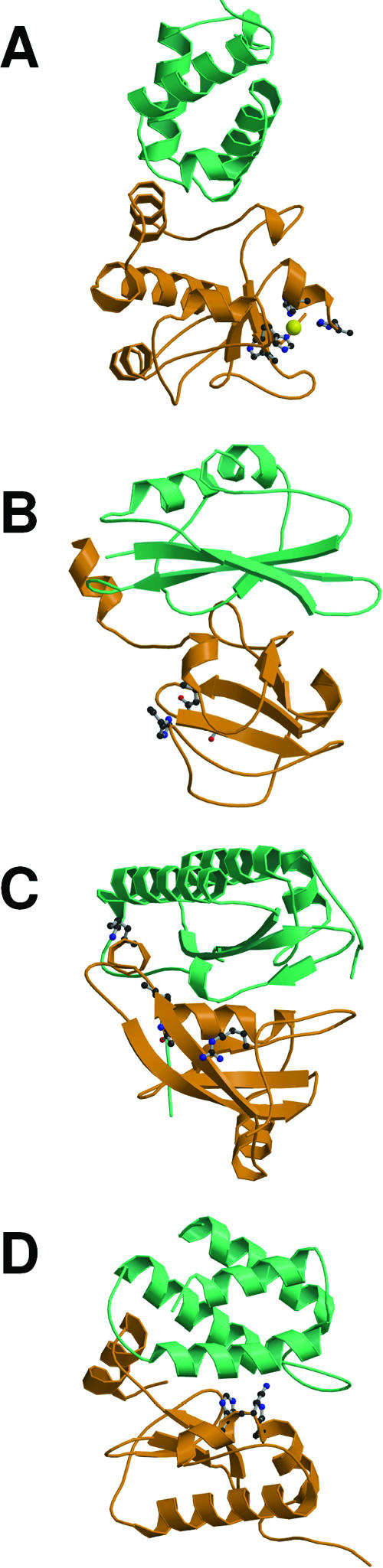
Structures of colicin nuclease domains bound to their cognate immunity proteins highlighting exosite (top) and active-site (bottom) binding by their neutralizing inhibitors. (A) ColE9 DNase-Im9 complex (PDB accession number 1BXI). (B) ColE3 rRNase-Im3 (PDB accession number 1E44). (C) ColE5 tRNase-Im5 (PDB accession number 2FHZ). (D) ColD tRNase-ImD (PDB accession number 1V74). Colicin nucleases are shown in orange, and their Im proteins are shown in cyan. Each structure identifies key active-site residues and, in the case of the DNase, the catalytic metal ion (see the text for details). Courtesy of Irina Grishkovskaya, reproduced with permission.
Although the entire colicin A structure remains unsolved, it appears that its translocation domain lacks a secondary structure as shown by circular dichroism studies and by the poor resolution of its heteronuclear nuclear magnetic resonance (NMR) spectrum (147). Structural disorder is found in all the N-terminal domains of group A colicins (colicin N [650], colicin E3 [607], and colicin E9 [125]) and also in colicins B and Ia, as judged by the absence of electron density for part of the 43 and 22 N-terminal residues, respectively. This suggests, as for other disordered proteins, a propensity of these domains for molecular interactions with different partners. The structure of the translocation domain of colicin E7, lacking the 60 N-terminal residues, has recently been found to superimpose with homologous regions of colicins E3 and B, suggesting similar mechanisms for the import of group A and group B colicins (114). According to protein flexibility, previous results demonstrated that the disordered N-terminal domains of colicins N and E9 are stabilized upon interactions with TolA and TolB, respectively (5, 632). In contrast, the disordered colicin A translocation domain has been shown to remain disordered upon TolA binding, but this interaction induces a disordered TolA structure. The global structure of TolA is maintained upon interaction with the isolated N-terminal domain of g3p (g3p-N1) (147). Contrary to the flexible colicin translocation domains, the g3p-N1 domain is a structured peptide of 68 residues, containing two disulfide bridges, formed by one N-terminal α-helix and five β-strands linked through glycine clusters to the N2 domain (the receptor binding domain) (Fig. 4). As suspected, the glycine clusters are not resolved in the crystal, while the N2 domain presents structural similarities with N1 and is formed by eight β-strands and one α-helix (420). Concerning the active domains of colicins, the molecular dynamic simulations obtained for the C-terminal domain of colicin E9 have demonstrated its flexibility and its ability to form channels in lipid bilayers (467), while previous results demonstrated the molten-globule state of the pore-forming domain of colicin A as an intermediate membrane insertion step (640). Surprisingly, no globular or flexible structure appears to be involved in the receptor binding of E colicins. Recent data indicate the critical role of the coiled-coil and loop-rigid regions of colicin E3 and of residues bound to the receptor (463). Thus, the N-terminal domain of colicin E3 present in the BtuB-colicin complex interacts with the outer membrane porin OmpF, allowing colicin to bind simultaneously two OM proteins (275, 371, 694). Obtaining further structural information on colicins interacting with their receptor and/or import partners is necessary for a complete understanding of the import of macromolecules across membranes.
COLICIN RECEPTION
Recognition of the Receptor at the Bacterial Cell Surface
Both group A and group B colicins target E. coli cells by interacting with specific outer membrane proteins (54, 68, 387). In group A, colicins A and E1 to E9 target the TonB-dependent vitamin B12 transporter BtuB (Table 1 and Fig. 3). Colicin K binds to the nucleoside transporter Tsx, while colicin U interacts with OmpA. All of these colicins require a second outer membrane protein for translocation (usually OmpF, but TolC is used by colicin E1), implying that the initial binding interaction serves to position the colicin on the surface of the cell. The primary receptor is not thought to participate in the translocation of the colicin across the outer membrane. The requirement for two outer membrane proteins is not absolute, however. BtuB can be bypassed by incubating cells in buffers of low osmotic strength. Under these conditions, pore-forming colicins from group A (but not nuclease colicins) can kill cells lacking BtuB, implying that OmpF can be used as the sole receptor (96). The only group A colicin requiring a single outer membrane protein under all conditions is colicin N, which uses OmpF for recognition and transport. All group A colicins also require some combination of Tol proteins for translocation across the outer membrane (see Translocation through the Outer Membrane below).
Group B colicins use the Ton system for transit, and most of them also bind to specific TonB-dependent transporters in the outer membrane. In these systems, colicin appears to be bound and transported by a single outer membrane protein. For example, ferric enterobactin receptor FepA recognizes and transports colicins B and D, ferrichrome receptor FhuA is used by colicin M, and colicins Ia and Ib require colicin I receptor Cir for recognition and transport. For these colicins, no secondary outer membrane protein that could function as a transporter has been identified (54; S. K. Buchanan, P. Lukacik, S. Grizot, R. Ghirlando, M. Ali, T. J. Barnard, K. S. Jakes, P. K. Kienker, and L. Esser, unpublished data). The only group B colicins thought to use two outer membrane proteins are colicins 5 and 10, which bind to Tsx and use TolC as a transport channel.
Binding to the Receptor
While the binding of colicins to receptors has been characterized by genetic, biochemical, and biophysical approaches (54, 68, 387), two crystal structures of outer membrane proteins in complex with their cognate colicins that illustrate specific, detailed binding interactions are now available. BtuB was cocrystallized with the 135-residue receptor binding domain of colicin E3, resulting in a 2.75-Å structure (371), and Cir was cocrystallized with a 102-residue receptor binding domain of colicin Ia (Buchanan et al., unpublished), yielding a structure to 2.5-Å resolution. Full-length structures of colicins E3 (607) and Ia (675) were also solved separately, which allowed the modeling of the presumed in vivo interaction between these full-length colicins and their corresponding receptors (371; Buchanan et al., unpublished). Colicin E3 belongs to group A, using BtuB as a cell surface receptor (i.e., the natural transport function of BtuB is not used here), whereas colicin Ia belongs to group B and uses Cir for both binding and transport. Therefore, one can compare these structures to discover similarities and differences between group A and group B colicin recognition by related outer membrane proteins.
Both BtuB and Cir are TonB-dependent outer membrane transporters that share a similar architecture. They naturally function to transport small molecules, in this case, vitamin B12 and linear catecholates, respectively, into the periplasm (199, 676). TonB-dependent transporters are composed of two domains (Fig. 5): a 22-stranded β-barrel spans the membrane, and a globular domain (formed from the N-terminal ∼150 residues of the protein) is inserted inside the barrel, extending from the periplasm to the extracellular surface. The globular domain is referred to as a “cork” or “plug” because it completely fills the barrel lumen, preventing the transport of even small molecules in the ground state. At the N terminus of the plug domain (protruding into the periplasm) is a region of five residues called the TonB box. This region interacts with TonB during the transport process, and structures of TonB in complex with BtuB and FhuA have recently been described (507, 588). The transport of small molecules requires a functional TonB-ExbB-ExbD complex and energy derived from the proton motive force (PMF) (530) (see Translocation through the Outer Membrane). No one knows exactly what occurs when a TonB-transporter complex is energized, but something must happen to the plug domain to create a transient pore in the outer membrane. The most likely hypotheses are that the plug domain undergoes a conformational rearrangement within the barrel to create a small channel (200) or that the plug domain is pulled partially or completely out of the barrel by TonB (194).
At the extracellular surface, the 22 β-strands of the transporter are connected by 11 long extracellular loops. The extracellular loops are very mobile and have been observed in crystal structures to undergo large conformational changes when the receptor domain binds to BtuB (198, 691; Buchanan et al., unpublished). Large movements have also been observed by spectroscopic methods (312). Not surprisingly, colicins E3 and Ia bind primarily to these long extracellular loops on their respective receptors, and binding specificity is conferred through the individual residues found in these loops (Table 2). However, the binding sites are composed very differently for colicins E3 and Ia, and receptors respond differently to colicin binding.
TABLE 2.
Receptor-colicin interactions for Cir-colicin Ia and BtuB-colicin E3a
| Loop | Receptor region(s)b
|
|||
|---|---|---|---|---|
| Cir | Colicin Ia | BtuBc | Colicin E3 | |
| Plug | R116 | E357 | ||
| L5 | N348 | E357 | ||
| L7 | S434 | D358 | ||
| L7 | R436 | D362, P353, N352, D350, E369 | ||
| L8 | I488 | A315 | ||
| L8 | S489 | T317 | ||
| L8 | R490 | T317, K319, F314, D358 | ||
| L8 | T491 | K319 | ||
| L8 | Y520 | R313 | ||
| Plug | N77 | M383 | ||
| L3 | Y249 | R388 | ||
| L3 | L260 | R388 | ||
| L5 | E350 | Q398, T402 | ||
| L7 | Y422 | Q398 | ||
| L7 | Y425 | W390 | ||
| L8 | Y466 | N376 | ||
| L8 | D468 | H380 | ||
| L9 | R517 | D381 | ||
For Cir-colicin Ia, all interactions are less than 3.2 Å apart; for BtuB-colicin E3, the interactions listed occur at distances of less than 3.4 Å. Note that Cir uses primarily multiple interactions from two extracellular loops (L7 and L8) to bind colicin Ia, whereas the colicin E3 binding site is more distributed on the BtuB extracellular surface, involving five extracellular loops (L3, L5, L7, L8, and L9).
In TonB-dependent transporters such as Cir and BtuB, there are 22 transmembrane β-strands connected by 11 extracellular loops such that L1 connects strands 1 and 2, L2 connects strands 3 and 4, etc. Each transporter also contains a 140- to 150-residue N-terminal domain designated as “plug” here and in the text.
Residue numbering for BtuB includes the signal peptide (as it does for Cir), according to Swiss-Prot conventions (http://ca.expasy.org/).
Figure 5A shows the structure of BtuB in complex with the 135-residue receptor binding domain (R135) of colicin E3 (371). The short loop connecting the two long α-helices in colicin E3 sits slightly above the natural cobalamin binding pocket, making contacts with five extracellular loops and a single residue from the BtuB plug domain (Table 2). In this structure, several loops contribute one or two interactions, forming a binding site that is dispersed around the BtuB β-barrel. The distributed nature of the binding site is reflected by the lack of measurable effects of single-residue changes in colicin E3, and even the double mutant M383A-W390A diminishes colicin toxicity by only 50- to 100-fold (607). Despite the weak individual interactions, the BtuB-R135 complex exhibits a dissociation constant (Kd) of 0.5 to 1 nM; the Kd for full-length colicin E3 is estimated to be 100 times smaller, reflecting tighter binding (371, 694). Colicin E3 buries 1,533 Å2 of its surface upon interacting with BtuB and binds at an angle of about 45° with respect to the lipid bilayer. When colicin E3 binds to BtuB, only small conformational changes are observed in BtuB, reflecting its role as a cell surface receptor and its (presumed) lack of participation in transport across the outer membrane.
In contrast, colicin Ia binds to Cir using very different interactions and evokes changes in Cir not seen in BtuB (Fig. 5B). In Table 2, it is immediately clear that Cir uses only two extracellular loops (loop 7 [L7] and L8) to form most of the binding interactions with colicin Ia. A single interaction is contributed from loop 5, and one is contributed from the plug domain, but 16 of the 18 observed hydrogen bonds come from loops 7 and 8 (Buchanan et al., unpublished). Another difference in BtuB-colicin E3 is that the majority of these interactions come from just two Cir residues: R436 from loop 7 and R490 from loop 8; each binds multiple colicin Ia residues (residue numbering for Cir and BtuB includes the signal peptide according to Swiss-Prot conventions [http://ca.expasy.org/]). Although these two arginine residues have not been studied to date, we predict that a double mutant would have a considerable effect on colicin Ia binding. The dissociation constant has been estimated to be 0.1 nM in vivo (357), similar to that measured for BtuB-colicin E3. The Cir-colicin Ia interaction buries 990 Å2 of surface area, which is somewhat less than that of BtuB-colicin E3. Interestingly, colicin Ia binds Cir to the lipid bilayer at an angle of about 45°, very similar to what is observed for BtuB-colicin E3.
A superposition of the two structures (Fig. 5C) shows that the two colicins approach their targets at similar angles but from opposite directions. Although no structure is available for Cir bound to its natural metal chelate, Fig. 5C shows that colicins Ia and E3 sit at approximately the same depth in the natural ligand binding pocket, so in this respect, receptor-colicin binding interactions are very similar. A major difference in interactions between Cir-colicin Ia and BtuB-colicin E3 is that Cir undergoes large conformational changes in two extracellular loops (L7 and L8) when colicin Ia binds (Buchanan et al., unpublished). Specifically, L7 and L8 move outward by 37° compared to Cir in its ground-state structure. This large movement, not seen for BtuB, may reflect the use of Cir as both cell surface receptor and transporter of colicin Ia.
At the periplasmic side, it has been proposed that substrate binding induces a rotation of residues 6 and 7 of the BtuB TonB box of 180°, allowing the interaction with TonB in the periplasm (see Transit through the Periplasm) (117). Recently, an in meso structure of BtuB was solved at 1.95 Å (114a). Comparison of this structure with previous BtuB structures (117) suggests that while the strands of the β-barrel are relatively rigid, the loop regions between strands are highly mobile. Differences in structures in the conformation of the TonB box and the amino-terminal 5 amino acids of BtuB also emerged and raise the question of whether, as postulated previously (117), a substrate-induced 180° rotation of TonB box residues 6 and 7 is a unique signaling event.
Competition with the Natural Ligand
Colicins exhibit the same type of receptor specificity that small molecules do. A number of colicins have been shown to compete with the corresponding natural ligand for receptor binding, indicating that the binding sites for colicins and metal chelates overlap to some extent. For example, the addition of ferric enterobactin to cells expressing FepA protects them from killing by colicin B (672); similar experiments were done using ferrichrome, FhuA, and colicin M (672); cobalamin, BtuB, and E-type colicins (158); or spermine, OmpF, and colicin N (57). However, the nature of competition between colicins and natural metal chelates differs: cobalamin can rescue colicin (A or E)-treated bacteria, suggesting that this metal chelate displaces a bound colicin from BtuB (83). Ferric enterobactin does not displace colicin B on cells, although FepA binds ferric enterobactin with a Kd of 24 nM, compared to a Kd of 185 nM for colicin B. This may be due to the very slow dissociation of colicin B from FepA (508).
While these experiments confirm competition for binding, they do not indicate the precise residues involved in binding colicins versus metal chelates. Currently, the only system for which this level of detail exists comes from structures of BtuB bound to either cobalamin (117) or colicin E3 (371). Of the 13 residues involved in cobalamin binding, 5 of these also participate in binding colicin E3: Y249, N296, T309, R517, and Y599. Instead of being distributed throughout the cobalamin binding pocket, the colicin E3 binding residues occur on one side of the pocket, near the region where two calcium ions are bound. Now that the structure of colicin Ia bound to Cir is known (Buchanan et al., unpublished), it will be interesting to determine the structure of Cir bound to a natural ligand to see if principles of colicin and small-molecule discrimination are conserved.
Colicin Cleavage at the Cell Surface
A number of laboratories have characterized the cleavage of colicins A, E1 to E4, Ia, and Ib at the outer membrane (44, 59, 95, 97, 447a). Colicin proteolysis was attributed to the outer membrane protease OmpT and resulted in fragmentation into a number of smaller products upon binding to the cell surface. An ompT deletion mutant does not degrade the targeted colicins, but the mutant remains sensitive to killing (97). Although colicin cleavage at the cell surface may occur in vivo, it does not seem to be necessary for killing. ompT deletion strains are killed by colicin Ia with the same efficiency as that of strains expressing ompT (Buchanan et al., unpublished), and no proteolysis of colicin Ia was observed when incubated with ompT deletion strains (S. K. Buchanan, unpublished results). However, some nuclease colicins undergo an essential cleavage reaction after outer membrane translocation, releasing the nuclease domain (157, 585). This mechanism is discussed in “Enzymatic Colicins” below.
Energy Requirements for Receptor Binding
Colicins bind to their target outer membrane proteins without energy input, as demonstrated for colicin M and FhuA. Colicin M adsorbs to cells under energy-depleted conditions but kills only when the cells are energized by glucose and respiration (50). The lack of an energy requirement for colicin binding can also be visualized in the crystal structures of BtuB-colicin E3 and Cir-colicin Ia, which show that the conformation of the receptor binding domain of each colicin changes very little upon receptor binding (in the case of Cir-colicin Ia, there is no detectable change in colicin conformation) (Buchanan et al., unpublished). The binding of colicin E9 to BtuB has been characterized by isothermal titration calorimetry, displaying a (favorable) large negative free energy of binding (275). These results are also supported by the ease of making stoichiometric receptor-colicin complexes in vivo or in vitro that can be purified by size exclusion chromatography (275; Buchanan et al., unpublished). All binding experiments indicate that an input of energy is not required for complex formation.
A longstanding assumption is that colicins must unfold during the translocation process, and this event would require energy. The crystal structures suggest that the translocation domain (required for cell penetration) is positioned high above the membrane when a colicin initially binds its receptor and must undergo a large conformational change to come into close proximity with the outer membrane (371; Buchanan et al., unpublished). The notion that unfolding facilitates killing has been shown experimentally: colicin A was shown to kill cells more quickly when denatured with urea than the natively folded protein (24), and disulfide bonds engineered into the pore-forming domain delayed translocation (165). Similarly, a disulfide-bonded colicin E9 mutant, linked near the end of its coiled coil (close to the nuclease domain), was inactive unless a reductant was present (513). Colicin unfolding would presumably require energy, but this is likely to occur sometime after binding to the receptor.
Release of Immunity
Colicins using nuclease domains to exert cytotoxicity are secreted with tightly bound immunity proteins to protect the host cell from self-destruction. One example is Im9, which binds the DNase domain of colicin E9 and displays a Kd for the complex of 10−16 M (666, 667) (see “Nuclease-specific immunity proteins” below). This tight binding presents a problem for colicin activity—when and how is the immunity protein released? Although unfolding is thought to occur at some point in the translocation mechanism and could potentially facilitate the release of the immunity protein, this does not seem to occur upon receptor binding. A small amount of unfolding was seen in the BtuB-colicin E3 crystal structure at the two ends of the coiled-coil (371), but isothermal titration calorimetry experiments on BtuB interacting with a colicin E9 disulfide-bonded mutant (513) and wild-type colicin E9 yielded almost identical results (275), suggesting that colicin E9 does not significantly unfold upon binding its receptor. Therefore, the immunity protein is probably released into the medium sometime after receptor binding. Likewise, a colicin E9-Im9 complex was able to bind to BtuB and recruit OmpF, forming a BtuB-OmpF-colicin E9 translocon, without displacement of Im9 (275). Recently, in vitro experiments with colicin E3 demonstrated that the cognate immunity protein can be removed without unfolding by using anion-exchange chromatography (698). These results suggest that unfolding is not necessarily required for immunity protein removal. A recent report showed that the release of the immunity protein of colicin E2 requires the presence of the translocation machinery, including the periplasmic and inner membrane Tol components (165a). The dissociation of the colicin E3-immunity complex is dependent on the translocation of the colicin N-terminal domain (662). The crystal structure of the colicin E3-immunity complex showed that the immunity protein interacts with both the catalytic and the translocation domains (607). The loss of contacts with the translocation domain during the translocation process might have weakened the colicin-immunity complex and increased the dissociation rate, leading to immunity dissociation. In contrast, in vitro experiments suggested that the release of the cognate immunity from the colicin E3-immunity complex allows the binding of colicin to the OmpF translocon (698). However, immunity protein release is achieved ∼20 min after binding to the outer membrane receptor, the time required for the enzymatic colicins to reach their target (165a, 364, 645).
Reception of Bacteriophages
Tol-dependent Ff filamentous bacteriophages adsorbs on F+ E. coli cells by interacting with the tra/trb-encoded F pilus. This step involves the central domain (N2) of the minor coat protein g3p located at the tip of the phage capsid (182, 234). This domain recognizes the major subunit of the F pilus, TraA (145). Treatment of cells with purified g3p-N2 (but not with g3p-N1, the translocation domain) prior to bacteriophage attachment delays the infection process. Further alanine-scanning studies showed that residues involved in pilus binding are located on the outer rim of the three-dimensional N2 domain (144). Phage attachment is mediated by two distinct domains of the TraA pilin located at both termini (211, 432). Differently, the IKe filamentous bacteriophage adsorbs on N pili (encoded by IncN plasmids) via the g3p protein. Interestingly, sequence comparisons of F- and N-specific g3p proteins show extensive sequence arrangements, explaining the host specificity (511). Ff virions displaying a g3pIKe protein at the surface of their capsid attach onto N pili (180, 181). However, the phage particle is not infectious, probably because of the unconcerted action of g3p with another capsid component. In Vibrio cholerae, CTX-phi bacteriophage infection requires the toxin-coregulated pilus. Adsorption is initiated upon binding of g3pCTX (OrfU) to the pilus, as elegantly shown using g3pfd and g3pCTX fusions. An E. coli-specific fd phage displaying the fusion comprising g3pCTX-N1N2 and g3pfd-N3 domains was shown to infect V. cholerae in a toxin-coregulated-pilus-dependent fashion (258). In Pseudomonas aeruginosa, filamentous bacteriophages Pf1 and Pf3 attach to type IV pili and RP4 IncP-encoded pili, respectively. The specificity of attachment resides in the essential minor coat protein g3p (270). Following phage absorption, it is though that pili retract to the cell surface upon bacteriophage binding (298) in a ATP-dependent process (684, 685), allowing the next step of penetration to proceed (25, 272, 323).
TonB-dependent bacteriophages adsorb at the cell surface on TonB-dependent receptors. T1 and φ80 phages interact with the FhuA receptor, and mutational studies have delineated the fixation site on a large external gating loop (residues 322 to 355) (70, 341, 342). Contrary to Tol-dependent bacteriophages, the viral protein responsible for host (or TonB-dependent receptor) recognition has yet to be identified.
COLICIN IMPORT
In the colicin field, the term import refers to the step by which parts of the colicin (including N-terminal and C-terminal domains) reach their final target after recognition of OM receptors. This step of the colicin mode of action requires passage through membranes (we will use the term “translocation,” such as for the transport of polypeptide through membranes) and through the aqueous periplasm (we will use the term “transit”). We will refer herein to import or translocation for the entire mechanism and will distinguish OM translocation, transit through the periplasm, and IM translocation. For enzymatic colicins, IM translocation refers to the step by which the C-terminal domain crosses the IM to the cytoplasm (developed in Colicin Activities). For pore-forming colicins, IM translocation refers to the movement of large sections of the colicin C-terminal domain (several helices) from the periplasmic side to the cytoplasmic side of the IM (developed in Colicin Activities).
TRANSLOCATION THROUGH THE OUTER MEMBRANE
Tol-Dependent Colicins
Although colicin translocation across the outer membrane is still poorly understood, it is clear that Tol-dependent (group A) colicin translocation likely occurs by a mechanism distinct from that of Ton-dependent (group B) colicins. For group A, all colicins except colicin N require two outer membrane proteins for cell entry: a cell surface receptor, which does not participate in translocation (for example, if a TonB-dependent receptor is used for binding, no interaction with TonB is required), and either OmpF or TolC for cell penetration. BtuB has been studied in detail. This TonB-dependent receptor binds colicins E1 to E9 and colicin A. In each case, a cotransporter is required, usually OmpF. A reasonable model for translocation across the outer membrane is as follows. Once BtuB binds colicin E9, OmpF (which is a trimer consisting of three β-barrels with three independent pores) is recruited to interact with a natively unstructured segment of the colicin's translocation domain, forming a BtuB-OmpF-colicin translocon (275). This portion of the translocation domain then enters the cell through an OmpF pore to recruit TolB (417). The colicin's cytotoxic domain (conferring nuclease activity), now in close proximity to an OmpF trimer, probably enters the periplasm through another OmpF pore (698). It has recently been suggested that the unstructured region of 37 amino acids after the TolB binding sequence (BBS) (see below) of colicin E3 requires at least ∼20 residues. It has been suggested that this length may allow the TolB binding sequence to cross the OM and to interact with TolB in the periplasm (583a).
While this mechanism could potentially apply to most group A colicins, colicin N requires only OmpF for binding and permeation. OmpF-PhoE and OmpF-OmpC hybrid proteins demonstrated that the specificity of colicin N resides in residues near the N terminus of OmpF (207, 630). This region was further defined as requiring residues 67 to 182 for receptor binding and residues 17 to 66 for translocation (179). The binding of colicin N to OmpF is rather weak compared to that of colicin Ia or E-type colicins, displaying a Kd of 2 × 10−4 M and a stoichiometry of 3 colicins per OmpF trimer (191, 193). The receptor binding domain of colicin N initially interacts with OmpF, but the pore-forming domain has also been shown to associate with OmpF (161). Translocation requires domains II and III of TolA and does not require TolB as many other group A colicins do. An OmpF colicin N-resistant mutant has been isolated in the internal loop L3, which contributes to lumen constriction (311). However, experiments using OmpF containing a tethered L3 loop inside the β-barrel suggest that the translocation pathway is not through the barrel lumen (13).
Colicin E1 does not use OmpF for translocation but rather uses the tunnel channel TolC (Table 1). Random mutagenesis of the tolC gene followed by screening on colicin E1 recently showed that mutations abolishing colicin E1 binding are exposed to the external medium. Interestingly, other mutations that affect residues buried into the channel do not interfere with colicin E1 binding but slow down its translocation (447a).
TonB-Dependent Colicins
Compared to group A colicins, much less is known about translocation across the outer membrane for colicins belonging to group B. All group B colicins, except for colicins 5 and 10, appear to use a single outer membrane protein for binding and transport. For instance, Cir appears to function as both a cell surface receptor and a TonB-dependent transporter for colicin Ia (Buchanan et al., unpublished). Colicin Ia binds to Cir at the cell surface, stabilizing Cir in an extracellular “open” conformation. As seen for colicins B (456) and M (517), functional TonB box sequences are required in both Cir and colicin Ia for killing to occur (Buchanan et al., unpublished). In all cases, there are probably two sequential interactions with TonB. A plausible mechanism is that the binding of colicin Ia initiates an interaction between the TonB box of Cir and TonB, mimicking small-molecule transport. Energy from the proton motive force could then open a transient pore in Cir by pulling the plug domain partially or completely out of the barrel. The translocation domain of colicin Ia could enter through the Cir pore by a mechanism that might involve the unfolding of the colicin. Once the translocation domain can access the periplasm, an interaction between the TonB box of colicin Ia and TonB, again (possibly) requiring energy from the proton motive force, would facilitate the entry of the pore-forming domain. Although the individual events and energy requirements are presently not well defined, the need for two functional TonB boxes, and the lack of a second outer membrane protein for transport, argues for a mechanism distinct from that of Tol-dependent colicins.
Although energy is not required to form a colicin-receptor complex, it is clearly required at later stages in translocation. For example, the transport of colicin B by FepA was monitored in whole cells using electron paramagnetic resonance spin labeling. If the cells were cooled to 4°C, were deprived of glucose, or did not express a functional TonB complex, colicin transport ceased (312). Experiments using TolQ point mutants to monitor TolQR-TonB cross talk suggested that most group B colicins require energy for translocation across the outer membrane. However, colicins 5 and 10 (group B colicins using Tsx and TolC for cell penetration) did not demonstrate an energy requirement, suggesting that group B colicins require energy to translocate across the outer membrane (if they depend upon a TonB-dependent transporter) but not for transit through the periplasm (E. Goemaere, L. Journet, I. Schalk, R. Lloubès, and E. Cascales, unpublished results).
TRANSIT THROUGH THE PERIPLASM
General Principles of Colicin Transit
In the colicin uptake pathway, the term “transit” concerns the process by which colicins pass the intermembrane compartment called periplasm. It is known that two domains of the colicins (the N-terminal translocation and the C-terminal activity domains) enter the periplasm. The N-terminal domain should trigger a mechanism at the end of which the C-terminal domain crosses the periplasm to reach its final target (the inner membrane or the cytosolic compartment). This ultimate step of transport is completely unknown, and a hypothesis has been proposed that will be discussed below (see “Speculative models for colicin translocation”). For the transit step, it has been shown that the N-terminal domain interacts with components of the cell envelope. This process is probably driven by a Brownian ratchet mechanism, using a hierarchy of sequential interactions with the components of the import machinery, rather than the use of cellular energy (37, 313, 387). Colicin N-terminal domains have parasitized two different and conserved complexes that are essential for the development of the bacterial cell: the Tol and TonB systems. These two systems present topological, structural, and functional similarities or homologies and are coupled to the PMF across the inner membrane, acting as energy transducers for outer membrane stability and active transport, respectively (413, 530). Tol-dependent colicins are classified in group A (colicins A, E1 to E9, K, N, and cloacin DF13), whereas TonB-dependent colicins (colicins B, D, Ia, Ib, and M) are part of group B (Table 1). These two groups were originally described to distinguish two exclusive groups of colicins presenting cross-resistance and thus to reflect two different modes of colicin action (133, 134). Later, these two groups were assigned to the two different translocation pathways.
Tol-Dependent Colicins
The Tol system. (i) Identification.
The Tol system is composed of five proteins that form a multiprotein complex in the cell envelope of most gram-negative bacteria. It involves a complex network of interactions, which have been defined and characterized by a combination of biochemical, structural, and genetic approaches (391, 413). Its physiological role is unclear, but the tol-pal genes have been shown to be essential in pathogens such as Pseudomonas aeruginosa, Haemophilus ducreyi, and enteropathogenic Escherichia coli O157 (146, 214, 611). Mutations within the Escherichia coli K-12 tol genes were identified in the late 1960s by their insensitivity to colicins (474, 475, 487). Later, these genes were isolated based on their involvement in outer membrane integrity and bacteriophage uptake (29, 389, 390, 392, 615, 616). Aside from their role during colicin and filamentous bacteriophage infection processes, the cellular function of the Tol proteins has yet to be determined. However, the pleiotropic phenotype of the tol mutants, the network of interaction of the Tol proteins, as well as the mode of regulation of the tol genes converge to indicate a role in cell envelope stability (see below).
(ii) Regulation.
The regulation of the tol genes is clearly linked to cell envelope stability. It has been demonstrated that the E. coli tol-pal genes are induced by the RcsC sensor of the two-component system RcsBC, a regulatory system involved in the regulation of the cps genes (encoding the biosynthesis machinery of cholanic acid, the major component of the capsula) in response to cell envelope stresses (118). Expression of the Pseudomonas aeruginosa tol genes is modulated by iron and growth phase and involves the Fur and RegA regulatory elements (163, 372).
(iii) Localization and topology.
The five tol genes (tolQ, tolR, tolA, tolB, and pal) encode proteins located in the cell envelope (Fig. 6A). The polytopic TolQ and the bitopic TolR and TolA proteins localized in the inner membrane. The TolQ protein has three membrane-spanning segments, with the N-terminal extremity located in the periplasm (38, 321, 651). TM segment 1 (TM1) is connected to TM2 by a large cytoplasmic loop. TolR and TolA are anchored in the inner membrane through a single TM with the N-terminal extremity located in the cytoplasm (321, 400, 472). Both possess a large periplasmic domain, which can be delineated into two subdomains (314, 400). The C-terminal domain of TolR, predicted to form an α-helix, associates peripherally with the IM (314). The two periplasmic domains of the TolA protein are separated by a stretch of glycine residues (400). Molecular modeling of the central domain predicts that it might consist of an elongated structure, which tethers the TolA anchor to the functional C-terminal domain. Indeed, this central domain, which is essential for TolA function (149, 576), presents a three-stranded coiled-coil structure and is therefore thought to cross the periplasm to interact with outer membrane components (152, 400, 674). This is indeed the case, since TolA interacts with major porins and the TolB-Pal complex (see below). The C-terminal domain, which constitutes the site of colicin binding, presents a more globular fold (148, 421, 677).
FIG. 6.
Localization and topologies of components of colicin import machineries. (A) Topologies and predicted transmembrane domains of the inner membrane TolA, TolQ, and TolR proteins. The periplasmic TolB and the outer membrane-anchored peptidoglycan-associated lipoprotein Pal are shown. The Pal peptidoglycan binding sequence and TolQRA transmembrane segment boundaries are indicated (39, 78, 119, 400, 472, 651). (B) Topologies and predicted transmembrane domains of TonB, ExbB, and ExbD. Two predictions for ExbB transmembrane domains have been made (320, 325). Those described by Karlsson et al. (325) are in parentheses. ext, external medium; peri, periplasm; cyto, cytoplasm.
TolB is a periplasmic protein that peripherally associates with the outer membrane (291). Pal is an abundant (∼25,000 copies per cell) (75) lipoprotein anchored to the outer membrane via its N-terminal acylated residue, and it interacts with the peptidoglycan layer through a conserved domain (residues 86 to 110) also found in the OmpA and MotB protein families (143, 218, 350, 393, 460).
Recent data suggest that all five Tol-Pal proteins localize to the constriction site during the division process (219a).
(iv) Three-dimensional structures.
The three-dimensional structures of three Tol subunits or domains (e.g., TolB, TolA C-terminal domain, and Pal) are available (Fig. 7). The structure of the TolA C-terminal domain has been solved by both crystallography and NMR, alone and in association with partner domains (148, 421, 553, 677). It forms a novel fold consisting of three antiparallel β-strands with four helical motifs (Fig. 7A). In the crystal structure of the P. aeruginosa TolA C-terminal domain (677), the C-terminal 43 residues of the central domain are present and form a long coiled-coil α-helical structure, as suspected from biochemical and biophysical data (152, 400). It is noteworthy that despite the absence of sequence homologies, the crystal structures of the TolA and TonB C termini showed a remarkable similarity, suggesting an evolutionary relationship (677).
FIG. 7.
TolA, TolB, and Pal crystal structures. (A) Ribbon diagrams of TolA, TolB, and Pal. The Pseudomonas aeruginosa TolA C-terminal domain (TolAIII) (677) (PDB accession number 1LRO; 1.80 Å) is shown. The α-helix indicated by the arrow corresponds to the C-terminal 43 residues of the central domain (TolAII). (B) The Escherichia coli TolB structure (1, 72) (PDB accession number 1CRZ; 1.95 Å) showing the D1 domain and the six-bladed D2 domain (β-propeller) forming the cavity. (C) The Escherichia coli Pal lipoprotein structure (C. Abergel and A. Walburger, unpublished data) (PDB accession number 10AP; 1.93 Å). The peptidoglycan binding helix is indicated with an arrow, whereas the lateral chain of the Tyr residue of the conserved TolA binding motif (SYGK) (Fig. 8B) is shown and highlighted by the pink circle.
The crystal structure of the periplasmic TolB protein has also been solved (1, 72). It is composed of two subdomains (called D1 and D2) in which D2 forms a six-bladed β-propeller, as originally predicted by bioinformatics (528). Each blade consists of twisted β-sheets that are radially arranged around a central tunnel (Fig. 7B). This β-propeller domain is also found in three-dimensional structures of β-lactamases, suggesting that TolB may participate in peptidoglycan metabolism or OM lipoprotein assembly. This hypothesis remains to be experimentally confirmed. Recently, structures of a truncated form of the Pal lipoprotein (lacking the 20 N-terminal amino acids, which are unstructured and nonessential for function) (78) alone (Protein Data Bank [PDB] accession number 1OAP) or in complex with a synthetic peptidoglycan precursor (PDB accession number 2AIZ) (506) have been determined.
(v) Interaction network.
In 1994, Guihard et al. showed that treatment of whole cells with purified colicin A increases the level of Tol proteins at discrete sites of the cell envelope by a similar ratio, suggesting that the Tol proteins interact to form a multiprotein complex with a precise stoichiometry (240). Since then, many interactions have been characterized within the Tol-Pal complex. First, biochemical and suppressive studies have shown that TolQ, TolR, and TolA form a complex in the inner membrane, interacting via their TM domains (149, 220, 314, 326, 394). TolR has further been shown to dimerize through its central periplasmic domain (314). The inner membrane complex composed of the TolQ-TolR-TolA subunits has a stoichiometry of 4:2:1 to 6:2:1 (79, 240). This estimation is compatible with cell numeration proposing 600 to 800 copies of TolA and 2,000 copies of TolR per cell (166, 401, 472) and the precise measurements of the homologous ExbB-ExbD-TonB stoichiometry (7:2:1) (264). Isnard et al. showed that TolB is peripherally associated with membranes in addition to its periplasmic location (291). Later, Bouveret et al. demonstrated that this is due to the interaction between TolB and Pal (40). Recently, the interaction between TolB and Pal has been measured with a Kd of 30 nM (417). Furthermore, it has been shown that TolB and Pal are part of a more complex network of interactions including the OM OmpA protein and the major murein lipoprotein Lpp (75, 119). The domains of interaction between TolB and Pal have been mapped by different approaches. TolB interacts with Pal through its β-propeller D2 domain (119, 550, 658), while Pal interacts with TolB through the peptidoglycan binding sequence (39, 78, 119, 550). Both TolB and the peptidoglycan interact with the same region of Pal, and competition experiments have shown that these two interactions are exclusive (39).
Finally, interactions connecting the two subcomplexes have been identified. TolA interacts independently with both TolB and Pal (77, 658) through its C-terminal domain. Binding to TolB requires the TolB D1 subdomain (164, 658), while mapping of the TolA binding sequence (ABS) of Pal has demonstrated the presence of a tetrapeptide sequence critical for the TolA-Pal interaction at the Pal C terminus (78). This ABS has recently been shown to be present in different TolA partners such as colicins A, N, and K as well as the minor capsid bacteriophage protein g3p (527), a result reminiscent of the TonB box described 20 years earlier (see below).
Another similarity with the TonB system (described below in the TonB-dependent colicin transit section) is the observation that the TolA protein has two distinct conformations (221). The TolA conformation is controlled and regulated by the PMF and the TolQ and TolR proteins (221). Interestingly, the “energized” conformation of TolA is the form that can interact with Pal (77), and it has been suggested that both TolQ and TolR proteins combine to form an ion-conducting channel, coupling ion flow through the channel to structural modifications of the TolQR TM helices. In turn, the TolQR TM would transduce a conformational signal to the TolA protein through a conserved “SHLS” motif present within the TolA TM that has been shown to be involved in the interaction with TolQ and to be critical for the conformational change (79, 221). This model has been proposed in view of the high degree of homology between TolQ and TolR transmembrane helix sequences with those of the MotA and MotB proteins, two components of the flagella, constituting the stator of PMF-driven molecular motors (79, 354). Residues regulating ion flow through the TolQR complex have been recently identified and may form an ion pathway (227a).
(vi) Physiological functions.
The tol-pal mutants present a pleiotropic phenotype linked to the outer membrane. They show a hypersusceptibility to toxic compounds, antibiotics (including large antibiotics such as vancomycin), detergents, and bile salts (29, 392, 533). The OM of tol mutants disaggregates and forms blebs, which are accompanied by the release of periplasmic components (28, 206, 392, 410, 452). Interestingly, the level of OM stability has been shown to be dependent on the IM proton motive force, which drives the TolA-Pal interaction (see reference 482). The Tol-Pal system is thus involved in the maintenance of cell envelope integrity. However, how these proteins fulfill this function is unclear and has yet to be determined. Because the exact function is not known, current research focuses on the link between Tol-Pal proteins and porins, lipopolysaccharides (LPS), or O-antigen biogenesis. It has been shown that the central domain of the TolA protein, as well as the TolB protein, interacts in vitro with major OM trimeric porins such as OmpF, OmpC, LamB, and PhoE in the presence of sodium dodecyl sulfate (150, 554). TolA and TolB do not interact with OmpA or the OM denaturated porins. This observation led those authors to propose that TolA and TolB might interact only with OM proteins requiring LPS for their assembly or their integration into the lipid bilayer. It has thus been proposed that SDS may mimic LPS in the in vitro interaction assay. Furthermore, isothermal titration calorimetry (ITC) experiments have shown that the TolA periplasmic domains do not interact with purified OmpF in the absence of SDS (546). The levels of OmpF in WT and tol mutant cells are reduced (392). However, the porins are correctly inserted in WT or tol cells (28, 137), suggesting a role in the kinetics of porin transport or insertion rather than a direct role in porin biogenesis.
The link between the Tol proteins and LPS has been demonstrated recently. tol mutants present defects in O-antigen insertion into the OM (214), and more recently, the role of the Tol-Pal proteins in LPS assembly has been restricted to O-antigens assembled through the Wzy-dependent pathway (652). However, O antigens are correctly polymerized in these mutants, suggesting that the Tol proteins interfere with the pathway at a late stage of the biogenesis process. It is noteworthy that both Tol proteins and newly synthesized LPS molecules localize at the Bayer contact sites where the OM and IM come together (18, 240, 290, 395).
Besides this putative function in the assembly or transport of OM components, the Tol-Pal proteins and their numerous interactions with OM stabilizing components (OmpA and the major murein lipoprotein Lpp) might connect inner and outer membranes and the peptidoglycan layer, thus playing an architectural role in the cell envelope. Finally, because of their particular cell localization at the division site (219a) and the formation of filamentous cells by tol mutants in high- and low-osmolarity media (457), a role in the division process might also exist. Localization of the Tol proteins is dependent on FtsN, the last protein recruited at the division site, suggesting a role for the Tol-Pal complex in the late stages of cell division (219a).
Interactions between Tol subunits and colicin translocation domains.
Different group A colicins use different subsets of the Tol proteins for their translocation. For example, colicins A, E2 to E9, and K require TolABQR; colicin E1 requires only TolAQ; and colicin N translocation needs the TolAQR proteins (Table 1). Interactions of colicins with different components of the import machinery were suspected and first demonstrated in 1991 (23). Since then, data have been accumulated to reach a higher level of understanding. However, even if the domains of interactions are now delineated, we still lack a detailed overview of the general mechanism of translocation.
Guihard et al. showed that the level of Tol proteins present at contact sites increases upon treatment of whole cells with colicin A (240). Quantification of the different subunits further showed that each Tol protein level increases with the same ratio, suggesting that the Tol proteins are recruited to these sites upon colicin reception or OM translocation. More indirect evidence of the involvement of Tol proteins in the colicin import process was the observation that the number of translocation sites per cell (measured in receptor bypass experiments) was similar to the number of TolA molecules (166, 240, 401).
The information necessary for the OM translocation and transit steps is contained in the N-terminal domain of the colicin. This has been shown by analyses of proteolysed fragments and truncated colicins or by the use of colicin chimeras constructed by the recombination of colicin domains from Tol- and TonB-dependent colicins. The properties of these hybrid colicins demonstrate that the specific translocation pathway used is defined by the N-terminal domain (14, 21, 22, 24, 496, 515). Concerning the colicin E1 N-terminal domain, it has been proposed, based on sequence comparisons, that it might be composed of two subdomains of 34 and 140 residues, respectively, in which the former constitutes the TolA binding domain while the latter is responsible for TolC recognition and binding (237, 516).
Other indirect evidence includes the use of periplasmic overproduction of colicin or Tol domains (41). Periplasmic production of the colicin A or colicin E3 translocation domains into wild-type cells renders cells tolerant to the action of group A colicins, suggesting that the N-terminal domains interact with machinery components, thus blocking the access of exogenous colicins. Furthermore, these cells present the characteristic tol mutant phenotype, suggesting that interactions within the Tol complex are displaced by the colicin N-terminal domains (42, 43). Conversely, the sites of interaction in the Tol subunits have been defined by a similar approach. Overproduction of the TolA C-terminal or the TolR central domains in the periplasm of WT cells renders cells tolerant to the action of exogenous colicins, suggesting a titration of colicin N-terminal domains protruding into the periplasm (314, 401).
(i) Interaction with the TolA protein.
The first direct evidence for an interaction between the translocation domain and TolA came from the work of Bénédetti et al., who demonstrated that both intact colicins A and E1 or their translocation domains, but not the group B colicin B translocation domain, interact with the TolA protein, using an overlay technique (or Far-Western blot) (23). They further showed that the C-terminal part of TolA is involved in these interactions. Later, these results were confirmed by in vitro techniques using the purified central and C-terminal TolA and colicin A and E1 N-terminal domains and a combination of overlay, gel shift, and surface plasmon resonance (SPR) (151). The dissociation constants have been estimated to be 0.2 to 0.6 μM and 0.4 μM for colicins A and E1 translocation domains, respectively (151) (Table 3). ITC experiments have shown that the TolA periplasmic domain interacts with either the whole colicin N or its N-terminal domain with dissociation constants of 18 μM and 1 μM, respectively (546), suggesting that the site of interaction with TolA is more accessible in the isolated N-terminal domain than with the entire colicin. The stoichiometry of binding of the three colicin N-terminal domains (A, E1, and N) to the TolA protein has been shown to be equimolar (151, 546). Binding assays with increasing ionic strength suggested that the interaction between the TolA C-terminal domain and the colicin A translocation domain is not driven only via electrostatic interactions (147). However, the mode of TolA binding and requirements might be different for distinct colicins, since TolA mutations or truncations have been shown to have different properties in colicin A or E1 uptake (576). This variability in the mode of recognition or binding of different colicins to the TolA subunit is exemplified by the report that tolA point mutations that conferred tolerance to colicin A but not to colicins E1, E2, E3, and K have been isolated (164). It is noteworthy that no interaction of TolA with any enzymatic colicins has been demonstrated. The TolA C-terminal domain is the site of interaction for three partners, TolB, Pal, and the N-terminal domain of colicins (23, 77, 151, 658). However, overlay experiments have shown that a TolB-colicin-TolA complex can form (43), while competition experiments have demonstrated that the N-terminal domain of colicins does not compete with Pal for TolA binding (527). These data suggest that the partners interact differently with TolA, probably by distinct regions. NMR studies have shown that the interaction between TolA and colicin A induces a significant structural change accompanied by an increased flexibility of the TolA C-terminal domain (147) (see below).
TABLE 3.
Affinity constants between components of the Tol import machinery and group A colicin and g3p constructsa
| Tol subunit | Ligand | Kd(μM) | Method(s) | Reference(s) |
|---|---|---|---|---|
| TolAII-III | ColN-T | 1.4 | ITC | 546 |
| TolAII-III | ColN-T | 2.3-3.7 | FS | 228, 546 |
| TolAII-III | ColN-TR | 8.0 | ITC | 546 |
| TolAII-III | ColN | 18.0 | ITC | 546 |
| TolAIII | ColN-T | 1.0 | ITC | 546 |
| TolAIII | ColN40-90b | 0.9 | SPR | 228 |
| TolAIII | ColN-T | 0.8, 1.4 | SPR | 5, 228 |
| TolAIII | ColA-T | 0.2-0.6 | SPR | 151, 248 |
| TolAIII | ColA-T | NB | ITC | 228, 248 |
| TolAIII | ColE1-T | 0.4 | SPR | 151 |
| TolB | TolAII-III | NB | ITC | 228 |
| TolB | TolAIII | 15.3 | SPR | 248 |
| TolB | Pal | 0.03 | ITC | 417 |
| TolB | Pal | 0.09d | ITC | 417 |
| TolB | ColN-T | NB | ITC | 228 |
| TolB | ColN | NB | ITC | 228 |
| TolB | ColA-T | 10.0 | ITC | 228 |
| TolB | ColA-T | 10.5 | SPR | 248 |
| TolB | ColE9/imE9 | 14.0 | SPR | 248 |
| TolB | ColE9/imE9 | 1.0 | ITC | 248 |
| TolB | ColE9 | 1.0-1.2 | SPR, ITC | 248, 417 |
| TolB | ColE9-TR | 1.7 | SPR, ITC | 248 |
| TolB | ColE9-T | 0.9 | ITC | 417 |
| TolB | ColE9-T | 0.08d | ITC | 417 |
| TolB | ColE932-47c | 0.9 | ITC | 417 |
| TolB | ColE3/ImE3 | 7.0 | SPR | 248 |
| TolB | ColE3 | 0.9 | SPR | 248 |
| TolRII-III | ColA-T | >1,000 e | SPR | 248 |
| TolRII-III | ColE3-T | NB | SPR | 248 |
| TolRII-III | ColE9-T | NB | SPR | 248 |
| TolRII-III | ColN | NB | ITC | 228 |
| TolAIII | g3p-N1N2 | 23 | SPR | 323 |
| TolAIII | g3p-N1 | 1-1.9 | SPR | 323 |
| TolAIII | g3p-N2 | NB | SPR | 323 |
| TolAII | g3p-N2 | 1.3-2.4 | SPR | 323 |
| TolRII-III | g3p-N1 | NB | ELISA | 553 |
Affinity constants (micromolar) were calculated from SPR, ITC, fluorescence spectroscopy (FS), or enzyme-linked immunosorbent assay (ELISA) studies. NB, no binding observed. T, translocation domain; TR, translocation and reception domains.
Peptide corresponding to residues 40 to 90 of colicin N.
Peptide corresponding to residues 32 to 47 of colicin E9.
Affinity constants calculated in the presence of 1 mM Ca2+.
An interaction was detectable at high protein concentrations but was not quantifiable.
(ii) Interaction with the TolB protein.
N-terminal domains of various colicins have been shown to interact with TolB. The first evidence came from the work of Bouveret et al., who demonstrated the interaction between TolB and the colicin E3 N-terminal domain by coimmunoprecipitation and in vitro cross-linking experiments (42). Moreover, the periplasmic expression of the colicin E3 N-terminal domain has been shown to displace and dissociate the TolB-Pal complex, suggesting that both TolB protein partners interact with the same region of TolB. This is indeed the case, since Walburger et al. confirmed the interaction between TolB and the colicin E3 translocation domain by yeast two-hybrid analysis and further delineated the TolB β-propeller domain as the site of colicin E3 and Pal binding (658). Similarly, the interaction between TolB and the colicin A translocation domain was demonstrated using a combination of cross-linking, copurification, and overlay experiments (43), while interactions between the TolB protein and the TolB-independent colicin E1 translocation domain were not observed by using the same approaches. Yeast two-hybrid experiments also showed that the colicin E9 N-terminal domain interacts with TolB. Here again, truncation studies have defined the β-propeller of TolB as the colicin binding site (72). Therefore, this domain is responsible for the interaction of TolB with both the Pal lipoprotein and the colicin N-terminal domain (550, 658), and it has been demonstrated recently that the formation of the colicin E9 N-terminal domain-TolB complex displaces the TolB-Pal interaction (417). An identical site of TolB binding for colicin and Pal was recently confirmed for colicin E9. It was shown that the binding of colicin E9 to TolB mimics the interaction of Pal with TolB. Interestingly, Pal undergoes localized conformational changes as a result of TolB binding. These rearranged residue positions are mimicked by the colicin translocation domain when it interacts with TolB (C. Kleanthous, unpublished results). The TolB-ColE7 translocation domain interaction has been demonstrated by coimmunoprecipitation (114). However, ITC studies showed an absence of an interaction of TolB with the colicin N translocation domain while interacting with a Kd of ∼10 μM to colicin A (228) (Table 3), a result in agreement with earlier in vivo experiments demonstrating that colicin N activity does not require the TolB subunit (192). Using surface plasmon resonance and ITC, the dissociation constant for the colicin E9 (devoid of immunity)-TolB interaction was measured at ∼1 μM (248), an affinity that is 10-fold higher than that of the colicin A-TolB interaction measured by ITC (228) (Table 3). When bound to its immunity protein, the affinity of colicin E9 for TolB decreases to ∼14 μM (248). A comparison of dissociation constants between TolB and the two partners, the translocation domain of colicin E9 (ColE9-T) and Pal (0.9 μM versus 0.03 μM), is not consistent with the displacement of the TolB-Pal interaction by the colicin E9 N-terminal domain during transit (417). However, identical measurements in the presence of Ca2+ give similar affinities for TolB-ColE9-T and TolB-Pal complexes (0.08 μM versus 0.09 μM) (417) (Table 3).
(iii) Interaction with the TolR and TolQ proteins.
The evidence for an interaction between TolR and group A colicin N-terminal domains came from in vivo and in vitro cross-linking experiments demonstrating that the TolR central domain (TolRII) interacts with colicin A and colicin E3 translocation domains. Furthermore, the group B colicin B and the TolR-independent group A colicin E1 translocation domains do not interact with TolR (313). Unexpectedly, Gokce et al. could not measure any TolRII-colicin N interaction using ITC, whereas the colicin A translocation domain-TolRII interaction has not been detected by yeast two-hybrid assays (228, 658). No interaction between TolQ-dependent colicins and the TolQ subunit has yet been demonstrated. However, as shown for the TolA protein, different TolQ regions or residues may be involved in the recognition of distinct colicins (E. Cascales and R. Lloubès, unpublished data).
Definition of Tol boxes.
Experiments to map the Tol binding sequences have been performed. The capacity to interact with TolA, TolB, or TolR involves distinct regions of the colicin N-terminal domain. However, both TolB and TolR binding sequences overlap, suggesting that the TolB-colicin and TolR-colicin interactions are mutually exclusive.
(i) TolA binding sequence.
The ABS was originally proposed to be located between residues 30 and 39 of the N-terminal domains of colicins E1, E2, and E3 (516). Mutations within the consensus pentapeptide sequence (DGSGW) within colicins E3 and E9 revealed a critical role for these residues in the transit process (187, 213). However, overlay experiments have previously shown that the deletion of a domain including this sequence has no effect on the TolA-colicin A interaction (23). Later, competition SPR experiments performed with a synthetic peptide corresponding to this consensus sequence showed that even with a 1,000 molecular ratio of peptide over the colicin A N-terminal domain, no decrease of TolA-colicin A interaction was measured (151). Similarly, the deletion of the putative pentapeptide TolA binding sequence in the colicin N-terminal domain does not alter its capacity to interact with TolA, as shown by ITC (546). These results suggest that the pentapeptide sequence is not required for TolA binding. Thus, the TolA binding sequence has been reconsidered, and the above consensus sequence has indeed been found to correspond to the TolB binding sequence (43). The TolA binding sequence was then mapped in the colicin A N-terminal domain (43), and further mutagenesis studies have narrowed the region of colicin A involved in binding to TolA (313). The ABS locates between residues 52 and 97 of colicin A (Fig. 8A and B). More recently, based on homologies with the proposed ABS in the Pal sequence (SYGT) (78), Pommier et al. demonstrated that an SYNT sequence, and particularly the tyrosine residue, within the colicin A translocation domain is important for binding to the TolA protein (527). Multiple alignments further showed that a similar motif is present in both colicin N (SYHIT) and colicin K (SYLT) translocation domains and might constitute a TolA box (Fig. 8B). Here again, it has to be noted that such a motif is not found in any of the enzymatic colicin translocation domains. Indeed, mutations within this putative ABS in the colicin N N-terminal domain have previously been shown to reduce or abolish (notably the conserved tyrosine Y62 residue) TolA binding activity (546). However, the ABS does not seem to be restricted to this short sequence but rather to a longer sequence that extends into the N-terminal side of the TolA box. Using a combination of alanine-scanning mutagenesis and surface plasmon resonance studies, Gokce et al. showed that the minimal colicin N TolA recognition sequence spans at least 27 residues including the SYHIT sequence (residues 40 to 66) (228). Besides the conserved tetrapeptide motif, the TolA recognition regions from distinct colicins do not present any sequence homologies (Fig. 8B). Here again, it is notable that even if tolA mutants are tolerant to enzymatic colicins, no direct interaction between TolA and enzymatic colicin N-terminal domains has been demonstrated, and no TolA binding sequence has been identified.
(ii) TolB binding sequence.
As described above, the BBS is located at the N-terminal extremity of the colicin translocation domains. This sequence is composed of eight residues, DGT(S)GWSSE, in which the WSSE motif is uniquely found in TolB-dependent colicins (43) (Fig. 8C). Because of its conservation, its size, and its high specificity, this sequence has been called the “TolB box.” As described previously, mutants affected in this motif in colicins E3 and E9 are inactive on sensitive cells (187, 213) and do not interact with TolB in in vitro or in vivo assays (42, 43, 72). The TolB box is part of an unstructured region, and it has recently been proposed that the flexible region after the TolB box of colicin E3 requires a minimal length, allowing the TolB box to cross the OM and interact with TolB in the periplasm (583a). Construction of truncated variants of colicin A delimited the BBS in the translocation domain between residues 7 and 20 (including the TolB box, residues 11 to 18) (43, 313). Here again, it has been demonstrated that the BBS is larger than previously described. The addition of unlabeled TolB protein to 15N-labeled ColE9 affects signals corresponding to residues 33 to 46 (including the TolB box, residues 35 to 42) (125), and the importance of the extended sequence has been confirmed by a combination of alanine scanning and SPR (248) (Fig. 8C). It has been suggested that this region forms β-turns, and recent data indeed confirm that it forms a loop penetrating within the TolB β-propeller (417) (Fig. 8D and 8E).
(iii) TolR binding sequence.
The region of colicin A involved in the interaction with the TolR protein has been defined. It is located at the extreme N terminus, between residues 7 and 12, thus overlapping the TolB binding sequence (313). No experimental data are available for other colicins.
Structural information on Tol-dependent translocation.
Data are available concerning structural modifications of colicins or Tol subunits during the transit step, suggesting that it is not a passive process involving sequential interactions with cell envelope components but rather that it is a dynamic process involving conformational changes of both the machinery components and the ligand. First, it has been shown that the colicin A central domain remains in an interaction with the BtuB receptor while both N- and C-terminal domains are translocated (24, 165, 513). This implies that the three domains are linked via flexible regions that are subject to large-scale structural modifications. This is indeed the case, since most colicins (except colicins N and B) showed elongated structures with extended coiled-coil α-helices (102, 607, 675) and have been shown to unfold during the translocation process (24). Based on calorimetric, spectroscopic, and crystallographic experiments, it has been proposed that colicin E1 unfolds by a process triggered by binding to the BtuB receptor (237, 371). This implies structural modifications starting from the receptor binding domain (237, 607). This suggestion correlates with the inactivity of oxidized forms of double-cysteine mutants that lock the colicin E9 reception domain without interfering with its capacity to interact with the BtuB receptor (513). Overall, these data suggest that the unfolding of the receptor binding domain is necessary for starting the translocation process. However, such a mechanism has been ruled out in the case of colicin E9, based on ITC experiments and thermodynamic considerations. It has been proposed that unfolding occurs after recruitment of the “translocon” (i.e., OmpF or TolC) upon receptor binding (275). The N-terminal domains of colicins have been postulated to be poorly structured regions based on the low amount of ordered secondary structures observed by circular dichroism, a weak dispersion of NMR signals, the absence of detection of corresponding electron density during X-ray crystallography studies, and high susceptibility to proteolytic degradation (6, 125, 132, 147, 496, 546, 607, 650). Biophysical studies have demonstrated that the TolB binding sequence of colicin E9 becomes more structured when in interaction with TolB (125, 248, 431, 632). Similarly, transition of the N-terminal domain of colicin N from a disordered state to a folded conformation is induced upon TolA C-terminal binding (5, 6). Conversely, the increased structural flexibility of the 15N-labeled 96 C-terminal residues of the TolA protein has been observed by NMR upon the addition of the unlabeled colicin A N-terminal domain without interfering with the main secondary structure elements (147). In vivo, Pommier et al. showed that the expression of the colicin A N-terminal domain in the periplasm of WT cells induces TolA degradation (527). Similarly, the exogenous addition of colicin A to the culture induces the same degradation pattern, an observation the correlates with the structural modification of the TolA molecule upon colicin A binding. This probably suggests that molecular recognition involves concerted binding and folding events that occur simultaneously. It has been further suggested that this might be done through “induced-fit” (e.g., recognition of the ligand induces a conformational modification and subsequent high-affinity binding) or “preexisting-equilibrium” (e.g., distinct structural conformations of the Tol subunit or colicin N-terminal domains exist, with one of them with high affinity for the ligand) mechanisms (148). However, no structural modifications of the TolA C-terminal domain have been observed upon binding of the colicin N N-terminal domain (5), thus reflecting different behaviors of TolA with different partners. Overall, these data suggest that the N-terminal segments of the group A colicin translocation domains are disordered in their native states and fold into an ordered structure when binding a Tol subunit. This recognition mechanism has been suggested to contribute to the specificity of the recognition event, to allow a protein to bind several target partners, and to provide large protein binding surfaces in relatively small regions (168, 241, 248, 362).
Hierarchy of contacts during transit.
Because the translocation of group A colicins does not need any energy input (see below) and requires interactions of the N-terminal domain with the different subunits of the Tol machinery, it has been suggested that these colicins transit the periplasm through a Brownian ratchet mechanism involving a sequential and unidirectional cascade of interactions. This type of mechanism has been demonstrated for the BiP-dependent translocation of the prepro-α-factor into the endoplasmic reticulum (449). In the case of colicins, the interaction of the N-terminal domain with one of the subunits would prevent the colicin from moving backwards. In such a mechanism, the hierarchical interactions should be driven by the affinity constants between the colicin N-terminal domain and the different Tol subunits. This implies that each interaction between the colicin N-terminal domain and a Tol protein would have a higher affinity than the previous interaction. Taken separately, the data reported so far indicate a transit route, but most of the data are not compatible all together. For example, the overlay technique has shown that both TolB-colicin A N-terminal domain and TolR-colicin A N-terminal domain complexes interact with TolA, but the latter does not interact with TolB. This suggests that there is formation of ternary complexes during the transit process and that the colicin would not pass from TolB to TolR or vice versa. This correlates with the data showing that TolR and TolB binding sequences overlap in the colicin A N-terminal domain. With regard to affinity constants, the colicin A N-terminal domain displays a higher affinity for TolA than for TolB and TolR, suggesting that if a Brownian ratchet mechanism is correct, the colicin should pass from TolR to TolB to TolA (Table 3). Indeed, an intact TolB-TolA complex seems to be necessary for an efficient transport mechanism. A mutation within the TolA C-terminal domain abolishing the formation of the TolA-TolB complex has been isolated and was further shown to be important for the transit of colicins A and E3 but not of colicin E1, which does not require TolB for its import (658). Taken together, these observations suggest that the transit pathway should be defined as sequential interactions of colicin A with TolB, TolA, and then TolR. The location of TolQ in this pathway cannot be hypothesized because of the lack of data about affinity constants or binding sequences. This scenario has recently been supported by the observation that the addition of colicin A in wild-type, tolR, or tolQ cells induces TolA degradation, while no degradation is observed upon treatment of tolB cells. These observations led to the conclusion that TolB probably acts upstream of TolA, which acts upstream of TolR and TolQ in the transit pathway (A. Barnéoud, R. Lloubès, and E. Cascales, unpublished results). How is this result compatible with affinity constants? The interaction between the colicin A N-terminal domain and the TolR protein appears to be weak, whereas the previous interaction is of high affinity. One hypothesis for explaining this discrepancy is that in vivo, TolA degradation might be accompanied by a decrease in the affinity of colicin A for the TolA protein, which will in turn interact with the next partner.
Kinetics of translocation.
The time required for the translocation of group A colicins has been determined experimentally. In the case of ionophoric colicins, potassium efflux induced by pore formation in the inner membrane occurs within 30 s posttreatment at 37°C (37, 178, 240). This reflects the time necessary for colicin binding at the cell surface (which is on the order of a second), colicin unfolding, translocation through both the outer membrane and the periplasm, and insertion into the cytoplasmic membrane. When the same experiments are repeated in receptor bypass experiments or when the number of receptors is nonlimiting, the time needed is shorter (∼10 to 15 s), demonstrating that binding and unfolding are fast events (178, 240). Further experiments have shown that this time is required for the translocation step (24, 37, 165, 178). In the case of nuclease colicins, measurements of the time necessary for release of the immunity protein (∼20 min for cloacin [364] and colicin E2 [165a]), for the transcriptional profiling of genes of the LexA-regulated SOS response (∼10 min for colicin E9) (663), or for the induction of a synthetic SOS promoter-lux reporter system (∼15 min for colicin E9) (645) have clearly shown that the kinetics are much slower than those of ionophoric colicins. Although the underlying causes for this difference are not known, it has been proposed that it might correspond to the time required for the dissociation of the tightly associated colicin-immunity complex and translocation through the inner membrane or the time necessary for the nuclease domain to damage sufficient DNA to induce the SOS response. However, using the SOS reporter assay, Vankemmelbeke et al. reported that the treatment of E. coli cells with colicin E9 devoid of its immunity does not decrease the time necessary for translocation (645).
TonB-Dependent Colicins
It seems that every field goes through phases where, at first, so little is known that any conjecture seems reasonable, the whole field can be satisfyingly described, and everyone knows how it works. Next, in a second phase, data are gathered, differences of opinion and a variety of models emerge, and because there is so much apparently contradictory information, the field is unsettled and in a state of relative confusion. In the final phase, new data allow the resolution of apparently contradictory old data, the field becomes relatively simple again, and everyone knows how it works (at least for a while). The TonB field appears to be in the exciting second phase, and this section is written with that idea in mind.
Gram-negative bacteria face a dilemma due to the presence of their characteristic OM. The OM is protective on one hand and yet restrictive in terms of the size of nutrients that can be acquired through diffusion across it on the other hand. In particular, iron binding siderophores and vitamin B12 are too large, too scarce, and too important to have their acquisition left to diffusion through the OM porin proteins that serve for the diffusion of most nutrients. Instead, high-affinity active transporters have evolved to capture and transport these important nutrients into the periplasmic space. Due to the subnanomolar affinities of the transporters for iron-binding siderophores or vitamin B12, energy is required for their transport across the OM. The OM is devoid of conventional types of energy such as access to ATP or a significant ion potential. Thus, the TonB system apparently evolved to energize active transport across the OM. Similarly, a group of protein toxins that can parasitize the TonB-dependent OM transporters to enter the cells also evolved. In E. coli, where they have been most well studied, these toxins are known as colicins. As described above, the colicins that use the TonB system to gain entry into E. coli are known as group B colicins.
Early studies from the Braun laboratory indicated that cytoplasmic, or IM, proton motive force (or something coupled to it) was required for the translocation of phages (and later of group B colicins) across the unenergized OM (247, 250) (see Energy requirements for phage DNA uptake below). The need for an energy-transducing system was ultimately filled by the TonB system. For additional information, refer to recent reviews of the TonB system (49, 351, 530, 531, 676).
The TonB system consists of an OM transporter and three known IM proteins, TonB, ExbB, and ExbD (Fig. 6B), as well as at least one mystery protein whose existence can be experimentally inferred (530). Interestingly, the three IM proteins all carry some degree of amino acid and functional similarity to the Tol system proteins TolA, TolQ, and TolR, respectively (173).
E. coli strains with mutations in tonB are tolerant to all group B colicins, meaning that the colicins can still bind reversibly to their OM transporters but cannot translocate into the periplasm (133). E. coli strains with mutations in exbB or exbD are not entirely tolerant to colicins but retain some slight sensitivity due to cross talk with the Tol system (52). Mutant strains lacking both exbB/D and tolQ/R are tolerant to group B colicins (48).
Killing by group B colicins provides an indirect measure of TonB system activity. Because the killing is measured by the ability of colicin dilutions to kill lawns of bacteria over several hours, these assays are fairly sensitive to low levels of TonB system activity (vitamin B12-dependent growth and φ80 spot titers are most sensitive to low levels of TonB, detecting the presence of as little as one TonB molecule per cell). On the other hand, colicin killing assays are also among least sensitive to variations in TonB activities, lacking the ability to discriminate clearly between ∼20% and 100% TonB activity. However, both transport assays and irreversible φ80 adsorption can discriminate in that range (377).
The TonB protein: physiological function and its role in colicin translocation. (i) The TonB amino terminus.
The TonB protein of Escherichia coli has 239 amino acids, 17% of which are prolines (529). It can be divided into roughly three domains, the amino (amino acids 1 to 65) and carboxy (amino acids 103 to 239) termini separated by a proline-rich domain (amino acids 66 to 102), although other authors make somewhat different boundaries for these domains (507). At the amino terminus, a standard hydrophobic signal sequence necessary for the Sec-dependent export of TonB to the IM is found. This signal sequence is not cleaved, thus providing an anchor in the IM (249, 522, 532) and leaving the majority of TonB to occupy the periplasmic space (Fig. 6B) (249, 563, 593). In the IM, TonB appears to interact directly with one or more of the three transmembrane domains of ExbB (310, 326, 381). Interestingly, ExbB also acts as a chaperone for TonB in the cytoplasm, presumably during assembly (325; T. Letain and K. Postle, unpublished observations). The hydrophobic signal anchor is most conveniently referred to as a transmembrane domain, although as will be apparent below, it probably never contacts lipids directly, and its actual conformation is unknown. When modeled as an α-helix, the sequence of the putative transmembrane domain (amino acids 12 to 32) has a conserved face, S16, H20, L27, and S31, that also characterizes TolA (349). A deletion-scanning analysis of the TonB transmembrane domain suggested that S16 and H20 as well as the register between them were important for TonB activity (379). Recently, a TonB transmembrane domain where every residue except S16 and H20 was simultaneously replaced with alanyl residues was, surprisingly, shown to be fully active. An S16A replacement in the wild-type transmembrane domain was also active, whereas an H20Y or H20A replacement in the wild-type transmembrane domain was inactive. In spite of the conserved SHLS motif, it was concluded that only the H20 side chain is important for TonB activity (377b). In a mutagenic study of the TolA transmembrane domain, S16A also retained activity (220).
Interestingly, the amino terminus and carboxy terminus of TonB can function independently. Only the ∼140 N-terminal amino acids (at the maximum) of TonB are required for a PMF-dependent conformational change in spheroplasts (380). TonB transmembrane domain mutations that inactivate TonB also prevent the PMF-dependent conformational response (379). By itself, the TonB carboxy terminus can bind to OM transporters, although it cannot transduce energy to them (278, 335).
(ii) The proline-rich region.
The proline-rich spacer domain [amino acids 70 to 102; (Glu-Pro)4, Ile-Pro-Glu-Pro-9 amino acid spacer-(Lys-Pro)6] serves to divide the TonB amino and carboxy termini. Structural studies suggest that it is in an extended conformation that could be as long as 100 Å (193). This domain is ∼40% proline and appears in vivo to function only to extend the reach of TonB from the cytoplasmic membrane to the OM. Although a peptide corresponding to the proline-rich spacer was shown to bind specifically to FhuA in vitro, in vivo, the proline-rich region can be almost entirely deleted without preventing TonB activity in Vibrio cholerae as well as E. coli (58, 382, 580). The existence of a proline-rich region is one of the hallmarks of the TonB protein in gram-negative bacteria, but within that region, the primary amino acid sequences diverge.
(iii) The TonB carboxy terminus.
The carboxy terminus of TonB (amino acids 103 to 239) is a site of interaction with the OM transporters. In vivo, the region from Asn159 through Pro164 makes direct contact with the TonB box (amino-terminal site of interaction with TonB) of the OM transporters BtuB and FecA and presumably others as well (67, 492). Interestingly, the group B colicins have TonB boxes (456, 517, 564) as well. Without the TonB box, colicin B cannot be translocated across the OM, although when it is shocked across the OM, it can still kill cells by depolarizing the IM (456). By analogy with the TonB-dependent OM transporter, TonB probably interacts directly with the colicin B TonB box, although this has not been tested directly. The colicin B crystal structure suggests a means by which sequential interactions of TonB, first with the TonB box of the colicin B receptor FepA and second with the colicin B TonB box, would be the translocation mechanism across the OM (267) (Fig. 9). For colicin Ia, which has a very different crystal structure for receptor and translocation domains, the means of translocation are not so obvious. Here, the putative receptor domain is spatially separate from the putative TonB box.
FIG. 9.
Model for colicin B transport across the outer membrane. In step 1, binding of colicin B (blue) to FepA (red) causes the FepA TonB box (A) to become periplasmically exposed. In step 2, TonB releases FepA globular domain bound to colicin B. The colicin B TonB box (B) can now bind TonB. In step 3, TonB releases colicin B into the periplasm. TonB is not shown on this model because its active configuration at the outer membrane is not certain. (Reprinted from reference 267 with permission from Blackwell Publishing.)
Colicin D is 96% identical to colicin B in the first 313 amino acids, the region that determines binding to the OM transporter FepA and translocation by the TonB system rather than the Tol system. Surprisingly, spontaneous TonB mutants Arg158Ser and Pro161Leu are uniquely resistant to colicin D, retaining sensitivity to colicin B as well as colicin M, colicin Ia, and bacteriophage φ80. When colicin B was given the colicin D TonB box in exchange, it did not cease to be toxic in the TonB Arg158Ser and Pro161Leu strains; similarly, when colicin D was given the colicin B TonB box in exchange, it did not become fully toxic. Taken together, these results suggest that TonB interacts with regions in addition to the TonB box (464).
Interestingly, there is an entire class of protein toxins, called microcins, that require TonB and yet have no TonB box (recently reviewed in reference 54). It seems reasonable that their small size eliminates the need for two TonB-mediated events, and their transport might be similar to that of siderophores. In another variation on a theme, colicins 5 and 10 have a TonB box but use the Tsx pore protein, which has no TonB box, as a receptor (47, 516). Thus, the TonB box appears to be the handle by which the globular domains of transporters and colicins alike are brought across the OM.
The mechanism by which bacteriophage T1 (from which TonB takes its mnemonic) and φ80 cross the OM clearly requires TonB, but here, the mechanism is not so clear (247). The recent genome sequence of phage T1 suggests a growing interest in this problem (562). T1 and φ80 host-range mutants can bypass the need for TonB (250). Recently, TonB-independent mutants in FhuA were isolated and characterized, although it is not clear why they became TonB independent (376). Further study of these host-range mutants has the potential to reveal much about the mechanism of the OM transporters.
(iv) The carboxy terminus in vitro.
When overexpressed, the carboxy terminus is the only region that is sufficiently protease resistant to be purified and examined in vitro. This frustrating behavior means that an in vitro analysis of full-length wild-type TonB has not yet been possible. Even when nearly full-length TonB (amino acids 33 to 239) is purified, only TonB amino acids 124 to 239 are recovered (507).
Nonetheless, several studies have generated some thought-provoking data about the in vitro behavior of the TonB carboxy terminus. Phage panning against the purified TonB carboxy terminus recovered library peptides corresponding to the periplasm-accessible surface of FhuA, BtuB, FecA, and FepA, including the TonB box and, surprisingly, regions interior to the luminal domains. Even more surprisingly, the phage-panning results suggested interactions with external loops of the OM transporters (74). Recently, NMR characterizations of the TonB carboxy terminus with the purified luminal domain of FepA suggested that the interaction leads to TonB disorder rather than order (510). It is not clear if these findings have in vivo correlates, but if they do, it suggests that TonB is a dynamic and structurally mobile protein.
It has been possible to crystallize the carboxy-terminal domain of TonB as a dimer (111, 348). Current structures of the carboxy terminus are perhaps best represented by the sole NMR structure of amino acids 152 to 239, which shares and summarizes elements of all the structures (509). An excellent review comparing the current structures has recently been published (676). The NMR structure is monomeric, with two α-helices on one side and four β-strands on the other. It undergoes conformational shifts in the presence of TonB box peptides that, when coupled with the importance of the region at Q160, suggest an interaction of the TonB box with the β-3 strand of TonB.
This prediction was borne out in the recent exciting cocrystallization of the TonB carboxy terminus with the OM transporters BtuB and FhuA in the presence of their ligands, vitamin B12 and ferricrocin, respectively (507, 676). In both crystals, TonB monomers (residues 153 to 233 and 158 to 235 in the BtuB/TonB and FhuA/TonB crystals, respectively) occupy approximately one-half of the periplasmic face of the transporters (Fig. 10). While the overall structure of the TonB carboxy terminus is similar to those observed previously, in both cocrystals, the TonB box of each transporter is recognized by the TonB β-3 strand (amino acids 226 to 232 in BtuB) and displaces the TonB β-4 strand (amino acids 234 to 239), which is disordered in both structures. This antiparallel interaction then additionally allows contact between the TonB box (directly visible in the BtuB structure and modeled in the FhuA structure) and the region around Q160 in TonB, previously identified as a site of interaction in vivo (67) (Fig. 11B). Indeed, the BtuB-TonB cocrystal provides a satisfying rationale for the phenotypes of BtuB TonB box mutant L8P, which inactivates BtuB by preventing interactions with TonB in vivo (67). In the TonB-BtuB structure, BtuB L8P, if present, would decrease hydrogen bonding to the TonB β-3 strand. Because the chemically different TonB suppressor mutants Q160L and Q160K restore low levels of BtuB activity (259), they must be able to compensate for this destabilization.
FIG. 10.
BtuB-TonB and FhuA-TonB crystal structures. (A) BtuB-TonB view from the periplasm. The BtuB barrel is in orange, vitamin B12 is shown as red spheres, the BtuB internal globular domain is in green, and the TonB carboxy terminus (amino acids 153 to 233) is in magenta, with the TonB box highlighted in blue. (B) BtuB-TonB side view from within the outer membrane. (Panels A and B are reprinted from reference 588 with permission of AAAS.) (C) FhuA-TonB side view from within the outer membrane. The FhuA barrel is in blue, the FhuA internal globular domain is in green, and the TonB carboxy terminus (amino acids 158 to 235) is in yellow. (D) FhuA-TonB view from the periplasm. TonB α-helices (α1 and α2) and β-strands are labeled, as are periplasmic turns in FhuA (T1 and T7 to T10), for reference. (Panels C and D are reprinted from reference 507 with permission of AAAS.)
Another interesting aspect of the cocrystals is that TonB covers only one-half of the periplasmic face of BtuB and FhuA. This leaves room for a second TonB carboxy terminus to bind, as has been observed in vitro (335). Since there is no second TonB box available in the transporter, it is not clear how this would be accomplished, although a second low-affinity site has been proposed (337). Alternatively, if the shuttle model (described below) is correct, it leaves room for the entire TonB protein to bind. Unfortunately, there are currently no definitive in vivo data on the oligomeric state of energized TonB at the OM to guide our thinking about this problem.
(v) What do the crystal/NMR structures represent?
TonB goes through at least three conformational changes during energy transduction. Currently, there is in vivo evidence for an uncharged conformation (prior to energization by ExbB/D), a charged (energized) conformation, and a discharged conformation that has already transduced energy to the OM transporter (380). Which, if any, of these forms do the crystal/NMR structures of TonB represent? Are there additional conformational changes that TonB goes through? On one hand, the interaction of TonB region 160 with the TonB box of BtuB in the cocrystal beautifully explains the phenotypes of TonB box mutants and is consistent with the in vivo disulfide cross-linking studies (67), suggesting that it is physiologically relevant. The fact that, in vivo, carboxy-terminal TonB fragments are competitively inhibitory of wild-type TonB at the OM transporters (278) has also been taken as evidence of the biological activity of the carboxy terminus (74). However, the observation that a TonB box pentapeptide is inhibitory in vivo suggests that competitive inhibition need not involve active forms (633). Any TonB fragments could theoretically competitively inhibit wild-type activity; indeed, this appears to be the basis of the dominant negative gene dosage effect that arises from TonB overexpression (434). Furthermore, unenergized forms of TonB clearly exist in vivo and in fact can form ligand-dependent complexes with FepA that are detectable by in vivo cross-linking (223, 592).
Perhaps the crystal/NMR structures represent one of several conformations in the energy transduction cycle. However, because they arise from TonB that has never been exposed to ExbB/D or the IM PMF, they theoretically could represent a form of TonB that is not physiologically relevant. The instances where the in vitro behavior of the purified carboxy-terminal TonBs is different than TonB behavior in vivo lend urgency to answering this question. For example, in vitro, a significant interaction between TonB and FhuA can occur in the absence of the ligand ferric ferricrocin (335), but in vivo, TonB apparently cannot transduce energy to nonliganded OM transporters (380). In addition, as noted above, the proline-rich domain can be deleted without inactivating TonB in vivo (382, 580), but in vitro, it is required for the formation of the 2:1 complex with FhuA believed to represent TonB at the OM (336).
(vi) The carboxy terminus in vivo.
Recent in vivo results suggest that at the OM, the TonB carboxy terminus is a dynamic domain that interacts with different transporters in different ways, possibly through a large aromatic cluster. Individual alanine replacements at five aromatic amino acids in the TonB carboxy terminus (F180, F202, W213, Y215, and F230), when expressed at chromosomal levels, give rise to substitution-specific profiles for sensitivity to group B colicins, bacteriophage φ80, and ferrichrome transport (223). For example, TonB F202A is resistant to colicin D but has only slightly decreased sensitivity to colicin Ia and can support ferrichrome transport at 95% of wild-type rates, while F180A has only slightly decreased sensitivity to colicin D, exhibits a substantial loss of sensitivity to colicin Ia, and can support ferrichrome transport at only 17% of wild-type rates. These differences in phenotypic profiles suggest that during translocation, the aromatic amino acids are involved in the differential recognition of transporters, group B colicins, or both. In addition, all possible combinations of two alanine replacements exhibit synergistic behavior, suggesting that they all interact with one another. This expectation was borne out when cysteine replacements at four out of five of the aromatic amino acids spontaneously formed disulfide-linked dimers in vivo (224).
How do these data fit with the crystal/NMR structures? In the crystal/NMR structures, the aromatic amino acid side chains are arranged in two buried clusters (F180, W213, and Y215 in one cluster and F202 and F230 in the other), thus rendering them unlikely to participate in transporter/colicin recognition (Fig. 12). The two clusters are also sufficiently distant from one another that the individual aromatic residues within them should not act synergistically between clusters, yet the genetic and biochemical data indicate that they do. The two aromatic clusters and their end-on aromatic stacking interactions were first identified in the 76-amino-acid crystal structure dimer (111); however, the relationships among the five aromatic amino acids have remained similar in subsequent monomer structures and the cocrystals. Thus, it seems probable that the crystal structures do not represent the energized form of TonB (676).
FIG. 12.
Space-filling model of the TonB NMR monomer (509). Backbone atoms are gray, aromatic side chains (F180, F202, W213, Y215, and F230) are yellow, and other side chains are magenta.
Although the oligomeric state of TonB at the OM is unknown, there is solid evidence for TonB dimers in the IM. First, the ability of ToxR-TonB hybrid proteins to activate the transcription of lacZ under the control of the cholera toxin promoter demonstrated ExbB/ExbD-dependent dimerization of TonB through the transmembrane domain. In contrast, the carboxy-terminal domain (residues 164 through 239) was capable of mediating dimerization as well but without the need for ExbB/ExbD, perhaps mimicking what occurs in the crystal/NMR structures (573). Second, with the exception of F180C, cysteine replacements at each of the carboxy-terminal aromatic residues form spontaneous disulfide-linked dimers that leave TonB confined to the IM. These data suggest that there is an obligatory dimeric intermediate conformation of TonB prior to its interaction with the OM. Here again, the significance of these disulfide-linked dimers is demonstrated by the requirement for ExbB/D and a wild-type amino-terminal transmembrane domain in order to form (224).
(vii) Models for TonB energy transduction.
There are currently two models under consideration for TonB energy transduction: a propeller model where TonB, remaining anchored in the IM, somehow pulls or twists the OM transporters to release their ligands, and a model where TonB shuttles to the OM to deliver conformationally stored potential energy (for recent discussions of the models, see references 530 and 676).
The propeller model was based largely on the first TonB crystal structure, a rigid, strand-exchanged dimer of 76 amino acids (111). Now that a looser dimeric crystal structure has been observed (92 amino acids) (348), as well as a monomeric NMR structure (88 amino acids) (509), the rationale for a strict propeller model has faded somewhat. In addition, the fact that the lack of IM ExbB/D and TolQ/R complexes can be overcome by the overexpression of TonB (48) suggests that a strictly pulling or twisting force originating in the IM complexes and transmitted along the length of TonB to the OM is unlikely. Nonetheless, mechanical aspects of the model are still attractive—how else might the internal globular domain be released from the barrel? Although two studies have reached different conclusions, it seems likely that the passage of large colicins will require the release of the internal globular domain into the periplasm (174, 183). The idea that a force perpendicular to the β-sheets of the globular domain would not have to be large to denature the globular domain out of the barrel is an attractive one (116). This model is weakened by the lack of candidates to anchor the TonB/ExbB/ExbD complex sufficiently that a force could be exerted.
A shuttling model for TonB-dependent energy transduction has been suggested based on finding TonB associated with either the IM or the OM in sucrose density gradient fractionations (397). It was subsequently shown that an engineered cysteine at the TonB amino terminus became accessible to thiol-specific reagents only under conditions where TonB could shuttle to the OM, suggesting that the amino terminus did pop out of the IM ExbB/D complex (378). In the shuttle model, TonB is converted by ExbB, ExbD, and the IM proton motive force to a form that conformationally stores potential energy, rather like setting a mouse trap. Energized TonB can then shuttle to the OM to deliver the energy (spring the mousetrap). One possibility is that the carboxy terminus serves to direct TonB to only those OM transporters that are ligand occupied by colicins, phages, or nutrients (380). The amino-terminal region of TonB would then transduce the energy to the OM transporters. This model is weakened by the lack of direct evidence for an interaction of the amino terminus with OM or periplasmic proteins. A further weakness of the shuttling model is that it does not address what TonB does once it gets to the OM. It could be that some mechanical force is necessary, or it could be that chemical signals are delivered, or both. There is still room for all the models to carry some truth.
New thoughts on an old protein.
Regardless of which model is correct, the TonB carboxy terminus clearly interacts directly and physically with the OM transporters in vivo (67, 377a, 492). The crystal structure of the TonB carboxy terminus bound to FhuA indicates that TonB residues 158 to 235 occupy only ∼25 Å of the 200-Å periplasmic space (Fig. 10C), raising the question of how the TonB carboxy terminus locates a ligand-bound OM transporter. This question deepens when the nonessential role of the potentially 100-Å proline-rich domain is considered (190). Because this proline-rich extension is not required for TonB to contact an OM transporter, the conformation of TonB that locates and binds to a ligand-bound OM transporter is almost certainly much different than that depicted by the cocrystal structures. In the crystal structure conformation, TonB lacking the proline-rich domain would be too short.
The “reach” of TonB across the periplasmic space could be extended if TonB has a semidisordered conformation. Consistent with this idea, the NMR structure of a monomeric TonB carboxy terminus shows a disordered region from residues 103 to 153. In addition, large regions of disorder are predicted from the TonB primary amino acid sequence, approximately from residues beginning at the edge of the amino-terminal transmembrane domain through residue 160 (omitting the proline-rich region) and approximately residues 180 to 220 (377b). There are several aspects of TonB behavior that parallel those of disordered proteins. Like disordered proteins, TonB consists of apparently independent modular domains (amino and carboxy termini) separated by a spacer rich in proline (among other amino acids) (reviewed in reference 170a). Signal transduction proteins often have regions of disorder that allow for great specificity of induced-fit binding while retaining the low affinity needed for reversible interactions (170a). Finally, disordered proteins are highly susceptible to proteolytic degradation outside their normal in vivo environment. It is tempting to speculate that the TonB carboxy terminus is induced or maintained in a disordered conformation by ExbB, ExbD (with ExbD as a chaperone), and the PMF, without which it would collapse into the ordered conformation characterized by the crystal structure.
The ExbB and ExbD proteins: physiological function and their role in colicin transit.
Far less is known about ExbB and ExbD than about TonB and its OM transporters. The current hypothesis is that these two integral IM proteins somehow harvest the energy from the IM proton motive force and use it to energize TonB. Since we do not yet know the details of what it means for TonB to be “energized,” it is challenging to study the mechanism of these two proteins. ExbB and ExbD belong to what is known as the MotA/TolQ/ExbB and MotB/TolR/ExbD families of proteins, respectively (48, 79, 699). TolQ/R somehow energizes the TonB analogue TolA (79, 221) and can to some degree (∼10%) also energize TonB in a process known as cross talk (46, 48, 52, 592) (see Cross complementation between Tol and TonB systems below). MotA/B energizes flagellar rotation (32). In all three cases, there is evidence that the activity of these proteins is coupled either directly or indirectly to the IM proton motive force. Consistent with that observation, these three sets of proteins have the highest degrees of similarity in their transmembrane domains (79, 531). In the absence of ExbB/D and TolQ/R, TonB expressed at chromosomal levels is inactive (48). Surprisingly, the overexpression of TonB under the same circumstances leads to low but detectable levels of activity (48), suggesting that the function of ExbB/D or TolQ/R is to overcome an energetic barrier that TonB/TolA can only rarely overcome on its own (380).
DNA sequences and topologies of ExbB and ExbD have been known for some time (173, 319, 320). ExbB appears to constitute a primary scaffolding protein since both TonB and ExbD are unstable in its absence (205). Careful quantitation indicates that under four different conditions, the ratios of TonB to ExbB to ExbD per cell are 1:7:2, respectively (264). Similar numbers have been obtained for Tol system paralogs (79). Consistent with those ratios, ExbB and ExbD can be cross-linked into dimers and trimers by in vivo formaldehyde cross-linking (265). Given the ratio of proteins in the energy transduction complex, it is most likely that TonB is buried somewhere in the middle of it, with the TonB “transmembrane domain” making direct contact with ExbB and ExbD but not lipids. Certainly, there are interactions between the TonB signal anchor and ExbB transmembrane domains, but the actual arrangement of TonB in the ExbB/D complex is unknown (381). TonB and ExbD can both bind to ExbB in vitro (51).
If the ExbB/D complex harvests the IM proton motive force, there might be a proton pathway. Two hypothetical pathways have been modeled based on the reasonable assumption that only the transmembrane domains are involved, the increasingly less reasonable assumption that conserved residues are functionally important, and the assumption that TonB, ExbB, and ExbD or ExbB and ExbD are present in ratios of 1:1:1 or 1:1, respectively, in each complex (699). Mutagenesis of appropriate Ser and Thr residues (T148, S155, and T181) in the second ExbB transmembrane domain suggests that neither pathway is important for energizing TonB (53). Also, in those studies, an Ala replacement at the highly conserved Glu176 in the third ExbB transmembrane domain rendered ExbB inactive. Interestingly, the Glu176Gln mutation had a much more negative effect on Tol system activities than on TonB system activities, whereas Glu173Gln in TolQ had a much more negative effect on TonB system activities.
Both Asp25Asn and Leu132Glu abolish ExbD activity, suggesting important roles for both the amino-terminal transmembrane domain and the periplasmic domain (51). Asp25 is also conserved in MotB and TolR. Beyond those two mutants, little is known about the domains or function of the ExbD protein.
Cross-Complementation between Tol and TonB Systems
Early genetic evidence suggested that TolQ and ExbB are somewhat interchangeable (48). Moreover, it has been shown that tolQ and tolR can replace exbB and exbD, albeit at low levels (∼10 to 20%), for TonB-dependent functions (46, 42). Braun demonstrated that colicins B and D, two group B colicins, are still active on exbBD mutants (48). Activities of group B colicins and cobalamin transport are completely abolished in a tolQR-exbBD mutant (46, 52), while the overexpression of tolQ and tolR in the exbBD mutants renders cells more sensitive to the action of these two colicins (52). Strikingly, expression of the P. aeruginosa tolQ gene (which cannot complement an E. coli tolQ mutant) has been shown to complement an E. coli exbB mutant (146). Similarly, the decreased level of activity of group A colicins, notably colicin E1, on the tolQR mutant is completely abolished in an exbBD-tolQR strain. In contrast, no cross-complementation between TolA and TonB has been observed, suggesting that these proteins regulate the specificity toward group A and group B colicins, respectively (52). However, the affinity of the TonB protein appears to be higher for ExbBD than for the TolQR complex (326). Reciprocally, TolA has a higher affinity for TolQR than for the ExbBD complex (Goemaere et al., unpublished).
Energy Dependence for Translocation
Among all the data collected on colicin translocation, few studies have been devoted to the energetic aspects. Following a report on the energy dependence of TonB-dependent bacteriophage entry (247) (see below), the group of Volkmar Braun demonstrated by competition experiments with FhuA-dependent T5 phages that the FhuA-dependent colicin M required energy to cross the OM (50). Data reported in the same study suggested that both colicin B and colicin Ib had similar energy requirements, whereas the Tol-dependent colicin E1 did not. Those authors suggested that since the irreversible absorption was energy dependent and TonB dependent, the ExbBD-TonB complex is likely to energize colicin uptake. In the case of colicin E1, it is intriguing that this colicin does not need TolR for its transit, thus reflecting its different behavior. However, Bourdineaud et al. demonstrated that the time necessary for potassium efflux upon colicin A treatment was not diminished in de-energized susceptible cells (37). Taken together, these data suggest that group A colicins might not need any energy to cross the cell envelope, whereas group B colicins do. Further results converge to the same results. First, colicin-dependent TolA degradation is observed in both energized and de-energized susceptible cells, demonstrating that group A colicins cross the OM to reach the TolA protein even under energy-depleted conditions (Barnéoud et al., unpublished). Second, it was shown that point mutations within the TolQR ion channel that abolish the PMF-dependent TolA-Pal interaction exhibit a wild-type activity towards group A colicin uptake (227a). When these mutants were used in cross-complementation experiments, they abolished both siderophore and group B colicin uptake. However, these results appear to be restricted to colicins using a TonB-dependent receptor for both cell binding and OM translocation, since the import of colicins 5 and 10 was not affected by these mutations (Goemaere et al., unpublished). Overall, these data suggest that group A colicins do not need energy to cross the cell envelope at any point, whereas group B colicins (except colicins 5 and 10) require energy provided by the ExbBD-TonB complex to cross the OM (see Transport through the Outer Membrane). In both case, energy does not appear to be involved in crossing the periplasm, and it is likely that both group A and group B colicins use a Brownian ratchet process.
Speculative Models for Colicin Translocation
From the data reviewed above, a speculative model for colicin translocation can be drawn (Fig. 9 and 13). Upon binding to the receptor on the bacterial cell surface, colicin undergoes a partial unfolding. The translocation domain is then translocated across the OM via the transporter-receptor channel in an energy- and TonB-dependent way (in the case of most group B colicins) or via a secondary β-barrel OM protein (for group A colicins and group B colicins 5 and 10). Subsequently, the translocation domain interacts in the periplasm with components of its transit machinery (TonB via the TonB box, ExbD, and ExbB for group B colicins and TolB, TolA, TolR, and probably TolQ by specific binding sequences for group A colicins). The immunity proteins of nuclease colicins are then released into the medium (165a). The cytotoxic domain is translocated through the OM and into the periplasm by an unknown mechanism. Dover et al. suggested that the pore-forming domain of colicin N might translocate through OmpF, following the same route as the N-terminal domain (161). Alternatively, the C-terminal domain might translocate through the OM bilayer by conformational changes induced by its interaction with the phospholipids (468, 469, 470) or at the interface between the porin trimer and the LPS leaflet. It has also been proposed that for group A colicins, interactions between the colicin N-terminal domain and the Tol machinery would displace Tol-Tol interactions (such as the TolB-Pal complex) (417) and then induce an OM defect and the subsequent entry of the cytotoxic domain (147). Nevertheless, the Tol proteins are not involved in the translocation of pore-forming C-terminal domains or in their insertion into the inner membrane (188). However, translocation of a fusion protein composed of the fd bacteriophage g3p N1 and N2 domains (which are necessary for both reception and TolAQR-dependent translocation) (see below) with the C-terminal RNase domain of the TolABQR colicin E3 showed a requirement for TolB, suggesting that TolB is needed for nuclease colicins to cross the IM (301). However, when the C-terminal domain has completed its translocation, the central domain is still bound to the receptor at the cell surface (24, 27, 489), and the N-terminal domain interacts with the Tol subunits (166).
FIG. 13.
Speculative models of colicin import. (A) Import of group A colicins. In stage 1, the colicin contacts the bacterial cell and binds to the outer membrane receptor by its central domain. In stage 2, the colicin partly unfolds and recruits the translocation machinery (e.g., the outer membrane translocon and the Tol complex). In stage 3, the colicin N-terminal domain translocates through the OM β-barrel and protrudes into the periplasm, where it interacts with the TolB subunit, thus dissociating the TolB-Pal complex. Stages 4 and 5 depict the Brownian ratchet mechanism. The colicin N-terminal domain dissociates from TolB to interact with TolA (stage 4) and then with TolQ and/or TolR (stage 5), probably after TolA degradation. In stage 6, the colicin C-terminal domain, carrying the lethal activity, is translocated by an unknown mechanism (?) and forms a pore in the inner membrane or translocates to the cytoplasm to degrade nucleic acids. The extracellular face is on the top, and the cytoplasm is on the bottom. A, TolA; A*, degradation product of TolA; B, TolB; P, Pal; Q, TolQ; R, TolR; Rec, OM receptor; T, OM translocon. Colicin is in red (t, translocation domain; r, reception domain; a, activity domain). (B) Import of group B colicins. In stage 1, the colicin contacts the bacterial cell and binds to a TonB-dependent gated channel by its central domain. In stage 2, the receptor recruits the TonB machinery through the colicin-induced receptor TonB box accessibility. In stage 3, following energy input by the ExbBD-TonB complex (yellow flash), the colicin N-terminal domain translocates through the OM and interacts with TonB by its own TonB box. Please note that two models are currently envisaged. The second model implies that TonB shuttles from the IM to the OM to release stored energy (see the group B colicin transit section). In stage 4, the colicin N-terminal domain dissociates from TonB to interact with ExbB and/or ExbD. In stage 5, the colicin C-terminal domain, carrying the lethal activity, is translocated by an unknown mechanism (?) to reach its final compartment. The extracellular face is on the top, and the cytoplasm is on bottom. B, ExbB; D, ExbD; Rec, TonB-dependent OM-gated channel. Colicin is in red (t, translocation domain; r, reception domain; a, activity domain). The TonB boxes of the receptor and the colicin are indicated by trapezoids. Another speculative model for group B colicin translocation is depicted Fig. 9.
Translocation of Phage DNA
Similarities between colicin and phage DNA translocation.
Some bacteriophages require the Tol or the TonB system to infect bacteria. Upon binding to conjugative F-pili receptors or a TonB-dependent gated receptor (see Colicin Reception), the bacteriophage particle or a part of it penetrates the cell envelope. Few data are available for TonB-dependent bacteriophages (T1 and φ80) (Table 1), although they require the ExbB, ExbD, and TonB proteins during the infection process (48, 247). In the case of Tol-dependent E. coli Ff bacteriophages (f1, fd, and M13), it has been shown that they require the products of the tolQ, tolR, and tolA genes but neither the TolB nor the Pal proteins (120, 565, 615, 616, 674). Similarly, CTX-phi bacteriophages that encode cholera toxin (659) infect Vibrio cholerae by parasitizing the TolQRA proteins (257, 258). As discussed in Colicin Reception, binding and translocation require the presence of the minor coat capsid protein g3p (or pIII), which is located at the tip of the bacteriophage particle (182, 234). Interestingly, like colicins, this protein is organized into three distinct domains separated by flexible glycine-rich linkers, each of them involved in a specific stage of the infection process (361, 443, 612) (see structural organization section). The functional features of the similar organizations between colicins and g3p proteins were discovered by the construction of active chimeras between the M13 g3p protein and colicin E3 (301). The g3p N-terminal domain (g3p-N1) interacts with the TolA subunit, whereas the central domain (g3p-N2) recognizes and binds the pilus tip. The function of the C-terminal N3 domain is less clear, but it anchors the g3p protein in the phage capsid and has been proposed to oligomerize and form channel of ∼8 Å in the inner membrane of infected cells, by which the DNA is thought to be injected into the bacterial cytoplasm (25, 182, 227). Furthermore, the TolQRA proteins have been shown to be essential for the insertion of the major coat protein g8p into the E. coli IM (121). However, this observation is probably indirect, since g8p insertion requires the translocation of the minor coat protein g3p to be completed. Unlike the Tol-dependent filamentous bacteriophages, the protein(s) involved in receptor recognition and translocation has not been identified for the TonB-dependent bacteriophages. In the case of the V. cholerae Tol-dependent bacteriophage CTX-phi, sequence alignments have demonstrated homologies between the CTX-phi OrfU (g3pCTX) and the fd g3p protein (271). Functional homologies between the two proteins have been demonstrated, since fd coliphages expressing g3pCTX-g3pfd protein fusions at their surface infect V. cholerae cells in an F-pilus- and TolA-dependent manner (258).
Interaction between Tol subunits and minor capsid phage translocation domains: a TolA box?
Like colicins, the first suggestion for an interaction between the g3p protein and a Tol subunit came from indirect observations. First, Lopez and Webster (418) showed that bacteriophages adsorb at discrete locations of bacteria corresponding to the contact sites between the IM and OM described by Bayer (18), to which the Tol proteins also localize (240). Second, the periplasmic expression of the g3p protein or the g3p-N1 domain in E. coli susceptible cells induces tolerance towards group A colicins, resistance to bacteriophage infection, sensitivity to deoxycholate, and the release of periplasmic proteins into the medium (33, 41, 527, 549, 702). These data suggest that the g3p protein interacts with at least one component of the Tol machinery, thus competing with natural partners or exogenous colicins or bacteriophages. Indeed, the g3p-N1 domain interacts with the C-terminal domain of TolA (120, 324, 527, 553). Heteronuclear NMR studies using labeled g3p-N1 and TolAIII domains and a costructure between the g3p-N1 domain and the C-terminal domain of the TolA protein have provided structural insights (421, 553). The interface involves a long α-helix of the TolA III domain. Two important features arise from this costructure (Fig. 14). First, the region of the g3p-N1 domain in interaction with TolAIII overlaps with the region in interaction with g3p-N2 (272, 420), suggesting that the interaction of g3p-N2 with the conjugative F pilus dissociates the g3p N1-N2 interaction, allowing the N1 domain to interact with TolA. Indeed, contrary to g3p-N1, g3p-N1-N2 does not interact with TolA (553). Second, the g3p interaction site involves a CYGT motif similar to the TolA-binding tetrapeptide motif found in the Pal lipoprotein and colicins A, K, and N (78, 527) (see TolA binding sequence above) (Fig. 8B). In the costructure, this motif is at the interface with the TolAIII molecule (421) (Fig. 14). Mutations within this motif affect interactions with the TolA subunit, and mutated g3p-N1 domains expressed in the periplasm of wild-type cells have no effect on colicin or bacteriophage uptake (527). A micromolar-range affinity between g3p-N1 and TolAIII was measured by SPR (323) (Table 3). Here again, similarly to colicins, the affinity decreases when g3p-N1N2 instead of g3p-N1 is used (323) (Table 3). Another similarity with colicins is the observation that the infection of susceptible cells with M13 bacteriophages provokes the degradation of the TolA protein. However, the degradation pattern is quite different, suggesting that proteolytic cleavage is not identical (Barnéoud et al., unpublished). Indeed, the addition of the purified g3p-N1 domain on labeled TolAIII molecules induces weak structural changes of the TolA C-terminal domain (148), demonstrating that if both colicins and g3p N-terminal domains interact with TolAIII, and if both interactions involve a similar tetrapeptide motif, the mode of interaction is likely to be different. Conversely, the addition of TolAIII on labeled g3p-N1 showed chemical shifts indicative of conformational modifications within the g3p molecule (148, 553). Competition between the g3p-N2 and the TolA C-terminal domains is suggested by NMR analyses showing that the addition of purified g3p-N2 on labeled g3p-N1 induces identical modifications (553). Interactions between the g3p-N2 domain and the central domain of TolA have been reported (323).
FIG. 14.
Crystal structure of g3p-N1 bound to TolAIII. The cocrystal structure of the M13 g3p translocation domain (g3p-N1, left) with the TolA C-terminal domain (TolAIII, right) (PDB accession number 1TOL; 1.85 Å) (421) is shown. The conserved TolA binding motif (CYGT) (Fig. 8B) of g3p-N1, located at the interface between the two domains, is shown in a ball-and-stick representation and is highlighted by a pink circle.
No interactions between the g3p protein (or another component of the phage particle) and the TolQ and TolR subunits have been reported so far. Interactions between g3p-N1 and TolR were detected neither in enzyme-linked immunosorbent assay experiments carried out with coated TolRII nor by NMR when TolRII was added to labeled g3p-N1 (553) (Table 3).
Energy requirements for phage DNA uptake.
No data are currently available for the energy requirements of Tol-dependent bacteriophage DNA uptake, unless their uptake requires both ATP and proton motive force (685). This energy is probably required for pilus retraction. Nevertheless, TonB-dependent bacteriophage uptake requires PMF (247). PMF is involved in OM translocation through TonB-dependent transporters acting as receptors, since it is necessary for bacteriophage irreversible adsorption (247). In V. cholerae, the entry of phage CTX-phi requires PMF (253).
Speculative models for phage DNA translocation.
A model for phage DNA translocation via the Tol system is depicted in Fig. 15. Phage infection is initiated by the binding of the g3p-N2 domain to the tip of the conjugative F pilus. This recognition and binding event is followed by the retraction of the pilus (298) and the dissociation of the N1 domain (553). Near the OM or in the periplasm, the g3p-N1 domain would interact with the C-terminal domain of TolA. Since the affinity of g3p-N1 for TolAIII is higher than that for g3p-N2, g3p cannot reassociate with g3p-N2 (553). It is interesting that N1-N2 domain assembly and dissociation are very slow events, probably allowing pilus retraction to occur (439, 440). Further steps are unclear, since no interaction has been demonstrated with the TolQ and TolR proteins, which are required for the infection process. The g3p-N1-TolAIII interaction would induce structural modifications within g3p-N3, allowing the entry phase of infection to proceed (25). The g3p-N3 domain then inserts into the inner membrane, oligomerizes, and forms a pore with a size compatible with the diameter of a single-stranded DNA molecule. This stage is probably followed by the disassembly of the capsid, the insertion of the major coat protein g8p in the membrane, and the subsequent translocation of the DNA through the g3p channel. The major force driving the DNA out of the capsid has been proposed to be the relief of the capsid internal pressure (398, 399) or alternatively that DNA uptake might be linked to its cellular replication (435).
FIG. 15.
Speculative model for Tol-dependent filamentous phage DNA translocation. The model is depicted in six consecutive stages. In stage 1, the filamentous bacteriophage contacts the bacterial cell and binds to the tip of an F pilus by its reception N2 domain (r). In stage 2, the interaction between g3p-N2 and the F pilus dissociates g3p-N1. In stage 3, the F pilus retracts and brings the g3p molecule into the periplasm, where it interacts through the translocation domain (t) with the TolA C-terminal domain (TolAIII). In stage 4, the g3p-N2 domain interacts with the TolA central domain (TolAII). In stage 5, the g3p-N3 domain that anchors the minor coat protein into the phage particle dissociates, inserts into the inner membrane, oligomerizes, and forms pores by which the single-stranded DNA (in red) translocates (stage 6) after disaggregation of the capsid by the insertion of the major coat g8p proteins into the membrane. The TolA degradation product observed upon the treatment of WT cells with M13 bacteriophage is represented (A*). Periplasm is on the top, and cytoplasm is on the bottom. The insert at stage 2 represents the electron microscopy picture of an M13 filamentous bacteriophage bound at an F pilus tip (bar, 100 nm). (Reprinted from reference 442 with permission.)
COLICIN ACTIVITIES
Pore-Forming Colicins
Judging from the marked similarities of the known crystal structures of colicin pore-forming domains, all of the pore-forming colicins ought to form pores in a similar way. The C-terminal domain of each colicin retains its ability to form channels when isolated from the rest of the protein, and it was just such an isolated domain, that of colicin A, that was the first domain of a colicin molecule to be solved by crystallography (502). It, and the four other pore domains that have now been solved (of colicins Ia, E1, N, and B) (177, 267, 650, 675), consists of a tightly packed bundle of 10 α-helices. Its one hydrophobic segment forms a helical hairpin (helices 8 [H8] and H9) sequestered from the aqueous phase by the other eight (mostly amphipathic) helices (Fig. 16). This is the structure of a compact, water-soluble protein, and the pathway of its transformation into a voltage-gated ion channel is far from obvious. Nevertheless, the ability of this domain to rearrange itself upon interaction with the inner membrane accounts for one of colicin's most striking qualities: despite its origin in the producing cell as a water-soluble, monomeric protein (102), it goes on to become, effectively, a membrane protein of the target cell. In what follows, we will consider some of the properties, many quite atypical, of the channel formed by this multifaceted protein.
The closed channel.
The discovery that purified, aqueous colicin can bind to pure lipid membranes and form voltage-dependent channels demonstrated that no target cell proteins were required for membrane insertion or channel formation (575), opening the way for colicins to be studied in various in vitro systems. The initial binding of colicin to the membrane, which is nearly irreversible under some conditions, was proposed to result from an umbrella-like structure in which the unique hydrophobic segment (helices 8 and 9) would be inserted into the membrane, where it could interact with the hydrophobic core of the bilayer, while the other eight helices, many of which are amphipathic, splay out onto the surface while retaining their secondary structure (502). The umbrella model was supported by fluorescence resonance energy transfer experiments on colicin A in a lipid vesicle system, which showed that the distance between the hairpin formed by H1/2 and the rest of the molecule increased by 10 to 15 Å in the bound state compared to the solution state (373). However, subsequent work failed to support some details of the umbrella model and led to the “penknife” model (Fig. 17A), in which it was postulated that only H1/2 moves substantially away from the rest of the channel-forming domain, which remains compact but sinks into the membrane to a depth sufficient to bury the (now exposed) hydrophobic hairpin (374). The penknife model was supported by disulfide bond engineering experiments in which a disulfide bond between two introduced cysteine residues was used to covalently link adjacent helices in the crystal structure (167). The only disulfide links that blocked the insertion of colicin A into 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (DOPG) vesicles (determined by a fluorescence quenching assay) were those that prevented H1 or H2 from moving away from the rest of the domain. However, time-resolved fluorescence resonance energy transfer and quenching experiments with colicin E1 (408, 696, 697) showed that initial unfolding at the membrane surface begins with a subtle rearrangement of H9/10, followed by the movement of the H1/2 hairpin away from the hydrophobic hairpin. Half a second later, helices 3 to 7 have moved out away from the hydrophobic hairpin. The final structure is reminiscent of the umbrella model in that the hydrophilic helices are all on the surface, but they are longer than the crystal structure helices, and they form a spiral rather than spokes (Fig. 17B). The data allow some flexibility in the arrangement of the helices, and in any case, such a structure would be expected to be rather mobile, which has been postulated to be a necessity for subsequent membrane insertion and channel formation (693). Luo et al. used NMR experiments to arrive at a similar structure for the closed state of colicin Ia (428). Interestingly, the maximum channel activity of colicin Ia correlated with conditions that maximized helix rigidity, suggesting that the helices insert into the bilayer as preformed units (688). Further complicating the picture, it has been proposed that colicins A and B interact with the membrane via a molten-globule intermediate in which the secondary structure is retained while the tertiary structure is lost (192, 640), whereas this is not the case with colicin E1 (577, 692) and colicin N (192). Presumably, the discrepancies among these models represent real differences among the colicins, despite their seductively similar crystal structures. However, the penchant of individual laboratories for focusing on particular colicins may contribute to the apparent dissimilarities.
FIG. 17.
Schematic representation of certain closed states. Closed colicin channels can adopt several membrane-bound conformations, two of which are illustrated. (A) Penknife model based on studies of colicin A (374). (B) Variant of the umbrella model, as proposed for colicin E1 (408). The figure is not meant to suggest that the secondary structure of the protein in these states replicates the crystal structure.
None of the data challenge the central role of the hydrophobic hairpin in directly contacting the lipid tails, for which there is independent evidence both in vivo with colicin E1 (608) and in vitro with colicin Ia. Experiments in planar bilayers (340) showed that the interhelical loop between H8 and H9 of colicin Ia channels is exposed to the solution on the trans side of the membrane in the open state and in at least some closed states (including channels which had never opened), thus demonstrating that the hydrophobic hairpin can adopt a transmembrane orientation in the closed state, before the channel opens.
The open channel.
Uncertainties about the structure(s) of the closed channel, which entail choosing among feasible models, pale in comparison to those about the open channel, for which no feasible models exist. It is clear that positive voltage applied to the closed channel leads to further insertion into the membrane and the formation of an ion channel with a conductance of a few pS under physiological conditions. However, several properties of the channel are irreconcilable with the available structural data. In particular, all evidence asserts that colicin channels are monomeric (reviewed in reference 596), and yet they form pores that appear to be too large to model with the available protein.
Even the entirety of the 10-helix pore domain would seem to be inadequate for the task, but persuasive evidence that rules out the use of a sizable part of it has emerged. For both colicins E1 and A, several helices worth of protein can be completely eliminated from the N terminus of the domain without preventing channel formation (16, 124, 408, 478). There remains some uncertainty about the precise location of the upstream edge of the channel, part of which results from the fact that some of the shorter fragments tested form channels with altered properties compared to the native protein and part of which may reflect true differences among the colicins. In any case, it seems clear that, at least for colicins A and E1, the first three helices can be safely discarded and that some or all of the next two (i.e., H4 and H5) may not be crucial, leaving only five to seven helices to form the pore. This basic inference is bolstered by a distinct set of experiments that looked at the geometry of the open channel in the membrane by mapping the exposure of biotin tags introduced at particular residues to soluble streptavidin in the bathing buffers. For both colicin Ia and colicin A, most or all of helices 2 through 5 were deduced to be fully exposed to the trans solution (Fig. 18), ruling them out as transmembrane elements required for the channel structure (544, 595).
FIG. 18.
Schematic representations of the transmembrane segments of the open colicin channel. Only the channel-forming domain is shown. The depicted arrangement of the protein segments within the membrane derives from experiments, but the secondary structure (shown as blue, numbered, α-helical cylinders) follows that of the crystal structure and is purely speculative. Likewise, the distortion of the lipid bilayer is shown only as an acknowledgment of the potential involvement of lipids in the structure and not as a specific model. (A) Colicin Ia. Helices 2 to 5 are translocated across the membrane during channel opening. The inset shows the electrophysiological record of colicin Ia channels in a voltage-clamped lipid bilayer. Two channels open at positive voltage and close at negative voltage. (B) Colicin A. The colicin A channel resembles that of colicin Ia, but helices 2 and 5 are thought to be incompletely translocated. The inset shows colicin A channels in a lipid bilayer.
The remaining helices (which, we remind the reader, have not been shown to retain their helical structure in the open-channel state) form a weakly selective pore of approximately 10 Å. A definitive value for this number remains elusive, but the lumen is surely larger than the well-known tetrameric voltage-gated channels and may be much larger. The lowest value reported for a diameter of a colicin channel is 7 Å for colicin Ia (360) using an assay based on the influence of nonelectrolytes on conductance. Perhaps more germane are a succession of experiments that measured the permeation of ions of various sizes. None of those experiments succeeded in identifying a monovalent ion that was too large to permeate (65, 66, 551). Tetraethyl ammonium (TEA), a cation of about 8 Å with tetrahedral symmetry, permeates easily. We recently found that tetra-kis-2-hydroxyethyl ammonium, which is the tetrahydroxyl derivative of TEA and thus is even larger and more hydrophilic, can go through the colicin A and colicin Ia channels (P. Kienker and S. Slatin, unpublished observations). (The transmembrane movement of molecules far larger than these monovalent ions is also catalyzed by colicin, but it is not likely that they cross by passing through the lumen in the same way as the smaller ions are presumed to permeate [discussed below].) The resolution of the too-little-protein/too-much-pore problem might be resolved if the protein was somehow recruiting lipid molecules to participate in the structure of the pore (594, 695). Such a mechanism has been suggested for certain peptides that form multimeric pores (405, 450, 451), but it is not yet clear how it could be applied to colicin. Recently, however, evidence in support of this proposal has been reported (605, 606).
A credible model of the open pore must account not only for the apparently large size of the lumen but also for the comparatively small conductance and, in the case of colicin A, for the anomalously high proton selectivity (338). All of the colicin channels exhibit a high selectivity of protons over other cations, but the PH+/PK+ of colicin A is so high (∼104 at neutral pH) that it is difficult to envision a conduction pathway in which the permeant ions do not interact strongly with the pore. How this may occur in a 10-Å lumen, with or without the participation of lipids, is unclear.
Channel gating and translocation.
All colicin channels are voltage dependent, opening at positive voltage and closing at negative voltage. The voltage dependence is steep, signifying that several formal charges cross the electrical field to sense the voltage, although only a few quantitative studies have been undertaken (124, 483), no doubt because of the complexity of the system: there are a multiplicity of rate constants, open states, and closed states. We presume that most of this complexity arises from the numerous partially inserted configurations of the protein, although some may be due to bound impurities, as was suggested by Cavard (89). The basic mechanism by which the channel gates is believed to be via the insertion of a large fraction of the protein into and across the membrane (598). In this model, positive voltage drives helices 2 through 5 across the membrane, leaving only four transmembrane segments (the two formed by the hydrophobic segment and one each from H1 and H6/7) to span the membrane. The hydrophobic hairpin, which can insert at zero potential (340), would precede the voltage-dependent steps, followed next by H6/7 and then by the translocated domain (H2 to H5), each section catalyzing the translocation of the next, by mechanisms not yet understood. Whatever the mechanism, it is quite indiscriminate, since surprisingly large molecules attached to the translocated domain can be translocated along with it (597). The simplest explanation would allow the translocated entities to go through the nascent pore, an idea whose feasibility depends on the size of the pore and the size of the translocated substrates. Kienker et al. tried to determine the size of the translocation pathway by presenting colicin Ia with a series of potential peptide cargoes of various sizes that were constrained to remain folded by disulfide bonds (339). They found that cargoes at least as large as 26 Å could be translocated, a figure that seems to be incompatible with the conductance of the stably open colicin pore. (The conductance of the colicin pore is similar to that of highly selective pores, such as gramicidin and the voltage-gated tetrameric channels; i.e., its conductance is “too small” for a pore that conducts TEA and not too large.) If the translocation “pore” is an open pathway across the membrane, it must be present only transiently.
The unexpected translocation machinery of the colicin channel has several interesting consequences. In the case of colicin Ia, the channel formed by the isolated C-terminal domain acquires a new configuration, since H1 is free to translocate along with H2 to H5, a transition that is prevented in the intact colicin by upstream domains not directly involved in channel formation (299). This new state of the channel has only three, rather than the usual four, transmembrane domains, and while its conductance is about sixfold smaller than that of the native channel, it is nevertheless a channel with properties not very unlike that of the whole protein (Fig. 19A and B). This raises the prospect that the lumen of the native channel is formed by only three transmembrane segments, with the fourth, formed by H1, serving to modify its conductance. Any alternative explanation demands that one-fourth of the pore's structural elements can be arbitrarily plucked from the channel without destroying it. Interestingly, the isolated C-terminal domain of colicin A also undergoes a conductance transition, but the new conductance state is only slightly smaller than the native state, and it is not the result of the translocation of H1 (595). Since the open-channel topology of native colicin A is similar to that of native colicin Ia, the implication is that the channels formed by its C-terminal domain are rather different (Fig. 19) and serves as a reminder that the fundamental features of these proteins that make them pore formers remain unexplained.
FIG. 19.
Schematic representation of the transmembrane segments of the open states of the isolated C-terminal domain of colicin Ia. (A) Normal conductance channel similar to the channel formed by the whole molecule. (B) Small conductance channel, the result of the translocation of H1. The inset shows the electrophysiological record showing the transition from the normal to the small conductance state. The membrane is clamped at +70 mV.
Immunity to pore-forming colicin.
The cell-killing potential of a pore-forming colicin is remarkable in that one molecule kills one cell. Equally remarkable is the protection system that operates at 104 to 107 times the concentration of colicin that would kill a nonimmune cell. This protective mechanism is achieved by a small polypeptide of 11 to 18 kDa, called the immunity protein, encoded by the same plasmid as colicin. The immunity proteins confer upon cells protection against the colicin they produce but not against heterologous colicins with identical modes of action, even those with considerable sequence similarity. Interestingly, immunity proteins are required to protect the cell against the action of exogenous colicin, probably produced by its neighbors, but are not required to protect it from internal colicin, since the polarity of the transmembrane potential is opposite to that required to open the pore.
(i) Immunity proteins interact with the pore-forming domain in the inner membrane.
The immunity proteins are constitutively expressed at a low level despite the high-level expression of the corresponding colicin (411) (see Colicin Synthesis above), and in order to identify the products of the immunity genes, it has been necessary to clone the genes in appropriate overexpression vectors (215, 433). Such techniques have made the demonstration that immunity proteins reside in the inner membrane possible (215, 229). Examination of DNA sequences of the immunity genes reveals that they fall into two prospective classes: the A type (proteins conferring immunity to colicins A, B, N, and U) and the E1 type (proteins conferring immunity to colicins E1, 5, K, 10, Ia, and Ib). These two types correspond to two natural classes of the pore-forming colicins based on the length of the hydrophobic segment. The topology of the colicin A, E1, U, and 5 immunity proteins (Cai, Cei, Cui, and Cfi) was determined by studies of these proteins fused to alkaline phosphatase or β-lactamase. Cai and Cui contain four transmembrane segments, and their N- and C-terminal regions are directed toward the cytoplasm (216, 519) (Fig. 20). Cei and Cfi have only three membrane-spanning segments; their N termini are directed toward the cytoplasm, and their C termini are located in the periplasm (514, 608, 609). The orientation of immunity proteins is in agreement with predictions based on the positive-charges rule, which describes the topological charge bias of cytoplasmic membrane proteins (482). These results suggest that the A-type immunity proteins have four transmembrane segments, whereas the E1-type proteins have three (Fig. 20).
FIG. 20.
Topographical models of (A) Cfi and (B) Cai in the cytoplasmic membrane of Escherichia coli. H1, H2, H3, and H4 denote the trans-membrane α-helices; L1, L2, and L3 denote loops; and T1 and T2 correspond to the N- and C-terminal ends, respectively.
In an attempt to identify the colicin domains involved in immunity recognition, domain exchanges between colicins Ia and Ib and between colicins A and E1 were performed. In all cases, the immunity protein recognized the C-terminal domain of its cognate colicin (21, 433). A similar conclusion was reached in experiments with the purified pore-forming domain of colicin E1. This peptide, devoid of its translocation and receptor-binding domains, killed osmotically shocked sensitive cells but did not kill immune cells treated in the same way (31). Based on those results, the specificity of colicins to their immunity proteins was localized to the C-terminal domain. However, the ability of a low copy number of immunity proteins to protect cells against high concentrations of exogenous colicins led to the hypothesis that immunity proteins may also interact with inner membrane components of the transit machinery, a speculation that subsequent work failed to support. Thus, active hybrid colicins in which the receptor and the translocation domains of two colicins were changed so as to direct the pore-forming domain of each colicin to the transit machinery of the other remain susceptible to their cognate immunity proteins (217, 701), suggesting that the transit machinery is not involved in the immune process. This was confirmed by introducing a prokaryote signal peptide downstream from the colicin A pore-forming domain (sp-pfColA). sp-pfColA has been found to be directed to the inner membrane via the periplasm and to form a functional channel specifically inhibited by its immunity protein by a mechanism independent of the tol genes (188). It was concluded that immunity proteins interact directly with their corresponding specific pore-forming domains in the inner membrane. It is thought that the lateral diffusion of immunity proteins in the membrane would ensure the rapid recognition of the pore-forming domain (217, 701).
(ii) Immune process involves intramembrane association.
The specificity of colicins with respect to immunity is determined by the C-terminal pore-forming domains. This raises questions as to the identity of the specificity determinant in the C-terminal domain. Several genetic studies of A- and E1-type colicins have designated transmembrane helices as the main determinants recognized by immunity proteins. Based on interchanging residues between homologous colicins (407, 514) and on screening immunity bypass mutants (701), the N terminus of helix 6 and the C terminus of helix 7 have been identified as being the immunity determinants of colicin E1, whereas for colicin 5, it is helix 6 that confers immunity. Therefore, the inactivation of E1-type colicins occurs via interactions between the voltage-gated region and the transmembrane helices of their immunity proteins. Studies on A-type colicins have designated another region as being the main determinant recognized by immunity proteins, the hydrophobic helical hairpin (31, 189, 477) and, in the case of ColU and ColB, the tip of the hydrophobic helical hairpin (519). Apparently, A- and E1-type colicins differ with regard to the membrane locations of the sites of interactions with the cognate immunity proteins. Does this difference reflect two different mechanisms of colicin inactivation? Based on the membrane location of the immune process and the identification of the colicin transmembrane helices involved in this process, it is clear that immunity proteins do not prevent the membrane insertion of their cognate colicins. It is reasonable that immunity proteins either prevent channel opening or block the channel once it is opened. Using a coimmunoprecipitation procedure, Espesset et al. showed that there is an interaction between the colicin A pore-forming domain bound to the cytoplasmic membrane and its immunity protein (189). This interaction did not require the channel to be in the open state but required the presence of the hydrophobic helical hairpin in the membrane (189, 477), indicating that A-type immunity proteins can bind colicins prior to pore formation. In the E1-type colicin, the situation seems to be more complex, since the transmembrane helices recognized by the immunity protein are located in the voltage-gated region of the colicin. Despite the uncertainties about the structure of the closed channel (see above), it is believed that the voltage-gated region remains at the membrane surface. According to these data, the helices recognized by the E1-type immunity proteins (H6 and H7) would be at the membrane surface and thus accessible only to the small periplasmic loop of the immunity protein, which has not been identified as being a crucial region for the colicin-immunity association. However, upon voltage gating, H6/7 adopts a transmembrane orientation, placing it proximal to the transmembrane helices of the immunity protein. The N terminus of H6 would be in the inner leaflet of the inner membrane, close to the cytoplasmic loop of the immunity protein, which has been shown, in the Col5 and Col10 systems, to contain colicin recognition determinants. Judging from those results, it is tempting to favor the idea that the A-type immunity proteins inactivate colicin in a closed state and that E1-type immunity proteins inactivate colicin in an open state or shortly before channels in the membrane are opened.
The genetic approach to the question of the colicin-immunity association has been extensively used and has yielded interesting insights. However, further progress may depend on being able to reconstitute the colicin-immunity complex in artificial vesicles or other membrane systems where it can be subjected to biophysical and biochemical techniques. Unfortunately, immunity function has yet to be reconstituted in an in vitro model system.
Enzymatic Colicins
Enzymatic colicins fall into two groups, those that function in the cytoplasm as nucleases and colicin M, which disrupts peptidoglycan biosynthesis in the periplasm. Underpinning the modes of action of these colicins are different translocation mechanisms and distinct modes of neutralization by immunity (Im) proteins. Each of these enzymatic groups is discussed in the following sections, with emphasis on the nuclease colicins that have been more widely studied.
Overview of nuclease colicins and their immunity proteins.
The majority of enzymatic colicins that have been described target phosphodiester bonds in the bacterial cytoplasm, eliciting cell death as either hydrolases or transferases. This varied group of enzymes target genomic DNA (DNases), 16S rRNA (rRNases), or tRNAs (tRNases) and, in several instances, are evolutionarily and/or structurally related to other enzyme families of differing biological functions. Enzymatic colicins begin their passage into cells through either the Tol or Ton system, with T-domain-swapping experiments showing that their dependence on these systems is often interchangeable, implying that colicin translocation across the OM is performed by a single generic mechanism. Nuclease colicins must further translocate across the IM, raising the question of how this is accomplished. The following section deals with nuclease colicin translocation across the IM. Thereafter is a discussion of the structural biology and biochemistry of colicin nucleases. The final section deals with colicin nuclease immunity proteins, summarizing their structure and function and the basis for their specific, high-affinity recognition of colicin cytotoxic domains. The sequence numberings shown in the following sections are generally those for full-length (Col) colicins, with reference to the equivalent residues in the isolated nuclease domains that are often used in the literature.
Nuclease transport across the inner membrane.
Once a nuclease colicin has been translocated across the OM and periplasm, the task faced by the cytotoxic domain is quite different from that of the pore-forming colicins, since the entire domain has to cross the cytoplasmic membrane. Since the nuclease domains are structurally unrelated to one another, their mechanism of translocation to the cytosol must be structure independent and likely to proceed by one of three routes: the enzymatic domains themselves might have the capacity for translocating across the membrane, possibly at the terminal stage of energized Tol/Ton-mediated import (“self-propulsion”); the domains are actively translocated from the periplasm by a host system that has yet to be identified (“retrotranslocation”); or there is a combination of these mechanisms. Recent progress in this area is summarized, although it is emphasized that this aspect of colicin biology remains poorly understood. Also reviewed is recent work suggesting that colicin nuclease domains are proteolytically processed either before or during translocation across the IM.
(i) Colicin nucleases associate with anionic phospholipids.
Evidence that colicin nucleases associate with phospholipids and that this partitioning into the lipid phase may represent the first step in translocation across the IM is accumulating. Early experiments by Escuyer et al. focused on the intact colicin E3-Im3 complex and the effects of low pH (mimicking the local pH at the surface of the inner membrane) and neutral detergents (186). They deduced that the toxin exposes a hydrophobic domain at a low pH (∼3.5), at which point the Im protein dissociates, with the nuclease domain partitioning into the aqueous phase and Im3 partitioning into the detergent phase. The difficulty in interpreting the effect of neutral detergents on intact colicins is that both the inner and outer membranes of E. coli carry net negative charges and that colicins are multidomain proteins, with some domains involved in interactions with receptors/translocation proteins that are spatially distinct from the cytotoxic domain. Changes in pH may induce structural changes that may have little physiological relevance, since these changes may not be experienced by the whole protein.
Once the cytotoxic nuclease is brought to the IM, it is conceivable that domains might begin their journey across the membrane by interacting first with the bilayer by analogy with pore-forming colicins and with bacterial toxins that target eukaryotic cells. Recent experiments have begun to address this question, focusing primarily on the effects of anionic phospholipid vesicles that mimic the E. coli inner membrane on the structural integrity of colicin DNase and rRNase domains by using intrinsic tryptophan fluorescence emission and circular dichroism spectroscopy as indicators of structure (468, 469). Two assumptions underpin these experiments: first, only the cytotoxic domain of these toxins approaches the IM; and second, the Im protein is lost at the stage of OM translocation and therefore is not bound to the nuclease as it approaches the cytoplasmic membrane. Those studies yielded essentially analogous results. E3 rRNase and E9 DNase domains, which are both basic proteins, interact strongly with anionic phospholipid vesicles, with binding being driven by electrostatic attraction. As a consequence, the association can be abolished either by high salt concentrations (>300 mM NaCl) or by decreasing the amount of negative charge on the vesicles, with no observed binding to neutral lipid vesicles. Not surprisingly, pure anionic phospholipid vesicles, such as DOPG, act as strong denaturants of colicin nuclease domains at pH 7, while lipid mixtures of DOPG and dioleoylphosphatidylcholine that mimic the charge state of the IM (∼25 to 30% negative charge) do not unfold the proteins but destabilize them significantly (468, 469). Importantly, the association and destabilization of both the rRNase and DNase domains of colicins are enhanced when such vesicles are acidified (pH ∼4), which may be more indicative of the situation in vivo.
The DNase and rRNase domains of nuclease colicins undergo similar structural changes when exposed to anionic phospholipid vesicles, even though they are structurally unrelated. The tertiary structure is largely abolished, but significant secondary structure remains, suggesting that colicin nuclease domains attain molten-globule-like states akin to those of some pore-forming colicins prior to channel formation (640). The level of destabilization/denaturation induced in rRNase and DNase domains by anionic phospholipid vesicles is much less than that induced by chemical denaturants, such as low pH or urea (468, 469). Nevertheless, pH-denatured domains interact strongly with anionic phospholipid vesicles, implying that interactions with the anionic surface are not dependent on the nuclease having a tertiary structure.
Further evidence that colicin nucleases interact with membrane surfaces has come from planar lipid bilayer experiments with colicin DNase domains, all of which exhibit random channel activity (467). The channels themselves are not responsible for cell killing, since active-site mutants of colicin DNases that are inactive as bacteriocins still have channel activity. Other colicin nucleases such as the rRNase domains do not show this activity, implying that this is either part of a mechanism of translocation specific to DNase colicins or just a manifestation of their partitioning into membranes, with random fluctuations occasionally causing a disruption in membrane continuity that generates ion conductance. The colicin E9 DNase channels are evident at low (nanomolar) protein concentrations, and although the activity is random, there appears to be a single-molecule-unit conductance of ∼100 pS (at pH 7 and 0.1 M NaCl) with rarer larger conductance states (800 to 1,000 pS) seemingly multiples of this unit size. Unlike the pore-forming colicins, E9 DNase channels are short-lived, with lifetimes of the order of milliseconds, and are not voltage dependent. DNA and metal ion binding has little effect on the ion channel activity, but Im protein binding abolishes the activity, consistent with the idea that the immunity protein blocks the ability of the DNase to associate with lipid membranes. The biological relevance of the colicin channel activity is emphasized by the effects of a single intramolecular disulfide bond that was engineered into the colicin E9 DNase domain. The disulfide, which did not affect endonuclease activity, abolished both channel activity and colicin cytotoxicity, effects that were reversed upon reduction with dithiothreitol (467). Interestingly, the disulfided protein still bound DOPG phospholipid vesicles and experienced structural changes similar to those of wild-type E9 DNase, showing that the formation of colicin DNase channels is subsequent to the structural changes induced by lipid binding (469).
(ii) Translocation across the IM and refolding.
At present, it is not known how colicin nucleases translocate across the cytoplasmic membrane. The association of colicin DNases and rRNase with anionic phospholipid surfaces suggests that at least part of the mechanism for translocation across the IM may involve “self-propulsion,” that is, the electrostatically driven association of the nuclease with the membrane. Whether such associations occur in vivo and whether they would be sufficient to allow the complete traversal of the IM are unclear. It is possible that a host system that might dislocate colicin nuclease domains into the cytosol from a membrane environment could be hijacked. An analogous process occurs in the uptake pathway of the ricin A chain in mammalian cells that is imported into the cell cytoplasm via the energy-dependent endoplasmic reticulum-associated degradation pathway (591). Moreover, like nuclease colicins, the ricin A chain is also destabilized by interactions with negatively charged phospholipids (135). Although such a retrotranslocation pathway has yet to be identified, possible candidates include reversal of the general secretory (Sec) or Tat pathways, which normally secrete protein into the periplasm in their unfolded and folded states, respectively. These pathways can, however, be ruled out, since E. coli mutants in both remain sensitive toward nuclease colicins (D. Walker and C. Kleanthous, unpublished observations). Assuming that a colicin nuclease is unfolded upon entry into the cytosol, the domain would have to refold in order to begin enzymatic digestion of cellular targets. This step is the most facile of the entry mechanisms, since there are many reports in the literature of colicin nuclease domains being refolded from chemical denaturants without the need for chaperones (491, 662, 668). How such unfolded domains escape proteolysis during folding, however, remains to be established.
(iii) Processing of colicin nuclease domains.
There have been a number of recent reports on the proteolytic processing of colicin nuclease domains prior to and/or concomitant with translocation across the cytoplasmic membrane. Those reports followed early work showing the susceptibility of nuclease colicins or related bacteriocins to proteolytic breakdown, but the link to the biological mechanism of import was less clear (97, 364). Recent work has sought to link proteolytic processing directly with import. de Zamaroczy et al. identified the IM leader peptidase LepB as being required for colicin D cytotoxicity by using a screen for colicin D mutants that were not receptor binding mutants (157). From this screen, those authors identified a mutant LepB (Asn274Lys) that was resistant to colicin D and that failed to release the nuclease domain in in vitro proteolytic cleavage assays. The mutant, which lies close to the LepB active site but does not impair native LepB function, retains sensitivity to other Tol-dependent nuclease colicins such as E2 and E3; consistent with this observation, no processing by LepB-containing extracts was seen for other enzymatic colicins (156). Processing of the colicin occurs at Lys607 and is protected by the binding of the immunity protein to the nuclease. Mutants at this and surrounding residues yielded poorly active colicins that were no longer processed properly. Some uncertainty persists regarding whether LepB itself is the protease, since purified LepB fails to cleave ColD, suggesting the involvement of additional factors.
The specific proteolytic excision of a nuclease domain has also been demonstrated for the DNase colicin E7 but by a mechanism distinct from that of colicin D. Following extended incubations of colicin with periplasmic extracts, Shi et al. identified a cleavage site in ColE7 between Lys446 and Arg447 that released the intact DNase and that was dependent on an EDTA-sensitive protease in the extracts (LepB is not EDTA sensitive) (585). This is a highly conserved region of DNase colicins positioned at the junction of the R-domain coiled coil and the nuclease. Mutants at Arg447 showed aberrant processing and weak colicin activity while retaining substantial DNase activity in vitro.
Structural biology of colicin nucleases and their modes of action.
E-group colicins comprise colicins E2 to E9 and D. E-group colicins are ∼60 kDa, while ColD is a 75-kDa toxin. The nuclease domains of these toxins are housed in the C-terminal 90 to 130 amino acids of the toxin, which can be liberated from the rest of the protein. Structures for all major classes of colicin nuclease-Im protein complexes are available (Fig. 21), and in some instances, the basic enzymological properties of the nuclease domains are known and their mechanisms of substrate binding and hydrolysis are characterized. The following sections highlight recent findings on these enzymes and their ability to cleave nucleic acids.
(i) Colicin DNases.
(a) H-N-H/ββα-Me enzymes by another name. DNase colicins are metal-dependent enzymes that randomly degrade DNA (309). Four DNase colicins have been reported: ColE2 was the first to be recognized (574), followed by E7, E8, and E9 (108, 126). The DNase domains of these colicins share ∼65% sequence identity. The structures of the E7 and E9 DNases, which have very similar three-dimensional structures, have been published (343, 346). The enzymes are composed of a central three-stranded antiparallel β-sheet surrounded by helices, with a concave active-site cleft, which is the DNA binding site (Fig. 21A and 22A). Interestingly, most of the amino acid sequence variation in this family of enzymes is confined to the immunity protein binding site, which lies adjacent to the conserved active site (344). The core of the colicin DNase active site is the so-called H-N-H motif, a ∼34-amino-acid sequence at the C terminus of the protein that resembles a distorted zinc finger of two β-strands and an α-helix, with a single metal ion sandwiched between them (343) (Fig. 21A). The motif contains four histidine residues, all of which are conserved in colicin DNases but only some of which are conserved in related enzyme families (where they are usually replaced by aspartic acid or asparagines that serve similar functions) (332). The most critical histidine is the invariant N-terminal residue of the motif H-N-H, most often part of a His-His or Asp-His dyad. This residue serves as a general base in the hydrolysis mechanism (see below). The central asparagine residue (H-N-H) has a structural role, forming a stabilizing backbone hydrogen bond across the motif. The C-terminal histidine (H-N-H) is a metal-binding residue. The H-N-H motif was first described in 1994 by Shub et al. and Gorbalenya; since then, over 500 enzymes have been reported to contain the motif, and in all cases, the enzymes are nucleases involved in a wide range of unrelated functions, including mobile intron homing, DNA repair, mammalian apoptosis, and DNA restriction (230, 332, 587).
FIG. 22.
The Im9 binding site partially overlaps the DNA binding site on the colicin E9 DNase. (A) Crystal structure of ColE9 H551A (equivalent to His103 in the isolated E9 DNase) bound to 8-mer dsDNA showing the bound catalytic Mg2+ ion (red sphere) in the context of the Im9 exosite (blue shading) (PDB accession number 1V14). The enzyme binds to the minor groove of the DNA, causing it to widen and bend. (B) Structural overlay showing the steric clash of the salt bridge between ColE9 Arg502 and Im9 Glu30 with DNA (shown as a molecular surface). In the DNA-bound structure, ColE9 Arg502 swings out of the way to form a salt bridge with ColE9 Asp499. ColE9 Arg502 is equivalent to Arg54 in the isolated E9 DNase domain. Courtesy of Maria Maté, reproduced with permission.
The H-N-H motif is most frequently associated with homing endonucleases, which are intron-encoded endonucleases that cleave intronless sites with high specificity (212). H-N-H-containing enzymes are one of four homing endonuclease families, the others being the LAGLIDAG, GIY-YIG, and His-Cys enzymes. The structures of the colicin E7 and E9 DNase domains were the first for H-N-H motif-containing enzymes (343, 346). It has also been shown that this structural motif is representative of a much wider group of nucleases that includes the His-Cys homing endonuclease family, now generally referred to as “ββα-Me” endo-nucleases (370). This superfamily includes homing enzyme I-PpoI from Physarum polycephalum, Serratia nuclease from Serratia marcescens, the periplasmic nuclease Vvn from Vibrio vulnificus, and the Holliday junction-resolving enzyme T4 endo VII (9). Hence, the basic active-site architecture of a colicin DNase is structurally and mechanistically related to a larger superfamily of nucleases distributed across all the major kingdoms of life.
(b) Metal dependence of colicin DNases and mechanism of DNA hydrolysis. Colicin DNases are thought to elicit cell death through the random destruction of the bacterial genome. This produces a nick in double-stranded DNA (dsDNA) that on its own is unlikely to be cytotoxic, since nicks can be fixed by the repair machinery of the cell. Cell death most likely results from a double-strand break(s) caused by the repeated cutting of the DNA.
The E9 DNase has been shown to cleave both double- and single-stranded DNA substrates in vitro to yield 5′-phosphate and 3′-OH products with the enzyme most active against dsDNA (524). The enzyme will also cleave single-stranded RNA but poorly. Interestingly, the enzyme preferentially cleaves dsDNA after thymine, and consistent with this, a recent structure of the E9 DNase bound to dsDNA shows that it is bound to a scissile phosphodiester adjacent to a thymine base (448). Optimal endonucleolytic activity of the colicin E9 DNase against dsDNA substrates requires high concentrations (>5 mM) of either Mg2+ or Ca2+ and is optimal at alkaline pHs (>8), properties that are shared with many other H-N-H/ββα-Me enzymes (524, 526). The ability to utilize either Mg2+ or Ca2+ is unusual for an endonuclease (for example, restriction enzymes are rarely able to substitute Ca2+ for Mg2+) and signifies a degree of promiscuity in which divalent cations can be accommodated within the active site. Indeed, colicin DNases can readily bind first-row transition metals such as Ni2+, Co2+, and Cu2+ (329, 525), although in the case of the E9 DNase, these metals influence substrate specificity, with the enzyme preferentially cleaving single-stranded DNA in the presence of transition metal ions (524). Recent structures of the E9 DNase bound to dsDNA have highlighted the metal adaptability of the H-N-H/ββα-Me motif, with structures of both Mg2+- and Zn2+-bound enzymes showing how the side chains of the motif rearrange to accommodate the octahedral and tetrahedral geometries, respectively, that each metal ion prefers (448).
Controversy still surrounds the identity of the physiological metal ion of colicin DNases. Chak and Yuan and coworkers maintained that colicin E7 DNase and therefore, by inference, other colicin DNases, since they have identical active sites, are zinc-dependent enzymes (see references 282 and 365 and references therein). The main arguments in favor of Zn2+ include the following: (i) when purified from overexpressing strains, the enzyme is predominantly loaded with Zn2+; (ii) Chak et al. reported that in their hands, the enzyme preferentially uses Zn2+ to cleave DNA substrates (365); (iii) single-transition metal ions were bound at the active sites of the E7 and E9 DNases (Zn2+ and Ni2+, respectively) in the originally published structures for these enzymes (343, 346), with structures also available for both enzymes bound to DNA and Zn2+ (159, 448); and (iv) isothermal titration calorimetry experiments have shown that zinc binds with the highest affinity (Kd in the nanomolar range) to all colicin DNases, while other first-row transition metals such as Ni2+ bind with μM affinity. In contrast, alkaline earth metal ions do not bind to colicin DNases, at least not in the absence of DNA (525).
There is no doubt that the H-N-H/ββα-Me motif of all colicin DNases binds transition metal ions (636). The question, however, is whether this is the physiological cofactor for DNA digestion. Several observations cast serious doubt on zinc or indeed any other transition metal ion as the cofactor in vivo. First, the Kleanthous laboratory reported that zinc does not support E9 DNase activity and supports only very weak activity of the E7 DNase (524, 525, 639). The E9 DNase is much more active against supercoiled plasmids in the presence of Mg2+ or Ca2+ than transition metal ions (526). Second, nearly all H-N-H/ββα-Me motif enzymes reported in the literature utilize Mg2+ or Ca2+ ions for catalysis and are often inhibited by zinc, which is the case for the E9 DNase (329). Finally, in a mutagenic screen of the E9 DNase active site, it was found that a subset of mutants that retained catalytic activity with transition metal ions had little or no activity with Mg2+ ions. These mutants were biologically inactive as colicins, indicating that Mg2+ must be the physiological metal ion (664).
Crystallographic studies on both the E7 and E9 DNases bound to DNA and metal ions have helped elaborate the basic mechanism by which these colicins hydrolyze DNA (159, 281, 448) but with the most detailed information coming from structural and biochemical work on the broader family of ββα-Me enzymes such as I-PpoI, Serratia nuclease, and Vvn (reviewed in reference 332). Those studies point to a general mechanism where the enzyme binds to the minor groove of substrate DNA, causing the groove to widen and bend, straining the scissile phosphate toward the single divalent cation (Mg2+ or Ca2+) within the H-N-H/ββα-Me motif (Fig. 22A). In the colicin E9 DNase, the Mg2+ ion is coordinated by a bridging and a nonbridging phosphate oxygen from the DNA and two histidine residues from the motif (His550 and His575, equivalent to His102 and His127 in the isolated DNase), with the remaining coordination sites presumed to be taken by water molecules. The Mg2+ ion serves to polarize the scissile phosphate, stabilize the phosphoanion transition state, and activate a water molecule that protonates the 3′-oxygen-leaving group (this latter point has yet to be demonstrated in colicins). ColE9 His551 (His103 in the isolated domain) of the catalytic motif serves as the general base in the activation of the hydrolytic water molecule for an in-line attack of the scissile bond. The 5′-phosphate product is also likely stabilized by interactions with a conserved arginine residue (ColE9 Arg453 or E9 DNase Arg5). Key to the distortion of the DNA duplex is the unyielding V-shaped architecture of the H-N-H/ββα-Me motif and a neighboring salt bridge (Arg544-Glu548 ColE9, equivalent to Arg96-Glu100 in the isolated E9 DNase) that is inserted directly into the minor groove, which together form a network of hydrogen bonds to the DNA phosphate backbone that straddle the metal site (448).
(ii) RNase colicins.
All RNase colicins (E3, E4, E5, E6, and D) elicit cell death through the inhibition of protein synthesis by cleaving specific phosphodiester bonds in RNA, and, as with most ribonucleases, the enzymes do not require metal ion cofactors. Colicin RNases fall into two distinct groups: those that cleave 16S rRNA (rRNases) (colicins E3, E4, and E6) and those that cleave the anticodon loops of particular tRNAs (tRNases) (colicins E5 and D). Structures are now available for enzymatic domains from both groups, and although all of them have folds that are recognizable as RNA binding proteins, there is no discernible structural similarity between them (Fig. 21B and C).
(a) rRNase colicins. One of the first enzymatic colicins to be isolated and characterized was colicin E3 by the laboratories of Holland, Boon, and Nomura in the early 1970s (36, 45, 581), with colicins E4 and E6, which share 80% sequence identity, being identified subsequently. The enzyme specifically attacks the 30S subunit of the bacterial ribosome, cleaving 16S rRNA towards the 3′ end between nucleotides A1493 and G1494. This region of the 30S subunit is one of the most critical in the ribosome, as the adjoining nucleotides A1492 and A1493 form the decoding center of the ribosomal A site. The significance of this target site has become particularly apparent through the recent crystal structures of the 30S ribosomal subunit from the Ramakrishnan laboratory (see reference 493 and references therein). Nucleotides 1492 and 1493 undergo a large conformational change during translation, interacting directly with, and sampling the dimensions of, the minor groove of the helix formed between a cognate tRNA-mRNA codon-anticodon duplex. However, it is still not clear which parts of the translational cycle are most affected by ColE3 enzymatic cleavage, with conflicting data appearing in the literature that have yet to be resolved (12, 618). There is also little information on the cleavage mechanism itself. The structure of the enzyme is comprised of a twisted central β-sheet with peripheral α-helices (73) (Fig. 21B). The active site has been mapped by mutagenesis, with mutants assayed in vitro by a transcription-translation assay and in vivo by toxicity tests from which three conserved residues were identified as being essential for catalytic activity, Asp510, His513, and Glu517 (equivalent to Asp55, His58, and Glu62 in the isolated E3 rRNase domain), which line a groove in the enzyme (661) (Fig. 21B). Asp510 and His513 are hydrogen bonded to each other, appearing to form a catalytic diad (73). Consistent with the mutagenesis data, modeling studies have placed these residues within interaction distance of the target nucleotides A1493/A1494 in the ribosome (26).
(b) tRNase colicins. Colicins E5 and D cleave single phosphodiester bonds in the anticodon loops of specific transfer RNAs, with this activity demonstrated in vivo and in vitro. Both colicin activities were discovered by Masaki and coworkers, who recognized that the absence of sequence identity with the rRNase colicins signaled that they likely had different targets, although the cellular outcome was the same (491, 629). They also noticed that while colicin E3 and colicin E5 both inhibit RNA-dependent protein synthesis supported by an E. coli cytoplasmic extract, only ColE3 inhibited poly(U)-directed incorporation of [14C]Phe, suggesting a different mode of action for ColE5. They went on to show that protein synthesis could be inhibited by both ColE5 and ColD but only if natural RNAs were used as templates. ColE5 was found to specifically cleave the anticodon loop of the natural tRNAs for His, Asn, Tyr, and Asp; in each case, cleavage occurred after the wobble position in the anticodon loop corresponding to a Q34-C35 linkage (491), where Q (queuine) is a hypermodified form of guanine. ColD was subsequently shown to cleave four isoaccepting arginine tRNAs between bases 38 and 39, corresponding to either an AG or an AC linkage within the anticodon loop (629).
ColE5 and ColD yield identical cleavage products of 5′-OH and a 2′,3′-cyclic phosphate intermediate (491, 629). Neither enzyme hydrolyzes phosphodiester bonds, but, rather, they act as phosphotransferases. Nevertheless, the cleavage chemistry indicates that these enzymes behave as classic ribonucleases such as RNase A and T1, albeit ones that are unable to complete the catalytic cycle by hydrolyzing the cyclic intermediate. The catalytic domains of ColE5 and ColD also have weak structural homology to RNase T1, implying a distant but common evolutionary origin. Their topologies are similar to those of other RNA binding proteins in that they are positively charged, crescent-shaped molecules and are composed predominantly of β-sheets (233, 406) (Fig. 21C and D). In contrast to the classic two-histidine acid-base mechanism of RNase A, neither colicin E5 nor colicin D has two essential histidines in its active site. Indeed, the colicin E5 tRNase domain does not contain any histidine residues at all, while colicin D has only one essential histidine (His611). Mutagenesis studies of both enzymes have, however, highlighted other important amino acids that are involved in RNA binding and/or catalysis. Lin et al. found that alanine mutants at Asp46 and Arg48 in the isolated E5 tRNase domain were compromised in terms of their ability to cleave model stem-loop mimics of tRNA anticodons (406). These residues are positioned on opposite sides of the presumed catalytic cleft, with Asp46 acting as a putative general base in the initial proton abstraction from the 2′-OH and Arg48 involved in transition state stabilization (406). That group also showed that queuine is in fact not required for cleavage by ColE5. Graille et al. found that in addition to His611 in ColD, Lys608 and Lys610 are also critical, most likely in stabilizing the formation of the trigonal bipyramidal transition state (233).
Nuclease-specific immunity proteins.
Soon after the discovery of the enzymatic colicin E3, it became apparent that the toxin was released from cells as a heterodimeric complex with a protective immunity protein (302, 590). Immunity towards colicin intoxication is an absolute requirement for colicin-producing bacteria, the immunity gene normally downstream of the colicin gene in the SOS-inducible colicin operon. Every nuclease colicin is released from cells in complex with its cognate immunity protein, helping to prevent the suicide of the producing organism before the toxin is released into the environment.
Nuclease-specific immunity proteins (Ims) are typically ∼10 kDa and inactivate colicin by binding to the C-terminal nuclease domain to form of a ∼71-kDa heterodimer. How the immunity protein is subsequently lost during colicin translocation into a cell is still the point of some uncertainty; one of the few reports in the literature, from Krone et al., showed, using Im-specific antibodies, that the Im protein for cloacin DF13 (a ColE3 rRNase homologue) is released at the cell surface (364) (see Colicin Reception). While this has yet to be demonstrated for nuclease colicins, it nonetheless appears that Im proteins do not play a role in import and that their release is likely to be a relatively minor aspect of cell entry. For example, DNase colicins are equally active against E. coli regardless of whether the Im protein is bound to the colicin or not (669).
The inhibition of colicin nucleases by immunity proteins has been the subject of intense biochemical and structural studies over the last 10 years that have shown that Im proteins are a structurally diverse group of nuclease inhibitors (Fig. 21). DNase-specific Im proteins and the immunity protein for ColD tRNase (ImD) are both four-helical bundle proteins (Fig. 21A and D), but their topologies are quite different (233, 498). The same is true of Im proteins specific for the ColE3 rRNase and ColE5 tRNase; both have α/β-folds but are otherwise unrelated (73, 427) (Fig. 21B and C). One of the great surprises from the structural work has been the realization that Im proteins inactivate colicin nucleases by one of two distinct mechanisms: either by binding directly to the nuclease active site (tRNase-specific Im proteins) (Fig. 21C and D) or by blocking access to the substrate binding site, leaving the active site open (DNase- and rRNase-specific Im proteins) (Fig. 21A and B). An important evolutionary ramification of these distinct modes of action is that sequence diversity has been possible at the interfaces of those enzyme-inhibitor complexes that avoid binding at the conserved active site. This diversification, which presumably explains why more DNase and rRNase colicin-Im protein complexes have been identified, has given rise to families of related protein complexes that have proven to be a valuable model system in protein-protein interaction studies (summarized below). DNase-specific immunity proteins have also been used as models in protein folding studies. Due to space constraints, this aspect of Im protein chemistry is not reviewed, but the reader is directed to recent publications from Capaldi et al. and Gsponer and coworkers (69, 238).
(i) tRNase-specific immunity proteins block the enzyme active site.
The tRNase-specific immunity proteins Im5 and ImD appear to inactivate colicins E5 and D by mimicking substrate RNA (Fig. 21C and D), similar to the classic inhibition mechanisms of other RNase inhibitors, such as the barstar inactivation of barnase (63, 233, 427). The continued uncertainty about their mode of action reflects the absence of structural information for the enzymes bound to nucleic acid. Nevertheless, it is clear that in both cases, the Im protein binds at the enzyme active site. There is a high degree of charge and shape complementarity in both complexes, with the negative charges of the Im proteins complementing the positive charges of the nuclease. The topologies of both active sites include a positively charged groove, which is deeper in the E5 enzyme. It is into these grooves that the specific Im proteins bind, with 2,425 and 1,920 Å2 of accessible surface area buried at the interfaces of the E5 tRNase-Im5 and D tRNase-ImD complexes, respectively (233, 427). The ColD tRNase active site is mostly but not completely occluded by ImD, with residues from two of the immunity protein helices (α2 and α3) interacting with the rim and sides of the groove. His611, the presumed general base in ColD, is completely buried at the interface with the Im protein, forming a hydrogen bond with Glu56 of ImD. Mutational analysis of the ImD interface residues have highlighted the importance of polar and charged residues in stabilizing the complex (233).
Binding of Im5 to the E5 tRNase does not cause a gross change in protein conformation, a general characteristic of all colicin nuclease-Im protein complexes. As with the ColD complex, critical E5 active-site residues identified by mutagenesis (E5 tRNase Asp46 and Arg48) are buried at the interface with the Im protein but do not interact directly with the Im protein. Mutational analysis of Im5 residues at the interface also highlighted the importance of charged residues that form hydrogen bond networks that stabilize the complex with the enzyme (e.g., Lys4 and Asp95).
(ii) DNase- and rRNase-specific immunity proteins are high-affinity exosite inhibitors.
In contrast to Im5 and ImD, immunity proteins that are specific for DNase (E2, E7, E8, and E9) and rRNase (E3, E4, and E6) colicins inactivate their target enzymes by binding at an exosite adjacent to the active-site cleft, with inhibition being the result of steric and electrostatic occlusion of their substrates, the bacterial genome and ribosome, respectively (344) (Fig. 21A and B). In the case of the E3 rRNase-Im3 complex, extensive mutagenesis of charged and polar residues, including Asp510 and His513 ColE3, has shown that no catalytic residues are buried at the interface by Im3 (607, 661). Mutagenesis of the E7 and E9 DNases as well as crystal structures of the enzymes bound to dsDNA also show that the Im protein does not contact essential catalytic groups, but, rather, the Im protein binding site overlaps the DNA binding site (343, 346) (Fig. 22A and B). The greatest variation in amino acid sequence discussed in both rRNase and DNase colicin enzyme complexes is localized at their inhibitor protein binding sites (344), which is particularly apparent in the DNase-Im protein complexes, where only one conserved colicin amino acid contacts the Im protein directly (the DNA-contacting residue ColE9 Arg502, equivalent to Arg54 in the E9 DNase) (Fig. 22B). In contrast, immunity proteins utilize both conserved and nonconserved residues to contact colicin DNases.
The rRNase domain of colicin E3 forms an extensive contact with Im3, burying over 2,550 Å2 of accessible surface area (73). The interface is formed between an exposed face of the Im3 four-stranded β-sheet with the β-strands of the rRNase and an adjoining N-terminal α-helix. As with other colicin nuclease-Im protein complexes, there is strong electrostatic and steric complementarity between the positively charged nuclease and negatively charged Im3. There is also a high degree of hydrophobic burial at the interface, with aromatic residues in both proteins making important contacts. Studies by Masaki et al. previously identified Cys47 of Im3 as being an important determinant of colicin specificity, differentiating between the rRNases of ColE3 and those of ColE6 (444). As expected, this residue is buried at the interface, although it does not appear to be contacted directly by E3 rRNase residues.
The principal point of contact of the DNase-specific Im protein with the enzyme is through helices II and III and includes interactions from an adjoining loop (343, 346). The Im protein, which does not undergo large structural changes, binds to a cleft on the enzyme constructed from a short α-helix, an extended β-strand, and two loops. Approximately 1,500 Å2 of accessible surface area is buried in both complexes and again shows a high degree of charge and steric complementarity. The E7 DNase-Im7 complex is more polar and contains more interprotein hydrogen bonds than the E9 DNase-Im9 complex, which is more hydrophobic, a reflection of the hydrophobic specificity contacts in the E9 complex. Kuhlmann et al. noticed that while the individual proteins within the complexes are similar to each other (the secondary structure elements of the DNases and Im proteins have root-mean-square deviations of ∼1.8 Å, respectively), the complexes are rotated by 19° with respect to the position of the Im protein, with the conserved residues of helix III being the axis of this rotation (369). The rigid-body rotation is central to the mechanism by which Im proteins “sample” the DNase surface for specificity contacts (see below). Consistent with the structural data, protein-engineering work from the Kleanthous laboratory has shown that DNase-specific immunity proteins exhibit “dual recognition,” wherein the conserved residues of helix III in the Im protein form a binding energy hotspot that anchors the Im protein to the DNase surface, with adjacent nonconserved residues in helix II being the principal determinants of protein-protein interaction specificity (402, 403, 404, 665).
There is extensive literature on the kinetics and thermodynamics of colicin DNase and rRNase nucleases binding their Im proteins (no information is currently available for tRNase-Im protein complexes). The first to be analyzed was Im9 binding the nuclease domain of ColE9. The Kd for the complex, obtained from the ratio of the individual dissociation and association rate constants (Kd = koff/kon), was determined to be 2.4 × 10−14 M at pH 7 and 25°C and in 200 mM NaCl, reducing it to 9.3 × 10−17 M in the absence of salt, with these values being essentially identical for the isolated domain and intact ColE9 (667). By comparison, Im3 binds the isolated nuclease domain of ColE3 more weakly (Kd = 1.4 × 10−12 M) than it does the full-length toxin (Kd = 1.4 × 10−14 M) (662). This increase in affinity is explained by additional interactions with Im3 (equivalent to ∼3 kcal/mol) from the T domain of the colicin (607). Since the ColE9-Im9 complex does not behave in the same way, this suggests that the “sandwiching” of Im proteins between the enzyme and T domain may not be a general feature of all nuclease colicin-Im protein complexes. Kinetic analysis has shown that Im protein binding is very rapid for the rRNase and DNase-Im protein complexes (kon ∼ 109 M−1 · s−1 in the absence of salt) and electrostatically driven. However, the kinetic mechanisms of binding of Im proteins to the nuclease domains of ColE3 and ColE9 differ: Im3 binding to the E3 rRNase involves a single bimolecular collision, whereas a conformational change occurs after the bimolecular collision between Im9 and the E9 DNase, which is the rigid-body rotation described above (331, 662).
An area that has been the focus of much work is the specificity of DNase colicin-Im protein complexes. Inhibitors such as barstar display little nuclease specificity because recognition involves binding to the conserved active site. Colicin-DNase immunity protein complexes also appear, at first glance, to lack specificity since every colicin DNase can be bound and inactivated in vitro by every immunity protein. However, in contrast to other nuclease-inhibitor systems, there are very large differences in the binding free energies of cognate and noncognate complexes, with cognate complexes generally 106- to 108-fold more stable than noncognate complexes (403, 404, 666). Of the four DNase colicins, E9 shows the broadest cross-reactivity, forming noncognate complexes with Im7 (Kd ∼ 10−4 M), Im8 (Kd ∼ 10−6 M), and Im2 (Kd ∼ 10−7 M) (404, 666). In general, however, noncognate colicin DNase-Im protein complexes display Kds of the order of 10−6 to 10−8 M, with this cross-reactivity being due to residues from helix III making sequence-independent interactions with the DNase surface through backbone hydrogen bonds and van der Waal's interactions (330, 369, 404). Li et al. also established the link between in vitro binding affinity and in vivo colicin cross-reactivity, showing that Im proteins need to bind with a Kd of <10−10 M in order to provide bacterial cells with full protection against a colicin DNase (404). The breadth of colicin DNase-Im protein binding affinities transcends the binding affinities of all known protein-protein interactions in the literature, which makes this an attractive system with which to address fundamental questions concerning specificity in protein-protein recognition.
Riley proposed that the evolution of novel colicin-immunity protein binding specificities occurs through a two-step process termed “diversifying selection” (see Colicin Evolution and Ecology) (557). The first step is envisaged to be a mutation within the immunity protein that broadens specificity, followed by a second mutation within the DNase that is selected for tighter binding to the mutant immunity protein. This is postulated to generate a “superkiller” colicin-immunity protein pair that is proposed to allow it to outcompete the ancestral pair. This evolutionary view of colicin DNase-Im protein complexes requires that Im proteins can display broadened colicin specificity. This hypothesis is consistent with the way in which Im proteins bind and recognize their cognate nuclease domains, which is through overlapping but nonidentical specificity sites. Binding involves the close interdigitation of conserved and variable amino acids of the Im protein (from helices III and II, respectively) with specificity sites of the DNase (344, 369). Three conserved Im residues, Asp51, Tyr54, and Tyr55 in Im9, form a generic hotspot; many of their interactions with the enzyme are mediated by conserved water molecules. The two conserved tyrosine side chains from the Im protein present a key specificity residue of the DNase to their own specificity residues displayed from the adjacent helix II (e.g., Leu33 in Im9 or Asp33 in Im2). These interactions are augmented by additional specificity contacts along helix II that are nonoverlapping. Importantly, the rigid body rotation centered on the conserved tyrosines of helix III allows the Im protein to sample the enzyme for specificity contacts (369). In this way, new Im protein specificities could emerge while the original specificity was retained.
Colicin M.
Colicin M is a Ton-dependent, 28-kDa colicin first described in the 1970s by Braun and colleagues (56). In contrast to all other enzymatic colicins, ColM (one of the smallest colicins known) is not released bound to its Im protein, which instead is located in the periplasm and anchored to the cytoplasmic membrane, where it is presumed that the cytotoxic activity of the toxin is expressed, close to the membrane surface. Unlike most other enzymatic colicins, ColM causes cell lysis that can be blocked by controlling the osmolarity of the growth medium. The cytotoxic activity of ColM is directed toward bacterial murein, inhibiting both peptidoglycan biosynthesis and LPS O-antigen synthesis (54, 252). Recent work has shown that ColM exerts its cytotoxic effect through the enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors (176). Both lipid I and lipid II peptidoglycan intermediates are digested by colicin M, with cleavage occurring between the lipid moiety and the pyrophosphoryl group.
COLICIN-LIKE BACTERIOCINS FROM OTHER BACTERIAL GENERA
Although colicins are the most studied bacterial toxins, Escherichia coli and related species (Citrobacter freundii for colicin A or Shigella boydii for colicin U) are not the only bacteria to produce bacteriocin to kill neighboring bacteria. Toxins with similar domain organizations and infection characteristics have been described for Pseudomonas pyogenes (pyocins), Enterobacteriaceae such as Enterobacter cloacae (cloacins), Yersinia pestis (pesticins), Klebsiella species (klebicins or klebocins), Serratia marcescens (marcescins, colicins L and 24), Photorhabdus luminescens (lumicins), and even the gram-positive bacterium Bacillus megaterium (megacins) (505). Bacteriocins such as lantibiotics, microcins, or other gram-positive peptide bacteriocins will not be discussed here, and we refer the reader to excellent recent reviews (154, 162, 256).
Pesticins
The major pesticin produced by Yersinia pestis was first shown to induce the formation of spheroplasts from E. coli or Yersinia cells (176, 246), which correlates with its muramidase activity (196, 653). It possesses an immunity protein localized in the periplasm, but its release is not dependent on a lysis protein (518, 547, 653). The pesticin gene is probably regulated by the SOS system, as suggested by the existence of a LexA box in its promoter region (547). The pesticin possesses a TonB box at the N terminus (518, 547) (Fig. 11A) and indeed requires the TonB-dependent FyuA receptor and the products of the exbB, exbD, and tonB genes for its translocation (197, 352, 548).
Klebicins or Klebocins
Klebicins or klebocins are bacteriocins produced by various Klebsiella strains upon treatment with mitomycin C, suggesting that their genes are under the control of the SOS system (115, 305). Klebicin A1 uses the ferric aerobactin receptor to bind the cell and is likely to be a TonB-dependent bacteriocin (127). Klebicin B-resistant mutants of K. pneumoniae are sensitive to klebicin A, suggesting that klebicins A and B use different pathways to penetrate the cell (305). The observations that the klebicin B sequence is homologous to DNase colicins and that klebicin A is blocked by the colicin E2 immunity protein suggest that both klebicins A and B function as nonspecific endonucleases (307, 560). Two other klebicins have been described recently. Based on protein sequence comparisons and phylogenetic analyses, it has been proposed that klebicin C might display rRNase activity, whereas klebicin D belongs to tRNase bacteriocins (113).
Pyocins
The bacteriocins produced by Pseudomonas species are called pyocins. Expression of the chromosomally encoded pyocin genes is induced by UV irradiation or mitomycin C treatment. Pyocins are released by bacterial lysis (probably using a bacteriophage-like lytic system) (476) and bind and kill susceptible cells (295, 315). Pyocins are classified into different types, such as R, F, and S pyocins, which present distinct features and behaviors (see reference 458). R-type pyocins present a rod-like structure resembling contractile bacteriophage tails, whereas S-type pyocins are more related to colicins for domain organization (255, 317, 476, 619). The phage-tail-like structure of R-type pyocins is not unique, since other genera produce similar bacteriocins (for example, serracin P from Serratia plymithicum) (293). R-type pyocins adsorb to LPS molecules at the cell surface (287, 454) and then penetrate through the outer membrane by a process resembling phage contraction (232) before killing the target bacteria by membrane depolarization (635). The S-type pyocins that have been characterized so far are nonspecific endonucleases. The C-terminal domains of pyocins AP41, S1, S2, and S3 share homologies with the C-terminal domains of klebicin B and colicins E2, E7, and E9 (169, 560, 570, 571, 572, 582), specific tRNase (pyocin S4, related to colicin E5) (504), or pore-forming colicins (pyocin S5 shares homologies with colicin Ia) (504). They are released in association with a cognate immunity protein (570, 571). S-type pyocins are organized into four domains arranged linearly: the N-terminal domain is involved in the recognition of the cell surface receptor, domain II has an unknown function and is dispensable for killing activity, the third domain is responsible for pyocin translocation and penetration, and the C-terminal domain carries the lethal activity (458). Pyocins S1, S2, and S3 probably belong to TonB-dependent bacteriocins because their highest activity is detected on susceptible cells grown under iron-limited conditions and because of their use of the ferripyoverdine receptor FpvA as a receptor (19, 136, 169, 494, 572, 602). In contrast, pyocin AP41 is more related to Tol-dependent colicins because it is inactive on Pseudomonas aeruginosa tolQRA cells (146, 273, 372). Chimeras between S-type pyocins and colicins have been constructed and indicate that both receptor binding and translocation domains are species specific (316, 318, 572).
Lumicins
The bacteriocin produced by the insect-pathogenic bacterium Photorhabdus luminescens, called lumicin, has been described recently. Like colicin, lumicin (which has sequence similarities with pyocin S3) (505) is encoded with an immunity protein and possesses a similar domain organization (584). However, this bacteriocin does not exhibit any bactericidal activity. Based on the observation that it kills insect hemocytes and induces actin cytoskeleton modifications, it has been proposed that it might have evolved to bind and kill eukaryotic cells (686).
Megacins
The SOS-inducible plasmid-encoded bacteriocins produced by the gram-positive bacterium Bacillus megaterium have features, such as regulatory and inhibition mechanisms, that are similar to those of colicins (269, 654, 655). Interestingly, megacin is unique in that it kills other B. megaterium cells by means of phospholipase activity (490).
COLICINS AND PHAGES AS LABORATORY TOOLS
For a number of years, colicins, Ff phages, and their specific DNA-encoding matrices corresponding to colicinogenic plasmids or to single-stranded circular DNA have been used for biotechnological and molecular biology applications. These applications exploit properties listed below, which are related to (i) bacterial containment, (ii) biosensing of genotoxic compounds, (iii) improvement in protein purification, (iv) production of outer membrane vesicles, (v) screening of biological processes, and (vi) Ff phage display. This broad range of applications is certainly not exhaustive and may increase in the future. Since colicin import and phage DNA entry require the Tol and Ton cell envelope machineries, deciphering the translocation processes may lead to the development of new classes of antibiotics.
Bacterial Containment
DNA vectors harboring the origin of replication of plasmid ColE1 are often used to develop multicopy plasmids. They were used to construct the first stable vectors that were able to propagate in bacteria, which were selected by colicin (10) or antibiotic markers (34). Moreover, colicin activity has been used for bacterial containment in order to control and stop the propagation of recombinant bacteria in the natural environment. The first trial was performed with Pseudomonas putida, using its biotechnological property to degrade toluene. This was achieved by inserting the colicin E3 gene into the P. putida chromosome under tightly regulated expression in order to induce colicin E3 in the absence of toluene; thus, bacteria are killed only when the toluene is totally degraded (473). This technique is promising for the production of conditional lethal suicide vectors. The bacterial containment by colicin E3 has been further developed to reduce gene spread. Thus, to ensure efficient bacterial containment, a dual system corresponding to colicin E3 and to the EcoRI restriction endonuclease has been designed. Both the colicin and EcoRI genes are cloned on the same plasmid but under the control of different regulatory signals for their additive effects. This dual-containment approach targeting both RNA and DNA ensures efficient programmed bacterial suicide (631).
Biosensing of Genotoxic Compounds
The genotoxicity of environmental substances can be measured by exploiting colicin operator sequences brought into focus for high-throughput screening and for in situ detection assays. All colicin operons described present a genetic organization that is tightly regulated by SOS promoters. To detect DNA-damaging agents, the first important experiments were carried out using the sfiA SOS promoter by monitoring an sfiA::lacZ operon fusion using colorimetric assays (545). Recently, a system using the SOS promoter/operator of colicin D fused to GFP was developed (488), exploiting the strong affinity due to the cooperative binding of LexA repressors on the tandem operators of colicin genes (415). Using this type of reporter system, those authors compared the various SOS promoters that were available and found that the colicin D promoter yields greater sensitivity together with a higher response level toward genotoxic substances (mitomycin C, nitrosoguanidine, nalidixic acid, and formaldehyde) than recA, sulA, or umuDC promoters.
Improvement in Protein Purification
The production of soluble proteins isolated from the periplasm or the cytoplasm often requires cell permeabilization steps prior to purification. To produce and recover stable and soluble proteins in the cell supernatant, the Tol system, colicins, and colicin lysis proteins have been exploited to induce protein release during cell growth. Secretion of proteins into the culture medium of tol-pal mutants has been shown to improve the recovery of soluble periplasmic proteins (109). To induce the tol phenotype in E. coli cells, the Tol-Pal-targeting technique was further modified to produce periplasmic colicin, g3p N-terminal domains, or soluble Tol domains (for a review, see reference 41) and was previously shown to be useful for preventing periplasmic protein aggregation by using the C-terminal domain of TolA (670). In other experiments, the inducible production of colicin lysis proteins allowed the recovery of recombinant periplasmic or cytoplasmic proteins in the extracellular medium (284, 327, 416, 644).
Production of Outer Membrane Vesicles
The proteins or peptides that interact with the Tol system have been used to induce the formation of outer membrane vesicles. The results clearly demonstrate that the N-terminal domains of group A colicins or of the g3p protein are sufficient to induce vesicle production in various bacterial species (260). The formation of outer membrane vesicles is a property of gram-negative bacteria, well documented over many years. For new developments, the reader is referred to recent reviews (30, 366). The production of blebs in tol mutants or the induction of their production in wild-type cells might be used to obtain and purify large amounts of vesicles from gram-negative bacteria. In addition to the general interest in understanding their role (host-pathogen interactions, transport of virulence factors, toxin delivery, etc.), vesicles are promising vaccine particles since they contain all the major bacterial antigens. Moreover, designing recombinant vesicles or using vesicles in the analyses of biological processes, for example, GFP-labeled vesicles directly observable by fluorescence microscopy (A. Bernadac, S. Pommier, M. Gavioli, and R. Lloubès, unpublished data), is a current challenge.
Screening of Biological Processes
Colicins have been used to obtain functional information on transport systems. Protein export across the inner membrane to the periplasmic space utilizes two different translocation pathways: the twin-arginine (tat) and the SecYEG (sec) systems. The secreted protein LasB from Pseudomonas aeruginosa has been fused to the pore-forming domain of colicin A. LasB follows a sec-dependent export pathway coupled with the type II secretion system formed by the Xcp machinery. The recombinant protein causes a lethal phenotype when produced in the periplasm of a P. aeruginosa xcp mutant, but in the presence of the intact Xcp machinery, the fusion is secreted as the LasB wild type and is not toxic at all. Suppressors of xcp mutants were then obtained following random chemical mutagenesis of the bacteria (656). Thus, targeting the lethal activity domain of colicin A to the periplasm is an attractive genetic tool for analyzing the type II secretion machinery. Similarly, using a recombinant pore-forming colicin fused with a tat-dependent signal sequence, it has been possible to screen mutants and identify genes required for tat secretion in P. aeruginosa (204). Such experiments have also been performed using E. coli to dissect the tat export machinery using a suicide vector encoding a fusion formed by the tat signal sequence and colicin V (292). In this case, colicin V (which belongs to the microcin family) was used as a pore-forming toxin. By using random Tn10 transposition mutagenesis with bacteria encoding tat-exported colicin V, a new inner membrane protein necessary for the colicin V import process was identified (219).
Phage Display and Role of the g3p Protein
The generation of new drugs has long involved searching among hundreds of thousands of compounds using well-defined in vitro assays chosen to mimic the desired in vivo activity of the new drug as closely as possible. New library methodologies now offer powerful alternatives by combining the generation of billions of compounds with a fast screening or selection procedure to identify the most interesting leading candidates. One of the most widely used library methodologies, called phage display, is based on the use of filamentous phages. In 1990, McCafferty et al. showed that antibody fragments could be displayed on the surface of filamentous phage particles by fusion of the antibody variable genes to one of the phage coat proteins (essentially g3p protein) (453). The antigen-specific phage antibodies could subsequently be enriched by multiple rounds of affinity selection, because the phage particle carries the gene encoding the displayed antibody. Phage display has proven to be a very powerful technique to display libraries containing millions or even billions of different epitopes for antibodies, peptides, or proteins (for a review, see reference 274). Although phage display has been extensively used, the mechanism of phage infection remains poorly understood (see “Translocation of Phage DNA” above).
COLICIN EVOLUTION AND ECOLOGY
Two-Step Process in Colicin Evolution
Colicins and other enteric bacteriocins, such as klebicins, remain the only bacteriocins for which detailed evolutionary investigations have been undertaken. Among the colicins, there are two main evolutionary lineages, which also distinguish the two primary modes of killing: pore formation and nuclease activity (557). Studies that include DNA and protein sequence comparisons (55, 555), surveys of DNA sequence polymorphisms in natural isolates (521, 561, 620), experimental evolution (28, 91), and mathematical modeling (5a) have revealed two primary modes of colicin evolution (621).
The more abundant pore former colicins share one or more regions with high levels of sequence similarity (Fig. 23). This patchwork of shared and divergent sequences suggests frequent recombination. The location of the different patches frequently corresponds to the different functional domains of the proteins. Such domain-based shuffling between bacteriocins is responsible for much of the variability observed among gram-negative bacteriocins.
An alternative mode of evolution is responsible for the current diversity of nuclease colicins. These colicins, which include both RNase- and DNase-killing functions, share a recent common ancestry. Their DNA sequences are quite similar, ranging from 50% to 97% sequence identity. However, many pairs of nuclease colicins have elevated levels of divergence in the immunity region (Fig. 24). To explain this pattern of divergence, Riley and collaborators proposed a two-step process of mutation and selection (555, 556, 621). The diversifying selection hypothesis posits the action of strong positive selection acting on mutations that generate novel immunity and killing functions (Fig. 25). The first event in this process is the occurrence of a mutation in the immunity gene resulting in a broadened immunity function. The resulting producer cell is now immune to the ancestral version of the colicin and has gained immunity to some number of similar colicins. This broadened immunity function increases the fitness of the producer strain in populations where multiple colicins are found, which is the case in all E. coli populations sampled to date (231, 559). A second mutation, this time in the colicin gene, is paired with the immunity mutation.
FIG. 25.
Survey of colicin production and resistance in E. coli. Over 400 strains were isolated from two populations of feral mice in Australia over a period of 7 months. The isolates were scored for colicin production and resistance. (a) Colicin production is abundant, with just under 50% of the strains producing eight distinct colicin types. col− represents nonproducer strains. (b) The majority of isolates are resistant to most co-occurring colicins. (c) A small proportion of the population is sensitive to co-occurring colicins.
This pair of mutations produces a novel colicin that is no longer recognized by the ancestral immunity protein. Thus, the possessor of the novel colicin will rapidly displace (by killing) the ancestral, formerly abundant bacteriocin-producing strain. This evolved colicin will ultimately be replaced by yet another novel colicin as the cycle repeats itself. This process results in a family of closely related proteins that have diverged most extensively in the region involved in the immunity binding and killing function, as seen for nuclease colicins (556).
Recently, several E2 colicins isolated from Australia suggest that diversifying recombination is not restricted to pore former colicins (620). Half of the E2 producers carry the characterized E2 plasmid. The other half of the E2 producers carry a recombinant plasmid with sequences derived from colicin E7 and the characterized E2 plasmid. These observations suggest that it is not the case that pore formers diversify only by means of recombination and that nuclease colicins diversify only by diversifying selection. The evolutionary process is more complex than the proposed simple dichotomy suggests.
Riley developed a model of colicin diversification that involves two phases (557). When rare, as is currently the case for most nuclease colicins, the occurrence of point mutations that alter immunity function may be the primary mode for generating novel bacteriocin phenotypes. Novel immunity and killing functions are rapidly selected since they allow a cell to avoid being killed by other bacteriocins or allow cells carrying them to displace their ancestors. These novel bacteriocins are then maintained until a new immunity or killing function emerges. When colicins are abundant, as is the case for many pore former colicins, domain swapping may become a more frequent mode of diversification. This “switch” in the evolutionary mechanism is due simply to the requirement for a set of bacteriocins to be abundant enough to serve as templates for recombination. Once abundant, recombination can more rapidly generate additional diversity.
We have only just begun to tap into the diversity of enteric bacteriocins. However, recent work suggests that similar evolutionary mechanisms may play a role in the diversification of other enteric bacteriocins. Sequence comparisons reveal that in several cases, enteric bacteriocins are chimeras of known gram-negative bacteriocins (560; M. A. Riley, C. M. Goldstone, and J. E. Wertz, unpublished data). For other enteric bacteriocins, the action of diversifying selection has been proposed (Riley et al., unpublished). Finally, some new enteric bacteriocins have no similarity with those characterized previously. A particularly interesting example of this latter observation is the recently described colicin Js (600). This plasmid-borne bacteriocin has a typical colicin gene cluster composition with toxin, immunity, and lysis genes. However, the organization of the gene cluster is unique in that the lysis gene is transcribed 5′ to the toxin gene. The genes themselves show no similarity to any known bacteriocin genes, and the encoded toxin is 94 amino acids, which is smaller than any other described colicin.
Bacteriocin-encoding plasmids, such as pColJs (which encodes colicin Js) and pKlebB (which encodes klebicin B), demonstrate another aspect of bacteriocin evolution (560, 600). These bacteriocin plasmids are chimeras with a plasmid “backbone” comprising replication and maintenance sequences typical of plasmids found in the bacteriocins' host species. In the case of pKlebB isolated from Klebsiella pneumoniae, the plasmid contains sequences similar to those of pNBL63 (680) and pJHCMW1 (153) isolated from Klebsiella oxytoca and K. pneumoniae, respectively, encoding plasmid maintenance functions. The sequence surrounding and comprising part of the klebicin B gene cluster shares similarity with colicin A and E9, originally isolated from E. coli (560). In the case of pColJs, the plasmid backbone is virtually identical to ColE1, whereas the DNA flanking the colicin Js gene cluster shows high similarity to pPCP1 from Yersinia pestis (285). The colicin Js gene cluster itself has a significantly lower G+C content (33.6%) than the rest of the plasmid (52.9%), indicating that it originated from yet a third source (600), perhaps even outside of the Enterobacteriaceae. This type of recombination, although not altering the bacteriocin genes proper, results in an increased host range. As we continue to explore bacteriocin diversity, our model of bacteriocin evolution will almost certainly become more elaborate and complex.
Role of Colicins in Promoting Microbial Diversity
Without question, colicins serve some function in microbial communities. This statement follows from the detection of bacteriocin production in all surveyed lineages of Bacteria. Equally compelling is the inference of strong positive selection acting on enteric bacteriocins. Such observations argue that these toxins play a critical role in mediating microbial population or community interactions. What remains in question is what, precisely, that role is.
Colicins may serve as anticompetitors, enabling the invasion of a strain into an established microbial community. They may also play a defensive role and act to prohibit the invasion of other strains or species into an occupied niche or limit the advance of neighboring cells. An additional role has recently been proposed for gram-positive bacteriocins in which they mediate quorum sensing. It is likely that whatever roles bacteriocins play, these roles change as components of the environment, both biotic and abiotic, change.
A theoretical and empirical base that has defined the conditions that favor the maintenance of toxin-producing bacteria in both population and community settings has been established. Almost exclusively, those studies have modeled the action of colicins. Chao and Levin showed that the conditions for invasion of a colicin producer strain were much broader in a spatially structured environment than in an unstructured one (112). In an unstructured environment with mass action, a small population of producers cannot invade an established population of sensitive cells. This failure occurs because the producers pay a price for toxin production, the energetic costs of plasmid carriage and lethality of production, but the benefits, the resources made available by killing sensitive organisms, are distributed at random. Moreover, when producers are rare, the reduction in growth rate experienced by the sensitive strain (owing to extra deaths) is smaller than the reduction felt by the producer (owing to its costs), and the producer population therefore becomes extinct. In a physically structured environment, such as that on the surface of an agar plate, the strains grow as separate colonies. Toxin diffuses out from a colony of producers, thus killing sensitive neighbors. The resources made available accrue disproportionately to the producing colony owing to its proximity, and therefore, killers can increase in frequency even when initially rare.
Recent modeling efforts have incorporated additional biological realities. Two such efforts introduced a third species, one that is resistant to the toxin but cannot itself produce the toxin (130, 334). Resistance can be conferred through mutations in either the binding site or the translocation machinery required for a bacteriocin to enter the target cell. The acquisition of an immunity gene will also confer resistance to its cognate bacteriocin. The authors of both studies reasonably assumed that there is a cost to resistance and that this cost is less than the cost of toxin production borne by the killer strain (195). Owing to this third member, pairwise interactions among the strains have the nontransitive structure of the childhood game of “rock-scissors-paper” (Table 4) (396). The producer strain beats the sensitive strain, owing to the toxin's effects on the latter; the sensitive strain beats the resistant strain because only the latter suffers the cost of resistance; and the resistant strain wins against the producer because the latter bears the higher cost of toxin production and release, while the former pays only the cost of resistance. In an unstructured environment, this game allows periodic cycles in which all three types coexist indefinitely but each with a fluctuating abundance. In a structured environment, this game permits a quasistable global equilibrium, one in which all three strains can persist with nearly constant global abundance (130).
TABLE 4.
Chemical warfare among microbes as a nontransitive, three-way game similar to the rock-scissors-paper game
| Strain below | Wins against | Loses against |
|---|---|---|
| Killer | Sensitive | Resistant |
| Sensitive | Resistant | Killer |
| Resistant | Killer | Sensitive |
Further effects of evolution were incorporated into the model described by Czárán et al. by allowing as many as 14 distinct systems of toxin production, sensitivity, and resistance along with the genetic processes of mutation and recombination that can alter these traits and their associations (130). The permutations of these systems permit the existence of several million different strains. From this additional complexity emerged two distinct quasiequilibrium conditions, the “frozen” and “hyperimmunity” states. In the frozen state, all the toxins are maintained globally, but most colonies are single-toxin producers; that is, each colony produces one toxin to which it is also immune. By contrast, in the hyperimmunity state, many colonies produce no toxin, many others make one, and still others produce several toxins, but only a few produce most of the available toxins. Resistance shows a different distribution, with all of the colonies being resistant to most or all of the toxins. Which of these two outcomes is obtained depends upon initial conditions. If the evolving system begins with the entire population being sensitive to all toxins, then the frozen state results. The hyperimmunity state is reached if the system starts with enough diversity that most colonies already have multiple killer and resistance traits.
Numerous surveys of colicin production in natural populations suggest that populations of E. coli may closely match predictions of the model described by Czárán et al. (231, 559). In E. coli, producer strains are found in frequencies ranging from 10% to 50%. Resistant strains are even more abundant and are found at frequencies from 50% to 98%. In fact, most strains are resistant to all cosegregating colicins. Finally, there is a small population of sensitive cells. Figure 25 provides a summary of phenotype distributions in a population of E. coli cells isolated from wild field mice in Australia (231). The model described by Czárán et al. predicts that this distribution of phenotypes results from the frequent horizontal transfer of resistance and the significant cost to colicin production (130). In other words, if a strain can gain resistance and lose production, they will, over time, just as was observed in the E. coli strains isolated from the field mouse population over the course of a summer (231).
CONCLUDING REMARKS
The numerous results presented in this report demonstrate that the study of colicins, once the model for studies of bacterial toxins, has significantly contributed to progress in a number of fields. The fascinating aspect of colicin is the specificity and the diversity of interactions which it undergoes with various proteins under the course of its action and during its production and release. A great number of these interactions with proteins of the different cell compartments has been demonstrated by genetic, biochemical, and structural investigations and analyzed in vivo and, sometimes, in vitro, but many of them have yet to be deciphered. The most intricate association undertaken by colicin is the association with its cognate immunity protein, which has been extensively studied in the case of nuclease colicins. The specific associations of colicins with the outer membrane protein receptors, the different proteins of the translocation machinery, and the inner membrane immunity proteins have also been well characterized, and their studies are progressing rapidly. Among the various colicin processes that warrant further investigations are mainly the dissociation of the complex nuclease colicin immunity protein, the physiological role of the Tol system, the entry of nuclease colicin into the cytoplasm, the transformation of pore-forming colicins from soluble proteins to membrane proteins, the structure of the voltage-gated channels that they form, and the mechanism of colicin export. But other fundamental questions have to be answered: why are colicins produced after DNA damage, and what is their role in the producing cells and in the environment? Much work has yet to be performed. In the next 80 years, many answers will be found, more colicins will be isolated, more colicin interactions will be known, and more extensive use of colicins and colicin lysis proteins in genetic engineering will be realized than at present, but other questions will surely arise, and colicin studies will certainly continue.
Acknowledgments
We are grateful to many colleagues and coworkers for helpful discussions and critical appraisals of the manuscript. Susan K. Buchanan thanks P. Lukacik (NIDDK/NIH) for making Fig. 5. Kathleen Postle thanks Ray Larsen for critical reading of the manuscript and past and present Postle laboratory members for their excellent contributions to our understanding. Colin Kleanthous thanks Irina Grishkovskaya (York) and Maria Maté (Madrid) for figures (Fig. 21 and 22) used in this review, Dan Walker and Khédidja Mosbahi (York) for comments on the manuscript, and Geoff Moore (Norwich) and Richard James (Nottingham) for a long and fruitful collaboration on colicins. Eric Cascales, Danièle Cavard, Denis Duché, and Roland Lloubès thank Patrick Chames for helpful discussions. We thank Muriel Masi, Rajeev Misra, Emilie Goemaere, and Aurélie Barnéoud for sharing unpublished results, anonymous reviewers for helpful comments and corrections, and authors of original articles for permissions. We apologize for any omissions in citations of primary reports owing to space limitations, although each one has been useful to build the great story of colicins. This review was planned to thank Danièle Cavard for her contribution in the world of colicin since 1966 at the occasion of her retirement.
Susan K. Buchanan is supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH. Kathleen Postle acknowledges grant support from the NIH and the National Science Foundation Cell Biology Program. Colin Kleanthous acknowledges funding for colicin research from the BBSRC and the Wellcome Trust. Margaret A. Riley acknowledges grants from the NIH (“Evolution of Antibiotic Resistance in Enteric Bacteria” and “Molecular Evolution of Enteric Bacteriocins”). Eric Cascales, Danièle Cavard, Denis Duché, and Roland Lloubès are supported by Centre National de la Recherche Scientifique.
Footnotes
This review is dedicated to Robert Kadner (1942-2005).
REFERENCES
- 1.Abergel, C., E. Bouveret, J. M. Claverie, K. Brown, A. Rigal, C. Lazdunski, and H. Bénédetti. 1999. Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution. Structure 7:1291-1300. [DOI] [PubMed] [Google Scholar]
- 2.Akutsu, A., H. Masaki, and T. Ohta. 1989. Molecular structure and immunity specificity of colicin E6, an evolutionary intermediate between E-group colicins and cloacin DF13. J. Bacteriol. 171:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altieri, M., J. L. Suit, M. L. J. Fan, and S. E. Luria. 1986. Expression of the cloned ColE1 kil gene in normal and Kilr Escherichia coli. J. Bacteriol. 168:648-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amati, P. 1964. Vegetative multiplication of colicinogenic factors after induction in Escherichia coli. J. Mol. Biol. 78:239-246. [DOI] [PubMed] [Google Scholar]
- 5.Anderluh, G., I. Gokce, and J. H. Lakey. 2004. A natively unfolded toxin domain uses its receptor as a folding template. J. Biol. Chem. 279:22002-22009. [DOI] [PubMed] [Google Scholar]
- 6.Anderluh, G., Q. Hong, R. Boetzel, C. MacDonald, G. R. Moore, R. Virden, and J. H. Lakey. 2003. Concerted folding and binding of a flexible colicin domain to its periplasmic receptor TolA. J. Biol. Chem. 278:21860-21868. [DOI] [PubMed] [Google Scholar]
- 7.Andreoli, P. M., and H. J. J. Nijkamp. 1976. Mutants of the CloDF13 plasmid in Escherichia coli with decreased bacteriocinogenic activity. Mol. Gen. Genet. 144:159-170. [DOI] [PubMed] [Google Scholar]
- 8.Andreoli, P. M., N. Overbeeke, E. Veltkamp, I. D. A. van Embden, and H. J. J. Nijkamp. 1978. Genetic map of the bacteriocinogenic plasmid CloDF13 derived by insertion of the transposon Tn901. Mol. Gen. Genet. 160:1-11. [DOI] [PubMed] [Google Scholar]
- 9.Aravind, L., K. S. Makarova, and E. V. Koonin. 2000. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28:3417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armstrong, K. A., V. Hershfield, and D. R. Helinski. 1977. Gene cloning and containment properties of plasmid ColE1 and its derivatives. Science 196:172-174. [DOI] [PubMed] [Google Scholar]
- 11.Asensio, C., J. C. Perez-Diaz, M. C. Martinez, and F. Baquero. 1976. A new family of low molecular weight antibiotics from enterobacteria. Biochem. Biophys. Res. Commun. 69:7-14. [DOI] [PubMed] [Google Scholar]
- 12.Baan, R. A., N. Naaktgeboren, R. van Charldorp, P. H. van Knippenberg, and L. Bosch. 1978. Consequences of a specific cleavage in situ of 16S ribosomal RNA for polypeptide chain intiation. Eur. J. Biochem. 87:131-136. [DOI] [PubMed] [Google Scholar]
- 13.Bainbridge, G., G. A. Armstrong, L. G. Dover, K. F. Whelan, and J. H. Lakey. 1998. Displacement of OmpF loop 3 is not required for the membrane translocation of colicins N and A in vivo. FEBS Lett. 432:117-122. [DOI] [PubMed] [Google Scholar]
- 14.Baty, D., M. Frenette, R. Lloubès, V. Geli, S. P. Howard, F. Pattus, and C. Lazdunski. 1988. Functional domains of colicin A. Mol. Microbiol. 2:807-811. [DOI] [PubMed] [Google Scholar]
- 15.Baty, D., M. Knibielher, H. Verheij, F. Pattus, D. Shire, A. Bernadac, and C. Lazdunski. 1987. Site-directed mutagenesis of the COOH-terminal region of colicin A: effect on secretion and voltage-dependent channel activity. Proc. Natl. Acad. Sci. USA 84:1152-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baty, D., J. Lakey, F. Pattus, and C. Lazdunski. 1990. A 136-amino-acid-residue COOH-terminal fragment of colicin A is endowed with ionophoric activity. Eur. J. Biochem. 189:409-413. [DOI] [PubMed] [Google Scholar]
- 17.Baty, D., R. Lloubès, V. Geli, C. Lazdunski, and S. P. Howard. 1987. Extracellular release of colicin A is non-specific. EMBO J. 6:2463-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayer, M. E. 1968. Areas of adhesion between wall and membrane of Escherichia coli. J. Gen. Microbiol. 53:395-404. [DOI] [PubMed] [Google Scholar]
- 19.Baysse, C., J. M. Meyer, P. Plesiat, V. Geoffroy, Y. Michel-Briand, and P. Cornelis. 1999. Uptake of pyocin S3 occurs through the outer membrane ferripyoverdine type II receptor of Pseudomonas aeruginosa. J. Bacteriol. 181:3849-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bazaral, M., and D. R. Helinski. 1968. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J. Mol. Biol. 36:185-194. [DOI] [PubMed] [Google Scholar]
- 21.Bénédetti, H., M. Frenette, D. Baty, M. Knibiehler, F. Pattus, and C. Lazdunski. 1991. Individual domains of colicins confer specificity in colicin uptake, in pore-properties and in immunity requirement. J. Mol. Biol. 217:429-439. [DOI] [PubMed] [Google Scholar]
- 22.Bénédetti, H., M. Frénette, D. Baty, R. Lloubès, V. Géli, and C. Lazdunski. 1989. Comparison of the uptake systems for the entry of various BtuB group colicins into Escherichia coli. J. Gen. Microbiol. 135:3413-3420. [DOI] [PubMed] [Google Scholar]
- 23.Bénédetti, H., C. Lazdunski, and R. Lloubès. 1991. Protein import into E. coli: colicins A and E1 interact with a component of their translocation system. EMBO J. 10:1989-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bénédetti, H., R. Lloubès, C. Lazdunski, and L. Letellier. 1992. Colicin A unfolds during its translocation in Escherichia coli cells and spans the whole cell envelope when its pore has formed. EMBO J. 11:441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett, N. J., and J. Rakonjac. 2006. Unlocking of the filamentous bacteriophage virion during infection is mediated by the C domain of pIII. J. Mol. Biol. 356:266-273. [DOI] [PubMed] [Google Scholar]
- 26.Ben-Zeev, E., R. Zarivach, M. Shoham, A. Yonath, and M. Eisenstein. 2003. Prediction of the structure of the complex between the 30S ribosomal subunit and colicin E3 via weighted-geometric docking. J. Biomol. Struct. Dyn. 20:669-675. [DOI] [PubMed] [Google Scholar]
- 27.Beppu, T., K. Kawabata, and K. Arima. 1972. Specific inhibition of cell division by colicin E2 without degradation of deoxyribonucleic acid in a new colicin sensitivity mutant of Escherichia coli. J. Bacteriol. 110:485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernadac, A., M. Gavioli, J. C. Lazzaroni, S. Raina, and R. Lloubès. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180:4872-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein, A., B. Rolfe, and K. Onodera. 1972. Pleiotropic properties and genetic organization of the tolA,B locus of Escherichia coli K-12. J. Bacteriol. 112:74-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bishop, L. J., E. S. Bjes, V. L. Davidson, and W. A. Cramer. 1985. Localization of the immunity protein-reactive domain in unmodified and chemically modified COOH-terminal peptides of colicin E1. J. Bacteriol. 164:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blair, D. F. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545:86-95. [DOI] [PubMed] [Google Scholar]
- 33.Boeke, J. D., P. Model, and N. D. Zinder. 1982. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol. Gen. Genet. 186:185-192. [DOI] [PubMed] [Google Scholar]
- 34.Bolivar, F., R. L. Rodriguez, M. C. Betlach, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. Ampicillin-resistant derivatives of the plasmid pMB9. Gene 2:75-93. [DOI] [PubMed] [Google Scholar]
- 35.Boon, T. 1971. Inactivation of ribosomes in vitro by colicin E3. Proc. Natl. Acad. Sci. USA 68:2421-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boon, T. 1972. Inactivation of ribosomes in vitro by colicin E3 and its mechanism of action. Proc. Natl. Acad. Sci. USA 69:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourdineaud, J. P., P. Boulanger, C. Lazdunski, and L. Letellier. 1990. In vivo properties of colicin A: channel activity is voltage dependent but translocation may be voltage independent. Proc. Natl. Acad. Sci. USA 87:1037-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bourdineaud, J. P., S. P. Howard, and C. Lazdunski. 1989. Localization and assembly into the Escherichia coli envelope of a protein required for entry of colicin A. J. Bacteriol. 171:2458-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouveret, E., H. Bénédetti, A. Rigal, E. Loret, and C. Lazdunski. 1999. In vitro characterization of peptidoglycan-associated lipoprotein (PAL)-peptidoglycan and PAL-TolB interactions. J. Bacteriol. 181:6306-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouveret, E., R. Derouiche, A. Rigal, R. Lloubès, C. Lazdunski, and H. Bénédetti. 1995. Peptidoglycan-associated lipoprotein-TolB interaction. A possible key to explaining the formation of contact sites between the inner and outer membranes of Escherichia coli. J. Biol. Chem. 270:11071-11077. [DOI] [PubMed] [Google Scholar]
- 41.Bouveret, E., L. Journet, A. Walburger, E. Cascales, H. Bénédetti, and R. Lloubès. 2002. Analysis of the Escherichia coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains. Biochimie 84:413-421. [DOI] [PubMed] [Google Scholar]
- 42.Bouveret, E., A. Rigal, C. Lazdunski, and H. Bénédetti. 1997. The N-terminal domain of colicin E3 interacts with TolB which is involved in the colicin translocation step. Mol. Microbiol. 23:909-920. [DOI] [PubMed] [Google Scholar]
- 43.Bouveret, E., A. Rigal, C. Lazdunski, and H. Benedetti. 1998. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol. Microbiol. 27:143-157. [DOI] [PubMed] [Google Scholar]
- 44.Bowles, L. K., and J. Konisky. 1981. Cleavage of colicin Ia by the Escherichia coli K-12 outer membrane is not mediated by the colicin Ia receptor. J. Bacteriol. 145:668-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowman, C. M., J. E. Dahlerg, T. Ikemura, J. Konisky, and M. Nomura. 1971. Specific inactivation of 16S ribosomal RNA induced by colicin E3 in vitro. Proc. Natl. Acad. Sci. USA 68:964-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradbeer, C. 1993. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol. 175:3146-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradley, D. E., and S. P. Howard. 1992. A new colicin that adsorbs to outer-membrane protein Tsx but is dependent on the tonB instead of the tolQ membrane transport system. J. Gen. Microbiol. 138:2721-2724. [DOI] [PubMed] [Google Scholar]
- 48.Braun, V. 1989. The structurally related exbB and tolQ genes are interchangeable in conferring tonB-dependent colicin, bacteriophage, and albomycin sensitivity. J. Bacteriol. 171:6387-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braun, V. 2003. Iron uptake by Escherichia coli. Front. Biosci. 8:1409-1421. [DOI] [PubMed] [Google Scholar]
- 50.Braun, V., J. Frenz, K. Hantke, and K. Schaller. 1980. Penetration of colicin M into cells of Escherichia coli. J. Bacteriol. 142:162-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braun, V., S. Gaisser, C. Herrman, K. Kampfenkel, H. Killman, and I. Traub. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braun, V., and C. Herrmann. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol. 8:261-268. [DOI] [PubMed] [Google Scholar]
- 53.Braun, V., and C. Herrmann. 2004. Point mutations in transmembrane helices 2 and 3 of ExbB and TolQ affect their activities in Escherichia coli K-12. J. Bacteriol. 186:4402-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84:365-380. [DOI] [PubMed] [Google Scholar]
- 55.Braun, V., H. Pilsl, and P. Groß. 1994. Colicins: structures, modes of actions, transfer through membranes, and evolution. Arch. Microbiol. 161:199-206. [DOI] [PubMed] [Google Scholar]
- 56.Braun, V., K. Schaller, and M. R. Wabl. 1974. Isolation, characterization, and action of colicin M. Antimicrob. Agents Chemother. 5:520-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bredin, J., V. Simonet, R. Iyer, A. H. Delcour, and J. M. Pages. 2003. Colicins, spermine and cephalosporins: a competitive interaction with the OmpF eyelet. Biochem. J. 376:245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brewer, S., M. Tolley, I. P. Trayer, G. C. Barr, C. J. Dorman, K. Hannavy, C. F. Higgins, J. S. Evans, B. A. Levine, and M. R. Wormald. 1990. Structure and function of X-Pro dipeptide repeats in the TonB protein of Salmonella typhimurium and Escherichia coli. J. Mol. Biol. 216:883-895. [DOI] [PubMed] [Google Scholar]
- 59.Brey, R. N. 1982. Fragmentation of colicins A and E1 by cell surface proteases. J. Bacteriol. 149:306-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brock, R. G. P., E. Brinkman, R. van Boxtel, A. C. P. A. Bekkers, H. Verheij, and J. Tommassen. 1994. Molecular characterization of enterobacterial pldA genes encoding outer membrane phospholipase A. J. Bacteriol. 176:861-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brunden, K. R., W. A. Cramer, and F. S. Cohen. 1984. Purification of a small receptor-binding peptide from the central region of the colicin E1 molecule. J. Biol. Chem. 259:190-196. [PubMed] [Google Scholar]
- 62.Reference deleted.
- 63.Buckle, A. M., G. Schreiber, and A. R. Fersht. 1994. Protein-protein recognition: crystal structural analysis of a barnase-barstar complex at 2.0 Å resolution. Biochemistry 33:8878-8889. [DOI] [PubMed] [Google Scholar]
- 64.Bullock, J. O., F. S. Cohen, J. R. Dankert, and W. A. Cramer. 1983. Comparison of the macroscopic and single channel conductance properties of colicin E1 and its COOH-terminal tryptic peptide. J. Biol. Chem. 258:9908-9912. [PubMed] [Google Scholar]
- 65.Bullock, J. O., and E. R. Kolen. 1995. Ion selectivity of colicin E1. III. Anion permeability. J. Membr. Biol. 144:131-145. [DOI] [PubMed] [Google Scholar]
- 66.Bullock, J. O., E. R. Kolen, and J. L. Shear. 1992. Ion selectivity of colicin E1. II. Permeability to organic cations. J. Membr. Biol. 128:1-16. [DOI] [PubMed] [Google Scholar]
- 67.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA 96:10673-10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao, Z., and P. E. Klebba. 2002. Mechanisms of colicin binding and transport through outer membrane porins. Biochimie 84:399-412. [DOI] [PubMed] [Google Scholar]
- 69.Capaldi, A. P., C. Kleanthous, and S. E. Radford. 2002. Im7 folding mechanism: misfolding on a path to the native state. Nat. Struct. Biol. 9:209-216. [DOI] [PubMed] [Google Scholar]
- 70.Carmel, G., and J. W. Coulton. 1991. Internal deletions in the FhuA receptor of Escherichia coli K-12 define domains of ligand interactions. J. Bacteriol. 173:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caro, L., and M. Schnos. 1966. The attachment of the male-specific bacteriophage F1 to sensitive strains of Escherichia coli. Proc. Natl. Acad. Sci. USA 56:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carr, S., C. N. Penfold, V. Bamford, R. James, and A. M. Hemmings. 2000. The structure of TolB, an essential component of the tol-dependent translocation system, and its protein-protein interaction with the translocation domain of colicin E9. Structure 8:57-66. [DOI] [PubMed] [Google Scholar]
- 73.Carr, S., D. Walker, R. James, C. Kleanthous, and A. M. Hemmings. 2000. Inhibition of a ribosome-inactivating ribonuclease: the crystal structure of the cytotoxic domain of colicin E3 in complex with its immunity protein. Structure 8:949-960. [DOI] [PubMed] [Google Scholar]
- 74.Carter, D. M., J. N. Gagnon, M. Damlaj, S. Mandava, L. Makowski, D. J. Rodi, P. D. Pawelek, and J. W. Coulton. 2006. Phage display reveals multiple contact sites between FhuA, an outer membrane receptor of Escherichia coli, and TonB. J. Mol. Biol. 357:236-251. [DOI] [PubMed] [Google Scholar]
- 75.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubès. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cascales, E., M. Gavioli, J. N. Sturgis, and R. Lloubès. 2000. Proton motive force drives the interaction of the inner membrane TolA and outer membrane Pal proteins in Escherichia coli. Mol. Microbiol. 38:904-915. [DOI] [PubMed] [Google Scholar]
- 78.Cascales, E., and R. Lloubès. 2004. Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol. Microbiol. 51:873-885. [DOI] [PubMed] [Google Scholar]
- 79.Cascales, E., R. Lloubes, and J. N. Sturgis. 2001. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42:795-807. [DOI] [PubMed] [Google Scholar]
- 80.Cavard, D. 1977. Role of ionic strength in colicin K adsorption. J. Gen. Microbiol. 99:13-18. [DOI] [PubMed] [Google Scholar]
- 80a.Cavard, D. 1991. Synthesis and functioning of the colicin E1 lysis protein: comparison with the colicin A lysis protein. J. Bacteriol. 173:191-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cavard, D. 1992. Colicin A and colicin E1 lysis proteins differ in their dependence on secA and secY gene products. FEBS Lett. 298:84-88. [DOI] [PubMed] [Google Scholar]
- 82.Cavard, D. 1994. Truncated mutants of the colicin A lysis protein are acylated and processed when overproduced. FEMS Microbiol. Lett. 117:169-174. [DOI] [PubMed] [Google Scholar]
- 83.Cavard, D. 1994. Rescue by vitamin B12 of Escherichia coli cells treated with colicins A and E allows measurement of the kinetics of colicin binding on BtuB. FEMS Microbiol. Lett. 116:37-42. [DOI] [PubMed] [Google Scholar]
- 84.Cavard, D. 1995. Role of DegP protease on levels of various forms of colicin A lysis protein. FEMS Microbiol. Lett. 125:173-178. [DOI] [PubMed] [Google Scholar]
- 85.Cavard, D. 1995. Effects of temperature and heat shock on the expression and action of the colicin A lysis protein. J. Bacteriol. 177:5189-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cavard, D. 1997. Role of the colicin A lysis protein in the expression of the colicin A operon. Microbiology 143:2295-2303. [DOI] [PubMed] [Google Scholar]
- 87.Cavard, D. 1998. Inhibition of colicin synthesis by the antibiotic globomycin. Arch. Microbiol. 171:50-58. [DOI] [PubMed] [Google Scholar]
- 88.Cavard, D. 2002. Assembly of colicin A in the outer membrane of producing Escherichia coli cells requires both phospholipase A and one porin, but phospholipase A is sufficient for secretion. J. Bacteriol. 184:3723-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cavard, D. 2002. Colicin A multimerizes when unfolded. Biochimie 84:485-488. [DOI] [PubMed] [Google Scholar]
- 90.Cavard, D. 2004. Role of Cal, the colicin A lysis protein, in two steps of colicin A release and in the interaction with colicin A-porin complexes. Microbiology 150:3867-3875. [DOI] [PubMed] [Google Scholar]
- 91.Cavard, D., D. Baty, S. P. Howard, H. M. Verheij, and C. Lazdunski. 1987. Lipoprotein nature of the colicin A lysis protein: effect of amino acid substitutions at the site of modification and processing. J. Bacteriol. 169:2187-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cavard, D., A. Bernadac, and C. Lazdunski. 1981. Exclusive localization of colicin A in cell cytoplasm of producing bacteria. Eur. J. Biochem. 119:125-131. [DOI] [PubMed] [Google Scholar]
- 93.Cavard, D., S. P. Howard, and C. Lazdunski. 1989. Functioning of the colicin A lysis protein is affected by Triton X-100, divalent cations and EDTA. J. Gen. Microbiol. 135:1715-1726. [DOI] [PubMed] [Google Scholar]
- 94.Cavard, D., S. P. Howard, R. Lloubès, and C. Lazdunski. 1989. High-level expression of the colicin A lysis protein. Mol. Gen. Genet. 217:511-517. [DOI] [PubMed] [Google Scholar]
- 95.Cavard, D., and C. Lazdunski. 1979. Interaction of colicin E4 with specific receptor sites mediates its cleavage into two fragments inactive towards whole cells. Eur. J. Biochem. 96:525-533. [DOI] [PubMed] [Google Scholar]
- 96.Cavard, D., and C. Lazdunski. 1981. Involvement of BtuB and OmpF proteins in binding and uptake of colicin A. FEMS Microbiol. Lett. 12:311-316. [Google Scholar]
- 97.Cavard, D., and C. Lazdunski. 1990. Colicin cleavage by OmpT protease during both entry into and release from Escherichia coli cells. J. Bacteriol. 172:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cavard, D., C. Lazdunski, and S. P. Howard. 1989. The acylated precursor form of the colicin A lysis protein is a natural substrate of the DegP protease. J. Bacteriol. 171:6316-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cavard, D., R. Lloubès, J. Morlon, M. Chartier, and C. Lazdunski. 1985. Lysis protein encoded by plasmid ColA-CA31. Gene sequence and export. Mol. Gen. Genet. 199:95-100. [DOI] [PubMed] [Google Scholar]
- 100.Cavard, D., J. Marotel-Shirmann, and E. Barbu. 1971. Réversion de la fixation des colicines. C. R. Acad. Sci. Paris 273:1167-1170. [PubMed] [Google Scholar]
- 101.Cavard, D., C. Rampini, E. Barbu, and J. Polonovski. 1968. Activité phospholipasique et autres modifications du métabolisme des phospholipides consécutives à l'action des colicines sur E. coli. Bull. Soc. Chim. Biol. 50:1455-1471. [PubMed] [Google Scholar]
- 102.Cavard, D., P. Sauve, F. Heitz, F. Pattus, C. Martinez, R. Dijkman, and C. Lazdunski. 1988. Hydrodynamic properties of colicin A. Existence of a high-affinity lipid-binding site and oligomerization at acid pH. Eur. J. Biochem. 172:507-512. [DOI] [PubMed] [Google Scholar]
- 103.Chai, T. J., and J. Foulds. 1977. Escherichia coli K12 tolF mutants: alterations in protein composition of the outer membrane. J. Bacteriol. 130:781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chai, T. J., and J. Foulds. 1979. Isolation and partial characterization of protein E, a major protein found in certain Escherichia coli K12 mutant strains: relationship to other outer membrane proteins. J. Bacteriol. 139:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chak, K. F., and R. James. 1984. Localization and characterization of a gene on the ColE3-CA38 plasmid that confers immunity to colicin E8. J. Gen. Microbiol. 130:701-710. [DOI] [PubMed] [Google Scholar]
- 106.Chak, K. F., and R. James. 1985. Analysis of the promoters for the two immunity genes present in the ColE3-CA38 plasmid using two new promoter probe vectors. Nucleic Acids Res. 13:2519-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chak, K. F., and R. James. 1986. Characterization of the ColE9-J plasmid and analysis of its genetic organization. J. Gen. Microbiol. 132:61-71. [DOI] [PubMed] [Google Scholar]
- 108.Chak, K. F., W. S. Kuo, F. M. Lu, and R. James. 1991. Cloning and characterization of the ColE7 plasmid. J. Gen. Microbiol. 137:91-100. [DOI] [PubMed] [Google Scholar]
- 109.Chames, P., J. Fieschi, and D. Baty. 1997. Production of a soluble and active MBP-scFv fusion: favorable effect of the leaky tolR strain. FEBS Lett. 405:224-228. [DOI] [PubMed] [Google Scholar]
- 110.Chan, P. T., H. Ohmori, J. Tomizaqa, and J. Lebowitz. 1985. Nucleotide sequence and gene organization of ColE1 DNA. J. Biol. Chem. 260:8925-8935. [PubMed] [Google Scholar]
- 111.Chang, C., A. Mooser, A. Pluckthun, and A. Wlodawer. 2001. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem. 276:27535-27540. [DOI] [PubMed] [Google Scholar]
- 112.Chao, L., and B. R. Levin. 1981. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl. Acad. Sci. USA 78:6324-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chavan, M., H. Rafi, J. Wertz, C. Goldstone, and M. A. Riley. 2005. Phage associated bacteriocins reveal a novel mechanism for bacteriocin diversification in Klebsiella. J. Mol. Evol. 60:546-556. [DOI] [PubMed] [Google Scholar]
- 114.Cheng, Y. S., Z. Shi, L. G. Doudeva, W. Z. Yang, K. F. Chak, and H. S. Yuan. 2006. High-resolution crystal structure of a truncated ColE7 translocation domain: implications for colicin transport across membranes. J. Mol. Biol. 356:22-31. [DOI] [PubMed] [Google Scholar]
- 114a.Cherezov, V., E. Yamashita, W. Liu, M. Zhalnina, W. A. Cramer, and M. Caffrey. 2006. In meso structure of the cobalamin transporter, BtuB, at 1.95 A resolution. J. Mol. Biol. 364:716-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chhibber, S., and D. V. Vadehra. 1987. Effect of various inducing agents on bacteriocin (klebocin) production by Klebsiella pneumoniae. Folia Microbiol. (Praha) 32:137-141. [DOI] [PubMed] [Google Scholar]
- 116.Chimento, D. P., R. J. Kadner, and M. C. Wiener. 2005. Comparative structural analysis of TonB-dependent outer membrane transporters: implications for the transport cycle. Proteins 59:240-251. [DOI] [PubMed] [Google Scholar]
- 117.Chimento, D. P., A. K. Mohanty, R. J. Kadner, and M. C. Wiener. 2003. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 10:394-401. [DOI] [PubMed] [Google Scholar]
- 118.Clavel, T., J. C. Lazzaroni, A. Vianney, and R. Portalier. 1996. Expression of the tolQRA genes of Escherichia coli K-12 is controlled by the RcsC sensor protein involved in capsule synthesis. Mol. Microbiol. 19:19-25. [DOI] [PubMed] [Google Scholar]
- 119.Clavel, T., P. Germon, A. Vianney, R. Portalier, and J. C. Lazzaroni. 1998. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol. Microbiol. 29:359-367. [DOI] [PubMed] [Google Scholar]
- 120.Click, E. M., and R. E. Webster. 1997. Filamentous phage infection: required interactions with the TolA protein. J. Bacteriol. 179:6464-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Click, E. M., and R. E. Webster. 1998. The TolQRA proteins are required for membrane insertion of the major capsid protein of the filamentous phage f1 during infection. J. Bacteriol. 180:1723-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Clowes, R. C. 1965. Transmission and elimination of colicin factors and some aspects of immunity to colicin E1 in Escherichia coli. Zentrabl. Bakteriol. Parasiten. Abt. I Orig. 196:152-157. [Google Scholar]
- 123.Cole, S. T., B. Saint-Joanis, and A. Pugsley. 1985. Molecular characterization of the colicin E2 operon and identification of its products. Mol. Gen. Genet. 198:465-472. [DOI] [PubMed] [Google Scholar]
- 124.Collarini, M., G. Amblard, C. Lazdunski, and F. Pattus. 1987. Gating processes of channels induced by colicin A, its C-terminal fragment and colicin E1, in planar lipid bilayers. Eur. Biophys. J. 14:147-153. [DOI] [PubMed] [Google Scholar]
- 125.Collins, E. S., S. B. Whittaker, K. Tozawa, C. MacDonald, R. Boetzel, C. N. Penfold, A. Reilly, N. J. Clayden, M. J. Osborne, A. M. Hemmings, C. Kleanthous, R. James, and G. R. Moore. 2002. Structural dynamics of the membrane translocation domain of colicin E9 and its interaction with TolB. J. Mol. Biol. 318:787-804. [DOI] [PubMed] [Google Scholar]
- 126.Cooper, P. C., and R. James. 1984. Two new colicins, E8 and E9, produced by a strain of Escherichia coli. J. Gen. Microbiol. 130:209-215. [DOI] [PubMed] [Google Scholar]
- 127.Cooper, P. C., and R. James. 1985. Three immunity types of klebicins which use the cloacin DF13 receptor of Klebsiella pneumoniae. J. Gen. Microbiol. 131:2313-2318. [DOI] [PubMed] [Google Scholar]
- 128.Cotter, P. D., C. Hill, and R. P. Ross. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777-788. [DOI] [PubMed] [Google Scholar]
- 129.Curtis, M. D., R. James, and A. Coddington. 1989. An evolutionary relationship between the ColE5-099 and the ColE9-J plasmids revealed by nucleotide sequencing. J. Gen. Microbiol. 135:2783-2788. [DOI] [PubMed] [Google Scholar]
- 130.Czárán, T. L., R. F. Hoekstra, and L. Pagie. 2002. Chemical warfare between microbes promotes biodiversity. Proc. Natl. Acad. Sci. USA 99:786-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dandeu, J. P., and E. Barbu. 1967. Purification de la colicine K. C. R. Acad. Sci. Paris 265:774-778. [PubMed] [Google Scholar]
- 131a.Dankert, J., S. M. Hammond, and W. A. Cramer. 1980. Reversal by trypsin of the inhibition of active transport by colicin E1. J. Bacteriol. 143:594-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dankert, J. R., Y. Uratani, C. Grabau, W. A. Cramer, and M. Hermodson. 1982. On a domain structure of colicin E1. A COOH-terminal peptide fragment active in membrane depolarization. J. Biol. Chem. 257:3857-3863. [PubMed] [Google Scholar]
- 133.Davies, J. K., and P. Reeves. 1975. Genetics of resistance to colicins in Escherichia coli K12: cross-resistance among resistance of group B. J. Bacteriol. 123:96-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Davies, J. K., and P. Reeves. 1975. Genetics of resistance to colicins in Escherichia coli K12: cross-resistance among resistance of group A. J. Bacteriol. 123:102-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Day, P. J., T. J. T. Pinheiro, L. M. Roberts, and J. M. Lord. 2002. Binding of ricin A-chain to negatively charged phospholipid vesicles leads to protein structural changes and destabilizes the lipid bilayer. Biochemistry 41:2836-2843. [DOI] [PubMed] [Google Scholar]
- 136.de Chial, M., B. Ghysels, S. A. Beatson, V. Geoffroy, J. M. Meyer, T. Pattery, C. Baysse, P. Chablain, Y. N. Parsons, C. Winstanley, S. J. Cordwell, and P. Cornelis. 2003. Identification of type II and type III pyoverdine receptors from Pseudomonas aeruginosa. Microbiology 149:821-831. [DOI] [PubMed] [Google Scholar]
- 137.de Cock, H., M. Pasveer, J. Tommassen, and E. Bouveret. 2001. Identification of phospholipids as new components that assist in the in vitro trimerization of a bacterial pore protein. Eur. J. Biochem. 268:865-875. [DOI] [PubMed] [Google Scholar]
- 138.de Graaf, F. K., H. G. D. Niekus, and J. Klootwijk. 1973. Inactivation of bacterial ribosomes in vivo and in vitro by cloacin DF13. FEBS Lett. 35:161-165. [DOI] [PubMed] [Google Scholar]
- 139.de Graaf, F. K., and B. Oudega. 1986. Production and release of cloacin DF13 and related colicins. Curr. Top. Microbiol. Immunol. 125:183-205. [DOI] [PubMed] [Google Scholar]
- 140.de Graaf, F. K., M. J. Stukart, F. C. Boogert, and K. Metselaark. 1978. Limited proteolysis of cloacin DF13 and characterization of the cleavage products. Biochemistry 17:1137-1142. [DOI] [PubMed] [Google Scholar]
- 141.Dekker, N., J. Tommassen, A. Lustig, J. P. Rosenbusch, and H. M. Verheij. 1997. Dimerization regulates the enzymatic activity of outer membrane phospholipase A. J. Biol. Chem. 272:3179-3184. [DOI] [PubMed] [Google Scholar]
- 142.Dekker, N., J. Tommassen, and H. M. Verheij. 1999. Bacteriocin release protein triggers dimerization of outer membrane phospholipase A in vivo. J. Bacteriol. 181:3281-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.De Mot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 144.Deng, L. W., and R. N. Perham. 2002. Delineating the site of interaction on the pIII protein of filamentous bacteriophage fd with the F-pilus of Escherichia coli. J. Mol. Biol. 319:603-614. [DOI] [PubMed] [Google Scholar]
- 145.Deng, L. W., P. Malik, and R. N. Perham. 1999. Interaction of the globular domains of pIII protein of filamentous bacteriophage fd with the F-pilus of Escherichia coli. Virology 253:271-277. [DOI] [PubMed] [Google Scholar]
- 146.Dennis, J. J., E. R. Lafontaine, and P. A. Sokol. 1996. Identification and characterization of the tolQRA genes of Pseudomonas aeruginosa. J. Bacteriol. 178:7059-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Deprez, C., L. Blanchard, F. Guerlesquin, M. Gavioli, J. P. Simorre, C. Lazdunski, D. Marion, and R. Lloubès. 2002. Macromolecular import into E. coli: TolA C-terminal domain changes conformation when interacting with the colicin A toxin. Biochemistry 41:2589-2598. [DOI] [PubMed] [Google Scholar]
- 148.Deprez, C., R. Lloubès, M. Gavioli, D. Marion, F. Guerlesquin, and L. Blanchard. 2005. Solution structure of the E. coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J. Mol. Biol. 346:1047-1057. [DOI] [PubMed] [Google Scholar]
- 149.Derouiche, R., H. Bénédetti, J. C. Lazzaroni, C. Lazdunski, and R. Lloubès. 1995. Protein complex within Escherichia coli inner membrane. TolA N-terminal domain interacts with TolQ and TolR proteins. J. Biol. Chem. 270:11078-11084. [DOI] [PubMed] [Google Scholar]
- 150.Derouiche, R., M. Gavioli, H. Bénédetti, A. Prilipov, C. Lazdunski, and R. Lloubès. 1996. TolA central domain interacts with Escherichia coli porins. EMBO J. 15:6408-6415. [PMC free article] [PubMed] [Google Scholar]
- 151.Derouiche, R., G. Zeder-Lutz, H. Bénédetti, M. Gavioli, A. Rigal, C. Lazdunski, and R. Lloubès. 1997. Binding of colicins A and E1 to purified TolA domains. Microbiology 143:3185-3192. [DOI] [PubMed] [Google Scholar]
- 152.Derouiche, R., R. Lloubès, S. Sasso, H. Bouteille, R. Oughideni, C. Lazdunski, and E. Loret. 1999. Circular dichroism and molecular modeling of the E. coli TolA periplasmic domains. Biospectroscopy 5:189-198. [DOI] [PubMed] [Google Scholar]
- 153.Dery, K. J., R. Chavideh, V. Waters, R. Chamorro, L. Tolmasky, and M. Tolmasky. 1997. Characterization of the replication and mobilization regions of the multiresistance Klebsiella pneumoniae plasmid pjhcmw1. Plasmid 38:97-105. [DOI] [PubMed] [Google Scholar]
- 154.Destoumieux-Garzon, D., J. Peduzzi, and S. Rebuffat. 2002. Focus on modified microcins: structural features and mechanisms of action. Biochimie 84:511-519. [DOI] [PubMed] [Google Scholar]
- 155.De Witt, W., and D. R. Helinski. 1965. Colicinogenic factor from a non-induced and a mitomycin C-induced Proteus strain. J. Mol. Biol. 13:692-703. [Google Scholar]
- 156.de Zamaroczy, M., and R. H. Buckingham. 2002. Importation of nuclease colicins into E. coli cells: endoproteolytic cleavage and its prevention by the immunity protein. Biochimie 84:423-432. [DOI] [PubMed] [Google Scholar]
- 157.de Zamaroczy, M., L. Mora, A. Lecuyer, V. Géli, and R. H. Buckingham. 2001. Cleavage of colicin D is necessary for cell killing and requires the inner membrane peptidase LepB. Mol. Cell 8:159-168. [DOI] [PubMed] [Google Scholar]
- 158.Di Masi, D. R., J. C. White, C. A. Schnaitman, and C. Bradbeer. 1973. Transport of vitamin B12 in Escherichia coli: common receptor sites for vitamin B12 and the E colicins on the outer membrane of the cell envelope. J. Bacteriol. 115:506-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Doudeva, L., H. Huang, K. C. Hsia, Z. Shi, C. L. Li, Y. Shen, Y. S. Cheng, and H. S. Yuan. 2006. Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+. Protein Sci. 15:269-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dougan, G., M. Saul, G. Warren, and D. Scherratt. 1978. A functional map of plasmid ColE1. Mol. Gen. Genet. 158:325-327. [Google Scholar]
- 161.Dover, L. G., L. J. A. Evans, S. L. Fridd, G. Bainbridge, E. M. Raggett, and J. H. Lakey. 2000. Colicin pore-forming domains bind to Escherichia coli trimeric porins. Biochemistry 39:8632-8637. [DOI] [PubMed] [Google Scholar]
- 162.Drider, D., G. Fimland, Y. Hechard, L. M. McMullen, and H. Prevost. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Duan, K., E. R. Lafontaine, S. Majumdar, and P. A. Sokol. 2000. RegA, iron, and growth phase regulate expression of the Pseudomonas aeruginosa tol-oprL gene cluster. J. Bacteriol. 182:2077-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dubuisson, J. F., A. Vianney, and J. C. Lazzaroni. 2002. Mutational analysis of the TolA C-terminal domain of Escherichia coli and genetic evidence for an interaction between TolA and TolB. J. Bacteriol. 184:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Duché, D., D. Baty, M. Chartier, and L. Letellier. 1994. Unfolding of colicin A during its translocation through the Escherichia coli envelope as demonstrated by disulfide bond engineering. J. Biol. Chem. 269:24820-24825. [PubMed] [Google Scholar]
- 165a.Duché, D., A. Frenkian, V. Prima, and R. Lloubès. 2006. Release of immunity protein requires functional endonuclease colicin import machinery. J. Bacteriol. 188:8593-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Duché, D., L. Letellier, V. Géli, H. Bénédetti, and D. Baty. 1995. Quantification of group A colicin import sites. J. Bacteriol. 177:4935-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Duché, D., M. W. Parker, J.-M. Gonzalez-Manas, F. Pattus, and D. Baty. 1994. Uncoupled steps of the colicin A pore formation demonstrated by disulfide bond engineering. J. Biol. Chem. 269:6332-6339. [PubMed] [Google Scholar]
- 168.Dunker, A. K., C. J. Brown, J. D. Lawson, L. M. Iakoucheva, and Z. Obradovic. 2002. Intrinsic disorder and protein function. Biochemistry 41:6573-6582. [DOI] [PubMed] [Google Scholar]
- 169.Duport, C., C. Baysse, and Y. Michel-Briand. 1995. Molecular characterization of pyocin S3, a novel S-type pyocin from Pseudomonas aeruginosa. J. Biol. Chem. 270:8920-8927. [DOI] [PubMed] [Google Scholar]
- 170.Durkacz, B. W., C. K. Kennedy, and D. J. Sherratt. 1974. Plasmid replication and the induced synthesis of colicins E1 and E2 in Escherichia coli. J. Bacteriol. 117:940-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170a.Dyson, H. J., and P. E. Wright. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6:197-208. [DOI] [PubMed] [Google Scholar]
- 171.Ebina, Y., F. Kishi, and A. Nakazawa. 1982. Direct participation of lexA protein in repression of colicin E1 synthesis. J. Bacteriol. 150:1479-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ebina, Y., and A. Nakazawa. 1983. Cyclic AMP-dependent initiation and ρ-dependent termination of colicin E1 gene transcription. J. Biol. Chem. 258:7072-7078. [PubMed] [Google Scholar]
- 173.Eick-Helmerich, K., and V. Braun. 1989. Import of biopolymers into Escherichia coli: nucleotide sequences of the exbB and exbD genes are homologous to those of the tolQ and tolR genes, respectively. J. Bacteriol. 171:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Eisenhauer, H. A., S. Shames, P. D. Pawelek, and J. W. Coulton. 2005. Siderophore transport through Escherichia coli outer membrane receptor FhuA with disulfide-tethered cork and barrel domains. J. Biol. Chem. 280:30574-30580. [DOI] [PubMed] [Google Scholar]
- 175.Elgat, M., and R. Ben-Gurion. 1969. Mode of action of pesticin. J. Bacteriol. 98:359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.El Ghachi, M., A. Bouhss, H. Barreteau, T. Touze, G. Auger, D. Blanot, and D. Mengin-Lecreulx. 2006. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J. Biol. Chem. 280:18689-18695. [DOI] [PubMed] [Google Scholar]
- 177.Elkins, P., A. Bunker, W. A. Cramer, and C. V. Stauffacher. 1997. A mechanism for toxin insertion into membranes is suggested by the crystal structure of the channel-forming domain of colicin E1. Structure 5:443-458. [DOI] [PubMed] [Google Scholar]
- 178.el Kouhen, R., A. Bernadac, and J. M. Pages. 1998. Colicin N interaction with sensitive Escherichia coli cells: in situ and kinetic approaches. Res. Microbiol. 149:645-651. [DOI] [PubMed] [Google Scholar]
- 179.el Kouhen, R., H. P. Fierobe, S. Scianimanico, M. Steiert, F. Pattus, and J. M. Pages. 1993. Characterization of the receptor and translocator domains of colicin N. Eur. J. Biochem. 214:635-639. [DOI] [PubMed] [Google Scholar]
- 180.Endemann, H., P. Bross, and I. Rasched. 1992. The adsorption protein of phage IKe. Localization by deletion mutagenesis of domains involved in infectivity. Mol. Microbiol. 6:471-478. [DOI] [PubMed] [Google Scholar]
- 181.Endemann, H., V. Gailus, and I. Rasched. 1993. Interchangeability of the adsorption proteins of bacteriophages Ff and IKe. J. Virol. 67:3332-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Endemann, H., and P. Model. 1995. Location of filamentous phage minor coat proteins in phage and in infected cells. J. Mol. Biol. 250:496-506. [DOI] [PubMed] [Google Scholar]
- 183.Endriss, F., M. Braun, H. Killmann, and V. Braun. 2003. Mutant analysis of the Escherichia coli FhuA protein reveals sites of FhuA activity. J. Bacteriol. 185:4683-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Eraso, J. M., and G. M. Weinstock. 1992. Anaerobic control of colicin E1 production. J. Bacteriol. 174:5101-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Eraso, J. M., M. Chidambaram, and G. M. Weinstock. 1996. Increased production of colicin E1 in stationary phase. J. Bacteriol. 178:1928-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Escuyer, V., P. Boquet, D. Perrin, C. Montecucco, and M. Mock. 1986. A pH-induced increase in hydrophobicity as a possible step in the penetration of colicin E3 through bacterial membranes. J. Biol. Chem. 261:10891-10898. [PubMed] [Google Scholar]
- 187.Escuyer, V., and M. Mock. 1987. DNA sequence analysis of three missense mutations affecting colicin E3 bactericidal activity. Mol. Microbiol. 1:82-85. [DOI] [PubMed] [Google Scholar]
- 188.Espesset, D., Y. Corda, K. Cunningham, H. Benedetti, R. Lloubes, C. Lazdunski, and V. Geli. 1994. The colicin A pore-forming domain fused to mitochondrial intermembrane space sorting signals can be functionally inserted into the Escherichia coli plasma membrane by a mechanism that bypasses the Tol proteins. Mol. Microbiol. 13:1121-1131. [DOI] [PubMed] [Google Scholar]
- 189.Espesset, D., D. Duché, D. Baty, and V. Geli. 1996. The channel domain of colicin A is inhibited by its immunity protein through direct interaction in the Escherichia coli inner membrane. EMBO J. 15:2356-2364. [PMC free article] [PubMed] [Google Scholar]
- 190.Evans, J. S., B. A. Levine, I. P. Trayer, C. J. Dorman, and C. F. Higgins. 1986. Sequence-imposed structural constraints in the TonB protein of E. coli. FEBS Lett. 208:211-216. [DOI] [PubMed] [Google Scholar]
- 191.Evans, L. J., A. Cooper, and J. H. Lakey. 1996. Direct measurement of the association of a protein with a family of membrane receptors. J. Mol. Biol. 255:559-563. [DOI] [PubMed] [Google Scholar]
- 192.Evans, L. J., M. L. Goble, K. A. Hales, and J. H. Lakey. 1996. Different sensitivities to acid denaturation within a family of proteins: implications for acid unfolding and membrane translocation. Biochemistry 35:13180-13185. [DOI] [PubMed] [Google Scholar]
- 193.Evans, L. J., S. Labeit, A. Cooper, L. H. Bond, and J. H. Lakey. 1996. The central domain of colicin N possesses the receptor recognition site but not the binding affinity of the whole toxin. Biochemistry 35:15143-15148. [DOI] [PubMed] [Google Scholar]
- 194.Faraldo-Gomez, J. D., G. R. Smith, and M. S. P. Sansom. 2003. Molecular dynamics simulations of the bacterial outer membrane protein FhuA: a comparative study of the ferrichrome-free and bound states. Biophys. J. 85:1406-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Feldgarden, M., and M. A. Riley. 1998. The phenotypic and fitness effects of colicin resistance in Escherichia coli K-12. Evolution 53:1019-1027. [DOI] [PubMed] [Google Scholar]
- 196.Ferber, D. M., and R. R. Brubaker. 1979. Mode of action of pesticin: N-acetylglucosaminidase activity. J. Bacteriol. 139:495-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ferber, D. M., J. M. Fowler, and R. R. Brubaker. 1981. Mutations to tolerance and resistance to pesticin and colicins in Escherichia coli φ. J. Bacteriol. 146:506-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 199.Ferguson, A. D., and J. Deisenhofer. 2004. Metal import through microbial membranes. Cell 116:15-24. [DOI] [PubMed] [Google Scholar]
- 200.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 201.Ferrer, S., M. B. Viejo, J. F. Guasch, J. Enfedaque, and M. Regué. 1996. Genetic evidence for an activator required for induction of colicin-like bacteriocin 28b production in Serratia marscescens by DNA-damaging agents. J. Bacteriol. 178:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fields, K. L., and S. E. Luria. 1969. Effects of colicins E1 and K on transport systems. J. Bacteriol. 97:57-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Fields, K. L., and S. E. Luria. 1969. Effects of colicins E1 and K on cellular metabolism. J. Bacteriol. 97:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Filloux, A., R. Voulhoux, B. Ize, F. Gérard, G. Ball, and L. F. Wu. 2002. Use of colicin-based genetic tools for studying bacterial protein transport. Biochimie 84:489-497. [DOI] [PubMed] [Google Scholar]
- 205.Fischer, E., K. Günter, and V. Braun. 1989. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J. Bacteriol. 171:5127-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fognini-Lefebvre, N., J. C. Lazzaroni, and R. Portalier. 1987. tolA, tolB and excC, three cistrons involved in the control of pleiotropic release of periplasmic proteins by Escherichia coli K12. Mol. Gen. Genet. 209:391-395. [DOI] [PubMed] [Google Scholar]
- 207.Fourel, D., C. Hikita, J. M. Bolla, S. Mizushima, and J. M. Pages. 1990. Characterization of ompF domains involved in Escherichia coli K-12 sensitivity to colicins A and N. J. Bacteriol. 172:3675-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fredericq, P. 1946. Sur la pluralité des récepteurs d'antibiose de E. coli. C. R. Soc. Biol. (Paris) 140:1189-1194. [Google Scholar]
- 209.Fredericq, P. 1964. Colicins and colicinogeny. Ann. Inst. Pasteur (Paris) 107:7-17. (In French.) [PubMed] [Google Scholar]
- 210.Fredericq, P., and M. Betz-Bareau. 1953. Genetic transfer of colicinogenic property in E. coli. C. R. Soc. Biol. (Paris) 147:1110-1112. [PubMed] [Google Scholar]
- 211.Frost, L. S., and W. Paranchych. 1988. DNA sequence analysis of point mutations in traA, the F pilin gene, reveal two domains involved in F-specific bacteriophage attachment. Mol. Gen. Genet. 213:134-139. [DOI] [PubMed] [Google Scholar]
- 212.Galburt, E. A., and B. L. Stoddard. 2002. Catalytic mechanisms of restriction and homing endonucleases. Biochemistry 41:13851-13860. [DOI] [PubMed] [Google Scholar]
- 213.Garinot-Schneider, C., C. N. Penfold, G. R. Moore, C. Kleanthous, and R. James. 1997. Identification of residues in the putative TolA box which are essential for the toxicity of the endonuclease toxin colicin E9. Microbiology 143:2931-2938. [DOI] [PubMed] [Google Scholar]
- 214.Gaspar, J. A., J. A. Thomas, C. L. Marolda, and M. A. Valvano. 2000. Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol. Microbiol. 38:262-275. [DOI] [PubMed] [Google Scholar]
- 215.Geli, V., D. Baty, and C. Lazdunski. 1988. Use of a foreign epitope as a “tag” for the localization of minor proteins within a cell: the case of the immunity protein to colicin A. Proc. Natl. Acad. Sci. USA 85:689-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Geli, V., D. Baty, F. Pattus, and C. Lazdunski. 1989. Topology and function of the integral membrane protein conferring immunity to colicin A. Mol. Microbiol. 3:679-687. [DOI] [PubMed] [Google Scholar]
- 217.Geli, V., and C. Lazdunski. 1992. An α-helical hydrophobic hairpin as a specific determinant in protein-protein interaction occurring in E. coli colicin A and B immunity systems. J. Bacteriol. 174:6432-6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gennity, J. M., and M. Inouye. 1991. The protein sequence responsible for lipoprotein membrane localization in Escherichia coli exhibits remarkable specificity. J. Biol. Chem. 266:16458-16464. [PubMed] [Google Scholar]
- 219.Gérard, F., N. Pradel, and L. F. Wu. 2005. Bactericidal activity of colicin V is mediated by an inner membrane protein, SdaC, of Escherichia coli. J. Bacteriol. 187:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219a.Gerding, M. A., Y. Ogata, N. D. Pecora, H. Niki, and P. A. J. de Boer. 17 January 2007, posting date. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer membrane invagination during cell constriction in Escherichia coli. Mol. Microbiol. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed]
- 220.Germon, P., T. Clavel, A. Vianney, R. Portalier, and J. C. Lazzaroni. 1998. Mutational analysis of the Escherichia coli K-12 TolA N-terminal region and characterization of its TolQ-interacting domain by genetic suppression. J. Bacteriol. 180:6433-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Germon, P., M. C. Ray, A. Vianney, and J. C. Lazzaroni. 2001. Energy-dependent conformational change in the TolA protein of Escherichia coli involves its N-terminal domain, TolQ, and TolR. J. Bacteriol. 183:4110-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ghosh, P., S. F. Mel, and R. M. Stroud. 1994. The domain structure of the ion channel-forming protein colicin Ia. Nat. Struct. Biol. 1:597-604. [DOI] [PubMed] [Google Scholar]
- 223.Ghosh, J., and K. Postle. 2004. Evidence for dynamic clustering of carboxy-terminal aromatic amino acids in TonB-dependent energy transduction. Mol. Microbiol. 51:203-213. [DOI] [PubMed] [Google Scholar]
- 224.Ghosh, J., and K. Postle. 2005. Disulphide trapping of an in vivo energy-dependent conformation of Escherichia coli TonB protein. Mol. Microbiol. 55:276-288. [DOI] [PubMed] [Google Scholar]
- 225.Gilson, L., H. K. Mahanty, and R. Kolter. 1987. Four plasmid genes are required for colicin V synthesis, export, and immunity. J. Bacteriol. 169:2466-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gilson, L., H. K. Mahanty, and R. Kolter. 1990. Genetic analysis of an MDR-like export system. EMBO J. 9:3875-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Glaser-Wuttke, G., J. Keppner, and I. Rasched. 1989. Pore-forming properties of the adsorption protein of filamentous phage fd. Biochim. Biophys. Acta 985:239-247. [DOI] [PubMed] [Google Scholar]
- 227a.Goemaere, E. L., E. Cascales, and R. Lloubès. 15. December 2006, posting date. Mutational analyses define helix organization and key residues of a bacterial membrane energy-transducing complex. J. Mol. Biol. doi: 10.1016/j.jmb.2006.12.020. [DOI] [PubMed]
- 228.Gokce, I., E. Raggett, Q. Hong, R. Virden, A. Cooper, and J. H. Lakey. 2000. The TolA-recognition site of colicin N. ITC, SPR and stopped-flow fluorescence define a crucial 27-residue segment. J. Mol. Biol. 304:621-632. [DOI] [PubMed] [Google Scholar]
- 229.Goldman, K., J. L. Suit, and C. Kayalar. 1985. Identification of the plasmid-encoded immunity protein for colicin E1 in the inner membrane of Escherichia coli. FEBS Lett. 190:319-323. [DOI] [PubMed] [Google Scholar]
- 230.Gorbalenya, A. E. 1994. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 3:1117-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gordon, D., M. A. Riley, and T. Pinou. 1998. Temporal changes in the frequency of colicinogeny in Escherichia coli from house mice. Microbiology 144:2233-2240. [DOI] [PubMed] [Google Scholar]
- 231a.Gould, J. M., and W. A. Cramer. 1977. Relationship between oxygen-induced proton efflux and membrane energization in cells of Escherichia coli. J. Biol. Chem. 252:5875-5882. [PubMed] [Google Scholar]
- 232.Govan, J. R. 1974. Studies on the pyocins of Pseudomonas aeruginosa: morphology and mode of action of contractile pyocins. J. Gen. Microbiol. 80:1-15. [DOI] [PubMed] [Google Scholar]
- 233.Graille, M., L. Mora, R. H. Buckingham, H. van Tilbeurgh, and M. de Zamaroczy. 2004. Structural inhibition of the colicin D tRNase by the tRNA-mimicking immunity protein. EMBO J. 23:1474-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Grant, R. A., T. C. Lin, W. Konigsberg, and R. E. Webster. 1981. Structure of the filamentous bacteriophage fl. Location of the A, C, and D minor coat proteins. J. Biol. Chem. 256:539-546. [PubMed] [Google Scholar]
- 235.Gratia, A. 1925. Sur un remarquable exemple d'antagonisme entre deux souches de colibacille. C. R. Soc. Biol. (Paris) 93:1040-1041. [Google Scholar]
- 236.Gratia, A., and P. Fredericq. 1946. Diversité des souches antibiotiques de Bacterium coli et étendue variable de leur champ d'action. C. R. Soc. Biol. (Paris) 140:1032-1033. [Google Scholar]
- 237.Griko, Y. V., S. D. Zakharov, and W. A. Cramer. 2000. Structural stability and domain organization of colicin E1. J. Mol. Biol. 302:941-953. [DOI] [PubMed] [Google Scholar]
- 238.Gsponer, J., H. Hopearuoho, S. B. M. Whittaker, G. R. Spence, G. R. Moore, E. Paci, S. E. Radford, and M. Vendruscolo. 2006. Determination of an ensemble of structures representing the intermediate state of the bacterial immunity protein Im7. Proc. Natl. Acad. Sci. USA 103:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Guasch, J. F., J. Enfedaque, S. Ferrer, D. Gargallo, and M. Regué. 1995. Bacteriocin 28b, a chomosomally encoded bacteriocin produced by most Serratia marscescens biotypes. Res. Microbiol. 146:477-483. [DOI] [PubMed] [Google Scholar]
- 240.Guihard, G., P. Boulanger, H. Bénédetti, R. Lloubès, M. Besnard, and L. Letellier. 1994. Colicin A and the Tol proteins involved in its translocation are preferentially located in the contact sites between the inner and outer membranes of Escherichia coli cells. J. Biol. Chem. 269:5874-5880. [PubMed] [Google Scholar]
- 241.Gunasekaran, K., C. J. Tsai, S. Kumar, D. Zanuy, and R. Nussinov. 2003. Extended disordered proteins: targeting function with less scaffold. Trends Biochem. Sci. 28:81-85. [DOI] [PubMed] [Google Scholar]
- 242.Guo, X., R. W. Harrison, and P. C. Tai. 2006. Nucleotide-dependent dimerization of the C-terminal domain of the ABC transporter CvaB in colicin V secretion. J. Bacteriol. 188:2383-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Guterman, S. K. 1973. Colicin B: mode of action and inhibition by enterochelin. J. Bacteriol. 114:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hakkaart, M. J. J., E. Veltkamp, and H. J. J. Nijkamp. 1981. Protein H encoded by plasmid CloDF13 involved in lysis of the bacterial host. I. Localization of the gene and identification and subcellular localization of the gene H product. Mol. Gen. Genet. 183:318-325. [DOI] [PubMed] [Google Scholar]
- 245.Hakkaart, M. J. J., E. Veltkamp, and H. J. J. Nijkamp. 1981. Protein H encoded by plasmid CloDF13 involved in lysis of the bacterial host. II. Functions and regulation of synthesis of the gene H product. Mol. Gen. Genet. 183:326-332. [DOI] [PubMed] [Google Scholar]
- 246.Hall, P. J., and R. R. Brubaker. 1978. Pesticin-dependent generation of somotically stable spheroplast-like structures. J. Bacteriol. 136:786-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hancock, R. E. W., and V. Braun. 1976. Nature of the energy requirement for the irreversible adsorption of bacteriophages T1 and φ80 to Escherichia coli. J. Bacteriol. 125:409-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Hands, S. L., L. E. Holland, M. Vankemmelbeke, L. Fraser, C. J. Macdonald, G. R. Moore, R. James, and C. N. Penfold. 2005. Interactions of TolB with the translocation domain of colicin E9 require an extended TolB box. J. Bacteriol. 187:6733-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hannavy, K., G. C. Barr, C. J. Dorman, J. Adamson, L. R. Mazengera, M. P. Gallagher, J. S. Evans, B. A. Levine, I. P. Trayer, and C. F. Higgins. 1990. TonB protein of Salmonella typhimurium. A model for signal transduction between membranes. J. Mol. Biol. 216:897-910. [DOI] [PubMed] [Google Scholar]
- 250.Hantke, K., and V. Braun. 1978. Functional interaction of the tonA/tonB receptor system in Escherichia coli. J. Bacteriol. 135:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hardy, K. G., G. G. Meynell, J. E. Dowman, and B. G. Spratt. 1973. Two major groups of colicinogenic factors: their evolutionary significance. Mol. Gen. Genet. 125:217-230. [DOI] [PubMed] [Google Scholar]
- 252.Harkness, R. E., and V. Braun. 1989. Colicin M inhibits peptidoglycan biosynthesis by interfering with lipid carrier recycling. J. Biol. Chem. 264:6177-6182. [PubMed] [Google Scholar]
- 253.Hase, C. C. 2003. Ion motive force dependence of protease secretion and phage transduction in Vibrio cholerae and Pseudomonas aeruginosa. FEMS Microbiol. Lett. 227:65-71. [DOI] [PubMed] [Google Scholar]
- 254.Havarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carries out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 255.Hayashi, T., H. Matsumoto, M. Ohnishi, S. Yokota, T. Shinomiya, M. Kageyama, and Y. Terawaki. 1994. Cytotoxin-converting phages, phi CTX and PS21, are R pyocin-related phages. FEMS Microbiol. Lett. 122:239-244. [DOI] [PubMed] [Google Scholar]
- 256.Hechard, Y., and H. G. Sahl. 2002. Mode of action of modified and unmodified bacteriocins from gram-positive bacteria. Biochimie 84:545-557. [DOI] [PubMed] [Google Scholar]
- 257.Heilpern, A. J., and M. K. Waldor. 2000. CTXphi infection of Vibrio cholerae requires the tolQRA gene products. J. Bacteriol. 182:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Heilpern, A. J., and M. K. Waldor. 2003. pIIICTX, a predicted CTXphi minor coat protein, can expand the host range of coliphage fd to include Vibrio cholerae. J. Bacteriol. 185:1037-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258a.Helinski, D. R. 1973. Chemistry and functions of colicins, p. 15-39. In L. P. Hager (ed.), Chemistry and functions of colicins. Academic Press, Inc., New York, NY.
- 259.Heller, K. J., R. J. Kadner, and K. Günter. 1988. Suppression of the btuB451 mutation by mutations in the tonB gene suggests a direct interaction between TonB and TonB-dependent receptor proteins in the outer membrane of Escherichia coli. Gene 64:147-153. [DOI] [PubMed] [Google Scholar]
- 260.Henry, T., S. Pommier, L. Journet, A. Bernadac, J. P. Gorvel, and R. Lloubès. 2004. Improved methods for producing outer membrane vesicles in gram-negative bacteria. Res. Microbiol. 155:437-446. [DOI] [PubMed] [Google Scholar]
- 261.Herschman, H. R., and D. R. Helinski. 1967. Purification and characterization of colicin E2 and colicin E3. J. Biol. Chem. 232:5360-5368. [PubMed] [Google Scholar]
- 262.Herschman, H. R., and D. R. Helinski. 1967. Comparative study of the events associated with colicin induction. J. Bacteriol. 94:691-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Hershield, V., H. W. Boyer, C. Yanovsky, M. A. Lovett, and D. R. Helinski. 1974. Plasmid ColE1 as a molecular vehicle for cloning and amplification of DNA. Proc. Natl. Acad. Sci. USA 71:3455-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Higgs, P. I., R. A. Larsen, and K. Postle. 2002. Quantitation of known components of the Escherichia coli TonB-dependent energy transduction system: TonB, ExbB, ExbD, and FepA. Mol. Microbiol. 44:271-281. [DOI] [PubMed] [Google Scholar]
- 265.Higgs, P. I., P. S. Myers, and K. Postle. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Hill, C., and I. B. Holland. 1967. Genetic basis of colicin E susceptibility in Escherichia coli. I. Isolation and properties of refractory mutants and the preliminary mapping of their mutations. J. Bacteriol. 94:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hilsenbeck, J. L., H. Park, G. Chen, B. Youn, K. Postle, and C. Kang. 2004. Crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution. Mol. Microbiol. 51:711-720. [DOI] [PubMed] [Google Scholar]
- 268.Hofinger, C., H. Karch, and H. Schmidt. 1998. Structure and function of plasmid pColD157 of enterohemorrhagic Escherichia coli O157 and its distribution among strains from patients with diarrhea and hemolytic-uremic syndrome. J. Clin. Microbiol. 36:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Holland, I. B. 1961. The purification and properties of megacin, a bacteriocin from Bacillus megaterium. Biochem. J. 78:641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269a.Holland, I. B. 1975. Physiology of colicin action. Adv. Microb. Physiol. 12:55-140. [Google Scholar]
- 270.Holland, S. J., C. Sanz, and R. N. Perham. 2006. Identification and specificity of pilus adsorption proteins of filamentous bacteriophages infecting Pseudomonas aeruginosa. Virology 345:540-548. [DOI] [PubMed] [Google Scholar]
- 271.Holliger, P., and L. Riechmann. 1997. A conserved infection pathway for filamentous bacteriophages is suggested by the structure of the membrane penetration domain of the minor coat protein g3p from phage fd. Structure 5:265-275. [DOI] [PubMed] [Google Scholar]
- 272.Holliger, P., and L. Riechmann. 1999. Crystal structure of the two N-terminal domains of g3p from filamentous phage fd at 1.9 A: evidence for conformational lability. J. Mol. Biol. 288:649-657. [DOI] [PubMed] [Google Scholar]
- 273.Holloway, B. W., H. Rossiter, D. Burgess, and J. Dodge. 1973. Aeruginocin tolerant mutants of Pseudomonas aeruginosa. Genet. Res. 22:239-253. [DOI] [PubMed] [Google Scholar]
- 274.Hoogenboom, H. R., and P. Chames. 2000. Natural and designer binding sites made by phage display technology. Immunol. Today 21:371-378. [DOI] [PubMed] [Google Scholar]
- 275.Housden, N. G., S. R. Loftus, G. R. Moore, R. James, and C. Kleanthous. 2005. Cell entry mechanism of enzymatic bacterial colicins: porin recruitment and the thermodynamics of receptor binding. Proc. Natl. Acad. Sci. USA 102:13849-13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Howard, S. P., D. Cavard, and C. Lazdunski. 1989. Amino acid sequence and length requirements for assembly and function of the colicin A lysis protein. J. Bacteriol. 171:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Howard, S. P., D. Cavard, and C. Lazdunski. 1991. Phospholipase A-independent damage caused by the colicin A lysis protein during its assembly into the inner and outer membranes of Escherichia coli. J. Gen. Microbiol. 137:81-89. [DOI] [PubMed] [Google Scholar]
- 278.Howard, S. P., C. Herrmann, C. W. Stratilo, and V. Braun. 2001. In vivo synthesis of the periplasmic domain of TonB inhibits transport through the FecA and FhuA iron siderophore transporters of Escherichia coli. J. Bacteriol. 183:5885-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Howard, S. P., M. Leduc, J. van Heijenoort, and C. Lazdunski. 1987. Lysis and release of colicin A in colicinogenic autolytic deficient Escherichia coli mutants. FEMS Microbiol. Lett. 42:147-151. [Google Scholar]
- 280.Howard, S. P., and L. Lindsay. 1998. In vivo analysis of sequence requirements for processing and degradation of the colicin A lysis protein signal peptide. J. Bacteriol. 180:3026-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hsia, K. C., K. F. Chak, P. H. Liang, Y. S. Cheng, W. Y. Ku, and H. S. Yuan. 2004. DNA binding and degradation by the HNH protein ColE7. Structure 12:205-214. [DOI] [PubMed] [Google Scholar]
- 282.Hsia, K. C., C. L. Li, and H. S. Yuan. 2005. Structural and functional insight into sugar-nonspecific nucleases in host defence. Curr. Opin. Struct. Biol. 15:126-134. [DOI] [PubMed] [Google Scholar]
- 283.Hsieh, S. Y., T. P. Ko, M. Y. Tseng, W. Ku, K. F. Chak, and H. S. Yuan. 1997. A novel role of ImmE7 in the autoregulatory expression of the ColE7 operon and identification of possible RNase active sites in the crystal structure of dimeric ImmE7. EMBO J. 16:1444-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Hsiung, H. M., A. Cantrell, J. Luirink, B. Oudega, A. J. Veros, and G. W. Becker. 1989. Use of the bacteriocin release protein in E. coli for excretion of human growth-hormone into the culture medium. Bio/Technology 7:267-271. [Google Scholar]
- 285.Hu, P., J. Elliot, P. McCready, E. Skowronski, J. Garnes, A. Kobayashi, R. R. Brubaker, and E. Garcia. 1998. Structural organization of virulence-associated plasmids of Yersinia pestis. J. Bacteriol. 180:5192-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Iijima, T. 1962. Studies on the colicinogenic factor in Escherichia coli K12. Induction of colicin production by mitomycin C. Bikens J. 5:1-8. [Google Scholar]
- 287.Ikeda, K., and F. Egami. 1969. Receptor substance for pyocin R. I. Partial purification and chemical properties. J. Biochem. (Tokyo) 65:603-609. [DOI] [PubMed] [Google Scholar]
- 288.Inukai, M., M. Takeuchi, K. Shimizu, and M. Arai. 1978. Mechanism of action of globomycin. J. Antibiot. 31:1203-1205. [DOI] [PubMed] [Google Scholar]
- 289.Isaacson, R. E., and J. Konisky. 1974. Studies on the regulation of colicin Ib synthesis. Antimicrob. Agents Chemother. 6:848-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ishidate, K., E. S. Creeger, J. Zrike, S. Deb, B. Glauner, T. J. MacAlister, and L. I. Rothfield. 1986. Isolation of differentiated membrane domains from Escherichia coli and Salmonella typhimurium, including a fraction containing attachment sites between the inner and outer membranes and the murein skeleton of the cell envelope. J. Biol. Chem. 261:428-443. [PubMed] [Google Scholar]
- 291.Isnard, M., A. Rigal, J. C. Lazzaroni, C. Lazdunski, and R. Lloubès. 1994. Maturation and localization of the TolB protein required for colicin import. J. Bacteriol. 176:6392-6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Ize, B., F. Gérard, M. Zhang, A. Chanal, R. Voulhoux, T. Palmer, A. Filloux, and L. F. Wu. 2002. In vivo dissection of the Tat translocation pathway in Escherichia coli. J. Mol. Biol. 317:327-335. [DOI] [PubMed] [Google Scholar]
- 293.Jabrane, A., A. Sabri, P. Compere, P. Jacques, I. Vandenberghe, J. Van Beeumen, and P. Thonart. 2002. Characterization of serracin P, a phage-tail-like bacteriocin, and its activity against Erwinia amylovora, the fire blight pathogen. Appl. Environ. Microbiol. 68:5704-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Jack, R. W., and G. Jung. 2000. Lantibiotics and microcins: polypeptides with unusual chemical diversity. Curr. Opin. Chem. Biol. 4:310-317. [DOI] [PubMed] [Google Scholar]
- 295.Jacob, F. 1954. Mutation of bacteriophage induced by irradiation of bacterial host alone before infection. C. R. Acad. Sci. 238:732-734. [PubMed] [Google Scholar]
- 296.Jacob, F., A. Lwoff, L. Siminovitch, and E. Wollman. 1953. Définition de quelques termes relatifs à la lysogénie. Ann. Inst. Pasteur (Paris) 84:222-224. [PubMed] [Google Scholar]
- 297.Jacob, F., L. Siminovitch, and E. Wollman. 1952. Sur la biosynthèse d'une colicine et sur son mode d'action. Ann. Inst. Pasteur (Paris) 83:295-315. [PubMed] [Google Scholar]
- 298.Jacobson, A. 1972. Role of F pili in the penetration of bacteriophage fl. J. Virol. 10:835-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Jakes, K. S., P. K. Kienker, and A. Finkelstein. 1999. Channel-forming colicins: translocation (and other deviant behaviour) associated with colicin Ia channel gating. Q. Rev. Biophys. 32:189-205. [DOI] [PubMed] [Google Scholar]
- 300.Jakes, K. S., and P. Model. 1979. Mechanism of export of colicin E1 and colicin E3. J. Bacteriol. 138:770-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Jakes, K. S., N. G. Davis, and N. D. Zinder. 1988. A hybrid toxin from bacteriophage f1 attachment protein and colicin E3 has altered cell receptor specificity. J. Bacteriol. 170:4231-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Jakes, K. S., and N. D. Zinder. 1974. Highly purified colicin E3 contains immunity protein. Proc. Natl. Acad. Sci. USA 71:3380-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Jakes, K. S., and N. D. Zinder. 1984. Plasmid ColE3 specifies a lysis protein. J. Bacteriol. 157:582-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Jakes, K. S., N. D. Zinder, and T. Boon. 1974. Purification and properties of colicin E3 immunity protein. J. Biol. Chem. 249:438-444. [PubMed] [Google Scholar]
- 305.James, R. 1988. Molecular cloning and purification of klebicin B. J Gen. Microbiol. 134:2525-2533. [DOI] [PubMed] [Google Scholar]
- 306.James, R., M. Jarvis, and D. F. Barker. 1987. Nucleotide sequence of the immunity and lysis region of the ColE9-J plasmid. J. Gen. Microbiol. 133:1553-1562. [DOI] [PubMed] [Google Scholar]
- 307.James, R., J. Schneider, and P. C. Cooper. 1987. Characterization of three group A klebicin plasmids: localization of their E colicin immunity genes. J. Gen. Microbiol. 133:2253-2262. [DOI] [PubMed] [Google Scholar]
- 308.James, R., C. Kleanthous, and G. R. Moore. 1996. The biology of E colicins: paradigms and paradoxes. Microbiology 142:1569-1580. [DOI] [PubMed] [Google Scholar]
- 309.James, R., C. N. Penfold, G. R. Moore, and C. Kleanthous. 2002. Killing of E. coli cells by E group nuclease colicins. Biochimie 84:381-389. [DOI] [PubMed] [Google Scholar]
- 310.Jaskula, J. C., T. E. Letain, S. K. Roof, J. T. Skare, and K. Postle. 1994. Role of the TonB amino terminus in energy transduction between membranes. J. Bacteriol. 176:2326-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Jeanteur, D., T. Schirmer, D. Fourel, V. Simonet, G. Rummel, C. Widmer, J. P. Rosenbusch, F. Pattus, and J. M. Pages. 1994. Structural and functional alterations of a colicin-resistant mutant of OmpF porin from Escherichia coli. Proc. Natl. Acad. Sci. USA 91:10675-10679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Jiang, X., M. A. Payne, Z. Cao, S. B. Foster, J. B. Feix, S. M. C. Newton, and P. E. Klebba. 1997. Ligand-specific opening of a gated-porin channel in the outer membrane of living bacteria. Science 276:1261-1264. [DOI] [PubMed] [Google Scholar]
- 313.Journet, L., E. Bouveret, A. Rigal, R. Lloubès, C. Lazdunski, and H. Bénédetti. 2001. Import of colicins across the outer membrane of Escherichia coli involves multiple protein interactions in the periplasm. Mol. Microbiol. 42:331-344. [DOI] [PubMed] [Google Scholar]
- 314.Journet, L., A. Rigal, C. Lazdunski, and H. Bénédetti. 1999. Role of TolR N-terminal, central, and C-terminal domains in dimerization and interaction with TolA and TolQ. J. Bacteriol. 181:4476-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Kageyama, M., K. Ikeda, and F. Egami. 1964. Studies of a pyocin. III. Biological properties of the pyocin. J. Biochem. (Tokyo) 55:59-64. [DOI] [PubMed] [Google Scholar]
- 316.Kageyama, M., M. Kobayashi, Y. Sano, T. Uozumi, and H. Masaki. 1992. Construction and characterization of chimeric proteins between pyocins and colicin E3, p. 503-504. In R. James, C. Lazdunski, and F. Pattus (ed.), Bacteriocins, microcins, and lantibiotics. Springer-Verlag, Berlin, Germany.
- 317.Kageyama, M., T. Shinomiya, Y. Aihara, and M. Kobayashi. 1979. Characterization of a bacteriophage related to R-type pyocins. J. Virol. 32:951-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Kageyama, M., M. Kobayashi, Y. Sano, and H. Masaki. 1996. Construction and characterization of pyocin-colicin chimeric proteins. J. Bacteriol. 178:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Kampfenkel, K., and V. Braun. 1992. Membrane topology of the Escherichia coli ExbD protein. J. Bacteriol. 174:5485-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Kampfenkel, K., and V. Braun. 1993. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 268:6050-6057. [PubMed] [Google Scholar]
- 321.Kampfenkel, K., and V. Braun. 1993. Membrane topologies of the TolQ and TolR proteins of Escherichia coli: inactivation of TolQ by a missense mutation in the proposed first transmembrane segment. J. Bacteriol. 175:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Kanoh, S., H. Masaki, S. Yajima, T. Ohta, and T. Uozomi. 1991. Signal peptide of the colicin E2 lysis protein causes host cell death. Agric. Biol. Chem. 55:1607-1614. [PubMed] [Google Scholar]
- 323.Karlsson, F., C. A. Borrebaeck, N. Nilsson, and A. C. Malmborg-Hager. 2003. The mechanism of bacterial infection by filamentous phages involves molecular interactions between TolA and phage protein 3 domains. J. Bacteriol. 185:2628-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Karlsson, F., A. C. Malmborg-Hager, and C. A. Borrebaeck. 2006. Escherichia coli TolA tolerates multiple amino-acid substitutions as revealed by screening randomized variants for membrane integrity and phage receptor function. FEMS Microbiol. Lett. 259:81-88. [DOI] [PubMed] [Google Scholar]
- 325.Karlsson, M., K. Hannavy, and C. F. Higgins. 1993. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol. Microbiol. 8:389-396. [DOI] [PubMed] [Google Scholar]
- 326.Karlsson, M., K. Hannavy, and C. F. Higgins. 1993. A sequence-specific function for the N-terminal signal-like sequence of the TonB protein. Mol. Microbiol. 8:379-388. [DOI] [PubMed] [Google Scholar]
- 327.Kato, C., T. Kobayashi, T. Kudo, T. Furusato, Y. Murakami, T. Tanaka, H. Baba, T. Oishi, E. Ohtsuka, M. Ikehara, et al. 1987. Construction of an excretion vector and extracellular production of human growth hormone from Escherichia coli. Gene 54:197-202. [DOI] [PubMed] [Google Scholar]
- 328.Kedzierska, S., A. Wawrzynow, and A. Taylor. 1996. The Rz1 gene product of bacteriophage λ is a lipoprotein localized in the outer membrane of Escherichia coli. Gene 168:1-8. [DOI] [PubMed] [Google Scholar]
- 329.Keeble, A. H., A. M. Hemmings, R. James, G. R. Moore, and C. Kleanthous. 2002. Multi-step binding of transition metals to the H-N-H endonuclease toxin colicin E9. Biochemistry 41:10234-10244. [DOI] [PubMed] [Google Scholar]
- 330.Keeble, A. H., N. Kirkpatrick, S. Shimizu, and C. Kleanthous. 2006. Calorimetric dissection of colicin DNase-immunity protein complex specificity. Biochemistry 45:3243-3254. [DOI] [PubMed] [Google Scholar]
- 331.Keeble, A. H., and C. Kleanthous. 2005. The kinetic basis for dual recognition in colicin endonuclease-immunity protein complexes. J. Mol. Biol. 352:656-671. [DOI] [PubMed] [Google Scholar]
- 332.Keeble, A. H., M. J. Maté, and C. Kleanthous. 2005. HNH endonucleases, p. 49-65. In M. Belfort, V. Derbyshire, B. L. Stoddard, and D. W. Wood (ed.), Homing endonucleases and inteins. Springer-Verlag, London, United Kingdom.
- 333.Kennedy, C. K. 1971. Induction of colicin production by high temperature or inhibition of protein synthesis. J. Bacteriol. 108:10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Kerr, B., M. A. Riley, M. Feldman, and B. Bohannan. 2002. Local dispersal and interaction promote coexistence in a real life game of rock-paper-scissors. Nature 418:171-174. [DOI] [PubMed] [Google Scholar]
- 335.Khursigara, C. M., G. De Crescenzo, P. D. Pawelek, and J. W. Coulton. 2004. Enhanced binding of TonB to a ligand-loaded outer membrane receptor. Role of the oligomeric state of TonB in formation of a functional FhuA-TonB complex. J. Biol. Chem. 279:7405-7412. [DOI] [PubMed] [Google Scholar]
- 336.Khursigara, C. M., G. De Crescenzo, P. D. Pawelek, and J. W. Coulton. 2005. Deletion of the proline-rich region of TonB disrupts formation of a 2:1 complex with FhuA, an outer membrane receptor of Escherichia coli. Protein Sci. 14:1266-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Khursigara, C. M., G. De Crescenzo, P. D. Pawelek, and J. W. Coulton. 2005. Kinetic analyses reveal multiple steps in forming TonB-FhuA complexes from Escherichia coli. Biochemistry 44:3441-3453. [DOI] [PubMed] [Google Scholar]
- 338.Kienker, P., S. L. Slatin, and A. Finkelstein. 2002. Colicin channels have a shockingly high proton permeability. Biophys. J. 82:555a.11806900 [Google Scholar]
- 339.Kienker, P. K., K. S. Jakes, R. O. Blaustein, C. Miller, and A. Finkelstein. 2003. Sizing the protein translocation pathway of colicin Ia channels. J. Gen. Physiol. 122:161-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Kienker, P. K., X.-Q. Qiu, S. L. Slatin, A. Finkelstein, and K. S. Jakes. 1997. Transmembrane insertion of the colicin Ia hydrophobic hairpin. J. Membr. Biol. 157:27-37. [DOI] [PubMed] [Google Scholar]
- 341.Killmann, H., R. Benz, and V. Braun. 1993. Conversion of the FhuA transport protein into a diffusion channel through the outer membrane of Escherichia coli. EMBO J. 12:3007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Killmann, H., G. Videnov, G. Jung, H. Schwarz, and V. Braun. 1995. Identification of receptor binding sites by competitive peptide mapping: phages T1, T5, and φ80 and colicin M bind to the gating loop of FhuA. J. Bacteriol. 177:694-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Kleanthous, C., U. C. Kuhlmann, A. J. Pommer, N. Ferguson, S. E. Radford, G. R. Moore, R. James, and A. M. Hemmings. 1999. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat. Struct. Biol. 6:243-252. [DOI] [PubMed] [Google Scholar]
- 344.Kleanthous, C., and D. Walker. 2001. Immunity proteins: enzyme inhibitors that avoid the active site. Trends Biochem. Sci. 26:624-631. [DOI] [PubMed] [Google Scholar]
- 345.Knibiehler, M., S. P. Howard, D. Baty, V. Geli, R. Lloubès, P. Sauve, and C. Lazdunski. 1989. Isolation and molecular and functional properties of the amino-terminal domain of colicin A. Eur. J. Biochem. 181:109-113. [DOI] [PubMed] [Google Scholar]
- 346.Ko, T. P., C. C. Liao, W. Y. Ku, K. F. Chak, and H. S. Yuan. 1999. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure 7:91-102. [DOI] [PubMed] [Google Scholar]
- 347.Köck, J., T. Öschläger, R. M. Kamp, and V. Braun. 1987. Primary structure of colicin M, an inhibitor of murein synthesis. J. Bacteriol. 169:3358-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Kodding, J., F. Killig, P. Polzer, S. P. Howard, K. Diederichs, and W. Welte. 2005. Crystal structure of a 92-residue C-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments. J. Biol. Chem. 280:3022-3028. [DOI] [PubMed] [Google Scholar]
- 349.Koebnik, R. 1993. The molecular interaction between components of the TonB-ExbBD-dependent and of the TolQRA-dependent bacterial uptake systems. Mol. Microbiol. 9:219. [DOI] [PubMed] [Google Scholar]
- 350.Koebnik, R. 1995. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol. Microbiol. 16:1269-1270. [DOI] [PubMed] [Google Scholar]
- 351.Koebnik, R. 2005. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13:343-347. [DOI] [PubMed] [Google Scholar]
- 352.Koebnik, R., A. J. Baumler, J. Heesemann, V. Braun, and K. Hantke. 1993. The TonB protein of Yersinia enterocolitica and its interactions with TonB-box proteins. Mol. Gen. Genet. 237:152-160. [DOI] [PubMed] [Google Scholar]
- 353.Kohiyama, M., and M. Nomura. 1965. DNA synthesis and induction of colicin E2 as studied with a temperature-sensitive mutant of colicinogenic E. coli strain. Zentrabl. Bakteriol. Parasiten. Abt. I Orig. 196:211-215. [Google Scholar]
- 354.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041-13050. [DOI] [PubMed] [Google Scholar]
- 355.Kolmar, H., P. R. Waller, and R. T. Sauer. 1996. The DegP and DegQ periplasmic endoproteases of Escherichia coli: specificity for cleavage sites and substrate conformation. J. Bacteriol. 178:5925-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Konings, R. N. H., P. M. Andreoli, E. Veltkamp, and H. J. J. Nijkamp. 1976. Clo DF13 plasmid deoxyribonucleic acid-directed in vitro synthesis of biologically active cloacin DF13 and Clo DF13 immunity protein. J. Bacteriol. 126:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356a.Konisky, J. 1978. The bacteriocins, p. 71-136. In L. N. Ornston and J. R. Sokatch (ed.), The bacteria, vol. 6. Bacterial diversity. Academic Press, Inc., New York, NY. [Google Scholar]
- 357.Konisky, J., and B. S. Cowell. 1972. Interaction of colicin Ia with bacterial cells. Direct measurement of Ia-receptor interaction. J. Biol. Chem. 247:6524-6529. [PubMed] [Google Scholar]
- 358.Konisky, J., and F. M. Richards. 1970. Characterization of colicin Ia and colicin Ib. Purification and some physical properties. J. Biol. Chem. 245:2972-2978. [PubMed] [Google Scholar]
- 359.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 360.Krasilnikov, O. V., J. B. Da Cruz, L. N. Yuldasheva, W. A. Varanda, and R. A. Nogueira. 1998. A novel approach to study the geometry of the water lumen of ion channels: colicin Ia channels in planar lipid bilayers. J. Membr. Biol. 161:83-92. [DOI] [PubMed] [Google Scholar]
- 361.Kremser, A., and I. Rasched. 1994. The adsorption protein of filamentous phage fd: assignment of its disulfide bridges and identification of the domain incorporated in the coat. Biochemistry 33:13954-13958. [DOI] [PubMed] [Google Scholar]
- 362.Kriwacki, R. W., L. Hengst, L. Tennant, S. I. Reed, and P. E. Wright. 1996. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. USA 93:11504-11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455-459. [DOI] [PubMed] [Google Scholar]
- 364.Krone, W. J. A., P. de Vries, G. Koningstein, A. J. R. de Jonge, F. K. de Graaf, and B. Oudega. 1986. Uptake of cloacin DF13 by susceptible cells: removal of immunity protein and fragmentation of cloacin molecules. J. Bacteriol. 166:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Ku, W. Y., Y. W. Liu, Y. C. Hsu, C. C. Liao, P. H. Liang, H. S. Yuan, and K. F. Chak. 2002. The zinc ion in the HNH motif of the endonuclease domain of colicin E7 is not required for DNA binding but is essential for DNA hydrolysis. Nucleic Acids Res. 30:1670-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Kuehn, M. J., and N. C. Kesty. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645-2655. [DOI] [PubMed] [Google Scholar]
- 367.Kuhar, I., J. P. M. van Putten, D. Zgur-Bertok, W. Gaastra, and B. J. A. M. Jordi. 2001. Codon-usage based regulation of colicin K synthesis by the stress alarmone ppGpp. Mol. Microbiol. 41:207-216. [DOI] [PubMed] [Google Scholar]
- 368.Kuhar, I., and D. Zgur-Bertok. 1999. Transcription regulation of the colicin K cka gene reveals induction of colicin synthesis by differential responses to environmental signals. J. Bacteriol. 181:7373-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Kuhlmann, U. C., A. J. Pommer, G. R. Moore, R. James, and C. Kleanthous. 2000. Specificity in protein-protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes. J. Mol. Biol. 301:1163-1178. [DOI] [PubMed] [Google Scholar]
- 370.Kühlmann, U. C., G. R. Moore, R. James, C. Kleanthous, and A. M. Hemmings. 1999. Structural parsimony in endonuclease active sites: should the number of homing endonuclease families be redefined? FEBS Lett. 463:1-2. [DOI] [PubMed] [Google Scholar]
- 371.Kurisu, G., S. D. Zakharov, M. V. Zhalnina, S. Bano, V. Y. Eroukova, T. I. Rokitskaya, Y. N. Antonenko, M. C. Wiener, and W. A. Cramer. 2003. The structure of BtuB with bound colicin E3 R-domain implies a translocon. Nat. Struct. Biol. 10:948-954. [DOI] [PubMed] [Google Scholar]
- 372.Lafontaine, E. R., and P. A. Sokol. 1998. Effects of iron and temperature on expression of the Pseudomonas aeruginosa tolQRA genes: role of the ferric uptake regulator. J. Bacteriol. 180:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Lakey, J. H., D. Baty, and F. Pattus. 1991. Fluorescence energy transfer distance measurements using site-directed single cysteine mutants: the membrane insertion of colicin A. J. Mol. Biol. 218:639-653. [DOI] [PubMed] [Google Scholar]
- 374.Lakey, J. H., D. Duché, J.-M. Gonzalez-Manas, D. Baty, and F. Pattus. 1993. Fluorescence energy transfer distance measurements: the hydrophobic helical hairpin of colicin A in the membrane bound state. J. Mol. Biol. 230:1055-1067. [DOI] [PubMed] [Google Scholar]
- 375.Langen, G. R., J. R. Harper, T. J. Silhavy, and S. P. Howard. 2001. Absence of the outer membrane phospholipase A suppresses the temperature-sensitive phenotype of Escherichia coli degP mutants and induces the Cpx and σE extracytoplasmic stress responses. J. Bacteriol. 183:5230-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Langenscheid, J., H. Killmann, and V. Braun. 2004. A FhuA mutant of Escherichia coli is infected by phage T1-independent of TonB. FEMS Microbiol. Lett. 234:133-137. [DOI] [PubMed] [Google Scholar]
- 377.Larsen, R. A., G. J. Chen, and K. Postle. 2003. Performance of standard phenotypic assays for TonB activity, as evaluated by varying the level of functional, wild-type TonB. J. Bacteriol. 185:4699-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377a.Larsen, R. A., D. Foster-Hartnett, M. A. McIntosh, and K. Postle. 1997. Regions of Escherichia coli TonB and FepA proteins essential for in vivo physical interactions. J. Bacteriol. 179:3213-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377b.Larsen, R. A., G. E. Deckert, K. A. Kastead, S. Devanathan, K. L. Keller, and K. Postle. 2007. His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 378.Larsen, R. A., T. E. Letain, and K. Postle. 2003. In vivo evidence of TonB shuttling between the cytoplasmic and outer membrane in Escherichia coli. Mol. Microbiol. 49:211-218. [DOI] [PubMed] [Google Scholar]
- 379.Larsen, R. A., and K. Postle. 2001. Conserved residues Ser(16) and His(20) and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J. Biol. Chem. 276:8111-8117. [DOI] [PubMed] [Google Scholar]
- 380.Larsen, R. A., M. G. Thomas, and K. Postle. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809-1824. [DOI] [PubMed] [Google Scholar]
- 381.Larsen, R. A., M. T. Thomas, G. E. Wood, and K. Postle. 1994. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (ΔV17) by a missense mutation in ExbB. Mol. Microbiol. 13:627-640. [DOI] [PubMed] [Google Scholar]
- 382.Larsen, R. A., G. E. Wood, and K. Postle. 1993. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol. Microbiol. 10:943-953. [DOI] [PubMed] [Google Scholar]
- 383.Lau, P. C. K., and J. A. Condie. 1989. Nucleotide sequence from the colicin E5, E6 and E9 operons: presence of a degenerate transposon-like structure in the ColE9-J plasmid. Mol. Gen. Genet. 217:269-277. [DOI] [PubMed] [Google Scholar]
- 384.Lau, P. C. K., M. A. Hefford, and P. Klein. 1987. Structural relatedness of lysis proteins from colicinogenic plasmids and icosahedral coliphages. Mol. Biol. Evol. 4:544-556. [DOI] [PubMed] [Google Scholar]
- 385.Reference deleted.
- 386.Lawrence, G. M. P., and R. James. 1984. Characterization of the ColE8 plasmid, a new member of the group E colicin plasmids. Gene 29:145-155. [DOI] [PubMed] [Google Scholar]
- 387.Lazdunski, C., E. Bouveret, A. Rigal, L. Journet, R. Lloubès, and H. Bénédetti. 1998. Colicin import into Escherichia coli cells. J. Bacteriol. 180:4993-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Lazzaroni, J. C., J. F. Dubuisson, and A. Vianney. 2002. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie. 84:391-397. [DOI] [PubMed] [Google Scholar]
- 389.Lazzaroni, J. C., N. Fognini-Lefebvre, and R. C. Portalier. 1986. Cloning of the lkyB (tolB) gene of Escherichia coli K12 and characterization of its product. Mol. Gen. Genet. 204:285-288. [DOI] [PubMed] [Google Scholar]
- 390.Lazzaroni, J. C., N. Fognini-Lefebvre, and R. Portalier. 1989. Cloning of the excC and excD genes involved in the release of periplasmic proteins by Escherichia coli K12. Mol. Gen. Genet. 218:460-464. [DOI] [PubMed] [Google Scholar]
- 391.Lazzaroni, J. C., P. Germon, M. C. Ray, and A. Vianney. 1999. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol. Lett. 177:191-197. [DOI] [PubMed] [Google Scholar]
- 392.Lazzaroni, J. C., and R. C. Portalier. 1981. Genetic and biochemical characterization of periplasmic-leaky mutants of Escherichia coli K-12. J. Bacteriol. 145:1351-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Lazzaroni, J. C., and R. Portalier. 1992. The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL). Mol. Microbiol. 6:735-742. [DOI] [PubMed] [Google Scholar]
- 394.Lazzaroni, J. C., A. Vianney, J. L. Popot, H. Bénédetti, F. Samatey, C. Lazdunski, R. Portalier, and V. Géli. 1995. Transmembrane alpha-helix interactions are required for the functional assembly of the Escherichia coli Tol complex. J. Mol. Biol. 246:1-7. [DOI] [PubMed] [Google Scholar]
- 395.Leduc, M., K. Ishidate, N. Shakibai, and L. Rothfield. 1992. Interactions of Escherichia coli membrane lipoproteins with the murein sacculus. J. Bacteriol. 174:7982-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Lenski, R. R., and M. A. Riley. 2002. Chemical warfare from an ecological perspective. Proc. Natl. Acad. Sci. USA 99:556-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Letain, T. E., and K. Postle. 1997. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in gram-negative bacteria. Mol. Microbiol. 24:271-283. [DOI] [PubMed] [Google Scholar]
- 398.Letellier, L., P. Boulanger, L. Plancon, P. Jacquot, and M. Santamaria. 2004. Main features on tailed phage, host recognition and DNA uptake. Front. Biosci. 9:1228-1339. [DOI] [PubMed] [Google Scholar]
- 399.Letellier, L., L. Plancon, M. Bonhivers, and P. Boulanger. 1999. Phage DNA transport across membranes. Res. Microbiol. 150:499-505. [DOI] [PubMed] [Google Scholar]
- 400.Levengood, S. K., W. F. Beye, Jr., and R. E. Webster. 1991. TolA: a membrane protein involved in colicin uptake contains an extended helical region. Proc. Natl. Acad. Sci. USA 88:5939-5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Levengood-Freyermuth, S. K., E. M. Click, and R. E. Webster. 1993. Role of the carboxyl-terminal domain of TolA in protein import and integrity of the outer membrane. J. Bacteriol. 175:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Li, W., C. A. Dennis, G. R. Moore, R. James, and C. Kleanthous. 1997. Protein-protein interaction specificity of Im9 for the endonuclease toxin colicin E9 defined by homologue-scanning mutagenesis. J. Biol. Chem. 272:22253-22258. [DOI] [PubMed] [Google Scholar]
- 403.Li, W., S. J. Hamill, A. M. Hemmings, G. R. Moore, R. James, and C. Kleanthous. 1998. Dual recognition and the role of specificity-determining residues in colicin E9 DNase-immunity protein interactions. Biochemistry 37:11771-11779. [DOI] [PubMed] [Google Scholar]
- 404.Li, W., A. H. Keeble, C. Giffard, G. R. Moore, R. James, and C. Kleanthous. 2004. Highly discriminating protein-protein interaction specificities in the context of a conserved binding energy hotspot. J. Mol. Biol. 337:743-759. [DOI] [PubMed] [Google Scholar]
- 405.Lin, J. H., and A. Baumgaertner. 2000. Stability of a melittin pore in a lipid bilayer: a molecular dynamics study. Biophys. J. 78:1714-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Lin, Y. L., Y. Elias, and R. H. Huang. 2005. Structural and mutational studies of the catalytic domain of colicin E5: a tRNA-specific ribonuclease. Biochemistry 44:10494-10500. [DOI] [PubMed] [Google Scholar]
- 407.Lindeberg, M., and W. A. Cramer. 2001. Identification of specific residues in colicin E1 involved in immunity protein recognition. J. Bacteriol. 183:2132-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Lindeberg, M., S. D. Zakharov, and W. A. Cramer. 2000. Unfolding pathway of the colicin E1 channel protein on a membrane surface. J. Mol. Biol. 295:679-692. [DOI] [PubMed] [Google Scholar]
- 409.Little, J. W., and D. W. Mount. 1982. The SOS regulatory system of Escherichia coli. Cell 29:11-22. [DOI] [PubMed] [Google Scholar]
- 410.Llamas, M. A., J. J. Rodriguez-Herva, R. E. Hancock, W. Bitter, J. Tommassen, and J. L. Ramos. 2003. Role of Pseudomonas putida tol-oprL gene products in uptake of solutes through the cytoplasmic membrane. J. Bacteriol. 185:4707-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Lloubès, R., D. Baty, and C. Lazdunski. 1986. The promoters of the genes for colicin production, release and immunity in the ColA plasmid: effects of convergent transcription and LexA protein. Nucleic Acids Res. 14:2621-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Lloubès, R., D. Baty, and C. Lazdunski. 1988. Transcriptional terminators in the caa-cal operon and cai gene. Nucleic Acids Res. 16:3739-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Lloubès, R., E. Cascales, A. Walburger, E. Bouveret, C. Lazdunski, A. Bernadac, and L. Journet. 2001. The Tol-Pal proteins of Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152:523-529. [DOI] [PubMed] [Google Scholar]
- 414.Lloubès, R., M. Granger-Schnarr, C. Lazdunski, and M. Schnarr. 1988. LexA repressor induces operator dependent DNA bending. J. Mol. Biol. 204:1049-1054. [DOI] [PubMed] [Google Scholar]
- 415.Lloubès, R., M. Granger-Schnarr, C. Lazdunski, and M. Schnarr. 1991. Interaction of a regulatory protein with a DNA target containing two overlapping binding sites. J. Biol. Chem. 266:2303-2312. [PubMed] [Google Scholar]
- 416.Lloubès, R., N. Vita, A. Bernadac, D. Shire, P. Leplatois, V. Géli, M. Frenette, M. Knibiehler, C. Lazdunski, and D. Baty. 1993. Colicin A lysis protein promotes extracellular release of active human growth hormone accumulated in Escherichia coli cytoplasm. Biochimie 75:451-458. [DOI] [PubMed] [Google Scholar]
- 417.Loftus, S. R., D. Walker, M. J. Maté, D. A. Bonsor, R. James, G. R. Moore, and C. Kleanthous. 2006. Competitive recruitment of the periplasmic translocation portal TolB by a natively disordered domain of colicin E9. Proc. Natl. Acad. Sci. USA 103:12353-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Lopez, J., and R. E. Webster. 1985. Assembly site of bacteriophage f1 corresponds to adhesion zones between the inner and outer membranes of the host cell. J. Bacteriol. 163:1270-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Lotz, W. 1978. Effect of guanosine tetraphosphate on in vitro protein synthesis directed by E1 and E3 colicinogenic factors. J. Bacteriol. 135:707-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Lubkowski, J., F. Hennecke, A. Pluckthun, and A. Wlodawer. 1998. The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p. Nat. Struct. Biol. 5:140-147. [DOI] [PubMed] [Google Scholar]
- 421.Lubkowski, J., F. Hennecke, A. Pluckthun, and A. Wlodawer. 1999. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Structure 7:711-722. [DOI] [PubMed] [Google Scholar]
- 422.Luirink, J., D. M. Clark, J. Ras, E. J. Veschoor, F. Stegehuis, F. K. DeGraaf, and B. Oudega. 1989. pCloDF13-encoded bacteriocin release proteins with shortened carboxyl-terminal segments are lipid modified and processed and function in release of cloacin DF13 and apparent host cell lysis. J. Bacteriol. 171:2673-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Luirink, J., F. K. DeGraaf, and B. Oudega. 1987. Uncoupling of synthesis and release of cloacin DF13 and its immunity protein by Escherichia coli. Mol. Gen. Genet. 206:126-132. [DOI] [PubMed] [Google Scholar]
- 424.Luirink, J., J. de Jong, A. J. van Putten, F. K. de Graaf, and B. Oudega. 1989. Functioning of a hybrid BRP-beta-lactamase protein in the release of cloacin DF13 and lysis of Escherichia coli cells. FEMS Microbiol. 49:25-31. [DOI] [PubMed] [Google Scholar]
- 425.Luirink, J., S. Mayashi, H. C. Wu, M. M. Kater, F. K. DeGraaf, and B. Oudega. 1988. Effect of a mutation preventing lipid modification on localization of the pCloDF13-encoded bacteriocin release protein and on release of cloacin DF13. J. Bacteriol. 170:4153-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Luirink, J., C. van der Sande, J. Tommassen, E. Veltkamp, F. K. DeGraaf, and B. Oudega. 1986. Effects of divalent cations and of phospholipase A activity on excretion of cloacin DF13 and lysis of host cells. J. Gen. Microbiol. 132:825-834. [DOI] [PubMed] [Google Scholar]
- 427.Luna-Chávez, C., Y. L. Lin, and R. H. Huang. 2006. Molecular basis of inhibition of the ribonuclease activity of colicin E5 by its cognate immunity protein. J. Mol. Biol. 358:571-579. [DOI] [PubMed] [Google Scholar]
- 428.Luo, W., X. Yao, and M. Hong. 2005. Large structure rearrangement of colicin Ia channel domain after membrane binding from 2D 13C spin diffusion NMR. J. Am. Chem. Soc. 127:6402-6408. [DOI] [PubMed] [Google Scholar]
- 428a.Luria, S. E. 1964. On the mechanisms of action of colicins. Ann. Inst. Pasteur (Paris) 107(Suppl.):67-73. [PubMed] [Google Scholar]
- 429.Luria, S. E. 1982. The mistaken identity of colicin A. J. Bacteriol. 149:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Lwoff, A., F. Jacob, E. Ritz, and M. Gage. 1952. Induction of bacteriophage production and of a colicin by peroxydes, ethyleneimines and halogenated alkylamines. C. R. Acad. Sci. 234:2308-2310. [PubMed] [Google Scholar]
- 431.Macdonald, C. J., K. Tozawa, E. S. Collins, C. N. Penfold, R. James, C. Kleanthous, N. J. Clayden, and G. R. Moore. 2004. Characterisation of a mobile protein-binding epitope in the translocation domain of colicin E9. J. Biomol. NMR 30:81-96. [DOI] [PubMed] [Google Scholar]
- 432.Manchak, J., K. G. Anthony, and L. S. Frost. 2002. Mutational analysis of F-pilin reveals domains for pilus assembly, phage infection and DNA transfer. Mol. Microbiol. 43:195-205. [DOI] [PubMed] [Google Scholar]
- 433.Mankovich, J. A., C. H. Hsu, and J. Konisky. 1986. DNA and amino acid sequence analysis of structural and immunity genes of colicins Ia and Ib. J. Bacteriol. 168:228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Mann, B. J., C. D. Holroyd, C. Bradbeer, and R. J. Kadner. 1986. Reduced activity of TonB-dependent functions in strains of Escherichia coli. FEMS Lett. 33:255-260. [Google Scholar]
- 435.Marco, R., S. M. Jazwinski, and A. Kornberg. 1974. Binding, eclipse, and penetration of the filamentous bacteriophage M13 in intact and disrupted cells. Virology 62:209-223. [DOI] [PubMed] [Google Scholar]
- 436.Markov, D., G. E. Christie, B. Sauer, R. Calendar, T. Park, R. Young, and C. Severinov. 2004. P2 growth restriction on an rpoC mutant is suppressed by alleles of the Rz1 homolog lysC. J. Bacteriol. 186:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Marotel-Shirmann, J., and E. Barbu. 1969. Quelques données sur la fixation des colicines. C. R. Acad. Sci. Paris 269:866-870. [PubMed] [Google Scholar]
- 438.Marotel-Shirmann, J., D. Cavard, L. Sandler, and E. Barbu. 1970. Temps nécessaire à une colicine fixée sur une bactérie pour déclencher des processus irréversibles. C. R. Acad. Sci. Paris 270:230-233. [PubMed] [Google Scholar]
- 439.Martin, A., and F. X. Schmid. 2003. Evolutionary stabilization of the gene-3-protein of phage fd reveals the principles that govern the thermodynamic stability of two-domain proteins. J. Mol. Biol. 328:863-875. [DOI] [PubMed] [Google Scholar]
- 440.Martin, A., and F. X. Schmid. 2003. The folding mechanism of a two-domain protein: folding kinetics and domain docking of the gene-3 protein of phage fd. J. Mol. Biol. 329:599-610. [DOI] [PubMed] [Google Scholar]
- 441.Martinez, M. C., C. Lazdunski, and F. Pattus. 1983. Isolation, molecular and functional properties of the C-terminal domain of colicin A. EMBO J. 2:1501-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Marvin, D. A., and B. Hohn. 1969. Filamentous bacterial viruses. Bacteriol. Rev. 33:172-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Marzari, R., D. Sblattero, M. Righi, and A. Bradbury. 1997. Extending filamentous phage host range by the grafting of a heterologous receptor binding domain. Gene 185:27-33. [DOI] [PubMed] [Google Scholar]
- 444.Masaki, H., A. Akutsu, T. Uozumi, and T. Ohta. 1991. Identification of a unique specificity determinant of the colicin E3 immunity protein. Gene 107:133-138. [DOI] [PubMed] [Google Scholar]
- 445.Masaki, H., and T. Ohta. 1985. Colicin E3 and its immunity genes. J. Mol. Biol. 182:217-227. [DOI] [PubMed] [Google Scholar]
- 446.Masaki, H., M. Toba, and T. Ohta. 1985. Structure and expression of the ColE2-P9 immunity gene. Nucleic Acids Res. 13:1623-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Mashburn, L. M., and M. Whiteley. 2005. Membrane vesicles traffic signals and facilitate group activities in a procaryote. Nature 437:422-425. [DOI] [PubMed] [Google Scholar]
- 447a.Masi, M., P. Vuong, M. Humbard, K. Malone, and R. Misra. 2007. Initial steps of colicin E7 import across the outer membrane of Escherichia coli. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 448.Maté, M. J., and C. Kleanthous. 2004. Structure-based analysis of the metal-dependent mechanism of H-N-H endonucleases. J. Biol. Chem. 279:34763-34769. [DOI] [PubMed] [Google Scholar]
- 449.Matlack, K. E., B. Misselwitz, K. Plath, and T. A. Rapoport. 1999. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell 97:553-564. [DOI] [PubMed] [Google Scholar]
- 450.Matsuzaki, K. 1998. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim. Biophys. Acta 1376:391-400. [DOI] [PubMed] [Google Scholar]
- 451.Matsuzaki, K., O. Murase, N. Fujii, and K. Miyajima. 1996. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry 35:11361-11368. [DOI] [PubMed] [Google Scholar]
- 452.McBroom, A. J., A. P. Johnson, S. Vemulapalli, and M. J. Kuehn. 2006. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.McCafferty, J., A. D. Griffiths, G. Winter, and D. J. Chiswell. 1990. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348:552-554. [DOI] [PubMed] [Google Scholar]
- 454.Meadow, P. M., and P. L. Wells. 1978. Receptor sites for R-type pyocins and bacteriophage E79 in the core part of the lipopolysaccharide of Pseudomonas aeruginosa PAC1. J. Gen. Microbiol. 108:339-343. [Google Scholar]
- 455.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Mende, J., and V. Braun. 1990. Import-defective colicin B derivatives mutated in the TonB box. Mol. Microbiol. 4:1523-1533. [DOI] [PubMed] [Google Scholar]
- 457.Meury, J., and G. Devilliers. 1999. Impairment of cell division in tolA mutants of Escherichia coli at low and high medium osmolarities. Biol. Cell 91:67-75. [PubMed] [Google Scholar]
- 458.Michel-Briand, Y., and C. Baysse. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499-510. [DOI] [PubMed] [Google Scholar]
- 459.Miyadai, H., K. Tanaka-Masuda, S. Matsuyama, and H. Tokuda. 2004. Effects of lipoprotein overproduction on the induction of DegP (HtrA) involved in quality control in the Escherichia coli periplasm. J. Biol. Chem. 279:39807-39813. [DOI] [PubMed] [Google Scholar]
- 460.Mizuno, T. 1979. A novel peptidoglycan-associated lipoprotein found in the cell envelope of Pseudomonas aeruginosa and Escherichia coli. J. Biochem. (Tokyo) 86:991-1000. [DOI] [PubMed] [Google Scholar]
- 461.Mock, M., and M. Schwartz. 1978. Mechanism of colicin E3 production in strains harboring wild-type or mutant plasmids. J. Bacteriol. 136:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Mock, M., and M. Schwartz. 1980. Mutations which affect the structure and activity of colicin E3. J. Bacteriol. 142:384-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Mohanty, A. K., C. M. Bishop, T. C. Bishop, W. C. Wimley, and M. C. Wiener. 2003. Enzymatic E-colicins bind to their target receptor BtuB by presentation of a small binding epitope on a coiled-coil scaffold. J. Biol. Chem. 278:40953-40958. [DOI] [PubMed] [Google Scholar]
- 464.Mora, L., N. Diaz, R. H. Buckingham, and M. de Zamaroczy. 2005. Import of the transfer RNase colicin D requires site-specific interaction with the energy-transducing protein TonB. J. Bacteriol. 187:2693-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Morlon, J., M. Chartier, M. Bidaud, and C. Lazdunski. 1988. The complete nucleotide sequence of the colicinogenic plasmid ColA. High extent of homology with ColE1. Mol. Gen. Genet. 211:231-243. [DOI] [PubMed] [Google Scholar]
- 466.Morlon, J., R. Lloubès, M. Chartier, J. Bonicel, and C. Lazdunski. 1983. Nucleotide sequence of promoter, operator and amino-terminal region of caa, the structural gene of colicin A. EMBO J. 2:787-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Mosbahi, K., C. Lemaitre, A. H. Keeble, H. Mobasheri, B. Morel, R. James, G. R. Moore, E. J. Lea, and C. Kleanthous. 2002. The cytotoxic domain of colicin E9 is a channel-forming endonuclease. Nat. Struct. Biol. 9:476-484. [DOI] [PubMed] [Google Scholar]
- 468.Mosbahi, K., D. Walker, R. James, G. R. Moore, and C. Kleanthous. 2006. Global structural rearrangement of the cell penetrating ribonuclease colicin E3 on interaction with phospholipid membranes. Protein Sci. 15:620-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Mosbahi, K., D. Walker, E. Lea, G. R. Moore, R. James, and C. Kleanthous. 2004. Destabilisation of the colicin E9 DNase domain by interaction with negatively charged phospholipids: implications for colicin translocation into bacteria. J. Biol. Chem. 279:22145-22151. [DOI] [PubMed] [Google Scholar]
- 470.Muga, A., J. M. Gonzalez-Manas, J. H. Lakey, F. Pattus, and W. K. Surewicz. 1993. pH-dependent stability and membrane interaction of the pore-forming domain of colicin A. J. Biol. Chem. 268:1553-1557. [PubMed] [Google Scholar]
- 471.Mulec, J., Z. Podlesek, P. Mrak, A. Kopitar, A. Ihan, and D. Zgur-Bertok. 2003. A cka-gfp transcriptional fusion reveals that the colicin K activity gene is induced in only 3 percent of the population. J. Bacteriol. 185:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Muller, M. M., A. Vianney, J. C. Lazzaroni, R. E. Webster, and R. Portalier. 1993. Membrane topology of the Escherichia coli TolR protein required for cell envelope integrity. J. Bacteriol. 175:6059-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Munthali, M. T., K. N. Timmis, and E. Diaz. 1996. Use of colicin E3 for biological containment of microorganisms. Appl. Environ. Microbiol. 62:1805-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Nagel de Zwaig, R. 1969. New class of conditional colicin-tolerant mutants. J. Bacteriol. 99:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Nagel de Zwaig, R., and S. E. Luria. 1967. Genetics and physiology of colicin-tolerant mutants of Escherichia coli. J. Bacteriol. 94:1112-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Nakayama, K., K. Takashima, H. Ishihara, T. Shinomiya, M. Kageyama, S. Kanaya, M. Ohnishi, T. Murata, H. Mori, and T. Hayashi. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38:213-231. [DOI] [PubMed] [Google Scholar]
- 477.Nardi, A., Y. Corda, D. Baty, and D. Duché. 2001. Colicin A immunity protein interacts with the hydrophobic helical hairpin of the colicin A channel domain in the Escherichia coli inner membrane. J. Bacteriol. 183:6721-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Nardi, A., S. Slatin, D. Baty, and D. Duché. 2001. The C-terminal half of the colicin A pore-forming domain is active in vivo and in vitro. J. Mol. Biol. 307:1293-1303. [DOI] [PubMed] [Google Scholar]
- 479.Nelson, F. K., S. M. Friedman, and G. P. Smith. 1981. Filamentous phage DNA cloning vectors: a noninfective mutant with a nonpolar deletion in gene III. Virology 108:338-350. [DOI] [PubMed] [Google Scholar]
- 480.Nijkamp, H. J. J., R. de Lang, A. R. Stuitje, P. J. M. van den Elzen, E. Veltkamp, and A. J. van Putten. 1986. The complete nucleotide sequence of the bacteriocinogenic plasmid CloDF13. Plasmid 16:135-160. [DOI] [PubMed] [Google Scholar]
- 481.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Nilsson, I., and G. von Heijne. 1990. Fine-tuning the topology of a polytopic membrane protein: role of positively and negatively charged amino acids. Cell 62:1135-1141. [DOI] [PubMed] [Google Scholar]
- 483.Nogueira, R. A., and W. A. Varanda. 1988. Gating properties of channels formed by colicin Ia in planar lipid bilayer membranes. J. Membr. Biol. 105:143-153. [DOI] [PubMed] [Google Scholar]
- 484.Nomura, M. 1963. Mode of action of colicins. Cold Spring Harbor Symp. Quant. Biol. 28:315-324. [Google Scholar]
- 485.Nomura, M. 1964. Mechanism of action of colicins. Proc. Natl. Acad. Sci. USA 52:1514-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Nomura, M., and M. Nakamura. 1962. Reversibility of inhibition of nucleic acids and protein sythesis by colicin K. Biochem. Biophys. Res. Commun. 7:306-309. [DOI] [PubMed] [Google Scholar]
- 487.Nomura, M., and C. Witten. 1967. Interaction of colicins with bacterial cells. III. Colicin-tolerant mutations in Escherichia coli. J. Bacteriol. 94:1093-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Norman, A., L. H. Hansen, and S. J. Sorensen. 2005. Construction of a ColD cda promoter-based SOS-green fluorescent protein whole-cell biosensor with higher sensitivity toward genotoxic compounds than constructs based on recA, umuDC, or sulA promoters. Appl. Environ. Microbiol. 71:2338-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Nose, K., and D. Mizuno. 1968. Degradation of ribosomes in Escherichia coli cells treated with colicin E2. J. Biochem. (Tokyo) 64:1-6. [DOI] [PubMed] [Google Scholar]
- 490.Ochi, T., K. Yano, M. Ozaki, Y. Higashi, and K. Inoue. 1971. Studies on immunity to megacin A: susceptibility of phospholipids to phospholipase A activity of megacin A. Biken J. 14:29-36. [PubMed] [Google Scholar]
- 491.Ogawa, T., K. Tomita, T. Ueda, K. Watanabe, T. Uozumi, and H. Masaki. 1999. A cytosolic ribonuclease targeting specific transfer RNA anticodons. Science 283:2097-2100. [DOI] [PubMed] [Google Scholar]
- 492.Ogierman, M., and V. Braun. 2003. Interactions between the outer membrane ferric citrate transporter FecA and TonB: studies of the FecA TonB box. J. Bacteriol. 185:1870-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Ogle, J. M., A. P. Carter, and V. Ramakrishnan. 2003. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28:259-266. [DOI] [PubMed] [Google Scholar]
- 494.Ohkawa, I., S. Shiga, and M. Kageyama. 1980. Effect of iron concentration in the growth medium on the sensitivity of Pseudomonas aeruginosa to pyocin S2. J. Biochem. (Tokyo) 87:323-331. [DOI] [PubMed] [Google Scholar]
- 495.Ohno-Iwashita, Y., and K. Imahori. 1980. Assignment of the functional loci in colicin E2 and E3 molecules by characterization of their proteolytic fragments. Biochemistry 19:652-659. [DOI] [PubMed] [Google Scholar]
- 496.Ohno-Iwashita, Y., and K. Imahori. 1982. Assignment of the functional loci in the colicin E1 molecule by characterization of its proteolytic fragments. J. Biol. Chem. 257:6446-6451. [PubMed] [Google Scholar]
- 497.Oka, A., N. Nomura, M. Morita, H. Sugisaki, K. Sugimoto, and M. Takanami. 1979. Nucleotide sequence of small ColE1 derivatives: structure of the regions essential for autonomous replication and colicin E1 immunity. Mol. Gen. Genet. 172:151-159. [DOI] [PubMed] [Google Scholar]
- 498.Osborne, M. J., A. L. Breeze, L.-Y. Lian, A. Reilly, R. James, C. Kleanthous, and G. R. Moore. 1996. Three dimensional solution structure and 13C NMR assignments of the colicin E9 immunity protein Im9. Biochemistry 35:9505-9512. [DOI] [PubMed] [Google Scholar]
- 499.Oudega, B., O. Mol, P. van Ulsen, F. Stegehuis, F. J. van der Wal, and J. Luirink. 1993. Escherichia coli SecB, SecA, and SecY proteins are required for expression and membrane insertion of the bacteriocin release protein, a small lipoprotein. J. Bacteriol. 175:1543-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Oudega, B., F. Stegehuis, G. J. van Tiel-Menkveld, and F. K. DeGraaf. 1982. Protein H encoded by plasmid CloDF13 is involved in excretion of cloacin DF13. J. Bacteriol. 150:1115-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Ozeki, H., B. A. D. Stocker, and H. de Margerie. 1959. Production of colicin by single bacteria. Nature 184:337-339. [DOI] [PubMed] [Google Scholar]
- 502.Parker, M. W., F. Pattus, A. D. Tucker, and D. Tsernoglou. 1989. Structure of the membrane pore-forming fragments of colicin A. Nature 337:93-96. [DOI] [PubMed] [Google Scholar]
- 503.Parker, M. W., J. P. Postma, F. Pattus, A. D. Tucker, and D. Tsernoglou. 1992. Refined structure of the pore-forming domain of colicin A at 2.4 A resolution. J. Mol. Biol. 224:639-657. [DOI] [PubMed] [Google Scholar]
- 504.Parret, A., and R. De Mot. 2000. Novel bacteriocins with predicted tRNase and pore-forming activities in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 35:472-473. [DOI] [PubMed] [Google Scholar]
- 505.Parret, A., and R. De Mot. 2002. Bacteria killing their own kind: novel bacteriocins of Pseudomonas and other gamma-proteobacteria. Trends Microbiol. 10:107-112. [DOI] [PubMed] [Google Scholar]
- 506.Parsons, L. M., F. Lin, and J. Orban. 2006. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry 45:2122-2128. [DOI] [PubMed] [Google Scholar]
- 507.Pawelek, P. D., N. Croteau, C. Ng-Thow-Hing, C. M. Khursigara, N. Moiseeva, M. Allaire, and J. W. Coulton. 2006. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312:1399-1402. [DOI] [PubMed] [Google Scholar]
- 508.Payne, M. A., J. D. Igo, Z. Cao, S. B. Foster, S. M. C. Newton, and P. E. Klebba. 1997. Biphasic binding kinetics between FepA and its ligands. J. Biol. Chem. 272:21950-21955. [DOI] [PubMed] [Google Scholar]
- 509.Peacock, R., A. M. Weljie, S. P. Howard, F. D. Price, and H. J. Vogel. 2005. The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J. Mol. Biol. 345:1185-1197. [DOI] [PubMed] [Google Scholar]
- 510.Peacock, R. S., V. V. Andrushchenko, A. R. Demcoe, M. Gehmlich, L. S. Lu, A. G. Herrero, and H. J. Vogel. 2006. Characterization of TonB interactions with the FepA cork domain and FecA N-terminal signaling domain. Biometals 19:127-142. [DOI] [PubMed] [Google Scholar]
- 511.Peeters, B. P., R. M. Peters, J. G. Schoenmakers, and R. N. Konings. 1985. Nucleotide sequence and genetic organization of the genome of the N-specific filamentous bacteriophage IKe. Comparison with the genome of the F-specific filamentous phages M13, fd and f1. J. Mol. Biol. 181:27-39. [DOI] [PubMed] [Google Scholar]
- 512.Penfold, C. N., C. Garinot-Schneider, A. M. Hemmings, G. M. Moore, C. Kleanthous, and R. James. 2000. A 76-residue polypeptide of colicin E9 confers receptor specificity and inhibits the growth of vitamin B12-dependent Escherichia coli 113/3 cells. Mol. Microbiol. 38:639-649. [DOI] [PubMed] [Google Scholar]
- 513.Penfold, C. N., B. Healy, N. G. Housden, R. Boetzel, M. Vankemmelbeke, G. R. Moore, C. Kleanthous, and R. James. 2004. Flexibility in the receptor-binding domain of the enzymatic colicin E9 is required for toxicity against Escherichia coli cells. J. Bacteriol. 186:4520-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513a.Phillips, S. K., and W. A. Cramer. 1973. Properties of the fluorescence probe response associated with the transmission mechanism of colicin E1. Biochemistry 12:1170-1176. [DOI] [PubMed] [Google Scholar]
- 514.Pilsl, H., and V. Braun. 1995. Evidence that the immunity protein inactivates colicin 5 immediately prior to the formation of the transmembrane channel. J. Bacteriol. 177:6966-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Pilsl, H., and V. Braun. 1995. Strong function-related homology between the pore-forming colicins K and 5. J. Bacteriol. 177:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Pilsl, H., and V. Braun. 1995. Novel colicin 10: assignment of four domains to TonB- and TolC-dependent uptake via the Tsx receptor and to pore formation. Mol. Microbiol. 16:57-67. [DOI] [PubMed] [Google Scholar]
- 517.Pilsl, H., C. Glaser, P. Gross, H. Killmann, T. Olschlager, and V. Braun. 1993. Domains of colicin M involved in uptake and activity. Mol. Gen. Genet. 240:103-112. [DOI] [PubMed] [Google Scholar]
- 518.Pilsl, H., H. Killmann, K. Hantke, and V. Braun. 1996. Periplasmic location of the pesticin immunity protein suggests inactivation of pesticin in the periplasm. J. Bacteriol. 178:2431-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Pilsl, H., D. Smajs, and V. Braun. 1998. The tip of the hydrophobic hairpin of colicin U is dispensable for colicin U activity but is important for interaction with the immunity protein. J. Bacteriol. 180:4111-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Pilsl, H., D. Smajs, and V. Braun. 1999. Characterization of colicin S4 and its receptor OmpW, a minor protein of the Escherichia coli outer membrane. J. Bacteriol. 181:3578-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Pinou, T., and M. Riley. 2001. Nucleotide polymorphism in microcin V plasmids. Plasmid 46:1-9. [DOI] [PubMed] [Google Scholar]
- 522.Plastow, G. S., and I. B. Holland. 1979. Identification of an Escherichia coli inner membrane polypeptide specified by a lambda-tonB transducing phage. Biochem. Biophys. Res. Commun. 90:1007-1014. [DOI] [PubMed] [Google Scholar]
- 523.Plate, C. A., and S. E. Luria. 1972. Stages in colicin K action, as revealed by the action of trypsin. Proc. Natl. Acad. Sci. USA 69:2030-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Pommer, A. J., S. Cal, A. H. Keeble, D. Walker, S. J. Evans, U. C. Kühlmann, A. Cooper, B. A. Connolly, A. M. Hemmings, G. R. Moore, R. James, and C. Kleanthous. 2001. Mechanism and cleavage specificity of the H-N-H endonuclease colicin E9. J. Mol. Biol. 314:735-749. [DOI] [PubMed] [Google Scholar]
- 525.Pommer, A. J., U. C. Kühlmann, A. Cooper, R. James, G. R. Moore, A. M. Hemmings, and C. Kleanthous. 1999. Homing-in on the role of transition metals in the HNH motif of colicin endonucleases. J. Biol. Chem. 274:27153-27160. [DOI] [PubMed] [Google Scholar]
- 526.Pommer, A. J., R. Wallis, R. Moore, R. James, and C. Kleanthous. 1998. Enzymological characterisation of the nuclease domain from the bacterial toxin colicin E9. Biochem. J. 334:387-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Pommier, S., M. Gavioli, E. Cascales, and R. Lloubès. 2005. Tol-dependent macromolecule import through the Escherichia coli cell envelope requires the presence of an exposed TolA binding motif. J. Bacteriol. 187:7526-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Ponting, C. P., and M. J. Pallen. 1999. A beta-propeller domain within TolB. Mol. Microbiol. 31:739-740. [DOI] [PubMed] [Google Scholar]
- 529.Postle, K., and R. F. Good. 1983. DNA sequence of the Escherichia coli tonB gene. Proc. Natl. Acad. Sci. USA 80:5235-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 531.Postle, K., and R. Larsen. 2004. The TonB, ExbB, and ExbD proteins, p. 96-112. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 532.Postle, K., and J. T. Skare. 1988. Escherichia coli TonB protein is exported from the cytoplasm without proteolytic cleavage of its amino terminus. J. Biol. Chem. 263:11000-11007. [PubMed] [Google Scholar]
- 533.Prouty, A. M., J. C. Van Velkinburgh, and J. S. Gunn. 2002. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J. Bacteriol. 184:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Pshennikova, E. S., M. N. Kolot, and I. A. Khmel. 1988. Structural and functional organization of the colicin operon of the ColD-CA23 plasmid. Mol. Biol. 22:1273-1278. [PubMed] [Google Scholar]
- 535.Pugsley, A. P. 1984. Genetic analysis of ColN plasmid determinants for colicin production, release, and immunity. J. Bacteriol. 158:523-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Pugsley, A. P. 1985. Escherichia coli K12 strains for use in the identification and characterization of colicins. J. Gen. Microbiol. 131:369-376. [DOI] [PubMed] [Google Scholar]
- 537.Pugsley, A. P. 1987. Nucleotide sequencing of the structural gene for colicin N reveals homology between the catalytic, C-terminal domains of colicins A and N. Mol. Microbiol. 1:317-325. [DOI] [PubMed] [Google Scholar]
- 538.Pugsley, A. P. 1988. The immunity and lysis genes of ColN plasmid pCHAP4. Mol. Gen. Genet. 211:335-341. [DOI] [PubMed] [Google Scholar]
- 539.Pugsley, A. P., and S. T. Cole. 1987. An unmodified form of the ColE2 lysis protein, an envelope lipoprotein, retains reduced ability to promote colicin E2 release and lysis of producing cells. J. Gen. Microbiol. 133:2411-2420. [DOI] [PubMed] [Google Scholar]
- 540.Pugsley, A. P., and J. P. Rosenbusch. 1981. Release of colicin E2 from Escherichia coli. J. Bacteriol. 147:186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Pugsley, A. P., and M. Schwartz. 1983. Expression of a gene in a 400-base-pair fragment of colicin plasmid ColE2-P9 is sufficient to cause host cell lysis. J. Bacteriol. 156:109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Pugsley, A. P., and M. Schwartz. 1984. Colicin E2 release: lysis, leakage or secretion? Possible role of a phospholipase. EMBO J. 3:2393-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Pugsley, A. P., M. Schwartz, M. Lavina, and F. Moreno. 1983. On the effect of ompR mutation on colicin E2 production. FEMS Microbiol. Lett. 19:87-92. [Google Scholar]
- 544.Qiu, X.-Q., K. S. Jakes, P. K. Kienker, A. Finkelstein, and S. L. Slatin. 1996. Major transmembrane movement associated with colicin Ia channel gating. J. Gen. Physiol. 107:313-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Quillardet, P., O. Huisman, R. d'Ari, and M. Hofnung. 1982. SOS chromotest, a direct assay of induction of an SOS function in Escherichia coli K-12 to measure genotoxicity. Proc. Natl. Acad. Sci. USA 79:5971-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Raggett, E. M., G. Bainbridge, L. J. Evans, A. Cooper, and J. H. Lakey. 1998. Discovery of critical TolA-binding residues in the bactericidal toxin colicin N: a biophysical approach. Mol. Microbiol. 28:1335-1343. [DOI] [PubMed] [Google Scholar]
- 547.Rakin, A., E. Boolgakowa, and J. Heesemann. 1996. Structural and functional organization of the Yersinia pestis bacteriocin pesticin gene cluster. Microbiology 142:3415-3424. [DOI] [PubMed] [Google Scholar]
- 548.Rakin, A., E. Saken, D. Harmsen, and J. Heesemann. 1994. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol. Microbiol. 13:253-263. [DOI] [PubMed] [Google Scholar]
- 549.Rampf, B., P. Bross, T. Vocke, and I. Rasched. 1991. Release of periplasmic proteins induced in E. coli by expression of an N-terminal proximal segment of the phage fd gene 3 protein. FEBS Lett. 280:27-31. [DOI] [PubMed] [Google Scholar]
- 550.Ray, M. C., P. Germon, A. Vianney, R. Portalier, and J. C. Lazzaroni. 2000. Identification by genetic suppression of Escherichia coli TolB residues important for TolB-Pal interaction. J. Bacteriol. 182:821-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Raymond, L., S. L. Slatin, and A. Finkelstein. 1985. Channels formed by colicin E1 in planar lipid bilayers are large and exhibit pH-dependent ion selectivity. J. Membr. Biol. 84:173-181. [DOI] [PubMed] [Google Scholar]
- 552.Reeves, P. 1968. Mode of action of colicins of types E1, E2, E3, and K. J. Bacteriol. 96:1700-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552a.Reeves, P. R. 1972. The bacteriocins. Springer-Verlag, New York, NY.
- 553.Riechmann, L., and P. Holliger. 1997. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell 90:351-360. [DOI] [PubMed] [Google Scholar]
- 554.Rigal, A., E. Bouveret, R. Lloubès, C. Lazdunski, and H. Bénédetti. 1997. The TolB protein interacts with the porins of Escherichia coli. J. Bacteriol. 179:7274-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Riley, M. 1993. Molecular mechanisms of colicin evolution. Mol. Biol. Evol. 10:1380-1395. [DOI] [PubMed] [Google Scholar]
- 556.Riley, M. A. 1993. Positive selection for colicin diversity in bacteria. Mol. Biol. Evol. 10:1048-1059. [DOI] [PubMed] [Google Scholar]
- 557.Riley, M. A. 1998. Molecular mechanisms of bacteriocin evolution. Annu. Rev. Genet. 32:255-278. [DOI] [PubMed] [Google Scholar]
- 558.Riley, M. A., L. Cadavid, M. S. Collett, M. N. Neely, M. D. Adams, C. M. Phillips, J. V. Neel, and D. Friedman. 2000. The newly characterized colicin Y provides evidence of positive selection in pore-former colicin diversification. Microbiol. 146:1671-1677. [DOI] [PubMed] [Google Scholar]
- 559.Riley, M. A., and D. M. Gordon. 1992. A survey of col plasmids in natural isolates of Escherichia coli and an investigation into the stability of col-plasmid lineages. J. Gen. Microbiol. 138:1345-1352. [DOI] [PubMed] [Google Scholar]
- 560.Riley, M. A., T. Pinou, J. E. Wertz, Y. Tan, and C. M. Valletta. 2001. Molecular characterization of the klebicin B plasmid of Klebsiella pneumoniae. Plasmid 45:209-221. [DOI] [PubMed] [Google Scholar]
- 561.Riley, M. A., Y. Tan, and J. Wang. 1994. Nucleotide polymorphism in colicin E1 and Ia plasmids from natural isolates of Escherichia coli. Proc. Natl. Acad. Sci. USA 91:11276-11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Roberts, M. D., N. L. Martin, and A. M. Kropinski. 2004. The genome and proteome of coliphage T1. Virology 318:245-266. [DOI] [PubMed] [Google Scholar]
- 563.Roof, S. K., J. D. Allard, K. P. Bertrand, and K. Postle. 1991. Analysis of Escherichia coli TonB membrane topology by use of PhoA fusions. J. Bacteriol. 173:5554-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Roos, U., R. E. Harkness, and V. Braun. 1989. Assembly of colicin genes from a few DNA fragments. Nucleotide sequence of colicin D. Mol. Microbiol. 3:891-902. [DOI] [PubMed] [Google Scholar]
- 565.Russel, M., H. Whirlow, T. P. Sun, and R. E. Webster. 1988. Low-frequency infection of F− bacteria by transducing particles of filamentous bacteriophages. J. Bacteriol. 170:5312-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Sabet, S. F., and C. A. Schnaitman. 1971. Localization and solubilization of colicin receptors. J. Bacteriol. 108:422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Sabet, S. F., and C. A. Schnaitman. 1973. Purification and properties of the colicin E3 receptor of Escherichia coli. J. Biol. Chem. 248:1797-1806. [PubMed] [Google Scholar]
- 568.Sabik, J. F., J. L. Suit, and S. E. Luria. 1983. cea-kil operon of the ColE1 plasmid. J. Bacteriol. 153:1479-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Salles, B., J. M. Weisemann, and G. M. Weinstock. 1987. Temporal control of colicin E1 induction. J. Bacteriol. 169:5028-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Sano, Y., and M. Kageyama. 1981. Purification and properties of an S-type pyocin, pyocin AP41. J. Bacteriol. 146:733-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Sano, Y., H. Matsui, M. Kobayashi, and M. Kageyama. 1993. Molecular structures and functions of pyocins S1 and S2 in Pseudomonas aeruginosa. J. Bacteriol. 175:2907-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Sano, Y., M. Kobayashi, and M. Kageyama. 1993. Functional domains of S-type pyocins deduced from chimeric molecules. J. Bacteriol. 175:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Sauter, A., S. P. Howard, and V. Braun. 2003. In vivo evidence for TonB dimerization. J. Bacteriol. 185:5747-5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Schaller, K., and M. Nomura. 1976. Colicin E2 is an endonuclease. Proc. Natl. Acad. Sci. USA 73:3989-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Schein, S. J., B. L. Kagan, and A. Finkelstein. 1978. Colicin K acts by forming voltage-dependent channels in phospholipid bilayer membranes. Nature 276:159-163. [DOI] [PubMed] [Google Scholar]
- 576.Schendel, S. L., E. M. Click, R. E. Webster, and W. A. Cramer. 1997. The TolA protein interacts with colicin E1 differently than with other group A colicins. J. Bacteriol. 179:3683-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Schendel, S. L., and W. A. Cramer. 1994. On the nature of the unfolded intermediate in the in vitro transition of the colicin E1 channel domain from the aqueous to the membrane phase. Protein Sci. 3:2272-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Schramm, E., J. Mende, V. Braun, and R. M. Kamp. 1987. Nucleotide sequence of the colicin B activity gene cba: consensus pentapeptide among TonB-dependent colicins and receptors. J. Bacteriol. 169:3350-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Schwartz, H. R., and D. R. Helinski. 1971. Purification and characterization of colicin E1. J. Biol. Chem. 246:6318-6327. [PubMed] [Google Scholar]
- 580.Seliger, S. S., A. R. Mey, A. M. Valle, and S. M. Payne. 2001. The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol. Microbiol. 39:801-812. [DOI] [PubMed] [Google Scholar]
- 581.Senior, B. W., and I. B. Holland. 1971. Effect of colicin E3 upon the 30S ribosomal subunit of Escherichia coli. Proc. Natl. Acad. Sci. USA 68:959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Seo, Y., and D. R. Galloway. 1990. Purification of the pyocin S2 complex from Pseudomonas aeruginosa PAO1: analysis of DNase activity. Biochem. Biophys. Res. Commun. 172:455-461. [DOI] [PubMed] [Google Scholar]
- 583.Shannon, R., and A. J. Hedges. 1973. Reversibility of the specific adsorption of colicin E2-P9 to cells of colicin-sensitive strains of Escherichia coli. J. Bacteriol. 116:1136-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583a.Sharma, O., and W. A. Cramer. 2007. Minimum length requirement of the flexible N-terminal translocation subdomain of colicin E3. J. Bacteriol. 189:363-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Sharma, S., N. Waterfield, D. Bowen, T. Rocheleau, L. Holland, R. James, and R. Ffrench-Constant. 2002. The lumicins: novel bacteriocins from Photorhabdus luminescens with similarity to the uropathogenic-specific protein (USP) from uropathogenic Escherichia coli. FEMS Microbiol. Lett. 214:241-249. [DOI] [PubMed] [Google Scholar]
- 585.Shi, Z., K. F. Chak, and H. S. Yuan. 2005. Identification of an essential cleavage site in ColE7 required for import and killing of cells. J. Biol. Chem. 280:24663-24668. [DOI] [PubMed] [Google Scholar]
- 586.Shirasu, K., P. Morel, and C. I. Kado. 1990. Characterization of the virB operon of an Agrobacterium tumefaciens Ti plasmid: nucleotide sequence and protein analysis. Mol. Microbiol. 4:1153-1163. [DOI] [PubMed] [Google Scholar]
- 587.Shub, D. A., H. Goodrich-Blair, and S. R. Eddy. 1994. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci. 19:402-404. [DOI] [PubMed] [Google Scholar]
- 588.Shultis, D. D., M. D. Purdy, C. N. Banachs, and M. Wiener. 2006. Outer membrane active transport: structure of the BtuB:TonB complex. Science 312:1396-1399. [DOI] [PubMed] [Google Scholar]
- 589.Sicard, N., and R. Devoret. 1962. Effets de la carence en thymine sur des souches d'Escherichia coli lysogènes K12 T− et colicinogènes 15T−. C. R. Acad. Sci. Paris 255:1417-1419. [PubMed] [Google Scholar]
- 590.Sidikaro, J., and M. Nomura. 1974. E3 immunity substance. J. Biol. Chem. 249:445-453. [PubMed] [Google Scholar]
- 591.Simpson, J. C., L. M. Roberts, K. Römisch, J. Davey, D. H. Wolf, and J. M. Lord. 1999. Ricin A chain utilises the endoplasmic reticulum-associated protein degradation pathway to enter the cytosol of yeast. FEBS Lett. 459:80-84. [DOI] [PubMed] [Google Scholar]
- 592.Skare, J. T., B. M. M. Ahmer, C. L. Seachord, R. P. Darveau, and K. Postle. 1993. Energy transduction between membranes—TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 268:16302-16308. [PubMed] [Google Scholar]
- 593.Skare, J. T., S. K. Roof, and K. Postle. 1989. A mutation in the amino terminus of a hybrid TrpC-TonB protein relieves overproduction lethality and results in cytoplasmic accumulation. J. Bacteriol. 171:4442-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Slatin, S. L. 1988. Colicin E1 in planar lipid bilayers. Int. J. Biochem. 20:737-744. [DOI] [PubMed] [Google Scholar]
- 595.Slatin, S. L., D. Duché, P. K. Kienker, and D. Baty. 2004. Gating movements of colicin A and colicin Ia are different. J. Membr. Biol. 202:73-83. [DOI] [PubMed] [Google Scholar]
- 596.Slatin, S. L., and P. Kienker. 2003. Colicin channels and protein translocation: parallels with diphtheria toxin, p. 102-131. In G. Menestrina (ed.), Pore forming peptides and protein toxins. Taylor and Francis, London, United Kingdom.
- 597.Slatin, S. L., A. Nardi, K. S. Jakes, D. Baty, and D. Duché. 2002. Translocation of a functional protein by a voltage-dependent ion channel. Proc. Natl. Acad. Sci. USA 99:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Slatin, S. L., X.-Q. Qiu, K. S. Jakes, and A. Finkelstein. 1994. Identification of a translocated protein segment in a voltage-dependent channel. Nature 371:158-161. [DOI] [PubMed] [Google Scholar]
- 599.Smajs, D., H. Pilsl, and V. Braun. 1997. Colicin U, a novel colicin produced by Shigella boydii. J. Bacteriol. 179:4919-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Smajs, D., and G. M. Weinstock. 2001. Genetic organization of plasmid ColJs, encoding colicin Js activity, immunity, and release genes. J. Bacteriol. 183:3949-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Smarda, J., J. Petrzelova, and B. Vyskot. 1987. Colicin Js of Shigella sonnei: classification of type colicin 7. Zentrabl. Bakteriol. Hyg. A 263:530-540. [DOI] [PubMed] [Google Scholar]
- 602.Smith, A. W., P. H. Hirst, K. Hughes, K. Gensberg, and J. R. Govan. 1992. The pyocin Sa receptor of Pseudomonas aeruginosa is associated with ferripyoverdin uptake. J. Bacteriol. 174:4847-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Snijder, H. J., I. Ubarretxena-Belandia, M. Blaauw, K. H. Kalk, H. M. Verheij, M. R. Egmond, N. Dekker, and B. W. Dijkstra. 1999. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 401:717-721. [DOI] [PubMed] [Google Scholar]
- 604.Snijder, W. B., L. J. B. Davis, P. N. Danese, C. L. Cosma, and T. J. Silhavy. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Sobko, A. A., E. A. Kotova, Y. N. Antonenko, S. D. Zakharov, and W. A. Cramer. 2004. Effect of lipids with different spontaneous curvature on the channel activity of colicin E1: evidence in favor of a toroidal pore. FEBS Lett. 576:205-210. [DOI] [PubMed] [Google Scholar]
- 606.Sobko, A. A., E. A. Kotova, Y. N. Antonenko, S. D. Zakharov, and W. A. Cramer. 2006. Lipid dependence of the channel properties of a colicin E1-lipid toroidal pore. J. Biol. Chem. 281:14408-14416. [DOI] [PubMed] [Google Scholar]
- 607.Soelaiman, S., K. Jakes, N. Wu, C. Li, and M. Shoham. 2001. Crystal structure of colicin E3: implications for cell entry and ribosome inactivation. Mol. Cell 8:1053-1062. [DOI] [PubMed] [Google Scholar]
- 608.Song, H. Y., F. S. Cohen, and W. A. Cramer. 1991. Membrane topography of ColE1 gene products: the hydrophobic anchor of the colicin E1 channel is a helical hairpin. J. Bacteriol. 173:2927-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Song, H. Y., and W. A. Cramer. 1991. Membrane topography of ColE1 gene products: the immunity protein. J. Bacteriol. 173:2935-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609a.Soong, B. W., S. Y. Hsieh, and K. F. Chack. 1994. Mapping of transcriptional start sites of the cea and cei genes. Mol. Gen. Genet. 243:477-481. [DOI] [PubMed] [Google Scholar]
- 610.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 611.Spinola, S. M., T. J. Hiltke, K. Fortney, and K. L. Shanks. 1996. The conserved 18,000-molecular-weight outer membrane protein of Haemophilus ducreyi has homology to PAL. Infect. Immun. 64:1950-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Stengele, I., P. Bross, X. Garces, J. Giray, and I. Rasched. 1990. Dissection of functional domains in phage fd adsorption protein. Discrimination between attachment and penetration sites. J. Mol. Biol. 212:143-149. [DOI] [PubMed] [Google Scholar]
- 613.Suit, J. L., M. L. J. Fan, J. F. Sabik, R. Labarre, and S. E. Luria. 1983. Alternative forms of lethality in mitomycin-induced bacteria carrying ColE1 plasmids. Proc. Natl. Acad. Sci. USA 80:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Suit, J. L., and S. E. Luria. 1988. Expression of the kil gene of the ColE1 plasmid in Escherichia coli Kilr mutants causes release of periplasmic enzymes and of colicin without cell death. J. Bacteriol. 170:4963-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Sun, T. P., and R. E. Webster. 1986. fii, a bacterial locus required for filamentous phage infection and its relation to colicin-tolerant tolA and tolB. J. Bacteriol. 165:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Sun, T. P., and R. E. Webster. 1987. Nucleotide sequence of a gene cluster involved in entry of E colicins and single-stranded DNA of infecting filamentous bacteriophages into Escherichia coli. J. Bacteriol. 169:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Suzuki, K., and K. Imahori. 1978. Preparation and characterization of an active fragment of colicin E3. J. Biochem. (Tokyo) 84:1021-1029. [DOI] [PubMed] [Google Scholar]
- 618.Tai, P. C., and B. D. Davis. 1974. Activity of colicin E3 treated ribosomes in initiation and in chain elongation. Proc. Natl. Acad. Sci. USA 71:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Takeya, K., Y. Minamishima, K. Amako, and Y. Ohnishi. 1967. A small rod-shaped pyocin. Virology 31:166-168. [DOI] [PubMed] [Google Scholar]
- 620.Tan, Y., and M. A. Riley. 1997. Nucleotide polymorphism in colicin E2 gene clusters: evidence for nonneutral evolution. Mol. Biol. Evol. 14:666-673. [DOI] [PubMed] [Google Scholar]
- 621.Tan, Y., and M. A. Riley. 1997. Positive selection and recombination: major molecular mechanisms in colicin diversification. Trends Ecol. Evol. 12:348-351. [DOI] [PubMed] [Google Scholar]
- 622.Teruda, M., T. Kuroda, S. Matsuyama, and H. Tokuda. 2001. Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 276:47690-47694. [DOI] [PubMed] [Google Scholar]
- 623.Thanassi, D. G., and S. J. Hultgren. 2000. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 12:420-430. [DOI] [PubMed] [Google Scholar]
- 624.Thomas, J. A., and M. A. Valvano. 1993. Role of tol genes in cloacin DF13 susceptibility of Escherichia coli K-12 strains expressing the cloacin DF13-aerobactin receptor IutA. J. Bacteriol. 175:548-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Tilby, M., I. Hindennach, and U. Henning. 1978. Bypass of receptor-mediated resistance to colicin E3 in Escherichia coli K-12. J. Bacteriol. 136:1189-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Timmis, K., F. Cabello, and S. N. Cohen. 1974. Utilization of two distinct modes of replication by a hybrid plasmid constructed in vitro from separate replicons. Proc. Natl. Acad. Sci. USA 71:4556-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Toba, M., H. Masaki, and T. Ohta. 1986. Primary structures of the ColE2-P9 and ColE3-CA-CA38 lysis genes. J. Biochem. 99:591-596. [DOI] [PubMed] [Google Scholar]
- 628.Toba, M., H. Masaki, and T. Ohta. 1988. Colicin E8, a DNase which indicates an evolutionary relationship between colicins E2 and E3. J. Bacteriol. 170:3237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 629.Tomita, K., T. Ogawa, T. Uozumi, K. Watanabe, and H. Masaki. 2000. A cytosolic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc. Natl. Acad. Sci. USA 97:8278-8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Tommassen, J., A. P. Pugsley, J. Korteland, J. Verbakel, and B. Lugtenberg. 1984. Gene encoding a hybrid OmpF-PhoE pore protein in the outer membrane of Escherichia coli K12. Mol. Gen. Genet. 197:503-508. [DOI] [PubMed] [Google Scholar]
- 631.Torres, B., S. Jaenecke, K. N. Timmis, J. L. Garcia, and E. Diaz. 2003. A dual lethal system to enhance containment of recombinant micro-organisms. Microbiology 149:3595-3601. [DOI] [PubMed] [Google Scholar]
- 632.Tozawa, K., C. J. Macdonald, C. N. Penfold, R. James, C. Kleanthous, N. J. Clayden, and G. R. Moore. 2005. Clusters in an intrinsically disordered protein create a protein-binding site: the TolB-binding region of colicin E9. Biochemistry 44:11496-11507. [DOI] [PubMed] [Google Scholar]
- 633.Tuckman, M., and M. S. Osburne. 1992. In vivo inhibition of TonB-dependent processes by a TonB box consensus pentapeptide. J. Bacteriol. 174:320-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 634.Uchimura, T., and P. C. K. Lau. 1987. Nucleotide sequences from the colicin E8 operon: homology with plasmid ColE2-P9. Mol. Gen. Genet. 209:489-493. [DOI] [PubMed] [Google Scholar]
- 635.Uratani, Y., and T. Hoshino. 1984. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J. Bacteriol. 157:632-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.van den Bremer, E. T. J., A. H. Keeble, W. Jiskoot, R. E. J. Spelbrink, C. S. Maier, A. van Hoek, A. J. W. G. Visser, R. James, G. R. Moore, C. Kleanthous, and A. J. R. Heck. 2004. Distinct conformational stability and functional activity of four highly homologous endonuclease colicins. Protein Sci. 13:1391-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.van den Elzen, P. J. M., R. N. H. Konings, E. Veltkamp, and H. J. J. Nijkamp. 1980. Transcription of bacteriocinogenic plasmid CloDF13 in vivo and in vitro: structure of the cloacin immunity operon. J. Bacteriol. 144:579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.van den Elzen, P. J. M., J. Maat, H. H. B. Walters, E. Veltkamp, and H. J. J. Nijkamp. 1982. The nucleotide sequence of the bacteriocin promoters of plasmids CloDF13 and ColE1: role of LexA repressor and cAMP in the regulation of promoter activity. Nucleic Acids Res. 10:1913-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.van den Heuvel, R. H. H., S. Gato, C. Versluis, C. Kleanthous, and A. J. R. Heck. 2005. Real-time monitoring of enzymatic DNA hydrolysis by electrospray ionisation mass spectrometry. Nucleic Acids Res. 33:U65-U71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.van der Goot, F. G., J. M. Gonzalez-Manas, J. M. Lakey, and F. Pattus. 1991. A molten-globule membrane insertion intermediate of the pore-forming domain of colicin A. Nature 354:408-410. [DOI] [PubMed] [Google Scholar]
- 641.van der Wal, F., J. Luirink, and B. Oudega. 1995. Bacteriocin release proteins: mode of action, structure and biotechnological application. FEMS Microbiol. Rev. 17:381-399. [DOI] [PubMed] [Google Scholar]
- 642.van der Wal, F., B. Oudega, M. M. Kater, C. M. ten Hagen-Jongman, F. K. de Graaf, and J. Luirink. 1992. The stable BRP signal peptide causes lethality but is unable to provoke the translocation of cloacin DF13 across the cytoplasmic membrane of Escherichia coli. Mol. Microbiol. 6:2309-2318. [DOI] [PubMed] [Google Scholar]
- 643.van der Wal, F., C. M. ten Hagen, B. Oudega, and J. Luirink. 1995. The stable bacteriocin release protein signal peptide, expressed as a separate entity, functions in the release of cloacin DF13. FEMS Microbiol. Lett. 131:173-177. [DOI] [PubMed] [Google Scholar]
- 644.van der Wal, F. J., C. M. ten Hagen-Jongman, B. Oudega, and J. Luirink. 1995. Optimization of bacteriocin-release-protein-induced protein release by Escherichia coli: extracellular production of the periplasmic molecular chaperone FaeE. Appl. Microbiol. Biotechnol. 44:459-465. [DOI] [PubMed] [Google Scholar]
- 645.Vankemmelbeke, M., B. Healy, G. R. Moore, C. Kleanthous, C. N. Penfold, and R. James. 2005. Rapid detection of colicin E9-induced DNA damage using Escherichia coli cells carrying SOS promoter-lux fusions. J. Bacteriol. 187:4900-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 646.van Putten, A. J., F. Stegehuis, P. M. P. van Bergen en Menengouwen, F. K. DeGraaf, and B. Oudega. 1988. Alterations in the carboxy-terminal half of cloacin destabilize the protein and prevent its export by Escherichia coli. Mol. Microbiol. 2:553-562. [DOI] [PubMed] [Google Scholar]
- 647.Varenne, S., D. Cavard, and C. Lazdunski. 1981. Biosynthesis and export of colicin A in Citrobacter freundii CA31. Eur. J. Biochem. 218:615-620. [DOI] [PubMed] [Google Scholar]
- 648.Varenne, S., M. Knibiehler, D. Cavard, J. Morlon, and C. Lazdunski. 1982. Variable rate of polypeptide chain elongation for colicins A, E2 and E3. J. Mol. Biol. 159:57-70. [DOI] [PubMed] [Google Scholar]
- 649.Varley, J. M., and G. J. Boulnois. 1984. Analysis of a cloned colicin Ib gene: complete nucleotide sequence and implications for regulation of expression. Nucleic Acids Res. 12:6727-6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Vetter, I. R., M. W. Parker, A. D. Tucker, J. H. Lakey, F. Pattus, and D. Tsernoglou. 1998. Crystal structure of a colicin N fragment suggests a model for toxicity. Structure 6:863-874. [DOI] [PubMed] [Google Scholar]
- 651.Vianney, A., T. M. Lewin, W. F. Beyer, Jr., J. C. Lazzaroni, R. Portalier, and R. E. Webster. 1994. Membrane topology and mutational analysis of the TolQ protein of Escherichia coli required for the uptake of macromolecules and cell envelope integrity. J. Bacteriol. 176:822-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652.Vines, E. D., C. L. Marolda, A. Balachandran, and M. A. Valvano. 2005. Defective O-antigen polymerization in tolA and pal mutants of Escherichia coli in response to extracytoplasmic stress. J. Bacteriol. 187:3359-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653.Vollmer, W., H. Pilsl, K. Hantke, J. V. Holtje, and V. Braun. 1997. Pesticin displays muramidase activity. J. Bacteriol. 179:1580-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Von Tersch, M. A., and B. C. Carlton. 1983. Megacinogenic plasmids of Bacillus megaterium. J. Bacteriol. 155:872-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Von Tersch, M. A., and B. C. Carlton. 1984. Molecular cloning of structural and immunity genes for megacins A-216 and A-19213 in Bacillus megaterium. J. Bacteriol. 160:854-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Voulhoux, R., A. Lazdunski, and A. Filloux. 2001. Colicin A hybrids: a genetic tool for selection of type II secretion-proficient Pseudomonas strains. EMBO Rep. 2:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Wai, S. N., B. Lindmark, T. Söderblom, A. Takade, M. Westermark, J. Oscarsson, J. Jass, A. Richter-Dahlfors, Y. Mizunoe, and B. E. Uhlin. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25-35. [DOI] [PubMed] [Google Scholar]
- 658.Walburger, A., C. Lazdunski, and Y. Corda. 2002. The Tol/Pal system function requires an interaction between the C-terminal domain of TolA and the N-terminal domain of TolB. Mol. Microbiol. 44:695-708. [DOI] [PubMed] [Google Scholar]
- 659.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 660.Waleh, N. S., and P. H. Johnson. 1985. Structural and functional organization of the colicin E1 operon. Proc. Natl. Acad. Sci. USA 82:8389-8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Walker, D., L. Lancaster, R. James, and C. Kleanthous. 2004. Identification of the catalytic motif of the microbial ribosome inactivating cytotoxin colicin E3. Protein Sci. 13:1603-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 662.Walker, D., G. R. Moore, R. James, and C. Kleanthous. 2003. Thermodynamic consequences of bipartite immunity protein binding to the ribonuclease colicin E3. Biochemistry 42:4161-4171. [DOI] [PubMed] [Google Scholar]
- 663.Walker, D., M. Rolfe, A. Thompson, G. R. Moore, R. James, J. C. Hinton, and C. Kleanthous. 2004. Transcriptional profiling of colicin-induced cell death of Escherichia coli MG1655 identifies potential mechanisms by which bacteriocins promote bacterial diversity. J. Bacteriol. 186:866-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Walker, D. C., T. Georgiou, A. J. Pommer, A. M. Hemmings, G. R. Moore, C. Kleanthous, and R. James. 2002. Mutagenic scan of the H-N-H motif of colicin E9: implications for the mechanistic enzymology of colicins, homing enzymes and apoptotic endonucleases. Nucleic Acids Res. 30:3225-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Wallis, R., K.-Y. Leung, M. J. Osborne, R. James, G. R. Moore, and C. Kleanthous. 1998. Specificity in protein-protein recognition: conserved Im9 residues are the major determinants of stability in the colicin E9 DNase-Im9 complex. Biochemistry 37:476-485. [DOI] [PubMed] [Google Scholar]
- 666.Wallis, R., K.-Y. Leung, A. Pommer, H. Videler, G. R. Moore, R. James, and C. Kleanthous. 1995. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 2. Cognate and noncognate interactions that span the mM-fM affinity range. Biochemistry 34:13751-13759. [DOI] [PubMed] [Google Scholar]
- 667.Wallis, R., G. R. Moore, R. James, and C. Kleanthous. 1995. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 1. Diffusion controlled association and femtomolar binding for the cognate complex. Biochemistry 34:13743-13750. [DOI] [PubMed] [Google Scholar]
- 668.Wallis, R., A. Reilly, K. Barnes, C. Abell, D. G. Campbell, G. R. Moore, R. James, and C. Kleanthous. 1994. Tandem overexpression and characterisation of the nuclease domain of colicin E9 and its cognate inhibitor protein Im9. Eur. J. Biochem. 220:447-454. [DOI] [PubMed] [Google Scholar]
- 669.Wallis, R., A. Reilly, A. J. Rowe, G. R. Moore, R. James, and C. Kleanthous. 1992. In vivo and in vitro characterization of overproduced colicin E9 immunity protein. Eur. J. Biochem. 207:687-695. [DOI] [PubMed] [Google Scholar]
- 670.Wan, E. W., and F. Baneyx. 1998. TolAIII co-overexpression facilitates the recovery of periplasmic recombinant proteins into the growth medium of Escherichia coli. Protein Expr. Purif. 14:13-22. [DOI] [PubMed] [Google Scholar]
- 671.Watson, R. J., T. Vernet, and L. P. Visentin. 1985. Relationships of the Col plasmids E2, E3, E4, E5, E6 and E7: restriction mapping and colicin genes fusions. Plasmid 13:205-210. [DOI] [PubMed] [Google Scholar]
- 672.Wayne, R., K. Frick, and J. B. Neilands. 1976. Siderophore protection against colicins M, B, V, and Ia in Escherichia coli. J. Bacteriol. 126:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 673.Weaver, C., A. H. Redborg, and J. Konisky. 1981. Plasmid-determined immunity of Escherichia coli K-12 to colicin Ia is mediated by a plasmid-encoded membrane protein. J. Bacteriol. 148:817-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674.Webster, R. E. 1991. The tol gene products and the import of macromolecules into Escherichia coli. Mol. Microbiol. 5:1005-1011. [DOI] [PubMed] [Google Scholar]
- 675.Wiener, M., D. Freymann, P. Ghosh, and R. M. Stroud. 1997. Crystal structure of colicin Ia. Nature 385:461-464. [DOI] [PubMed] [Google Scholar]
- 676.Wiener, M. C. 2005. TonB-dependent outer membrane transport: going for Baroque? Curr. Opin. Struct. Biol. 15:394-400. [DOI] [PubMed] [Google Scholar]
- 677.Witty, M., C. Sanz, A. Shah, J. G. Grossmann, K. Mizuguchi, R. N. Perham, and B. Luisi. 2002. Structure of the periplasmic domain of Pseudomonas aeruginosa TolA: evidence for an evolutionary relationship with the TonB transporter protein. EMBO J. 21:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 677a.Wooldridge, K. G., and P. H. Williams. 1991. Sensitivity of Escherichia coli to cloacin DF13 involves the major outer membrane protein OmpF. J. Bacteriol. 173:2420-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678.Wormald, M. R., A. R. Merrill, W. A. Cramer, and R. J. Williams. 1990. Solution NMR studies of colicin E1 C-terminal thermolytic peptide. Structural comparison with colicin A and the effects of pH changes. Eur. J. Biochem. 191:155-161. [DOI] [PubMed] [Google Scholar]
- 679.Wu, H. C. 1996. Biosynthesis of lipoproteins, p. 1005-1014. In F. C. Neidhardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol 1. ASM Press, Washington, DC. [Google Scholar]
- 680.Wu, S., K. Dornbusch, G. Kronvall, and M. Norgren. 1999. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type β-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC β-lactamase. Antimicrob. Agents Chemother. 43:1350-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681.Yamada, M., Y. Ebina, T. Miyata, T. Nagazawa, and A. Nakazawa. 1982. Nucleotide sequence of the structural gene for colicin E1 and predicted structure of the protein. Proc. Natl. Acad. Sci. USA 79:2827-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Yamada, M., and A. Nakazawa. 1984. Factors necessary for the export process of colicin E1 across the cytoplasmic membrane of Escherichia coli. Eur. J. Biochem. 140:249-255. [DOI] [PubMed] [Google Scholar]
- 683.Yamaguchi, K., F. Yu, and M. Inouye. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53:423-432. [DOI] [PubMed] [Google Scholar]
- 684.Yamamoto, M., S. Kanegasaki, and M. Yoshikawa. 1980. Effects of temperature and energy inhibitors on complex formation between Escherichia coli male cells and filamentous phage fd. J. Gen. Microbiol. 119:87-93. [DOI] [PubMed] [Google Scholar]
- 685.Yamamoto, M., S. Kanegasaki, and M. Yoshikawa. 1981. Role of membrane potential and ATP in complex formation between Escherichia coli male cells and filamentous phage fd. J Gen. Microbiol. 123:343-349. [DOI] [PubMed] [Google Scholar]
- 686.Yang, G., A. J. Dowling, U. Gerike, R. H. Ffrench-Constant, and N. R. Waterfield. 2006. Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J. Bacteriol. 188:2254-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.Yang, C. C., and J. Konisky. 1984. Colicin V-treated Escherichia coli does not generate membrane potential. J. Bacteriol. 158:757-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 688.Yao, X. L., and M. Hong. 2006. Effects of anionic lipid and ion concentrations on the topology and segmental mobility of colicin Ia channel domain from solid-state NMR. Biochemistry 45:289-295. [DOI] [PubMed] [Google Scholar]
- 689.Yokota, N., T. Koruda, S. Matsuyama, and H. Tokuda. 1999. Charaterization of the LolA-LolB system as the general lipoprotein localization mechanism of E. coli. J. Biol. Chem. 274:30995-30999. [DOI] [PubMed] [Google Scholar]
- 690.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 691.Yue, W. W., S. Grizot, and S. K. Buchanan. 2003. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J. Mol. Biol. 332:353-368. [DOI] [PubMed] [Google Scholar]
- 692.Zakharov, S. D., and W. A. Cramer. 2002. Colicin crystal structures: pathways and mechanisms for colicin insertion into membranes. Biochim. Biophys. Acta 1565:333-346. [DOI] [PubMed] [Google Scholar]
- 693.Zakharov, S. D., and W. A. Cramer. 2002. Insertion intermediates of pore-forming colicins in membrane two-dimensional space. Biochimie 84:465-475. [DOI] [PubMed] [Google Scholar]
- 694.Zakharov, S. D., and W. A. Cramer. 2004. On the mechanism and pathway of colicin import across the E. coli outer membrane. Front. Biosci. 9:1311-1317. [DOI] [PubMed] [Google Scholar]
- 695.Zakharov, S. D., E. A. Kotova, Y. N. Antonenko, and W. A. Cramer. 2004. On the role of lipid in colicin pore formation. Biochim. Biophys. Acta 1666:239-249. [DOI] [PubMed] [Google Scholar]
- 696.Zakharov, S. D., M. Lindeberg, and W. A. Cramer. 1999. Kinetic description of structural changes linked to membrane import of the colicin E1 channel protein. Biochemistry 38:11325-11332. [DOI] [PubMed] [Google Scholar]
- 697.Zakharov, S. D., M. Lindeberg, Y. Griko, Z. Salamon, G. Tollin, F. G. Prendergast, and W. A. Cramer. 1998. Membrane-bound state of the colicin E1 channel domain as an extended two-dimensional helical array. Proc. Natl. Acad. Sci. USA 95:4282-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 698.Zakharov, S. D., M. V. Zhalnina, O. Sharma, and W. A. Cramer. 2006. The colicin E3 outer membrane translocon: immunity protein release allows interaction of the cytotoxic domain with OmpF porin. Biochemistry 45:10199-10207. [DOI] [PubMed] [Google Scholar]
- 699.Zhai, Y. F., W. Heijne, and M. H. Saier, Jr. 2003. Molecular modeling of the bacterial outer membrane receptor energizer, ExbBD/TonB, based on homology with the flagellar motor, MotAB. Biochim. Biophys. Acta 1614:201-210. [DOI] [PubMed] [Google Scholar]
- 700.Zhang, N., and R. Young. 1999. Complementation and characterization of the nested Rz and Rz1 reading frames in the genome of bacteriophage lambda. Mol. Gen. Genet. 262:659-667. [DOI] [PubMed] [Google Scholar]
- 701.Zhang, Y. L., and W. A. Cramer. 1993. Intramembrane helix-helix interactions as the basis of inhibition of the colicin E1 ion channel by its immunity protein. J. Biol. Chem. 268:10176-10184. [PubMed] [Google Scholar]
- 702.Zinder, N. D. 1973. Resistance to colicins E3 and K induced by infection with bacteriophage f1. Proc. Natl. Acad. Sci. USA 70:3160-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]