Abstract
Insertion sequences (ISs) can constitute an important component of prokaryotic (bacterial and archaeal) genomes. Over 1,500 individual ISs are included at present in the ISfinder database (www-is.biotoul.fr), and these represent only a small portion of those in the available prokaryotic genome sequences and those that are being discovered in ongoing sequencing projects. In spite of this diversity, the transposition mechanisms of only a few of these ubiquitous mobile genetic elements are known, and these are all restricted to those present in bacteria. This review presents an overview of ISs within the archaeal kingdom. We first provide a general historical summary of the known properties and behaviors of archaeal ISs. We then consider how transposition might be regulated in some cases by small antisense RNAs and by termination codon readthrough. This is followed by an extensive analysis of the IS content in the sequenced archaeal genomes present in the public databases as of June 2006, which provides an overview of their distribution among the major archaeal classes and species. We show that the diversity of archaeal ISs is very great and comparable to that of bacteria. We compare archaeal ISs to known bacterial ISs and find that most are clearly members of families first described for bacteria. Several cases of lateral gene transfer between bacteria and archaea are clearly documented, notably for methanogenic archaea. However, several archaeal ISs do not have bacterial equivalents but can be grouped into Archaea-specific groups or families. In addition to ISs, we identify and list nonautonomous IS-derived elements, such as miniature inverted-repeat transposable elements. Finally, we present a possible scenario for the evolutionary history of ISs in the Archaea.
INTRODUCTION
Archaea, members of the third domain of life, are prokaryotic organisms that can be divided into two major groups: the Crenarchaeota and the Euryarchaeota. This division, based on small-subunit rRNA phylogeny, is also strongly supported by comparative genomics. A number of genes present in euryarchaeal genomes are missing altogether in crenarchaeota and vice versa (28). Recent studies have suggested the existence of a third phylum: the Nanoarchaeota (37). However it has been suggested that Nanoarchaeota may be representatives of a quickly evolving euryarchaeal lineage (7). Many new groups of as-yet-uncultured archaea have been detected by PCR amplification of 16S rRNA from environmental samples. These include seawater, sediments, tidal flats and lakes, and the human gut and buccal cavity (76). Archaea should therefore no longer be considered simply as extremophiles (18).
Like those of the other two domains of life, the Bacteria and Eukarya, members of the prokaryotic Archaea can carry a large number and variety of transposable elements within their genomes. These are principally insertion sequences (ISs) and miniature inverted-repeat transposable elements (MITEs) (8), although at least one active composite transposon has been documented (92) and other similar structures have been identified (see “Compound transposons, bits, and pieces,” below). ISs are short specific segments of DNA up to 2 kbp long. They carry one or two open reading frames (ORFs) encoding the enzyme that catalyzes their movement, the transposase (Tpase), generally (but not always) flanked by short terminal inverted repeats (IRs). IS insertion often results in the duplication of a short target sequence that flanks the insertion (direct repeat [DR]) (12). MITEs are nonautonomous ISs deleted for part or all of the Tpase ORF but retaining both ends, while composite transposons are structures in which a DNA segment is flanked by two copies of a given IS.
Little is known about the transposition behavior of the majority of these mobile genetic elements in archaea. This is certainly due to the limitation of genetic systems available for their analysis and to the extreme conditions (temperature, pressure, pH, and salinity) required for the growth of those archaea so far analyzed. Data from the available sequenced genomes suggests that, as among bacteria, the distribution of ISs is somewhat “haphazard,” with certain species exhibiting very few or no IS copies while others carry many (see “Genome comparisons: IS distribution, abundance, and geographical variations,” below). It is clear that the variety of archaeal ISs approximates that of bacteria rather than the limited types recognized at present in eukaryotes (8). However, apart from a survey compiled several years ago (8) before the availability of a significant number of archaeal genome sequences, no systematic and coherent comparison of archaeal and bacterial ISs is available. Since the transposition characteristics of a variety of bacterial ISs are known (14), such a comparison would provide a useful starting point for exploring transposition activity in archaea and the impact of mobile genetic elements on archaeal genome structure.
NOMENCLATURE
One major task that must be confronted initially is that of nomenclature. Apart from ISs originally identified in the extreme halophiles, named ISH, the more recently identified archaeal ISs have been distinguished by their appearance in the major archaeal divisions, the Crenarchaeota (ISC) and Euryarchaeota (ISE). For these individual ISs, the distinction ISC or ISE is followed by a number corresponding to the length, in base pairs (8). This, of course, obscures the relationship between IS derivatives that differ in length by deletion or insertion of one or a few base pairs and also inflates the number of apparently different ISs.
In the present review, we provide an updated survey of archaeal IS elements and include an analysis of their distribution and of their relationship to bacterial and eukaryotic ISs. Except for certain IS names already published (principally those of the halophiles and Sulfolobales), we adhere to the system of nomenclature used at present for ISs of Bacteria, namely, the first letter of the genus, in uppercase, and the first two letters of the species name, in lowercase (12; also see www-is.biotoul.fr). This is similar to the nomenclature system used for restriction enzymes. It renders more transparent the phylogenetic relationships between highly related ISs that differ simply in overall length. These designations have been included as the principal name in the ISfinder database (www-is.biotoul.fr). Any names previously used are also included in the database as synonyms to facilitate retrieval. We assign IS names only for those where we can identify the IS ends. In all other cases, we assume that the copies are only partial, and only the identification number of the corresponding transposase ORF is given.
At the time of writing, the public databases included the entire sequences of 28 archaeal genomes (23 euryarchaeotes and 5 crenarchaeotes). For operational simplicity, to avoid inundating the ISfinder database with specific names, we have adopted the use of “isoforms,” as first suggested by Ohtsubo et al. (57). We (arbitrarily) define isoforms as being sequences that are 98% similar at the protein level and/or more than 95% similar at the DNA level. Moreover, we also point out those previously published ISs that were given different names according to length but that are effectively identical to, or are isoforms of, other ISs. We have not yet systematically addressed the extensive accumulating data from environmental sequencing projects, although certain ISs have been identified and included in ISfinder.
IS elements were identified by manual reiterative BLAST analysis using an E value cutoff of 10−3. Tpase alignments were performed with CLUSTALX and refined by eye. To infer phylogenetic relationships, we performed preliminary analyses to assess the different subgroups of large families by neighbor joining using MUST.3.0 (68). TribeMCL (23) was also applied to confirm the clustering of all ISs into the various families and subgroups. Sequences belonging to different subgroups of a single family were then treated separately by maximum likelihood, using PROML (Phylip, version 3.6 [26]) with the Jones-Taylor-Thornton amino acid substitution matrix.
IS DISTRIBUTION IN ARCHAEA COMPARED TO BACTERIA AND EUKARYA
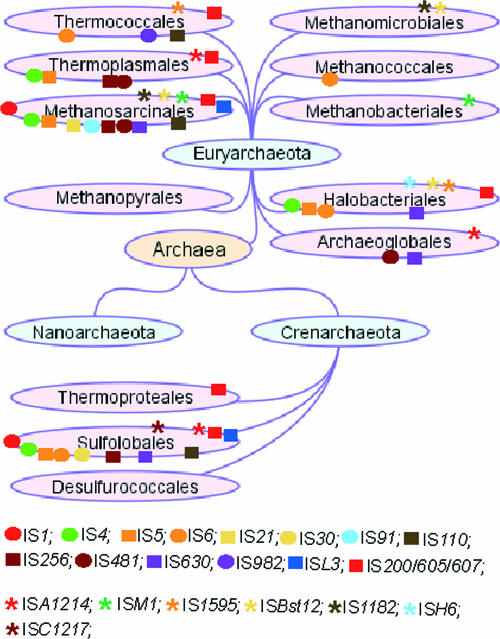
An overview of the results of database searches is presented in Fig. 1 and Table 1. IS elements are classified into families according to genetic organization, the relationship between their Tpases, and the sequences of their ends (12). The division into superfamilies, families, groups, and subgroups is relatively subjective and will change with time. A family can be defined as a closely related group with strong conservation of the catalytic site (identical spacing between the key residues and the presence of additional conserved residues within the catalytic domain; see “IS families and the nature of the catalytic site,” below), conservation of organization and expression signals (e.g., frameshifting), and a clear relationship between the IRs over their entire length. Examples of such large and closely knit families include IS3, IS21, IS30, IS481, and IS630. Not all IS groups are so coherent. Two such diverse groupings have been identified in prokaryotes: the IS4 and IS5 superfamilies (12). These are growing considerably, and the relationships within these superfamilies continue to evolve as additional members are identified. IS630 has also been included in a less well defined grouping with eukaryotic elements such as mariner and Tc. This has been referred to as a superfamily.
FIG. 1.
Comparison of IS families in archaea. The figure shows the distribution of IS families among the different archaeal phyla. The tree is from NCBI (http://www.ncbi.nlm.nih.gov/sutils/genom_tree.cgi). The color code for IS families is included within the figure beneath the phylogenetic tree. Stars represent emerging groups or families.
TABLE 1.
IS content of archaeal genomesa
The S. solfataricus genome is presently undergoing reannotation in ISfinder. The MITEs in the genomes of M. jannaschii, S. solfataricus, and S. tokodaii have not yet been fully annotated. Color coding is as follows: Sulfolobales, red; Thermoplasmatales, magenta; halophiles, green; methanogens, blue; “other,” orange.
Figure 1 shows the distribution of different IS families within the Archaea. The most striking feature here is that most of the archaeal ISs fall into families found in the Bacteria (present in the ISfinder database). Three Archaea-specific groups, ISA1214, ISC1217, and ISH6, have emerged in these studies. On the other hand, archaeal genomes lack elements from the IS1380 family and, moreover, several widespread bacterial IS families such as IS3, IS1182, IS21, IS91, IS30, and IS982 have few archaeal members. However, since the sequences of only 28 archaeal genomes were available, compared to more than 325 bacterial genomes, it is possible that the numbers of archaeal ISs from known families are underestimated. Conversely, we cannot rule out the existence of additional Archaea-specific ISs presenting limited or no obvious similarities with those from Bacteria (see “Emerging groups, orphans, waifs, and strays,” below).
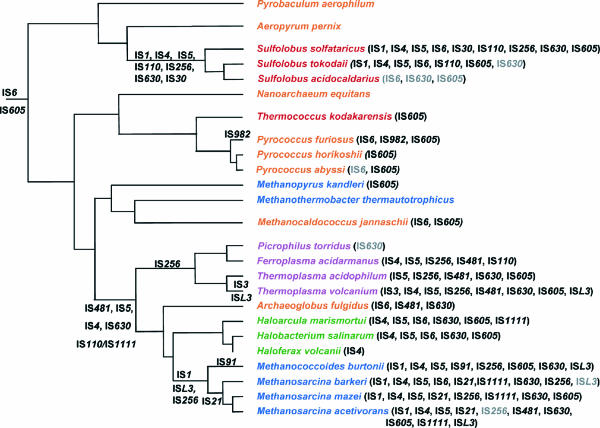
The distribution of ISs in archaeal genomes is very “patchy” (Fig. 1). Four phyla, comprising the Halobacteriales, Sulfolobales, Methanosarcinales, and Thermoplasmatales, monopolize more than 90% of archaeal ISs (Table 1). No ISs were identified in the Nanoarchaeota, the Desulfurococcales, the Methanomicrobiales, the Thermoproteales, or the Methanobacteriales, and only one or two families in the Methanococcales or the Methanopyrales. However, these lineages are represented by only one or two completely sequenced genomes, and this limited information may introduce some bias, as was initially the case for bacterial Mycoplasma species (www-is.biotoul.fr).
It is worth noting that archaeal ISs resemble bacterial ISs rather than those identified in eukaryotes. No elements with significant similarity to the nine currently recognized eukaryote DNA transposon superfamilies could be identified. These include notably the mariner/Tc (distantly related to the IS630 family) and the P (from Drosophila) families, which are structurally close to bacterial ISs; elements such as the CACTA or the hAT (e.g., hobo, Ac, and Tam) families (mainly recovered in plants and insects), Merlin (related to IS1016), Mutator (distantly related to IS256 family members), PIF/Harbinger (distant relatives of some IS5 family members), piggyback, and Transbib (12, 70); or to the helitrons (40), a family related to bacterial IS91 and identified in plants, fungi, and diverse animals (14). Extensive BLAST searches seeded with such sequences revealed no detectable homologies in the archaeal genomes. This is perhaps surprising in view of the fact that Archaea have important similarities to Eukarya, notably enzymes involved in DNA replication (47). Since it seems unlikely that eukaryal “ISs” were originally present in these genomes and were subsequently specifically deleted, this implies that any lateral transfer of transposable elements occurred between Bacteria and Archaea but not between Archaea and Eukarya.
In the light of the important differences between bacterial and archaeal replication systems, it is interesting to note the presence of members of the IS1, IS3, and IS256 families within archaeal genomes. Bacterial members of these families are thought to transpose by a mechanism involving a replication step to eject a circular IS transposition copy from the donor site, which then serves as a transposition intermediate (78). In the case of the IS3 family member IS911, this process has been shown to depend on the DnaG primase (22). Interestingly, each archaeal genome usually contains two types of primase: a dimeric eukaryotic-like primase (44) and a DnaG-like enzyme that shares the Toprim domain with bacterial DnaG (2).
However, recent biochemical analyses have demonstrated that the DnaG-like primase in Archaea may be involved in RNA processing and degradation rather than in DNA metabolism (25). The presence of these ISs in Archaea therefore implies that the replication step may be taken in charge by the host (Eukarya-like) replication system.
TRANSPOSITION IN THE ARCHAEA: HISTORICAL PERSPECTIVE
Spontaneous Mutation in the Extreme Halophiles
One of the earliest descriptions of IS element activity in archaea stemmed from the observation of an unusually high spontaneous mutation rate in Halobacterium salinarium (previously called H. halobium). Depending on the phenotypic marker observed (gas vacuole or bacterio-opsin genes), this was found to range between 10−2 and 10−4 in an aerobically grown culture which had undergone approximately 20 generations of growth (67). In the case of the gas vacuole genes, mutation was generally associated with the insertion of additional DNA at one of two specific places. Reversion of the mutation was often accompanied by loss of the inserted DNA, a characteristic of IS mutagenesis in the Bacteria. These “pregenomic” studies were facilitated by the fact that the H. salinarium genome could be physically separated into two fractions according to AT/GC content, and that the relatively AT rich fraction carried the genes of interest often as part of plasmids (66). Much of this and further work was done with wild strains of H. salinarium carrying various plasmids or megaplasmids such as pHH1, pHH2, pGRB1, or pNCR100 (54, 66).
The exceptional genome plasticity revealed by these studies was further reinforced by experiments establishing that strains of both H. salinarium and the related Halobacterium volcanii generally carry a large number of repeated elements. These were divided into several families by Southern hybridization. The elements appeared to be highly mobile, were associated with chromosome rearrangements, and were found both clustered and dispersed over the genome (79).
A collection of repeated sequences resembling bacterial ISs was subsequently assembled in H. salinarium with either gas vacuole or plasmid-carried purple membrane genes used as targets. Several of these have been isolated more than once and have received different names. Importantly, since the majority of these ISs were isolated as novel insertions, they therefore represent active copies.
ISH1.
The 1,118-bp ISH1 was isolated as an insertion into the bacteriorhodopsin (bop) gene. Its sequence revealed imperfect terminal inverted repeats of 9 bp and flanking 8-bp direct target repeats. These features are characteristic signatures of IS elements in Bacteria. The element was named ISH1 (84). The single ORF predicts a protein of 270 amino acids (aa) with a clear DDE catalytic motif (see “IS families and the nature of the catalytic site,” below), relating the Tpase to those of the majority of transposable elements presently identified. Further examination (12) placed ISH1 in the rather disperse IS5 family (see “IS families in the archaeal genomes,” below). Many isolates of ISH1 appeared to have inserted into the same site (5′-AGTTATTG-3′) of the bop gene but could do so in both orientations. This indicates relatively high target site specificity. Southern blot analysis revealed multiple ISH1 copies, ranging from one to more than five, in different halobacterial strains (84).
Moreover, analysis of one insertion mutant revealed a single additional ISH1-specific restriction fragment compared to its wild-type parent. This increase in copy number led to the supposition that ISH1 transposes by a replicative mechanism (84).
Evidence from Northern blots also showed that ISH1 was actively transcribed in these strains with a rough correlation between RNA band intensity and IS copy number. However, in view of the numerous regulatory mechanisms adopted by ISs to limit their activity (53), this does not necessarily mean that the Tpase is produced at comparative relative levels.
ISH2.
Examination of additional bop mutants revealed several other repeated sequences distinguishable by size. The most frequently observed was ISH2, only 521 bp long and carrying 19-bp terminal inverted repeats flanked by target duplications of 10 or 20 bp (17) and occasionally 11 bp (64). Although three potential ORFs were detected (ORF I, 80 codons; ORF II, 64; ORF III, 59), we have been unable to identify a typical Tpase catalytic motif (see “IS families and the nature of the catalytic site,” below). The majority of insertion mutations in the bop gene were caused by the elements ISH1 and ISH2. Unlike ISH1, ISH2 showed multiple insertion sites in the gene (17).
ISH2 was present in multiple copies in various H. salinarium strains, and, more recently, four additional copies were identified in the Halobacterium plasmid pNRC100 (54). The IS is clearly capable of transposition but is probably not an autonomous transposon. However, ISH2 shares nearly perfect terminal homology (but no internal homology) with an apparently complete IS, ISH26 (ISH8; see below). ISH2 transposition may therefore be driven in trans by the ISH26 Tpase.
ISH3/ISH27/ISH51.
Remarkably, 20% of H. salinarium PHH4 colonies were found to carry IS insertions into a resident pHH4 plasmid (16, 63). Among these, ISH27 was isolated as a major source of mutation. This group of ISs belongs to the IS4 family. They are 1,398 bp (ISH27-1) or 1,389 bp (ISH27-2 and ISH27-3) long and generate 5-bp target repeats (63) rather than the 3-bp repeats proposed for the identical ISH3 (16). They also include terminal IRs of 16 bp. Two ISH27-1-specific transcripts were observed in the pHH4 plasmid-carrying strain. One of these exhibited a size expected for a full ISH27 transcript (∼1,200 nucleotides [nt]), while the other was significantly shorter (∼650 nt). This could reflect regulation at the transcriptional or posttranscriptional level.
ISH27 is the generic name for three related ISs. Although closely related, these are not isoforms by our definition. At the nucleotide level, ISH27-1 is more similar to H. volcanii ISH51-1, ISH51-2, and ISH51-3 (88% DNA identity) than to ISH27-2 and ISH27-3 (80% identity). There are more than 20 copies of ISH51 in the H. volcanii genome (36). ISH27 was also observed to have undergone an amplification following storage of the host strain over a period of several years at 4°C (63). Further studies to determine the factors involved in this process would be interesting.
ISH8/ISH26.
ISH8/ISH26 was isolated as an insertion mutation of the gvp operon (gas vesicle proteins, Vac−) (31). ISH8, also a member of the IS4 family, is 1,402 bp long, carries 18-bp IRs, and generates 10-bp DRs. Its DNA sequence is 94% identical to that of ISH26. Copies of ISH8 were also found in the H. salinarium plasmid pNRC100.
A 70-kbp AT-rich island of H. salinarium was identified and proven to carry copies of ISH1, ISH2, and an IS-like sequence, ISH26, together with copies of an additional 10 repeated sequences, most of which were not characterized (62).
ISH26 was also isolated as an insertional inactivation of the bop gene. There are four ISH26 copies on pHH1 and four copies on the chromosome of H. salinarium PHH1 (65). ISH26 was described as harboring two overlapping ORFs. Although the first ORF has significant similarity with the putative Tpases of other IS4 family members (for example, 26% identity to IS231W over a 143-aa overlap), the second ORF has only very limited similarity, in the region of the conserved E residue (see “IS families and the nature of the catalytic site,” below). Detailed analyses suggest, however, that the introduction of several frameshifts would significantly increase this similarity. The first ORF is very closely related to the N-terminal end of the Tpase of ISH8. Like ISH27, ISH26 copies constitute a group of related, but not identical, elements (63).
ISH11.
ISH11, from H. salinarium, was observed as an insertion into plasmid pGRB1. It is 1,068 bp long, with 15-bp terminal IRs, and was flanked by 7-bp direct target repeats (43). It exhibits a single long ORF of 334 aa. ISH11 has been tentatively grouped within the IS427 cluster of the IS5 family. Two copies are present in pNRC100 of Halobacterium sp. strain NRC-1.
ISH23/ISH50.
ISH23/ISH50 is one of the least-frequent causes of insertion mutations in the bop gene (64). There are two ISH23 copies in H. salinarium NRC817.
ISH23 is flanked by 29-bp imperfect IRs and by a 9-bp direct target repeat. It is very similar (but not identical) to ISH50, an IS isolated as an insertion into the Halobacterium plasmid pNRC (93). ISH50 is 996 bp long, with terminal IRs of 23/29 bp and 8-bp flanking direct target repeats. It encodes a potential 273-aa Tpase and belongs to a newly defined family containing both archaeal and bacterial members (L. Gagnevin and P. Siguier, unpublished data) (see “Emerging groups, orphans, waifs, and strays,” below). The first and last 200 bp of ISH23 were found to be identical to those of ISH50 and, although ISH23 and ISH50 differ by at least two restriction sites and appear to generate either 9- or 8-bp target duplications, they are assumed be isoforms of the same IS (65).
ISH24.
Another infrequent insertion into the bop gene, ISH24, is 3,000 bp long, including two terminal IRs of 14 bp, and is flanked by 7-bp direct target repeats. The sequence of this element became available subsequent to the sequencing of the megaplasmid pNRC100 of H. salinarum. It was renamed ISH7 (54). ISH7 encodes two large ORFs. The second displays some weak and local similarities with the C-terminal parts of IS4 element Tpases. No clear DDE motif in ISH24 could be detected from this partial alignment.
ISH25.
The short 588-nt sequence of ISH25 is sometimes associated with ISH27 insertion, but it appears unlikely to be a simple IS, as no putative ORF can be found.
ISH28.
ISH28 was also isolated from a bop mutant (62). Its nucleotide sequence was revised (91). It is 938 bp long, with 16-bp terminal IRs, and carries an ORF of 828 bp. It is flanked by 8-bp direct target repeats. The putative Tpase protein is 49% similar to that of ISH1, a member of the IS5 family.
ISH28 has also been engineered to generate composite transposons, which are efficient tools for mutagenesis of Haloarcula hispanica and other halophilic organisms (92). This element showed little target sequence specificity but was biased toward target regions with a lower G+C content. Of 20 insertions characterized, 18 generated DRs of 8 bp, while the remaining 2 had DRs of 9 bp.
Collectively, these results clearly demonstrate the major role played by transposable elements in shaping the halophilic genome.
Transposition in Sulfolobus
Although most of the earliest exploratory studies in archaeal transposition were carried out with halobacteria due to the high level of transposon-mediated genome rearrangements in this model system, other archaea have received some attention. The 2.99-Mb Sulfolobus solfataricus genome is estimated to contain nearly 350 intact mobile elements (82). An early report (1) described the serendipitous isolation of an S. solfataricus IS, ISC1041 (named according to its length), which was related to the bacterial IS30 family of elements.
Like halobacterial species, S. solfataricus also exhibits a relatively high spontaneous mutation rate (52). These studies used 5-fluoro-orate resistance as a screen for uracil auxotrophs (pyrE and pyrF). Mutations were obtained at frequencies of between 10−4 and 10−5, significantly lower than in the halobacteria but at least 10-fold higher than for other members of the Sulfolobus genus. PCR analysis of several auxotrophic mutants revealed that all carried insertions ranging from 1 to 1.4 kbp. Similar auxotrophs of the related Sulfolobus acidocaldarius failed to show such insertions. Seven S. solfataricus mutants were analyzed in more detail and proved to carry insertions. These were named according to their individual lengths, in base pairs: ISC1058 (three examples), ISC1359 (two examples), and ISC1439 (one example). One example, of 1,147 bp, was closely related to, and presumably a deletion derivative of, a 1217-bp element previously isolated as an insertion of ISC1217 (13-bp IRs, 6-bp DRs) into a β-galactosidase gene (80). All four ISs show similarities to members of the IS4 or IS5 family: their putative Tpases include both the D · N · G/A-Y/F and Y · R · E · K motifs characteristic of these DDE families (see “IS families and the nature of the catalytic site,” below).
Additional active ISs have since been isolated (6), also with 5-fluoroorate resistance used as a screen. Several different, newly isolated, Sulfolobus strains from Siberia and the western United States were analyzed. As judged by the 99% nucleotide identities in the pyrB, pyrF, or pyrE gene, these appeared to be conspecific strains. Seven distinct ISs were isolated following PCR amplification across the mutated gene. Again, these were named for their lengths, in nucleotides.
In order of size they include ISC735, a member of the IS6 family with a single ORF, 18-bp IRs, and 8-bp DRs; ISC796, a member of the IS1 family with only a single reading frame, 21-bp IRs, and 8-bp DRs; ISC1057 and ISC1058b, related to ISC1058 and members of the IS5 family, with 88 to 93% shared nucleic acid identities, 20-bp IRs interrupted (“hyphenated”) by a hexanucleotide, and 8-bp DRs; ISC1205, related to ISC1217, with 17- to 20-bp IRs and 4- to 7-bp DRs; ISC1290, a member of the IS5 family, with 34-bp IRs and 5-bp DRs; and ISC1926, a member of the IS200/IS605 group, with the corresponding two characteristic ORFs. ISC1926 is an isoform of ISC1913 in the sequenced genome of S. solfataricus. In addition to these entire ISs, the authors also detected an insertion of a short 128-bp fragment with terminal inverted repeats similar to those of ISC1058. This sequence corresponds to a typical MITE (see “MITEs, MICs, and solo IRs,” below).
Transposition in Other Archaea
IS6-mediated gene rearrangements have been observed in the pyrococci (45, 95). These involve deletions (24), chromosome rearrangements (21, 45), and insertional inactivation (e.g., by insertion of ISpfu3 into napA in P. woesei [39]).
ISM1 was identified in a cloning study of the Methanobrevibacter smithii purE and proC genes (32). This has a typical IS structure, is distantly related to the ISL3 family, and is present in about 10 copies in M. smithii.
No data concerning transposition or the effects of transposable elements are available for other archaeal phyla, including important groups carrying numerous ISs such as the Methanosarcinales and Thermoplasmatales.
REGULATION OF TRANSPOSITION
Although regulation has not been addressed experimentally in any detail in the Archaea, in principle, many of the systems which regulate transposition activity in the Bacteria (53) might be expected to operate in the Archaea. These would include control at the level of gene expression (transcription initiation, translation initiation and elongation, translational or transcriptional frameshifting, and mRNA stability) and activity (Tpase stability, intervention of host proteins). Some studies have suggested that certain archaeal elements may be regulated by small, noncoding RNAs (ncRNAs) or by translational readthrough.
Lost in Transcription: ncRNAs in S. solfataricus
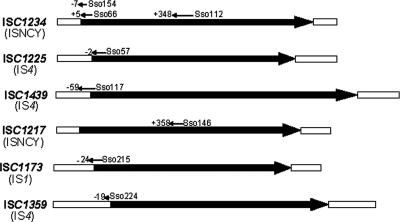
Interestingly, in a recent study designed to identify small, ncRNAs (87), 8 of the 57 ncRNAs identified proved to be complementary to mRNAs encoding various Tpases. These include ISC1173, ISC1217, ISC1225, ISC1234, ISC1359, and ISC1439 (Fig. 2). In the case of the most abundant S. solfataricus IS, ISC1234, one ncRNA would overlap the Tpase initiation codon. This is reminiscent of a regulatory mechanism observed for the bacterial IS10 (81, 83), where a short RNA, RNAout, is transcribed from a promoter, Pout, located close to the left end of the element and is complementary to the RNA. The complementarity between the mRNA and RNAout regulates Tpase expression by sequestering translation initiation signals. Two other ncRNAs were found to be complementary to sequences internal to the Tpase gene. The function of these internal ncRNAs is not yet clear. They could mask internal expression signals, interfere with the expression of full-length Tpase, or influence mRNA stability. Similar ncRNAs, complementary to the mRNA translation initiation signals, were also identified for ISC1439 and ISC1173, while internally complementary ncRNAs were identified for ISC1225 and ISC1217.
FIG. 2.
Noncoding RNA. The ISs are drawn to scale. The black arrows represent the length of the Tpase ORF. The open boxes represent the noncoding regions of the ISs. The noncoding RNA names from reference 72 are shown, together with their beginnings and ends (in bases from the first base of the Tpase coding sequence). The directions of transcription are shown.
In the case of ISC1217, the ncRNA complementary to an internal Tpase sequence proved to be a mixed population of identical size but carrying small nucleotide substitutions. Interestingly ISC1217 exists in several isoforms, some of which include nucleotide changes in this region. The ncRNA population was composed of examples carrying each of the isoform sequences. Finally, an ncRNA complementary to the upstream, nontranslated region of ISC1359 was identified.
Further studies are essential to determine the exact role of these ncRNAs in regulation of Tpase expression. As pointed out by Tang et al. (87), regulation at the posttranscriptional level would be an efficient strategy for S. solfataricus, since mRNAs in this organism have unusually long half-lives (4).
Lost in Translation: Translational Readthrough in Methanosarcina?
Methylamine methyltransferases are important in the production of methane by archaeal methanogens. Paul et al. (60) identified an in-frame amber codon (TAG) in the trimethylamine methyltransferase genes of both M. barkeri and M. thermophila. However, at least in the case of M. barkeri, abundant quantities of the full-length protein could be obtained and it appeared that the TAG codon was read as Lys. Moreover, all three copies of a dimethylamine methyltransferase gene were also shown to carry in-frame TAG codons. In addition, analysis of the M. mazei genome has identified seven methyltransferase genes of this type and a relatively large number of in-phase TAG termination codons within other genes. The additional genes include 18 that encode Tpases. M. barkeri encodes 58 tRNA genes, an unusually high number. This complement includes a putative amber suppressor tRNA (20). It is therefore possible that amber suppression leads to translational readthrough that regulates transposition activity in these cases.
IS FAMILIES AND THE NATURE OF THE CATALYTIC SITE
As stated above, Tpases can be classified according to the nature of their catalytic site. This defines the chemistry used in the transposition reactions. At present, five types have been identified. These are the DDE Tpases, the major recognized group; Y and S Tpases, related to the tyrosine (Y) and serine (S) site-specific recombinases; and Y2 enzymes, which share many characteristics of the rolling circle replicases (for reviews, see references 13 and 15). A fifth type, resembling the DNA relaxases associated with bacterial conjugation, has been identified more recently (77, 88). Only members of the DDE, serine, and relaxase classes of transposon have as yet been identified in Archaea.
The DDE Enzymes
Arguably the major transposon class encodes Tpases called DDE Tpases. The amino acids Asp (D)-Asp (D)-Glu (E) coordinate divalent metal ions necessary for DNA cleavage and joining involved in transposon movement. Additional conserved residues can also be observed. In particular, a basic K or R is often present at a distance of seven residues on the C-terminal side of the characteristic E. This places it on the same side of an α helix as the conserved E but two turns farther toward the C terminus.
Several groups of additional conserved amino acids, designated N1, N2, N3, and C1 encompass the D (N2), D (N3), and E (C1) regions in the IS4 family (74). These have been expanded to the motifs DDT, DREAD, and YREK respectively (73).
DDE enzymes ensure cleavage of the terminal phosphodiester bonds at the 3′ end of the transposon strand, which will be finally transferred into the target DNA site (transferred strand). Transposons and ISs using such enzymes generally carry imperfect IRs at their ends, including one or several Tpase binding sites. The ends of ISs (terminal IRs) that have adopted this transposition chemistry are generally the simplest. They can often be divided into two domains: a Tpase binding domain, an internal sequence of 10 to 15 bp, and a catalytic domain composed of the terminal 2 to 4 bp required for cleavage and strand transfer. DDE enzymes generally generate a characteristic direct duplication of target DNA flanking the insertion. This type of IR structure is conserved in the archaeal ISs but is generally more complicated in the Eukarya.
The Serine Enzymes
The serine enzymes are related to the site-specific recombinases involved in the resolution of cointegrate molecules, the final step in transposition of the Tn3 class of bacterial transposons. The Tpase of these elements generates a cointegrate or replicon fusion in which fused donor and target replicons are separated by directly repeated copies of the transposon at each junction. The serine recombinase intervenes by catalyzing site-specific recombination between the two transposon copies, separating the replicons and thereby completing transposition. Serine recombinases are so named because they use a serine residue as the nucleophile in DNA strand cleavage and generate covalent enzyme-DNA intermediates. In the single case analyzed, IS607 from Helicobacter pylori, the S Tpase generates a circular transposon intermediate (N. Grindley, personal communication), which presumably then undergoes integration into a target molecule.
The Relaxase Enzymes
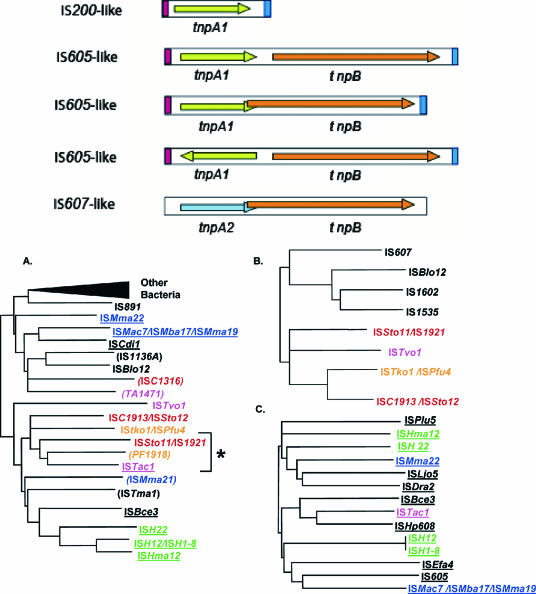
The relaxase enzymes represent a newly recognized class of generally small (∼150-aa) Tpases. They use a single tyrosine (Y) residue as a nucleophile in DNA cleavage and generate covalent Y-DNA substrate intermediates. The structures of two enzymes, the bacterial IS608 and an isoform of ISSto1 (from S. tokodaii [ISfinder]), have been solved (46, 77). They exhibit a structural topology close to that of the Rep and Relaxase proteins. Transposons using this type of Tpase do not carry terminal IRs and do not generate the small flanking direct target repeats generally produced by transposons with DDE Tpases. Instead, these Tpases bind to extensive subterminal secondary structural motifs and cleave at a fixed but distant position (88). They also use a defined tetra- or pentanucleotide as a target sequence and require this sequence for further transposition.
IS FAMILIES IN THE ARCHAEAL GENOMES
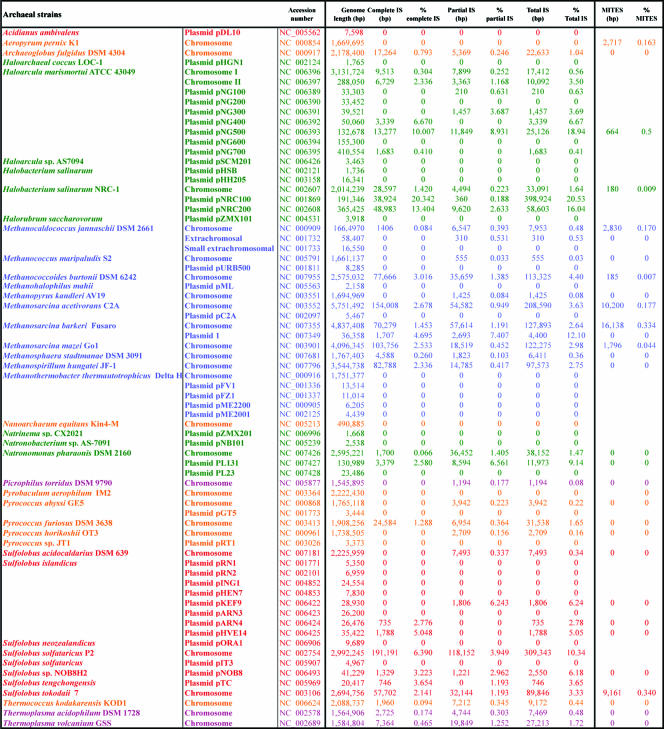
We have analyzed both the fully sequenced archaeal genomes and all partial sequences deposited in the public databases as of June 2006, with a few subsequent additions. The results are summarized in Fig. 1 and in Tables 1, 2 and 3. The archaeal genomes analyzed are listed in Table 1 together with the IS content. Table 2 lists the individual ISs in family groups and indicates their copy number, the presence of complete and partial copies, and the presence of MITEs. Table 3 lists the different types of MITE observed. Below, we present a more detailed description of the distribution and characteristics of each family. Where appropriate, we have included a tree for each IS family which relates the archaeal and eubacterial members. We have color-coded the origins of the ISs from Bacteria, Sulfolobales, Thermoplasmatales, halophiles, methanogens, and others throughout the figures and in Table 1. We have also included, where appropriate, diagrams of the organization of ISs of given families. We have not included this type of diagram for families whose members are simple and for which both bacterial and archaeal members are very similar (e.g., IS481) or for families whose members are extremely heterogeneous (e.g., IS4 and IS5 families) and which are at present undergoing extensive reanalysis. In addition, due to the limited number of members of some families in the Archaea, we have not included an individual figure for these families.
TABLE 2.
ISs identified in archaeal genomesb
| IS name or gene identifier | Accession no.a | Length (bp) | IR (bp) | DR (bp) | No. of ORFs | Host | No. of copies
|
Presence of MITEs | |
|---|---|---|---|---|---|---|---|---|---|
| C | P | ||||||||
| DDE transposons | |||||||||
| IS1 family (Fig. 3) | |||||||||
| ISC1173a | NC_002754 | 1,173 | ∼50 | 8 | 1 | S. solfataricus | 5 | 27 | |
| ISSto7 | NC_003106 | 1,174 | ∼50 | 8 | 1 | S. tokodaii | 2 | 12 | X |
| ISC796 | AY671943 | 796 | 21 | 8 | 1 | Sulfolobus sp. | 1 | ||
| ISSto9 | NC_003106 | 796 | 21 | 9 | 1 | S. tokodaii | 5 | 18 | |
| ISC1174 | NC_002754 | 1,174 | 49/50 | 9 | 1 | S. solfataricus | 2 | ||
| ISMac16 | NC_003552 | 740 | 24 | 8 | 2 | M. acetivorans | 16 | 7 | |
| ISMma7 | NC_003901 | 740 | 24 | 9 | 2 | M. mazei | 1 | 1 | |
| ISMba2 | NC_007355 | 740 | 24 | 8 | 2 | M. barkeri | 5 | ||
| ISMbu3 | NC_007955 | 741 | 15 | 8 | 2 | M. burtonii | 7 | 3 | |
| IS3 family | |||||||||
| TVN0691/92 | NC_002689 | T. volcanium | |||||||
| TVN0865/67 | NC_002689 | T. volcanium | |||||||
| IS4 family (Fig. 4) | |||||||||
| ISH8 subgroup | |||||||||
| ISH2 | NC_002607 | 521 | 19 | 10 | 1 | Halobacterium sp. | 4 | ||
| NC_001869 | Halobacterium sp. pNRC100 | 4 | |||||||
| NC_002608 | Halobacterium sp. pNRC200 | 5 | |||||||
| ISH26 | X04832 | 1,384 | 19 | 11 | 2 | H. salinarium PHH1 | |||
| ISH5 | NC_001869 | 1,442 | 19/20 | 10 | 1 | Halobacterium sp. pNRC100 | 2 | ||
| NC_002608 | Halobacterium sp. pNRC200 | 2 | 3 | ||||||
| ISH8 | NC_002607 | 1,402 | 16/18 | 10 | 1 | Halobacterium sp. | 5 | ||
| NC_001869 | Halobacterium sp. pNRC100 | 6 | |||||||
| NC_002608 | Halobacterium sp. pNRC200 | 10 | |||||||
| ISHma1 | NC_005125 | 1,403 | 17/19 | 10, 11 | 1 | H. marismortui chr I | 1 | 2 IRs | |
| NC_006397 | H. marismortui chr II | 1 | 2 | ||||||
| NC_006392 | H. marismortui pNG400 | 1 | |||||||
| NC_006393 | H. marismortui pNG500 | 1 | 5 | ||||||
| ISMba1 | NC_007355 | 1,539 | 11/13 | 0 | 1 | M. barkeri | 1 | ||
| ISMba6 | NC_007355 | 1,453 | 19/22 | 10 | 1 | M. barkeri | 4 | 28 | |
| NC_003552 | M. acetivorans | 1 | 4 + 35 IRs | ||||||
| NC_003901 | M. mazei | 3 IRs | |||||||
| M. thermophila | 1 IR | ||||||||
| ISMhu6 | NC_007796 | 1,781 | 14/16 | 0-10 | 1 | M. hungatei | 5 | 1 | |
| ISMhu9 | NC_007796 | 1,080 | 12 | 4 | 1 | M. hungatei | 7 | 1 | |
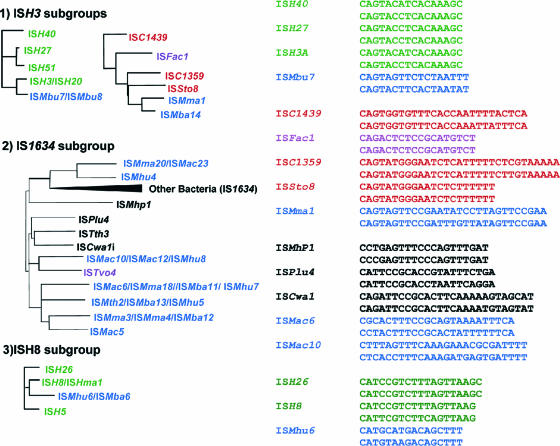
| IS1634 subgroup | |||||||||
| ISMac5 | NC_003552 | 1,719 | 19/20 | 6 | 1 | M. acetivorans | 7 | 2 | |
| ISMac6 | NC_003552 | 1,713 | 18/24 | 6 | 1 | M. acetivorans | 4 | 1 | |
| ISMac10 | NC_003552 | 1,926 | 18/26 | 5 | 1 | M. acetivorans | 2 | 1 | |
| ISMac12 | NC_003552 | 1,674 | 22/26 | 6 | 1 | M. acetivorans | 3 | ||
| ISMac23 | NC_003552 | 1,593 | 13 | 6 | 1 | M. acetivorans | 3 | ||
| ISMba11 | NC_007355 | 1,718 | 19/23 | 6 | 1 | M. barkeri | 4 | 7 | X |
| ISMba12 | NC_007355 | 1,729 | 18/20 | 6 | 1 | M. barkeri | 1 | 10 | X |
| ISMba13 | NC_007355 | 1,706 | 18/20 | 6 | 1 | M. barkeri | 2 | 6 | |
| ISMma3 | NC_003901 | 1,719 | 14/16 | 7 | 1 | M. mazei | 2 | ||
| ISMma4 | NC_003901 | 1,719 | 17/20 | 6 | 1 | M. mazei | 3 | 4 | |
| ISMma18 | NC_003901 | 1,718 | 18/23 | 6 | 1 | M. mazei | 2 | ||
| NC_003552 | M. acetivorans | 1 | |||||||
| NC_007355 | M. barkeri | 1 | |||||||
| ISMma20 | NC_003901 | 1,593 | 13/16 | 6 | 1 | M. mazei | 3 | 2 | |
| ISMhu4 | NC_007796 | 1,816 | 19/22 | 5 | 1 | M. hungatei | 6 | 0 | |
| ISMhu5 | NC_007796 | 1,719 | 18/21 | 5 | 1 | M. hungatei | 5 | 0 | |
| ISMhu7 | NC_007796 | 1,713 | 16/17 | 6 | 1 | M. hungatei | 1 | 3 | |
| ISMhu8 | NC_007796 | 1,762 | 10/14 | 6 | 1 | M. hungatei | 1 | 1 | |
| ISMth2 | 1,723 | 16/19 | 6 | 1 | M. thermophila | ||||
| ISFac6 | 1,511 | 16/18 | ND | 1 | F. acidarmanus | ||||
| ISTvo4 | NC_002689 | 1,525 | 25/31 | 6 | 1 | T. volcanium | 1 | ||
| ISArch8 | AY714833 | 1,923 | 20/24 | 0 | 1 | Uncultured archeon | |||
| ISH3 subgroup | |||||||||
| ISC1225 | NC_002754 | 1,225 | 17 | 4 | 1 | S. solfataricus | 9 | 30 | |
| ISC1200 | NC_002754 | 1,200 | 16/17 | 0 | 1 | S. solfataricus | 2 | 3 | |
| ISC1359 | NC_002754 | 1,359 | 29/31 | 4 | 1 | S. solfataricus | 9 | 10 | |
| ISC1439A | NC_002754 | 1,439 | 19/20 | 9 | 1 | S. solfataricus | 25 | 3 | |
| ISC1439B | NC_002754 | 1,439 | 21/24 | 9 | 1 | S. solfataricus | 4 | 3 | |
| ISSto8 | NC_003106 | 1,361 | 21 | 0 | 1 | S. tokodaii | 3 | 11 | |
| ISSto14 | NC_003106 | 1,227 | 16 | 4 | 1 | S. tokodaii | 1 | 1 | |
| ISH20 | NC_006397 | 1,374 | 15 | 0 | Pseudo | H. marismortui chr II | 1 | ||
| NC_006393 | H. marismortui pNG500 | 2 | X | ||||||
| ISH27 | X54432 | 1,398 | 16 | 5 | 1 | H. salinarium PHH1 | |||
| ISH3 | NC_002607 | 1,389 | 15/16 | 5 | 1 | Halobacterium sp. | 5 | ||
| NC_001869 | Halobacterium sp. pNRC100 | 6 | 1 | ||||||
| NC_002608 | H. sp pNRC200 | 10 | 3 | ||||||
| ISH40 | iso ISH27 | ||||||||
| ISH51 | XO4389 | 1,370 | 15/16 | 5 | Haloferax volcanii | ||||
| ISFac1 | 1,477 | 18 | ND | 2 | F. acidarmanus | ||||
| ISMma1 | NC_003901 | 1,500 | 12 | 7 | 1 | M. mazei | 5 | ||
| ISMba14 | NC_007355 | 1,503 | 12 | 0 | 1 | M. barkeri | 1 | 1 | |
| ISMbu7 | NC_007955 | 1,248 | 14/17 | 5 | 1 | M. burtonii | 7 | 4 | |
| ISMbu8 | NC_007955 | 1,247 | 13/17 | 5 | 1 | M. burtonii | 1 | 3 | |
| IS701 subgroup | |||||||||
| ISMba8 | NC_007355 | 1,646 | 19/24 | 1 | M. barkeri | 2 | 4 | ||
| Partial ISs | |||||||||
| NP1942A | NC_007426 | N. pharaonis | |||||||
| MM0877 | NC_003901 | M. mazei | |||||||
| MM0878 | NC_003901 | M. mazei | |||||||
| Mhun_1523 | NC_007796 | M. hungatei | |||||||
| TVN0693 | NC_002689 | T. volcanium | |||||||
| TVN0868 | NC_002689 | T. volcanium | |||||||
| TVN1465 | NC_002689 | T. volcanium | |||||||
| IS5 family (Fig. 5) | |||||||||
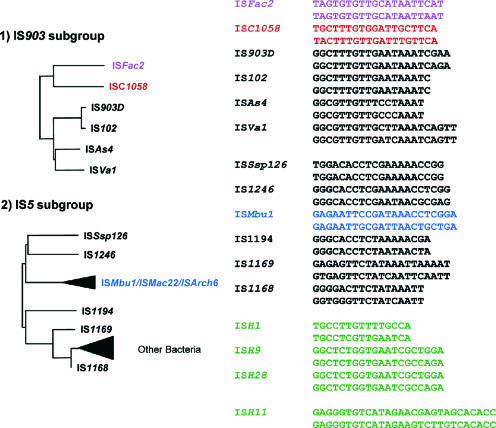
| IS903 subgroup | |||||||||
| ISC1058 | NC_002754 | 1,058 | 15/19 | 9 | 1 | S. solfataricus | 12 | 5 | |
| rmB0094 | NC_006397 | H. marismortui chr II | |||||||
| MM1429 | NC_003901 | M. mazei | |||||||
| Mbar_A1398/99 | NC_007355 | M. barkeri | |||||||
| Mbar_A2202 | NC_007355 | M. barkeri | |||||||
| TVN0139 | NC_002689 | T. volcanium | |||||||
| TVN0587 | NC_002689 | T. volcanium | |||||||
| IS1031 subgroup | |||||||||
| ISMac15 | NC_003552 | 886 | 15/17 | 3 | 1 | M. acetivorans | 2 | 20 | |
| IS427 subgroup | |||||||||
| ISMac11 | NC_003552 | 869 | 15/17 | 3 | 1 | M. acetivorans | 4 | 14 | X |
| ISMma12 | NC_003901 | 875 | 16/17 | 3 | 1 | M. mazei | 1 | 3 | |
| ISMba5 | NC_007355 | 872 | 15/17 | 3 | 1 | M. barkeri | 1 | 8 | X |
| ISMba19 | NC_007355 | 858 | 15 | 0 | 2 | M. barkeri | 3 | 31 | |
| IS5 subgroup | |||||||||
| ISMac22 | NC_003552 | 1,156 | 15/20 | 2 | 1 | M. acetivorans | 1 | 4 | |
| NC_007355 | M. mazei | 1 | |||||||
| ISMbu1 | NC_007955 | 1,141 | 17/22 | 4 | 1 | M. burtonii | 22 | 7 | |
| ISArch6 | AY714845 | 1,260 | 15/19 | 4 | 1 | Uncultured archeon | |||
| TVN1409 | NC_002689 | T. volcanium | 1 | ||||||
| ISH1 subgroup | |||||||||
| ISH9 | NC_002608 | 938 | 16 | 0 | 1 | Halobacterium sp. pNRC200 | 2 | ||
| NC_001869 | Halobacterium sp. pNRC100 | 2 | |||||||
| NC_002607 | Halobacterium sp. | X | |||||||
| ISH28 | X59158 | 938 | 16 | H. salinarium | 1 | ||||
| ISH1 | NC_002607 | 1,118 | 8/9 | 8 | 1 | Halobacterium sp. | 1 | ||
| NC_006396 | H. marismortui chr I | 1 | |||||||
| S78775 | H. salinarium | 1 | |||||||
| ISHma8 | NC_006396 | 937 | 16/21 | 8 | 1 | H. marismortui chr I | 1 | ||
| ISH19 | NC_006393 | 936 | 17/21 | 7 | 1 | H. marismortui pNG500 | 2 | ||
| NC_006395 | H. marismortui pNG700 | 1 | |||||||
| ISHma9 | NC_006396 | 921 | 10/11 | 0 | 1 | H. marismortui chr I | 1 | 1 | |
| ISHma10 | NC_006392 | 935 | 14/15 | 8 | 1 | H. marismortui pNG400 | 1 | ||
| ISHma11 | NC_006393 | 927 | 14/15 | 10 | 1 | H. marismortui pNG500 | 1 | ||
| ISNph4 | NC_007428 | 927 | 13/14 | 0 | 1 | N. pharaonis PL131 | 1 | ||
| NC_007426 | N. pharaonis | 1 IR | |||||||
| Sulfolobus subgroup | |||||||||
| ISSto3 | NC_003106 | 1,317 | 24/26 | 4 | 1 | S. tokodaii | 6 | 11 | X |
| ISC1234 | NC_002754 | 1,228 | 24/28 | 2 | 1 | S. solfataricus | 15 | 3 | |
| ISC1290 | NC_002754 | 1,286 | 34/36 | 0 | 1 | S. solfataricus | 3 | ||
| ISC1212 | NC_002754 | 1,213 | 19/24 | 4 | 2 | S. solfataricus | 10 | 5 | |
| IS5 orphans | |||||||||
| ISMbu10 | NC_007955 | 789 | 16/17 | 0 | 1 | M. burtonii | 2 | 19 + 29 | X |
| NC_003552 | M. acetivorans | 4 IRs | |||||||
| NC_003901 | M. mazei | 2 IRs | |||||||
| NC_007355 | M. barkeri | 1 IR | |||||||
| ISH11 | NC_002607 | 1,068 | 15 | 0-10 | 1 | Halobacterium sp. | 2 | ||
| NC_001869 | Halobacterium sp. pNRC100 | 2 | |||||||
| NC_002608 | Halobacterium sp. pNRC200 | 3 | |||||||
| ISHma6 | NC_006397 | 1,069 | 24/28 | 8 | 1 | H. marismortui chr II | 3 | ||
| NC_006393 | H. marismortui pNG500 | 1 | 4 + 1 IR | ||||||
| NC_007428 | N. pharaonis PL131 | 1 | |||||||
| NC_007426 | N. pharaonis | 1 | |||||||
| ISMba15 | NC_007355 | 1,251 | 11/16 | 0 | 1 | M. barkeri | 12 | ||
| NC_003901 | M. mazei | 1 | |||||||
| ISMhu10 | NC_007796 | 1,248 | 11 | 10 | 1 | M. hungatei | 1 | 2 | |
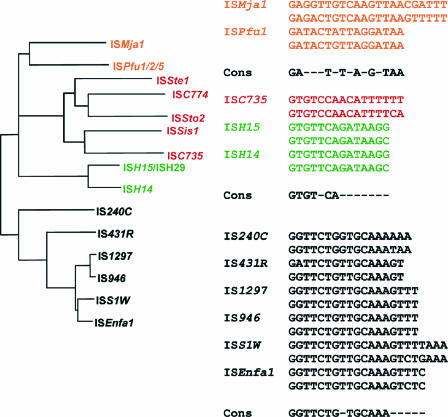
| IS6 family (Fig. 6) | |||||||||
| ISC735 | AY671942 | 735 | 15 | ND | 1 | Sulfolobus sp. | 1 | ||
| NC_002754 | S. solfataricus | 3 | |||||||
| ISC774 | NC_002754 | 778 | 16 | ND | 1 | S. solfataricus | 2 | 10 | |
| NC_007181 | S. acidocaldarius | 2 IRs | |||||||
| ISSto2 | NC_003106 | 851 | 33/34 | 5 | 1 | S. tokodaii | 4 | >13 | |
| NC_007181 | S. acidocaldarius | 1 | |||||||
| ISSte1 | NC_005969 | 746 | 16 | 8 | 1 | S. tokodaii pTC | 1 | ||
| ISSis1 | NC_006424 | 735 | 18 | 8 | 1 | S. islandicus pARN4 | 1 | ||
| ISH14 | NC_006396 | 696 | 14/15 | 8 | 1 | H. marismortui chr I | 1 | ||
| ISH15 | NC_006396 | 697 | 15/17 | 8 | 1 | H. marismortui chr I | 2 | ||
| NC_006393 | 9 | H. marismortui pNG500 | 1 | ||||||
| NC_007426 | N. pharaonis | IR | |||||||
| ISH17 | NC_006393 | 745 | 14 | 8 | 1 | H. marismortui pNG500 | 2 | 2 | |
| NC_006395 | H. marismortui pNG700 | 1 | |||||||
| NC_006397 | H. marismortui chr II | 1 | 1 | ||||||
| NC_002608 | Halobacterium sp. pNRC200 | 1 | |||||||
| NC_002607 | Halobacterium sp. | IR | |||||||
| ISH29 | NC_002608 | 697 | 16/17 | 8 | 1 | Halobacterium sp. pNRC200 | 1 | 4 | |
| NC_001869 | Halobacterium sp. pNRC100 | 4 | |||||||
| NC_002607 | Halobacterium sp. | 1 | |||||||
| ISNph1 | NC_007426 | 697 | 15/17 | 0 | 1 | N. pharaonis | 1 | ||
| ISMja1 | NC_000909 | 703 | 19/22 | 1 | M. jannaschii | 2 | 1 | X | |
| NC_001732 | M. jannaschii ECE | 3 | |||||||
| ISPfu1 | NC_003413 | 781 | 15/16 | 8 | 1 | P. furiosus | 8 | 1 | |
| NC_000868 | P. abyssi | 1 | |||||||
| ISPfu2 | NC_003413 | 782 | 15/16 | 0-8 | 1 | P. furiosus | 11 | 1 | |
| ISPfu5 | NC_003413 | 779 | 15/16 | 9 | 1 | P. furiosus | 4 | ||
| AF0138 | NC_000917 | A. fulgidus | |||||||
| AF0895 | NC_000917 | A. fulgidus | |||||||
| Mbar A0568 | NC_007355 | M. barkeri | |||||||
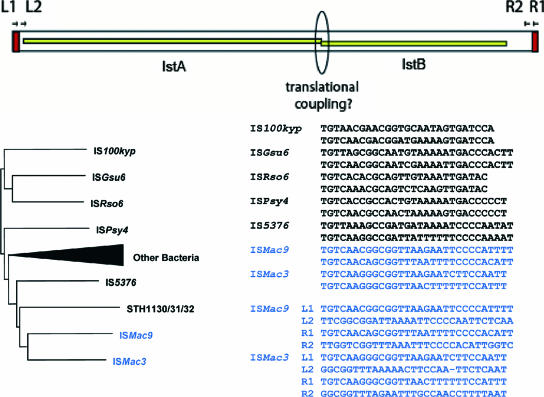
| IS21 family (Fig. 7) | |||||||||
| ISMac3 | NC_003552 | 2,199 | 16 | 4 | 2 | M. acetivorans | 11 | 1 | |
| NC_003901 | M. mazei | 1 | 1 | ||||||
| ISMac9 | NC_003552 | 2,199 | 21/26 | 4 | 2 | M. acetivorans | 7 | 1 | |
| Mbar A2360 | NC_007355 | M. barkeri | |||||||
| IS30 family | |||||||||
| ISC1041 | U85710 | 1,038 | 16/18 | S. solfataricus | |||||
| IS110 family (Fig. 8) | |||||||||
| ISC1190 | NC_002754 | 1,180 | 0 | 6 | 1 | S. solfataricus | 10 | 22 | |
| ISC1229 | NC_002754 | 1,232 | 0 | 0 | 1 | S. solfataricus | 2 | 7 | |
| ISC1228 | NC_002754 | 1,228 | 0 | 0 | 1 | S. solfataricus | 5 | 4 | |
| ISC1491 | NC_002754 | 1,403-1,494 | 0 | 0 | 1-2 | S. solfataricus | 4 | 3 | |
| ISSto4 | NC_003106 | 1,484 | 0 | 0 | 1 | S. tokodaii | 6 | 2 | |
| ISSto5 | NC_003106 | 1,191 | 0 | 0 | 1 | S. tokodaii | 3 | 4 | |
| ISSto6 | NC_003106 | 1,246 | 0 | 0 | 1 | S. tokodaii | 2 | 0 | |
| ISMac14 | NC_003552 | 1,534 | 9/12 | 3 | 1 | M. acetivorans | 4 | 23 | |
| ISMma5 | NC_003901 | 1,522 | 10/15 | 3 | 1 | M. mazei | 8 | 26 | |
| ISMba7 | NC_007355 | 1,522 | 0 | 0 | 1 | M. barkeri | 1 | ||
| ISMba20 | NC_007355 | 1,519 | 0 | 0 | 1 | M. barkeri | 1 | ||
| ISH18 | NC_006393 | 1,548 | 0 | 0 | 1 | H. marismortui pNG500 | 1 | 1 | |
| ISFac9 | 1,487 | 0 | 0 | 1 | F. acidarmanus | ||||
| Mhun 0755 | NC_007796 | M. hungatei | |||||||
| IS256 family (Fig. 9) | |||||||||
| ISC1250 | NC_002754 | 1,261 | 26/27 | 9 | 1 | S. solfataricus | 1 | 2 | |
| ISC1257 | NC_002754 | 1,257 | 1 | S. solfataricus | 1 | 12 | |||
| ISC1332 | NC_002754 | 1,332 | 19/23 | 9 | 1 | S. solfataricus | 1 | ||
| NC_006493 | Sulfolobus sp. pNOB8 | 1 | |||||||
| ISMma16 | NC_003901 | 1,268 | 21/25 | 8 | 1 | M. mazei | 4 | 2 | |
| NC_003552 | M. acetivorans | 6 | |||||||
| ISMba9 | NC_007355 | 1,268 | 21/27 | Pseudo | M. barkeri | 1 | 8 | ||
| ISMbu6 | NC_007955 | 1,252 | 22/27 | 8 | 1 | M. burtonii | 4 | ||
| ISFac7 | 1,254 | 31/39 | 8 | 1 | F. acidarmanus | ||||
| ISFac8 | 1,291 | 23/31 | 1 | F. acidarmanus | |||||
| ISTac2 | NC_002578 | 1,154 | 13/23 | Pseudo | T. acidophilum | ||||
| TVN0870 | NC_002689 | T. volcanium | |||||||
| TVN1468 | NC_002689 | T. volcanium | |||||||
| IS481 family (Fig. 10) | |||||||||
| ISMac4 | NC_003552 | 1,029 | 21/25 | 15 | 1 | M. acetivorans | 19 | 21 | |
| ISA0963-1 | NC_000909 | 963 | 20/24 | 15 | 1 | A. fulgidus | 1 | 3 | |
| ISA0963-2 | NC_000909 | 963 | 21/25 | 15 | 1 | A. fulgidus | 1 | ||
| ISA0963-3 | NC_000909 | 963 | 24/31 | 15 | 1 | A. fulgidus | 4 | ||
| ISA0963-7 | NC_000909 | 963 | 20/26 | 0 | 1 | A. fulgidus | 1 | ||
| ISFac5 | 972 | 23/27 | ND | 1 | F. acidarnus | ||||
| ISTvo3 | NC_002689 | 951 | 0 | 1 | T. volcanium | 1 | |||
| Ta1408 | NC_002578 | T. acidophilum | |||||||
| IS630 family (Fig. 11) | |||||||||
| ISHma2 | NC_006396 | 1,153 | 16/22 | TA | 1 | H. marismortui chr I | 1 | ||
| ISH16 | NC_006393 | 1,148 | 20/25 | TA | 1 | H. marismortui pNG500 | 2 | 2 | |
| NC_002607 | Halobacterium sp. | 1 | |||||||
| ISC1048 | NC_002754 | 1,044 | 18/22 | TA | 1 | S. solfataricus | 10 | 4 | |
| NC_003106 | S. tokodaii | IR | |||||||
| NC_007181 | S. acidocaldarius | IR | |||||||
| ISC1078 | NC_002754 | 1,086 | 30/31 | TA | 1 | S. solfataricus | 8 | 6 | |
| ISC1395 | NC_002754 | 1,392 | 87/92 | TA | 1 | S. solfataricus | 3 | 13 | |
| NC_003106 | S. tokodaii | 3 IR | |||||||
| ISMma6 | NC_003901 | 1,093 | 20/29 | TA | 2 | M. mazei | 1 | ||
| ISMma8 | NC_003901 | 1,176 | 23/30 | 4 | 2 | M. mazei | 1 | ||
| NC_007355 | M. barkeri | 6 IR | |||||||
| ISMma9 | NC_003901 | 1,200 | 15/22 | 4 | 2 | M. mazei | 6 | 1 | |
| NC_007355 | M. barkeri | 3 | |||||||
| ISMma10 | NC_003901 | 1,171 | 23/30 | 4 | 2 | M. mazei | 2 | 4 | |
| ISMma17 | NC_003901 | 1,075 | 22/27 | TA | 2 | M. mazei | 2 | ||
| NC_003552 | M. acetivorans | 1 | |||||||
| NC_007355 | M. barkeri | 1 | |||||||
| ISMac13 | NC_003552 | 1,284 | 19/21 | 4 | 1 | M. acetivorans | 1 | 2 | X |
| ISMac17 | NC_003552 | 1,170 | 20/26 | 4 | 1 | M. acetivorans | 3 | ||
| ISMac18 | NC_003552 | 1,200 | 15/23 | 4 | 2 | M. Acetivorans | 2 | ||
| ISMba3 | NC_007355 | 1,169 | 18/27 | 4 | 3 | M. barkeri | 1 | ||
| ISMba10 | NC_007355 | 1,095 | 18/24 | TA | 2 | M. barkeri | 1 | ||
| ISMth1 | 1,188 | 20/25 | 4 | 2 | M. thermophila | 1 | |||
| ISA1083-1 | NC_000917 | 2 | A. fulgidus | 1 | |||||
| ISA1083-2 | NC_000917 | 2 | A. fulgidus | 2 | |||||
| ISArch3 | AY714829 | 1,077 | 20/27 | TA | 1 | Uncultured archeon | |||
| ISArch4 | AY714840 | 1,212 | 19/24 | 4 | 2 | Uncultured archeon | |||
| rmAC1575 | NC_006396 | H. marismortui chr I | 1 | ||||||
| Mbur_0848 | NC_007955 | M. burtonii | |||||||
| rmAC1575 | NC_006396 | H. marismortui chr I | |||||||
| TVN1411 | NC_002689 | 1 | T. volcanium | ||||||
| PTO1017 | NC_005877 | P. torridus | |||||||
| PTO0855 | NC_005877 | P. torridus | |||||||
| PTO1049 | NC_005877 | P. torridus | |||||||
| IS982 family (Fig. 12) | |||||||||
| ISPfu3 | NC_003413 | 933 | 15/16 | 10 | 1 | P. furiosus | 5 | ||
| ISL3 family | |||||||||
| ISMac21 | NC_003552 | 1,327 | 22/26 | 8 | 1 | M. acetivorans | 2 | 14 | |
| NC_007355 | M. barkeri | 13 | |||||||
| NC_003901 | M. mazei | 1 | |||||||
| ISMbu4 | NC_007955 | 1,274 | 22/27 | 1 | M. burtonii | 2 | 3 | ||
| ISArch5 | AY714859 | 1,285 | 19/26 | 1 | Uncultured archeon | 1 | |||
| TVN0466 | NC_002689 | T. volcanium | |||||||
| TVN0686 | NC_002689 | T. volcanium | |||||||
| TVN0687 | NC_002689 | T. volcanium | |||||||
| TVN0688 | NC_002689 | T. volcanium | |||||||
| Non-DDE transposons | |||||||||
| IS91 family | |||||||||
| ISMbu9 | NC_007955 | 1,281 | 1 | M. burtonii | 2 | ||||
| IS200/IS605/IS607 family (Fig. 13) | |||||||||
| IS200 subgroup | |||||||||
| ISMma21 | NC_003901 | 725 | 0 | 0 | 1 | M. mazei | 9 | 3 | X |
| ISMba16 | NC_007355 | 728 | 0 | 0 | 1 | M. barkeri | 5 | 7 | X |
| ISMba18 | NC_007355 | 624 | 0 | 0 | 1 | M. barkeri | 4 | 2 | X |
| NC_003552 | M. acetivorans | 0 | 11 | X | |||||
| NC_003901 | M. mazei | 0 | 2 | X | |||||
| NP4630A | NC_007426 | N. pharaonis | |||||||
| IS605 subgroup | |||||||||
| ISC1476 | NC_002754 | ND | 0 | 0 | 2 | S. solfataricus | 2 | ||
| ISSto1 | NC_003106 | 1,793 | 0 | 0 | 2 | S. tokodaii | 7 | 3 | X |
| NC_007181 | S. acidocaldarius | 7 | |||||||
| ISSis2 | NC_006425 | 1,788 | 0 | 0 | 2 | S. islandicus pHVE14 | 1 | ||
| ISH12 | NC_002607 | 1,899 | 0 | 0 | 2 | Halobacterium sp. | 1 | 2 | |
| NC_001869 | Halobacterium sp. pNRC100 | 1 | |||||||
| NC_002608 | Halobacterium sp. pNRC200 | 1 | 3 | ||||||
| NC_006391 | H. marismortui pNG300 | 1 | |||||||
| ISH1-8 | Iso-ISH12 | ||||||||
| ISH22 | NC_002607 | 1,726 | 0 | 0 | 2 | Halobacterium sp. | 1 | ||
| NC_007427 | N. pharaonis PL131 | 1 | |||||||
| ISHma7 | NC_006396 | 2,008 | 0 | 0 | 2 | H. marismortui chr I | 1 | ||
| ISHma12 | NC_006393 | 1,672 | 0 | 0 | 2 | H. marismortui pNG500 | 1 | ||
| ISMac7 | NC_003552 | 1,711 | 0 | 0 | 2 | M. acetivorans | 3 | 14 | |
| ISMma19 | NC_003901 | 1,680 | 0 | 0 | 2 | M. mazei | 1 | 6 | X |
| ISMma22 | NC_003901 | 1,658 | 0 | 0 | 2 | M. mazei | 2 | 0 | |
| ISMba17 | NC_007355 | 1,708 | 0 | 0 | 2 | M. barkeri | 6 | 14 | |
| NC_007349 | M. barkeri plasmid 1 | 1 | 2 | ||||||
| ISTac1 | NC_002578 | 1,571 | 0 | 0 | 2 | T. acidophilum | 1 | 3 | |
| ISTvo5 | NC_002689 | 1,790 | 0 | 0 | 2 | T. volcanium | 1 | 2 | |
| Mbar_A2836 | NC_007355 | M. barkeri | |||||||
| TVN0750 | NC_002689 | T. volcanium | |||||||
| TK0931/32 | NC_006624 | T. kodakarensis | |||||||
| NP3908A/10A | NC_007424 | N. pharaonis | |||||||
| NP4810A/12A | NC_007424 | N. pharaonis | |||||||
| IS607 subgroup | |||||||||
| ISC1926 | AY671948 | 1,925 | 0 | 0 | 2 | Sulfolobus sp. L100 | 1 | ||
| ISC1913 | NC_002754 | 1,913 | 0 | 0 | 2 | S. solfataricus | 2 | 4 | |
| ISC1904 | NC_002754 | 1,903 | 0 | 0 | 2 | S. solfataricus | 7 | 9 | |
| ISC1778 | NC_002754 | Iso-ISC1904 | S. solfataricus | ||||||
| ISSto11 | NC_003106 | 1,727 | 0 | 0 | 2 | S. tokodaii | 1 | ||
| ISSto12 | NC_003106 | 1,956 | 0 | 0 | 2 | S. tokodaii | 2 | 6 | X |
| ISSto13 | NC_003106 | 1,928 | 0 | 0 | 2 | S. tokodaii | 1 | X | |
| IS1921 | X56616 | 1,922 | 0 | 0 | 2 | A. ambivalens | |||
| ISTko1 | NC_006624 | 1,960 | 0 | 0 | 2 | T. kodakarensis | 1 | ||
| ISPfu4 | NC_003413 | 1,961 | 0 | 0 | 2 | P. furiosus | 1 | 1 | |
| ISTvo1 | NC_002689 | 1,897 | 0 | 0 | 2 | T. volcanium | 1 | 3 | |
| SSO0836 | NC_002754 | S. solfataricus | |||||||
| Saci_2022/23 | NC_007181 | S. acidocaldarius | |||||||
| PF1985/86 | NC_003413 | P. furiosus | |||||||
| PAB2076/77 | NC_000868 | P. abyssi | |||||||
| TK1841/42 | NC_006624 | T. kodakarensis | |||||||
| MJ0012m/14 | NC_000909 | M. jannaschii | |||||||
| Single orfB elements | |||||||||
| ISC1316 | NC_002754 | 1,315-1,323 | 0 | 0 | 1 | S. solfataricus | 12 | 7 | |
| SSO0008 | NC_002754 | S. solfataricus | |||||||
| SSO0794 | NC_002754 | S. solfataricus | |||||||
| SSO0801 | NC_002754 | S. solfataricus | |||||||
| SSO0842 | NC_002754 | S. solfataricus | |||||||
| ST0152 | NC_003106 | S. tokodaii | |||||||
| ST0998 | NC_003106 | S. tokodaii | |||||||
| ST1777/1778 | NC_003106 | S. tokodaii | |||||||
| ST2008 | NC_003106 | S. tokodaii | |||||||
| ST2431 | NC_003106 | S. tokodaii | |||||||
| Saci_0269 | NC_007181 | S. acidocaldarius | |||||||
| Saci_1941 | NC_007181 | S. acidocaldarius | |||||||
| ORF355/245 | NC_006422 | S. islandicus pKEF9 | |||||||
| ORF191/309 | AJ748324 | S. islandicus pHVE14 | |||||||
| rmAC0534 | NC_006396 | H. marismortui chr I | 1 | ||||||
| rmAC1085 | NC_006396 | H. marismortui chr I | 1 | ||||||
| rmAC1559 | NC_006396 | H. marismortui chr I | 1 | ||||||
| rmAC1565 | NC_006396 | H. marismortui chr I | 1 | ||||||
| rmAC1588 | NC_006396 | H. marismortui chr I | 1 | ||||||
| rmAC1676 | NC_006396 | H. marismortui chr I | 1 | ||||||
| pNG3034 | NC_006391 | H. marismortui pNG300 | 1 | ||||||
| pNG5130 | NC_006393 | H. marismortui pNG500 | 1 | ||||||
| pNG5139 | NC_006393 | H. marismortui pNG500 | 1 | ||||||
| VNG0013C | NC_002607 | Halobacterium sp. | |||||||
| VNG0026G | NC_002607 | Halobacterium sp. | |||||||
| VNG0042G | NC_002607 | Halobacterium sp. | |||||||
| VNG2652H | NC_002607 | Halobacterium sp. | |||||||
| VNG6381H | NC_002608 | Halobacterium sp. pNRC200 | |||||||
| VNG6361G | NC_002608 | Halobacterium sp. pNRC200 | |||||||
| NP0294A | NC_007426 | N. pharaonis | |||||||
| NP0460A | NC_007426 | N. pharaonis | |||||||
| NP0706A | NC_007426 | N. pharaonis | |||||||
| NP0974A | NC_007426 | N. pharaonis | |||||||
| NP1210A | NC_007426 | N. pharaonis | |||||||
| NP1696A | NC_007426 | N. pharaonis | |||||||
| NP1714A | NC_007426 | N. pharaonis | |||||||
| NP1802A | NC_007426 | N. pharaonis | |||||||
| NP1814A | NC_007426 | N. pharaonis | |||||||
| NP1856A | NC_007426 | N. pharaonis | |||||||
| NP1890A | NC_007426 | N. pharaonis | |||||||
| NP2130A | NC_007426 | N. pharaonis | |||||||
| NP2528A | NC_007426 | N. pharaonis | |||||||
| NP2942A | NC_007426 | N. pharaonis | |||||||
| NP3036A | NC_007426 | N. pharaonis | |||||||
| NP3392A | NC_007426 | N. pharaonis | |||||||
| NP3590A | NC_007426 | N. pharaonis | |||||||
| NP3712A | NC_007426 | N. pharaonis | |||||||
| NP4280A | NC_007426 | N. pharaonis | |||||||
| NP4358A | NC_007426 | N. pharaonis | |||||||
| NP4546A | NC_007426 | N. pharaonis | |||||||
| NP4572A | NC_007426 | N. pharaonis | |||||||
| NP4634A | NC_007426 | N. pharaonis | |||||||
| NP4636A | NC_007426 | N. pharaonis | |||||||
| NP4742A | NC_007426 | N. pharaonis | |||||||
| NP4782A | NC_007426 | N. pharaonis | |||||||
| NP5054A | NC_007426 | N. pharaonis | |||||||
| NP5060A | NC_007426 | N. pharaonis | |||||||
| NP61114A | NC_007427 | N. pharaonis PL131 | |||||||
| NP6134A | NC_007427 | N. pharaonis PL131 | |||||||
| NP6142A | NC_007427 | N. pharaonis PL131 | |||||||
| NP6150A | NC_007427 | N. pharaonis PL131 | |||||||
| NP6178A | NC_007427 | N. pharaonis PL131 | |||||||
| NP6224A | NC_007427 | N. pharaonis PL131 | |||||||
| NP6268A | NC_007427 | N. pharaonis PL131 | |||||||
| MK0605 | NC_003551 | M. kandleri | |||||||
| MJ0751 | NC_000909 | M. jannaschii | |||||||
| MJ1635 | NC_000909 | M. jannaschii | |||||||
| MM0579 | NC_003901 | M. mazei | |||||||
| MM0766 | NC_003901 | M. mazei | |||||||
| Mbur_0800 | NC_007955 | M. burtonii | |||||||
| Mbur_2016 | NC_007955 | M. burtonii | |||||||
| Mbur_2253 | NC_007955 | M. burtonii | |||||||
| Mbur_1650 | NC_007955 | M. burtonii | |||||||
| Mbur_2248 | NC_007955 | M. burtonii | |||||||
| Mbur_A0319 | NC_007355 | M. barkeri | |||||||
| Mbar_A1230 | NC_007356 | M. barkeri | |||||||
| Mbar_A3217 | NC_007355 | M. barkeri | |||||||
| Mbar_A1973 | NC_007355 | M. barkeri | |||||||
| Mbar_B3751 | NC_007349 | M. barkeri plasmid 1 | |||||||
| Msp_1478 | NC_007681 | M. stadtmanae | |||||||
| Ta0381 | NC_002578 | T. acidophilum | 1 | ||||||
| Ta1471/72 | NC_002578 | T. acidophilum | 1 | ||||||
| TVN0006 | NC_002689 | T. volcanium | |||||||
| TVN0248 | NC_002689 | T. volcanium | |||||||
| TVN0323 | NC_002689 | T. volcanium | |||||||
| TVN0952 | NC_002689 | T. volcanium | |||||||
| TVN1187 | NC_002689 | T. volcanium | |||||||
| TVN0712/13/14 | NC_002689 | T. volcanium | |||||||
| TVN0770 | NC_002689 | T. volcanium | |||||||
| TVN0903 | NC_002689 | T. volcanium | |||||||
| PF0759/760 | NC_003413 | P. furiosus | |||||||
| PF1015 | NC_003413 | P. furiosus | |||||||
| PF1609 | NC_003413 | P. furiosus | |||||||
| PF1918 | NC_003413 | P. furiosus | |||||||
| PH0585 | NC_000961 | P. horikoshii | |||||||
| PH0630 | NC_000961 | P. horikoshii | |||||||
| PAB1802 | NC_000868 | P. abyssi | |||||||
| PAB1452 | NC_000868 | P. abyssi | |||||||
| TK0298 | NC_006624 | T. kodakarensis | |||||||
| TK0495 | NC_006624 | T. kodakarensis | |||||||
| TK0850 | NC_006624 | T. kodakarensis | |||||||
| Emerging families, orphans, waifs, and strays | |||||||||
| ISA1214 family | |||||||||
| ISA1214-1 | NC_000917 | 1,214 | 18/21 | 9-12 | 2 | A. fulgidus | 5 | ||
| ISA1214-6 | NC_000917 | 1,214 | 14/19 | 8 | 2 | A. fulgidus | 1 | ||
| ISFac3 | 1,270 | 17 | 9 | 2 | F. acidarmanus | ||||
| ISTvo2 | NC_002689 | 1,201 | 14/18 | 0 | 2 | T. volcanium | 1 | ||
| ISC1043 | NC_002754 | 1,043 | 13/14 | 0 | S. solfataricus | 1 | 7 | ||
| TVN1041 | NC_00268 | T. volcanium | |||||||
| MJ0362 | NC_000909 | M. jannaschii | |||||||
| MMP0468 | NC_005791 | M. maripaludis | |||||||
| MMP0751 | NC_005791 | M. maripaludis | |||||||
| Mhun 2372 | NC_007796 | M. hungatei | |||||||
| ISM1 group | |||||||||
| ISM1 | XO2587 | 1,381 | 33/34 | 8 | 1 | M. smithii | |||
| ISMst1 | NC_007681 | 1,529 | 24/26 | 8 | 1 | M. stadtmanae | 3 | 2 | |
| ISMbu2 | NC_007955 | 1,310 | 25 | 1 | M. burtonii | 2 | 2 | ||
| ISMma11 | NC_003901 | 1,619 | 19/24 | 9 | 1 | M. mazei | 6 | X | |
| ISMac19 | NC_003552 | 1,624 | 19/24 | 8 | 1 | M. acetivorans | 8 | 7 | |
| ISMba4 | NC_007355 | 1,621 | 20/24 | 8 | Pseudo | M. barkeri | 1 | 14 | X |
| IS1595 group | |||||||||
| ISH4 | NC_002607 | 1,004 | 23/29 | 8 | 1 | Halobacterium sp. | 1 | ||
| NC_001869 | Halobacterium sp. pNRC100 | 1 | |||||||
| ISH50 | XO1584 | 996 | 23/29 | 8 | 1 | H. salinarium | 1 | ||
| ISHma4 | NC_006392 | 1,001 | 8 | 1 | H. marismortui pNG400 | 1 | |||
| NC_006396 | H. marismortui chr I | 1 | |||||||
| NC_006397 | H. marismortui chr II | 1 | |||||||
| ISNph2 | NC_007426 | 1,003 | 21/25 | 8 | 1 | N. pharaonis | 1 | ||
| NC_007427 | N. pharaonis PL131 | 1 | |||||||
| PAB2064 | NC_000868 | P. abyssi | 1 | ||||||
| PH1854 | NC_000961 | P. horikoschii | 1 | ||||||
| NP6042A | NC_007427 | Pseudo | N. pharaonis PL131 | 1 | |||||
| ISBst12 group | |||||||||
| ISH10 | NC_002607 | 1,584 | 16/18 | 8 | 1 | Halopacterium sp. | 1 | ||
| NC_001869 | Halobacterium sp. pNRC100 | 2 | |||||||
| NC_002608 | Halobacterium sp. pNRC100 | 2 | |||||||
| ISH10B | NC_002607 | 1,629 | 14/15 | 8 | 1 | Halobacterium sp. | 1 | ||
| ISMac8 | NC_003552 | 1,603 | 13/15 | 8 | 1 | M. acetivorans | 3 | 2 | |
| ISMhu3 | NC_007796 | 1,727 | 9/12 | 0 | 2 | M. hungatei | 2 | 2 | |
| ISMma13 | NC_003901 | 1,534 | 20/28 | 8 | 1 | M. mazei | 4 | 1 | |
| ISMma14 | NC_003901 | 1,529 | 22/30 | 8 | 1 | M. mazei | 9 | ||
| ISMma15 | NC_003901 | 1,560 | 17/19 | 8 | 1 | M. mazei | 2 | ||
| ISMbu5 | NC_007955 | 1,696 | 18/19 | 8 | 1 | M. burtonii | 9 | 10 | |
| ISArch7 | CR937010 | 1,839 | 20/24 | 8 | 2 | Uncultured archeon | |||
| Mhun1220 | NC_007796 | M. hungatei | 1 | ||||||
| TVN0684 | NC_002689 | T. volcanium | |||||||
| IS1182 family | |||||||||
| ISMac1 | NC_003552 | 1,668 | 27/38 | 4 | 1 | M. acetivorans | 5 | 4 | |
| ISMac2 | NC_003552 | 1,707 | 35/41 | 4 | 1 | M. acetivorans | 1 | ||
| ISMac20 | NC_003552 | 1,723 | 26/29 | Pal | 1 | M. acetivorans | 5 | 8 | |
| NC_007355 | M. barkeri | 11 | |||||||
| NC_003901 | M. mazei | 4 | |||||||
| ISMma2 | NC_003901 | 1,669 | 30/39 | 5 | 1 | M. mazei | 3 | ||
| ISMhu1 | NC_007796 | 1,742 | 15/17 | 4 | 1 | M. hungatei | 7 | 2 | |
| ISMhu2 | NC_007796 | 1,760 | 14/16 | 4 | 1 | M. hungatei | 18 | 1 | |
| ISArch1 | AY714861 | 1,736 | 16/17 | 4 | 1 | Uncultured archeon | |||
| ISArch2 | AY714861 | 1,924 | 14 | 4 | 1 | Uncultured archeon | |||
| ISH6 family | |||||||||
| ISH6 | NC_002607 | 1,448 | 26/27 | 8 | 1 | H. salinarium | 1 | ||
| NC_002608 | Halobacteria sp. pNRC200 | 1 | |||||||
| ISHs1 | M38315 | 1,449 | 24/27 | 8 | 1 | H. salinarium S1 | 1 | ||
| ISHma5 | NC_006393 | 1,449 | 23/27 | 8 | 1 | H. marismortui pNG500 | 1 | ||
| NC_006389 | H. marismortui pNG100 | 1 | |||||||
| ISNph3 | NC_007427 | 1,449 | 26/27 | 0 | N. pharaonis PL131 | 1 | |||
| AF0120 | NC_000909 | A. fulgidus | 1 | ||||||
| AF0828 | NC_000909 | A. fulgidus | 1 | ||||||
| ISC1217 family | |||||||||
| ISC1217 | NC_002754 | 1,147 | 10/13 | 6 | 1 | S. solfataricus | 11 | 11 | |
| ISC1205 | AY671946 | 1,205 | 17/18 | ND | 1 | Sulfolobus sp. L00 24 | 1 | ||
| ISSto10 | NC_003106 | 1,145 | 14/23 | 7 | 1 | S. tokodaii | 3 | 17 | X |
| Orphans | |||||||||
| ISH7 | AF016485 | 3,302 | 14/19 | 4 | Halobacterium sp. | ||||
| ISMbu11 | NC_007955 | 1,278 | 18 | 5 | 1 | M. burtonii | 6 | 3 | |
| Others | |||||||||
| Ta1443 | NC_002578 | T. acidophium | |||||||
| MMP0766 | NC_005791 | M. maripaludis | |||||||
Where no accession number is given, the genome has not been sequenced completely. Abbreviations: Pal, palindrome; C, complete copies; P, partial copies; ND, not determined; chr, chromosome.
For IS630, the target duplication is shown as a TA dinucleotide. Pseudo, pseudogene.
TABLE 3.
Archaeal MITEs
| Organism | MITE | IS family | Length (bp) | Copy no. (partial) | IR (bp) | DR (bp) | Potential partner IS |
|---|---|---|---|---|---|---|---|
| S. solfataricus | SM1 | S630 | 79 | 40 | 23 | 2 | ISC1048 |
| SM2 | ISC1217 | 184 | 25 | 16 | 6 | ISC1217 | |
| SM3A/SM3B | IS5 | 131 | 44 | 24 | 9 | ISC1058 | |
| SM4 | IS1 | 164 | 34 | 27 | 8 | ISC1173 | |
| SM7 | IS1 | 330 | 10 | 13 | 0 | ISC1173 | |
| S. tokodaii | SM1 | IS630 | 79 | 1 | 23 | 2 | ISC1048a |
| SM2 | ISC1217 | 184 | 36 | 16 | 6 | ISSto10 | |
| SM5 | IS6 | 212 | 7 | 15 | 0 | ISC774 | |
| SM6 | IS1 | 127 | 8 | 12 | 0-10 | ISC794 | |
| IS1 | 315 | 1 | 14 | 0 | ISSto7 | ||
| 317 | 1 | 14 | |||||
| SMA | IS605 | 355 | 9 | 4 | 0 | ISSto1 | |
| IS607 | 274 | 1 | 0 | 0 | ISSto12 | ||
| IS607 | 266-271 | 3 | 0 | 0 | ISSto13 | ||
| IS5 | 329-332 | 4 | ISSto3 | ||||
| Halobacterium sp. NRC | IS5 | 180 | 19-23 | 8 | ISH9 | ||
| H. marismortui | IS4 | 277 | 1 | ISH20 | |||
| 287 | 1 | ||||||
| M. mazei | ISM1 | 199 | 3 (1) | 24 | 8 | ISMma11 | |
| IS200 | 248 | 2 | 0 | 0 | ISMma21 | ||
| IS200 | 209 | 1 | 0 | 0 | ISMba18 | ||
| IS605 | 206 | 1 | 0 | 0 | ISMma19 | ||
| M. barkeri | IS4 | 241 | 18 (13) | 18-22 | 3 | ISMba11 | |
| IS4 | 278 | 8 (3) | 21-22 | 6 | ISMba12 | ||
| IS5 | 131-152 | 3 | 3 | ISMba5 | |||
| ISM1 | 193-302 | 7 (11) | 24 | 8-9 | ISMba4 | ||
| IS200 | 272 | 7 | 0 | 0 | ISMba16 | ||
| 248 | 1 (1) | ||||||
| 195 | 1 | ||||||
| IS200 | 209 | 3 (1) | 0 | 0 | ISMba18 | ||
| 55 | 5 | ||||||
| 174 | 2 | ||||||
| 178 | 1 | ||||||
| M. acetivorans | IS5 | 130-131 | 30 (10) | 15-18 | 0-3 | ISMac11 | |
| IS630 | 172 | 15 (1) | 21 | 0 | ISMac13 | ||
| IS4 | 241 | 9 (2) | 18-22 | 3 | ISMba11 | ||
| IS200 | 209 | 4 (5) | 0 | 0 | ISMba18 | ||
| 65 | 1 | ||||||
| M. jannaschii | IS6 | 358-360 | 8 | 0 | 0 | ISMja1 | |
| MJRE1 | 108-115 | 59 | 17 | 0 | |||
| MJRE2 | 96-99 | 69 | 16 | 0 | |||
| MJRE3 | 101-105 | 13 | 15 | 0 | |||
| M. burtoni | IS5 | 185 | 1 | 18 | 0 | ISMbu10 | |
| A. pernix | 306-310 | 8 (1) | 0 | 0 |
The corresponding IS element is absent from the host.
IS1
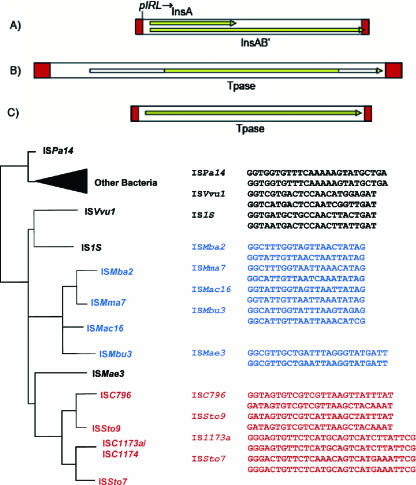
The IS1 family (Fig. 3) was thought to be restricted to the Enterobacteriaceae, but examples were subsequently found in several cyanobacteria. Bacterial IS1 family members are short (700 to 800 bp), bordered by highly conserved 15- to 24-bp IRs, and they generate 8- or 9-bp DRs on insertion. They generally carry two reading frames, insA and insB′ (Fig. 3A, top), although bacterial derivatives that carry only a single long frame (ISAba3 from Acinetobacter baumannii and possibly ISPa14 from Pseudomonas aeruginosa) have now been identified. However, these have yet to be demonstrated as active. The Tpase termination codon is often located within the distal IR. Expression of the IS1 family Tpases generally occurs by a programmed −1 translational frameshift between the two consecutive ORFs. This fuses the product of the upstream frame insA with that of the downstream frame (insAB′) to generate the Tpase as a fusion protein, InsAB′, which includes a catalytic DDE motif. InsAB′ also exhibits a zinc finger and a helix-turn-helix motif known to be important for Tpase binding (56, 89). InsA acts as a repressor, which binds to the IRs and regulates IS1 expression from the promoter partly included in the left end (IRL).
FIG. 3.
IS1 members. Shown is the phylogeny of the IS1 family and comparison of a representative set of terminal IRs. The top panel shows the general organization of members of this family. Red boxes indicate the terminal IRs. Yellow (or white) boxes within the larger IS box indicate ORFs (see the text). (A) Organization of the “classical” bacterial IS1. pIRL indicates the promoter, which drives Tpase synthesis. This class includes those from the archaeal methanogens. (B) The longer of the two Sulfolobus groups carries more-extensive IRs and N- and C-terminal extensions (white boxes) to the Tpase compared to the classical IS1 and the shorter Sulfolobus class. (C) Shorter Sulfolobus class. IRs are approximately the length of those found in the classical IS1 organization. The various Archaea have been color coded as follows for clarity: Sulfolobales, red; methanogens, blue. Bacteria are indicated in black.
Four IS1 members have been identified in the genomes of different Sulfolobus species. ISC1173a (S. solfataricus) and ISSto7 (S. tokodaii) (Fig. 3B, top) are closely related, as are ISC796 (Sulfolobus sp.) and ISSto9 (S. tokodaii) (Fig. 3C, top). Under our operational nomenclature, neither ISC1173a and ISSto7 nor ISSto9 and ISC796 are isoforms. Nevertheless the two pairs are phylogenetically closely related (91% and 84% amino acid identity, respectively). S. tokodaii carries both full-length and solo ISSto7 IRs, together with two complete small ISSto7-derived MITE-like elements (see “MITEs, MICs, and solo IRs,” below) with sizes of 315 and 317 bp. ISC796 is present as a single copy in Sulfolobus sp. and as several fragmented copies in S. solfataricus. There are both complete and partial copies of ISSto9 in S. tokodaii, as well as solo IRs.
All four Sulfolobus elements carry only a single long reading frame (although one ISSto9 copy appears to be degenerate, with an 8-bp deletion generating two ORFs). Although there is no ORF equivalent to insA, an upstream equivalent to InsA may be produced in these single ORF elements. This could occur, for example, by proteolysis of the larger Tpase or by frameshifting to create the smaller protein, as in Escherichia coli for dnaX (5).
ISC1173a and ISSto7 are significantly longer (1,173 and 1,174 bp) than other family members, with IRs of approximately 50 bp, over twice the length of other members of the family. Moreover, the Tpase is larger than that of ISC796, ISSto9, and other members of the family (∼340 aa compared to ∼240 aa) due to an 80-aa N-terminal extension and a 40-aa C-terminal extension (Fig. 3B, top). Both ISC796 and ISSto9 are 796 bp long, with IRs of 21 bp (Fig. 3C, top). DNA alignments show that the long and short ISs and the MITEs are clearly derived from a common ancestor, but their exact relationship is at present unclear.
Four additional IS1 family members, organized as a canonical eubacterial IS1 (Fig. 3A, top), are present in the Methanosarcinales: ISMac16 (Methanosarcina acetivorans); ISMma7 (M. mazei, M. barkeri, and Methanococcoides burtonii), ISMba2 (M. barkeri), and ISMbu3 (Methanococcoides burtonii). ISMac16, ISMma7, and ISMba2 are 740 bp long, with 24-bp IRs and 8- or 9-bp DRs. ISMbu3 (741 bp; 8-bp DRs) has IRs of only 15 bp. In contrast to the Sulfolobus IS1 members, these all carry the expected two ORFs. They are closely related elements, with 84 to 89% identity with respect to ISMac16. Inspection of their nucleic acid sequence reveals an appropriately placed stretch of eight A residues and raises the possibility that the Tpase is produced by transcriptional rather than translational frameshifting (3; O. Fayet, personal communication).
The Tpases of these elements are related to that of ISMae3 of the cyanobacterium Microcystis aeruginosa (Fig. 3; 89) and less closely to diverse IS1 elements of the γ-Proteobacteria, including IS1X and IS1S from E. coli and ISVvu1 from Vibrio vulnificus. The DDE catalytic motif and surrounding amino acid residues are also typical of this family. Finally, the terminal 23 to 30 bp are very similar to the IRs of the γ-proteobacterial and cyanobacterial IS1 elements and terminate with a highly conserved 5′-GGNNNTG (CANNNCC-3′). Where identified, the site of insertion is A+T rich.
IS3
The large IS3 family is widely distributed among Bacteria and forms an extremely coherent and highly related family characterized by lengths of between 1,200 and 1,550 bp; related terminal IRs of 20 to 40 bp terminating with 5′-TG… CA-3′; DRs of between 3 and 5 bp; two consecutive, partially overlapping reading frames, orfA and orfB, from which two proteins are expressed; and a strongly conserved DDE motif closely related to that of retroviral integrases. The product of the upstream frame, OrfA, acts as a regulatory protein, while the Tpase, OrfAB, is generated by programmed translational frameshifting as in IS1 (for a review, see reference 78).
A single, distantly related degenerate element has been identified in Thermoplasma volcanium (TVN0865/67 and TVN0691/92). Blast searches revealed a relationship with diverse bacterial IS3 elements such as ISAca1 of Acinetobacter calcoaceticus, ISSod2 of Shewanella oneidensis, and ISPg5 of Porphyromonas gingivalis. Multiple alignments of these reading frames suggested that TVN0865 and TVN0691 are truncated copies of the OrfA frame and that TVN0867 and TVG0898533 represent truncated versions of the OrfB frame lacking the first catalytic aspartic acid (D). The spacing between the second catalytic aspartic acid (D) and glutamic acid (E) is conserved (35 aa), and an arginine (R) is present 7 aa after the glutamic acid (E). No IRs or DRs could be found for these two archaeal elements. T. volcanium therefore apparently carries only partial copies of IS3 elements.
IS4
The IS4 superfamily (Fig. 4) (see Addendum in Proof) forms a vast, widespread, and extremely heterogeneous group of ISs in numerous prokaryote lineages. Previously it had been divided into five groups: IS231, IS4Sa, IS10, IS50, and IS1549 (12). However, as a result of an increasing number of ISs, much of this grouping is no longer appropriate and a reassessment is at present being undertaken. At present, Tribe analysis generates seven clusters. Three of these can be included in an IS4 superfamily. The four remaining clusters appear to define new emerging families (D. de Palmenaer and J. Mahillon, personal communication). Archaeal ISs are found in three distinct clusters. The ISH8 subgroup, included in the IS4 superfamily, is limited to the Archaea. The second group belongs to the emerging IS1634 family, while the third group, ISH3, which is also limited to the Archaea, forms a separate cluster.
FIG. 4.
IS4 members. Shown is the phylogeny of the different subgroups of the IS4 family and comparison of a representative set of terminal IRs. The various Archaea have been color coded as follows for clarity: Sulfolobales, red; Thermoplasmatales, magenta; halophiles, green; methanogens, blue. Bacteria are indicated in black.
ISH8 subgroup.
The ISH8 subgroup includes ISH26-1 and ISH26 from H. salinarium; ISH5, ISH8, and ISH8A to ISH8E from Halobacterium sp. and plasmids pNRC100 and pNRC200; ISHma1 from H. marismortui chromosomes I and II and plasmids pNG400 and pNG500; ISMba1 from M. barkeri; ISMba6 from M. barkeri and M. acetivorans; and ISMhu6 and ISMhu9 from M. hungatei. In addition, solo IRs of ISMba6 are found in M. acetivorans, M. barkeri, M. mazei, and M. thermophila. The ISH8 subgroup includes a 5′-CAT-3′ triad at the ends of the IR.
ISH26 was described as harboring two overlapping ORFs. Although the first has significant similarity to the putative Tpases of other IS4 family members (26% identity with IS231W over a 143-aa overlap), the second has only very limited similarity (in the region of the conserved E residue). Detailed analyses indicate, however, that several frameshifts could significantly increase this similarity. The first ORF is very closely related to the N-terminal end of the Tpase of ISH8. A reevaluation of the ISH26 DNA sequence is needed to clarify this issue.
It is interesting to note that all five copies of ISH5 are interrupted by ISH11 at an identical position. This suggests that the entire interrupted IS is capable of autonomous transposition.
IS1634 subgroup.
The IS1634 subgroup includes both bacterial and archaeal members. All archaeal members except ISFac6, from the incompletely sequenced F. acidarmanus, and ISTvo4, from T. volcanium, are restricted to methanogens. These include ISMac5, ISMac6, ISMac10, ISMac12, and ISMac23 from M. acetivorans; ISMba11, ISMba12, and ISMba13 from M. barkeri; ISMma3, ISMma4, and ISMma20 from M. mazei; ISMma18 from M. mazei, M. acetivorans, and M. barkeri; ISMhu4, ISMhu5, ISMhu7, and ISMhu8 from M. hungatei; and ISMth2 from M. thermophila. ISMba11 and ISMba12 also give rise to MITE derivatives (Table 3). An additional IS, ISArch8, has been identified in an uncultured environmental archaeon.
The IRs appear to be similar and begin with 5′CA or 5′CC. Short DRs generally of 5 or 6 bp are also present, but no similarities can be distinguished. Their presence, largely restricted to Methanosarcinales, could indicate horizontal acquisition of these elements from bacterial species by a common Methanosarcinales ancestor.
ISH3 subgroup.
The Archaea-specific subgroup ISH3 forms a separate cluster in Tribe analysis and can be further subdivided into two phylogenetic subgroups with BLAST. It includes ISH27 (an isoform of ISH40) from H. salinarium; ISH51 from Haloferax volcanii; ISH20 from Haloarcula marismortui; ISH3 from the Halobacterium sp. chromosome, pNRC100, and pNRC200; ISFac1 in the unfinished genome of Ferroplasma acidarmanus; ISC1200, ISC1225, ISC1359, and ISC1439A and ISC1439B (76% identity with ISC1439A) from S. solfataricus; ISSto8 and ISSto14 from S. tokodaii; ISMma1 from M. mazei; ISMba14 from M. barkeri and M. burtonii; and ISMbu7 and ISMbu8 from M. burtonii. ISMba14 was reconstructed in silico because it is interrupted by ISMba11. The ISH3 subgroup shares a conserved terminal 5′-CAG-3′ trinucleotide.
IS701 subgroup.
At present the IS701 cluster, which has emerged as a group separate from the IS4 family, contains a single example from the Archaea, ISMba8 (M. barkeri).
IS5
The IS5 superfamily (Fig. 5) is also a relatively heterogeneous group which had been divided into six or seven subgroups (12). It includes sequences from a large variety of Archaea. As is the case for the IS4 family, the IS5 family grouping is no longer appropriate and a reassessment is at present being undertaken. Archaeal IS5 elements are present in four of the bacterial groups (IS903, IS5, IS1031, and IS427). There are also two Archaea-specific groups (ISH1 and a Sulfolobus-specific group) and five IS5-related ISs that do not fall into any of these groups.
FIG. 5.
IS5 members. Shown is the phylogeny of the different subgroups of the IS5 family and comparison of a representative set of terminal IRs. The various Archaea have been color coded as follows for clarity: Sulfolobales, red; Thermoplasmatales, magenta; halophiles, green; methanogens, blue. Bacteria are indicated in black.
IS903 subgroup.
The IS903 subgroup includes two archaeal elements (Fig. 5): ISC1058 from S. solfataricus and ISFac2 in the unfinished genome of F. acidarmanus. Two short and partial copies of an IS903-related element are also found in the genome of T. volcanium (TVN0139, TVN0587). These are closely related to ISs from the γ-Proteobacteria (IS903D and IS102 of E. coli, ISAs4 from Aeromonas sp., and ISVa1 from Vibrio species). The IRs of this subgroup are very homogeneous despite the fact that the very terminal “catalytic” base pairs are different from the 5′-GGC-3′ consensus of the bacterial elements. They all carry a motif, TGTTG, common to the bacterial ISs between nt 6 and 10. All exhibit DRs with a length of 9 bp, as expected for this group, but no similarities between them are evident. Related partial copies are present in H. marismortui chromosome II (rrnB0094), M. mazei (MM1429), and M. barkeri (Mbar_A1398/99, Mbar_A2202).
IS5 subgroup.
The IS5 subgroup (Fig. 5) includes ISMbu1 (M. burtonii), ISMac22 (M. acetivorans), and ISArch6 (from an uncultured archaeon). Three complete copies of ISMbu1 carry an in-phase insertion of 52 bp, which introduces a termination codon. Four complete copies also carry an additional tandem left end of 97 bp. A possible MITE derivative of ISMac22 was also identified. A fragment of an IS related to IS1194 can also be found in T. volcanium (TVN1409, TVN1410) and another in T. acidophilum (ID: Ta0379). ISMbu1 is related to IS1246 (Pseudomonas species) and ISSsp126 (Sphingomonas sp.). The IRs of this subgroup are heterogeneous. ISMbu1 have long DRs (14 bp), with no similarities to bacterial DRs.
IS1031 subgroup.
Only a single example of this group, ISMac15 (M. acetivorans), has been identified.
IS427 subgroup.
Four archaeal ISs have been identified in this subgroup: ISMac11, ISMma12 (M. mazei), ISMba5, and ISMba19 (M. barkeri). ISMac11- and ISMba5-related MITEs have also been identified.
The halophilic subgroup ISH1.
The halophilic subgroup ISH1 includes ISH1 and two isoelements, ISH9 and ISH28, together with ISH19, ISHma8, ISHma9, ISHma10, ISHma11, and ISNph4. Where present, DRs are between 7 and 10 bp. A single ISH9 MITE derivative was also identified.
The Sulfolobus subgroup.
Several elements in the genome of S. solfataricus (ISC1212, ISC1234, and ISC1290) are annotated as IS5 family members (8). These, together with ISSto3 from S. tokodaii, show only very weak similarities to other IS5 elements and also vary significantly among themselves. Moreover, the spacing of the DDE catalytic motifs does not align with that of other IS5 family members. MITE derivatives of ISSto3 have been identified.
IS5 orphans.
Several elements that display only weak similarities with the other IS5 elements are also present in both archaeal methanogens and halophiles. We have identified ISMba15 (M. barkeri), ISMhu10 (M. hungatei), and ISMbu10 (M. burtonii). ISMbu10-related MITEs and numerous solo IRs were also identified. Solo IRs are also found in M. acetivorans, M. mazei, and M. barkeri. Two related ISs are also present in the halophiles: ISH11 (Halobacterium sp. plasmids pNRC100 and pNRC200) and ISHma6 (H. marismortui pNG500 and N. pharaonis chromosome II and pL131).
IS6
All bacterial members of the IS6 family (Fig. 6) carry short, related (15- to 20-bp) terminal IRs and generally create 8-bp DRs. No marked target selectivity has been observed. The putative Tpases are very closely related, with identity levels ranging from 40 to 94%. A single ORF is transcribed from a promoter at the left end and stretches across almost the entire IS. There is a strongly conserved DDE motif. Transposition of these elements is presumably accompanied by replication, since IS6 family members appear to give rise exclusively to replicon fusions (cointegrates) in which the donor and target replicons are separated by two directly repeated IS copies. Following cointegration, a resolution step would be required to separate donor and target replicons transferring a copy of the transposon to the target replicon. In contrast to members of the Tn3 family, which encode a specific enzyme, a site-specific recombinase, recombination between the directly repeated ISs necessary for this separation occurs by homologous recombination and requires a recombination-proficient host (12). IS6 family elements are abundant in archaea and cover almost all of the traditionally recognized archaeal lineages (methanogens, halophiles, thermoacidophiles, and hyperthermophiles (Fig. 1 and 6; Table 1). Fourteen IS6 members could be identified. Phylogenetically, these can be divided into three groups present in the halophiles, the sulfolobales, and the pyrococcales/methanosarcinales.
FIG. 6.
IS6 members. Shown is the phylogeny of the IS6 family and comparison of a representative set of terminal IRs. The various Archaea have been color coded as follows for clarity: Sulfolobales, red; halophiles, green; “other,” orange. Bacteria are indicated in black.
Three closely related elements were found in the halophiles: ISH14, ISH15, and ISH29. ISH14 is 75% identical to ISH15 and is present as a single copy in H. marismortui. ISH29 is present as a single copy in Halobacterium sp. plasmid pNRC200. In addition, an ISH29-related structure composed of 15 bp and 35 bp of one end flanking a 15-kb DNA segment in direct repeat is present in two identical copies in pNRC100 and in pNCR200. These are in an inverted orientation on both plasmids. ISH15 is found in the plasmid pNRG500 of H. marismortui and in Halobacterium sp. An additional sequence less related to these, ISH17, was found in H. marismortui plasmids pNG500 and pNG700 and chromosome II. One partial copy is also present in Halobacterium sp. and in the plasmid pNRC200. A single copy of another member, ISNph1, was found in Natronomonas pharaonis.
Five different members were identified in the Sulfolobales: ISC735, ISC774, ISSto2, ISSte1, and ISSis1. ISC735 is indicated as a single copy in Sulfolobus sp. (AY671942). There are also three degenerate copies (with rearrangements and deletions within the IS) in S. solfataricus. S. solfataricus also carries full and partial (mostly solo IRs) copies of ISC774, while S. acidocaldarius carries only two IRs. ISSto2 is present in four complete copies, three of which carry different mutations in one IR and at least 13 partial copies. ISSte1 is present in a single copy in Sulfolobus tengchongensis plasmid pTC. Finally, ISSis1 is present in a single copy in Sulfolobus islandicus plasmid pARN4.
Methanocaldococcus jannaschii carries ISMja1 (ISE703) in two complete and one partial copy in the genome and three partial copies in the large extrachromosomal element. In addition, eight small elements of 358 to 360 bp resembling MITEs were identified (see “MITES, MICs, and solo IRs,” below).
Only a single partial copy of an IS6 family member could be identified in the Methanosarcina genus (M. barkeri Mbar_A0568).
The hyperthermophilic P. furiosus carries another three closely related elements, ISPfu1, ISPfu2, and ISPfu5, while P. abyssi carries a partial iso-ISPfu1 copy. Isoforms of these ISs are present in P. woesei and in a wide range of Pyrococcus strains.
Finally, two partial copies of an IS6-like element are present in the genome of Archaeoglobus fulgidus (AF0138, AF0895).
These archaeal elements form a monophyletic group related to bacterial ISs from Firmicutes: IS240 (Bacillus sp.), IS431 (Staphylococcus aureus), IS1297 (Leuconostoc mesenteroides), ISS1W (Lactococcus lactis), and ISEnfa1 (Enterococcus faecalis). Most carry DRs of 8 bp, but no clear sequence similarities can be observed in the DRs or surrounding sequences either between different ISs or copies of the same IS. The IRs of the archaeal IS6 members are quite variable compared to those of the bacterial members and might be divided into two subgroups. They generally terminate with 5′-GT or 5′-GA, as opposed to the 5′-GG found in Bacteria (Fig. 6). The bacterial and archaeal IRs clearly fall into different groups. The large phylogenetic distribution of IS6 family members in the Archaea and the monophyly of the IS6 archaeal group (in agreement with the IR resemblances) suggest that these elements were ancestrally present in archaea rather than being recently acquired by lateral gene transfer from bacteria.
IS21
The IS21 family (Fig. 7) is homogenous and widespread (Fig. 7, top) in Bacteria, characterized by a large size of between 2 to 2.5 kb. Members carry two ORFs (a long upstream frame, istA, and a shorter downstream frame, istB) and are bordered by long IRs beginning with 3′-CA. They can carry multiple repeated sequences within their ends (including part of the terminal IRs), which are possibly Tpase binding sites. Insertion results in a DR of 4 bp or, more frequently, 5 bp. The arrangement of the two ORFs suggests that translational coupling could occur. IstA, the Tpase, carries a potential helix-turn-helix motif together with a motif related to the DDE signature. IstB, a regulatory protein, includes a relatively well conserved potential NTP binding domain (30).
FIG. 7.
IS21 members. Shown is the phylogeny of the IS21 family and comparison of a representative set of terminal IRs. The top panel shows the organization of a typical IS21 family member. Red boxes, terminal IRs; yellow boxes, ORFs (IstA is the Tpase, and IstB is a regulatory protein). The overlap between these ORFs is indicated as a possible control region where translational coupling might be required for expression of istB. The terminal repeated sequences L1, L2, R1, and R2 are indicated by small arrows. The Archaea have been color coded as follows for clarity: methanogens, blue. Bacteria are indicated in black.
Two different IS21 elements, ISMac3 and ISMac9, are present in the M. acetivorans genome. An interrupted full copy and a partial copy of an ISMac3 isoform were also identified in M. mazei. A partial copy of another IS21 family IS is also present in M. barkeri (Mbar_A2360). These two closely related elements encode two overlapping ORFs. Phylogenetically, they are closely related to bacterial IS21 members IS5376 (Bacillus stearothermophilus), ISPsy4 (Pseudomonas syringae), ISRso6 (Ralstonia solanacearum), ISGsu6 (Geobacter sulfurreducens), and IS100kyp (Yersinia pseudotuberculosis). Both bacterial and archaeal ISs are characterized by very homogeneous IRs beginning with 5′-TGTNAA-3′. Like the bacterial elements, both ISMac3 and ISMac9 carry multiple repeated sequences at their ends (Fig. 7). Short DRs of 4 bp are observed, with no apparent similarities. ISMac3 and ISMac9 encode two overlapping ORFs (the catalytic DDE motifs are encoded by the first, istA). Interestingly, the phylogeny of this family based on the Tpase (IstA) (Fig. 7) is almost the same as that based on IstB. The restricted distribution of IS21 in Archaea and the close similarities of ISMac3 and ISMac9 with bacterial IS21 family members suggest that these two elements derive from lateral gene transfers of an element primarily present in Bacteria.
IS30
Members of the IS30 family are characterized by lengths of between 1,000 bp and 1,200 bp, a single ORF of between 293 and 383 codons and spanning almost the entire length, a well conserved DD(33)E motif, somewhat heterogeneous terminal IRs in the range of 20 to 30 bp, and 2- or 3-bp DRs.
ISC1041 (S. solfataricus MT-4) is the unique archaeal member of this family. It encodes a single ORF with 93% identity in DNA sequence with ISAba125 from Acinetobacter baumannii and is also closely related to ISPst1 and IS1394 of Pseudomonas species. Unlike other IS30 elements, ISC1041 carries only short and very imperfect IRs (18 bp.) As the genome sequence of S. solfataricus MT-4 has not been determined, very little genomic information is available for this element. However, the absence of IS30 elements in the sequenced species of Sulfolobus is compatible with the idea that ISC1041 had been laterally and recently acquired in strain MT-4.
IS110
Members of the IS110 family (Fig. 8) show important differences from most of the other IS families. They generally possess no IRs, show little similarity in their ends, and do not usually create DRs. They can be divided into two subgroups (59): IS110 and IS1111. These were defined on the basis of Tpase similarities and by the fact that IS1111 subgroup members carry short IRs of approximately 12 bp located at a short distance (3 to 7 bp) from the physical ends of the element. Those of the IS110 subgroup do not; however, the Tpases of these subgroups form a single cluster in Tribe analysis.
FIG. 8.
IS110/IS1111 members. Shown is the phylogeny of the IS110/IS1111 family. The various Archaea have been color coded as follows for clarity: Sulfolobales, red; Thermoplasmatales, magenta; halophiles, green; methanogens, blue. Bacteria are indicated in black.
IS110 subgroup.
Six elements belonging to the IS110 subgroup could be identified in Sulfolobus genomes: ISC1190, ISC1228, ISC1229, and ISC1491 in S. solfataricus and ISSto4, ISSto5, and ISSto6 in S. tokodaii. No examples were detected in S. acidocaldarius. These Sulfolobus ISs (Fig. 8), like their bacterial counterparts, do not carry internal IRs. ISC1190 and ISSto5, ISC1229 and ISSto6, and ISC1491 and ISSto4 are closely related. The complete ISC1190 copies are flanked by 6-bp DRs (CTCCTT). The Tpases of all ISC1491 copies are split into two or three parts by insertions, frameshifts, or translation termination codons, and, in every example, ISC1491 is associated with ISC1385 (IS630 family), either as an insertion into a full ISC1385 copy or into ISC1385 ends. Four of the five ISC1228 copies are interrupted by insertions of ISC1048 (IS630) or ISC1359 (IS4).
One example, ISFac9, was also identified in Ferroplasma acidarmanus.
These elements form a monophyletic group that is distantly related to diverse bacterial IS110 elements from proteobacteria such as ISPpu10 (Pseudomonas putida), ISNgo3 (Neisseria gonorrhoeae), and ISIMb1 (Moraxella sp.) but also from IS elements of Thermus thermophilus such as IS1000A and IS1000B.
IS1111 subgroup.
Several members of the IS1111 subgroup are also found in the methanogens: ISMma5 (M. mazei), ISMac14 (M. acetivorans), and ISMba7 and ISMba20 (both from M. barkeri). These closely related elements display some significant similarities with other bacterial IS1111 elements, mainly in the C-terminal region of the Tpase. On the other hand, ISMma5 and ISMac14 display very short and typical DRs of 3 bp, together with typical IS1111 internal IRs. Another element, ISH18, was identified in pNG500 of H. marismortui and in partial copy in M. hungatei (Mhun_0755).
IS256
The IS256 family (Fig. 9) is characterized by related inverted terminal repeats of between 24 and 41 bp; DRs of 8 bp, or sometimes 9 bp; possible internal IRs close to the ends; and a single long ORF carrying a potential DDE motif with a spacing of 112 residues between the second D and E residues, together with a correctly placed K/R residue. This homogenous family is widely represented in bacteria (12).
FIG. 9.
IS256 members. Shown is the phylogeny of the IS256 family and comparison of a representative set of terminal IRs. The various Archaea have been color coded as follows for clarity: Sulfolobales, red; Thermoplasmatales, magenta; methanogens, blue. Bacteria are indicated in black.
There are three elements present in the genome of S. solfataricus: ISC1250, ISC1332, and, ISC1257. The latter is present as 1 complete copy interrupted by ISC1439 and 12 partial copies. A single ISC1332 copy is also located in pNOB8 (Sulfolobus sp.).
Three different family members have also been identified in the Methanosarcinales. These are ISMma16 (M. mazei and partial isocopies in M. acetivorans), ISMba9 (one copy with several termination codons and phase changes and partial copies in M. barkeri), and ISMbu6 (M. burtonii).
The Thermoplasmatales also carry three distinct members: ISFac7 and ISFac8, from Ferroplasma acidarmanus, and ISTac2, a single copy carrying several termination codons in Thermoplasma acidophilum. Short and closely related partial copies can also be found in the genome of T. volcanium (TVN0870, TVN1468).
These are all phylogenetically linked to IS256 elements of the Firmicutes (Fig. 9): IS905A (Lactococcus lactis), IS1310 (Enterococcus sp.), IS1191 (Streptococcus thermophilus), ISLdl12 (Lactococcus delbrueckii), and IS1201 (Lactobacillus helveticus). With the exception of ISFac8, the archaeal IS256 elements form a monophyletic group in the phylogeny, suggesting little or no transfer of these between archaea and bacteria.
All are characterized by relatively highly conserved IRs beginning with 5′-GG or 5′-GA and carrying blocks of conserved sequence throughout. Most archaeal members appear to generate an 8- or 9-bp DR, although no sequence similarities between these DRs are apparent.
IS481
The very homogenous IS481 family (Fig. 10) carries a single ORF and is distantly related to the IS3 family (12). There are three euryarchaeal members (Fig. 10). Several elements annotated as ISMac4 were found in the genome of M. acetivorans. Several copies “share” their DRs (the flanking 3 bp of target DNA at one end of an ISMac4 copy can be found at the opposite end of another copy), suggesting homologous recombination between the two IS copies. Elements belonging to this family are also present in the genome of A. fulgidus, where four distinct ISs coexist: ISA0963-1, ISA0963-2, ISA0963-3, and ISA0963-7. Two distinct IS481 elements were also found in thermoplasmatale genomes: ISFac5 in F. acidarmanus and ISTvo3 in T. volcanium. A partial IS copy (Ta1408) was also identified in T. acidophilum.
FIG. 10.
IS481 members. Shown is the phylogeny of the IS481 family and comparison of a representative set of terminal IRs. The various Archaea have been color coded as follows for clarity: Thermoplasmatales, magenta; methanogens, blue; “other,” orange. Bacteria are indicated in black.
These elements are characterized by highly conserved IRs of between 24 nt and 26 nt, but no clear conservation was found between their DRs (15 bp). They form a monophyletic group in the tree (Fig. 10) positioned as a sister group of a diverse set of bacterial elements belonging to Proteobacteria (ISVch1, Vibrio cholerae; ISCc3, Caulobacter crescentus), to Actinobacteria (ISSco2, Streptomyces coelicolor; ISMav2, Mycobacterium avium), and to Firmicutes (ISVi1, Spiroplasma virus). Like bacterial IS481 elements, the IRs terminate with a conserved 5′-TGT; in addition (but unlike the bacterial elements), they are highly conserved overall. Except for ISTvo3, whose IRL is degenerate, the IRs include a large subset of the consensus sequence TGTNNTNTCCCNAAATTA.
IS630
The IS630 family (Fig. 11) is widespread in bacteria and appears related to eukaryotic elements such as Tc1 or Tc3 of Caenorhabditis elegans or mariner in insects and a variety of animals. Typically, they include one ORF and generate a conserved TA as DRs (12). Several members are found in both the Crenarchaeota and the Euryarchaeota (Fig. 11).
FIG. 11.
IS630 members. Shown is the phylogeny of the IS6 family and comparison of a representative set of terminal IRs. The various Archaea have been color coded as follows for clarity: Sulfolobales, red; halophiles, green; methanogens, blue; “other,” orange. Bacteria are indicated in black.
Three different elements can be distinguished in the S. solfataricus genome: ISC1048, ISC1078, and ISC1395. A single ISC1048 solo IR is present in S. tokodaii and in S. acidocaldarius, and three solo IRs of ISC1395 are present in S. tokodaii. Two ISC1048 copies carry a single ORF, while this is split by mutation in the eight other copies. Similarly, three copies of ISC1078 carry a single ORF, while in the others it is split into two at different positions. Moreover, while a single copy carries the expected IR sequence (Fig. 11), all other copies appear to carry variations in the first dinucleotide of the IR of both ends. The Tpase of two complete copies of ISC1395 is also split by mutation into two ORFs, while in the third copy it is split by insertion of ISC1491 (IS110 family). There are few similarities between the IRs of these three ISs.
In the case of the Euryarchaeota, ISHma2 is present in one complete copy on chromosome I of H. marismortui. A closely related element, ISH16, is found as two complete copies and two partial copies in pNG500 of H. marismortui and one partial copy in Halobacterium sp. It encodes a single polypeptide, has IRs of 25 bp, and generates a typical TA dinucleotide DR. A more distant but partial element (rrnAC1575) is also present on chromosome I of H. marismortui.
We have identified 11 distinguishable IS630 family members in the Methanosarcina group: ISMac13, ISMac17, ISMac18, ISMma6 ISMma8, ISMma9, ISMma10, ISMma17, ISMba3, ISMba10, and ISMth1. All except ISMac13 and ISMac17 carry two ORFs. ISMma6 (M. mazei) is related to the Sulfolobus and halophilic elements. Several of these elements are mutated and presumably nonfunctional. The single complete ISMac13 (M. acetivorans) copy is interrupted by insertion of a 1,695-bp sequence present six times elsewhere in the genome (see “Compound transposons, bits, and pieces,” below). ISMac13-related MITEs can also be identified (see “MITES, MICs, and solo IRs,” below). In addition to a single complete copy in M. mazei, six solo 22-bp IRs of ISMma8 are present in M. barkeri. One of the two ISMma17 copies is interrupted by insertion of ISMma11. All three copies of ISMac17 include a stop codon (TAG) at the same location in the Tpase ORF, while that of the single ISMba3 copy is degenerate and distributed over three ORFs (but see “Lost in translation,” above).
The presence of two ORFs in ISMac18, ISMma6, ISMma8, ISMma9, ISMma10, ISMma17, ISMba10, and ISMth1 raises the possibility that expression involves translational or transcriptional frameshifting. Indeed, ISMth1 (Methanosaeta thermophila) carries an extended stretch of 21 A's, representing a potential frameshifting site.
Finally, Archaeoglobus fulgidus carries two IS630 family members, ISA1083-1 ISA1083-2. We were unable to identify terminal IRs for these elements.
We also note that TVN1411 of T. volcanium and PTO1017, PTO0855, and PTO1049 of Picrophilus torridus encode partial Tpases with some similarities to that of IS630. Several examples from uncultured archaea have also been identified (Table 2 and www-is.biotoul.fr).
Phylogenetic analyses (Fig. 11) show that there are two subgroups and that the archaeal and bacterial elements are interspersed in both, suggesting multiple transfer events between these two domains.
IS982
The heterogeneous IS982 family (Fig. 12) was initially discovered in Lactococcus (94) and was recently identified in other Firmicutes as well as several γ-Proteobacteria and Bacteroidetes species. The two from Bacteroidetes form a distinct subgroup. Members are characterized by lengths of between 950 and 1,200 bp, with similar terminal IRs of between 18 and 35 bp, generally beginning with 5′-ACCC; DRs of 7 bp or 9 bp; and a single ORF capable of specifying a protein of between 271 and 313 aa, with a possible DDE motif lacking the conserved downstream K/R residue.
FIG. 12.

IS982 members. Shown is a comparison of a representative set of terminal IRs of the IS982 family. The archaeal IS is indicated in orange. Bacterial ISs are shown in black.
A single element, ISPfu3, can be found in archaea in the genome of P. furiosus. It displays some weak but significant similarities with IS982 elements from the Bacteroidetes group: ISSra1 of Riemerella anatipestifer and IS1187 of Bacteroides fragilis. However, there are not sufficient similarities with other IS982 elements to generate a phylogeny of the family. Nevertheless, ISPfu3 displays some specific characteristics of IS982 elements: highly conserved termini starting with 5′-ACCC and the absence of a K/R residue downstream of the DDE motif. ISPfu3 appears to generate a 10-bp DR, which, like other members of the family, is rich in AT although no sequence similarities with other IS982 members are apparent.
ISL3
Bacterial members of the ISL3 family range in size from 1,150 bp to 1,550 bp. They carry closely related IRs of between 15 and 39 bp and generate a DR of 8 bp. One long ORF that gives potential proteins of about 400 to 440 aa is present, showing good alignment particularly in the C-terminal half (12). Archaeal members of this family include ISMac21 (2 complete and 14 partial copies in M. acetivorans, 13 partial isocopies in M. barkeri and 1 in M. mazei); ISMbu4 (at least 2 complete copies in M. burtonii); ISArch5 (from an uncultured archaeon), which carries an in-phase stop codon at position 101; and several partial elements in T. volcanium.
Non-DDE Transposons: the IS91 Group
Members of the IS91 family have no significant terminal IRs or DRs. They carry a long Tpase ORF whose potential products are related to a family of replication proteins of bacteriophages and small gram-positive plasmids, which propagate by a rolling-circle replication mechanism. The similarity is localized to a consensus of four motifs (42). These Tpases are more related to the φX174 gpA protein than to the plasmid Rep protein family. Similar types of transposon have been identified in eukaryotic genomes (40).
A single IS91 family element can be found in an archaeal genome, ISMbu9 in M. burtonii.
Non-DDE Transposons: the IS200/IS605/IS607 Group
The IS200/IS605/ IS607 group (Fig. 13) forms a heterogeneous assembly of ISs characterized by a complex organization (12, 88). Generally, they carry two ORFs: tnpA (the Tpase) and tnpB (of unknown function). Note that TnpB is not required for transposition. They are not bordered by IRs and do not generate DRs on insertion (41). However, several elements encode only a single ORF (tnpA or tnpB) and very few carry short IRs. Two nonhomologous Tpases encoded by tnpA can be discerned, and they form two subgroups: the IS605 (tnpA1) subgroup and the IS607 (tnpA2) subgroup (Fig. 13). That from IS607 resembles an S-site-specific recombinase (29), while the other (from IS605) resembles a relaxase (involved in plasmid transfer) or RCR protein (involved in rolling-circle replication of bacterial plasmids and single-strand bacteriophages) (77). The ends of elements associated with tnpA1 exhibit significant potential secondary structures that are important for Tpase recognition (15, 88).
FIG. 13.
IS200/IS605/IS607 members. The top panel shows the organization of different members of this group. The direction of gene expression is indicated by the arrows. Yellow, tyrosine Tpase TnpA1; blue arrowed boxes, serine Tpase TnpA2; orange arrowed boxes, TnpB frame of unknown function. The left and right ends of the transposons containing TnpA1 are shown in magenta and blue, respectively. These are not IRs but include potential secondary structures. (A) Phylogeny based on orfB of the IS200/IS605/IS607 family. (B) Phylogeny based on orfA of the IS607 family (orfA1). (C) Phylogeny based on orfA of the IS605 family (orfA2). IS608 elements are underlined, single orfB elements are indicated between brackets, and the asterisk indicates the mosaic construction of the elements of this family (see the text). The various Archaea have been color coded as follows for clarity: Sulfolobales, red; Thermoplasmatales, magenta; halophiles, green; methanogens, blue; “other,” orange. Bacteria are indicated in black.
Numerous ISs related to the IS200/IS605/IS607 group are present in archaeal genomes. A few include only tnpA1, and many encode only tnpB. Each IS200/IS605-related element inserts in an oriented way 3′ to a conserved tetra- or pentanucleotide, and we define the left end of the IS as that proximal to these sequences.
IS200 subgroup.
As in the Bacteria, isolated copies of tnpA1 could be observed but, except for a partial copy in N. pharaonis (NP4630A), are limited to the methanogens (ISMma21, ISMba16, and ISMba18), and all have corresponding MITE derivatives in their host genomes. We have yet to search for and identify equivalent isolated copies of tnpA2.
IS605-related elements.
Members of the IS605 subgroup carry a copy of tnpA1 together with tnpB. Three different members were identified in the Sulfolobales, ISC1476 (S. solfataricus), ISSto1 (present in S. tokodaii and as partial copies in S. acidocaldarius), and ISSis2 (S. islandicus plasmid pHVE14). A recent publication reporting the structure of a TnpA1 protein from S. solfataricus implied that the corresponding gene was an isolated copy (46), but this element is associated with an overlapping tnpB annotated as ISC1476 (8). MITE derivatives of ISSto1, including both left and right ends with respective potential secondary structures, are also present in S. tokodaii.
The halobacteria carry four distinct ISs: ISH12 and ISH1-8, “iso” copies of the same elements (Halobacterium sp. chromosome and plasmids pNRC100 and pNRC200 and H. marismortui plasmid pNG300); ISH22 (Halobacterium sp. and N. pharaonis plasmid pL131); ISHma7 (chromosome I of H. marismortui); and ISHma12 (H. marismortui plasmid pNG500).
Four different members were also identified in the methanogens: ISMac7 (M. acetivorans), ISMma19 and ISMma22 (M. mazei), and ISMba17 (M. barkeri chromosome and plasmid 1). MITE derivatives of ISMma19 are also present in the M. mazei genome.
Two ISs are present in the Thermoplasmatales, ISTac1 (T. acidophilum) and ISTvo5 (T. volcanium), and additional partial copies are found in T. volcanium (TVN0750), T. kodakarensis (TK0931/32), M. barkeri (Mbar_A2836), and N. pharaonis (NP3908A/10A and NP4810A/12A).
IS607-related elements.
Several archaeal elements encoding two overlapping ORFs can be assigned to the IS607 subgroup on the basis of the nature of orfA. The Sulfolobus genomes carry six such elements: ISC1904 and ISC1913, from S. solfataricus; ISC1926 from Sulfolobus sp. strain L00 11; and ISSto11, ISSto12, and ISSto13, from S. tokodaii. ISC1926 is closely related to ISC1913. An additional IS, IS1921, was also identified some time ago in the sulfolobale Acidianus ambivalens (A. ambivalens = Desulfurolobus ambivalens). MITE derivatives of ISSto12 and ISSto13 are also present in the S. tokodaii genome.
The pyrococcal group contains a single complete element present in the genomes of T. kodakarensis (ISTko1) and P. furiosus (ISPfu4). Partial ISs are also present in the T. kodakarensis genome (TK1841/1842), P. furiosus (PF1985/86), and P. abyssi (PAB2076/2077).
T. volcanium carries ISTvo1, whose TnpA2 shows only weak similarities with bacterial IS607 TnpA2, but TnpB shows extensive similarities with IS605 sequences.
Finally, a partial IS is found in M. jannaschii (MG0012m/14) and in S. acidocaldarius (Saci_2022/23).
Single orfB elements.
It is not yet clear even in the Bacteria that isolated copies of orfB, lacking the Tpase encoded by orfA1 or orfA2, are active in transposition or whether their transposition can be activated in trans by the Tpase of related IS200/IS605 elements in the same genome.
Most archaeal genomes carry isolated copies of orfB. These include S. solfataricus (ISC1316); S. tokodaii, S. acidocaldarius, and S. islandicus plasmids pEF9 and pHVE14; H. marismortui chromosome I and plasmids pNG300 and pNG500; Halobacterium sp. chromosome and pNRC200; N. pharaonis chromosome and pL131; M. kandleri; M. jannaschii; M. mazei; M. burtonii; M. barkeri chromosome and plasmid 1; M. stadtmanae; T. acidophilum; T. volcanium; P. furiosus; P. horikoshii; P. abyssi; and T. kodakarensis. At least some of these isolated orfB copies are flanked by short DNA regions including the expected secondary structural features necessary for mobilization. In addition, several copies of individual ends, not associated with either orfA or orfB, can be detected.
In Sulfolobus, the 12 complete but isolated TnpB genes are flanked by regions exhibiting potential secondary structures similar to those observed for other members of the family carrying both TnpA and TnpB. These have been called ISC1316. The left end, defined as the end upstream from the TnpB translation initiation signals, exhibits a short AT-rich sequence consistent with target sequences identified for other members of the family. The right end terminates with TCAC (compared to TCAA found with IS608). These extremities differ in sequence from the complete ISs (with both TnpA and TnpB) in the same genome, suggesting that they are probably not mobilizable by the TnpA genes encoded by these complete IS copies. A single copy of a left end is also present in this genome.
Phylogenetic distribution.
To determine the relationship within this IS group, we have analyzed the phylogeny of orfB, which is universally present in archaeal IS605/IS200 elements (Fig. 13A), and two distinct phylogenies for each orfA (Fig. 13B and C).
While orfB is universally present in archaeal IS605 elements, the distribution of the two nonhomologous orfA frames appears complex. Phylogenetic analysis of orfA1 (IS605 [Fig. 13B]) and orfA2 (IS607 [Fig. 13C]) and of the combined orfB frames from both groups was used to assess the relationship between different members of this complex group. In the orfB phylogeny (Fig. 13A), single orf elements of both IS605 and IS607 subgroups are interspersed and many bacterial sequences are intermingled with the archaeal sequences, suggesting a complex evolutionary history for orfB. The phylogeny of orfA1 shows that archaeal and bacterial IS605 elements are intermixed (Fig. 13B), suggesting several events of gene transfer between archaea and bacteria. On the other hand, orfA2 of archaeal IS607 is monophyletic (Fig. 13C), favoring a hypothesis of vertical transmission of these genes. Taken together, these observations could indicate the existence of diverse recombination events between divergent IS copies, leading to a mosaic construction of such elements. For example, replacement of an orfA of IS607 by an orfA of IS605, and vice versa, by nonhomologous recombination or with microhomologies would appear to have occurred frequently. As several highly similar orfB genes can be found alone or with orfA, it is likely that loss and gain of an orfA gene has occurred.
EMERGING GROUPS, ORPHANS, WAIFS, AND STRAYS
Small groups of closely related elements with a more distant relationship to the families in the ISfinder database appear frequently. These groups often represent members of a new family that is subsequently validated by the appearance of additional members. Three such emerging groups can be discerned in archaea: the ISA1214 family (A. fulgidus) and elements distantly related to ISL3 and IS66.
ISA1214-Related Elements
Members of the ISA1214 family are limited to the Archaea. A BLAST search revealed virtually no homology with other IS families or related elements in bacterial genomes. These elements are between 1,043 bp and 1,270 bp in length (with a deletion of 25 aa in the Tpase of ISA1214) and are flanked by IRs of >20 bp. They carry a terminal 5′-GG dinucleotide and several small additional sequence signatures; generate DRs of various lengths, up to 12 bp; and generally carry a long ORF encoding the Tpase (∼320 aa) and a short ORF (47 to 74 aa) of unknown function. These are arranged in a nonoverlapping divergent configuration, with the small ORF located upstream of the Tpase. In addition, the small ORF can be found in at least four isolated copies: one in T. volcanium (TVN1041) and M. jannaschii (MJ0362) and two in Methanococcus maripaludis (MMP0468 and MMP0751). The first element discovered in this family was ISA1214, from A. fulgidus (six complete copies: five are identical and the other is more distantly related [see reference 8 and www-is.biotoul.fr). It is closely related to ISFac3 of F. acidarmanus and to an element in T. volcanium, ISTvo2. An element was also identified in the genome of S. solfataricus and named ISC1043 (12). Reannotation has shown that the Tpase of the single complete copy of this element carries many termination codons. Seven partial copies are also found in this genome. Alignment of the Tpase of these five elements did not clearly identify a catalytic DDE motif, and it is possible that these Tpases use a different chemistry for transposition.
ISL3-Related Elements
Several elements distantly related to the ISL3 family (E value, >10−4) are observed in archaeal genomes. These can be divided into two groups: ISM1 (restricted to Archaea; could be considered an Archaea-specific ISL3 subgroup) and IS1595 (which is very distant from the ISL3 family).
ISM1 group.
Members of the ISM1 group are slightly longer (by 100 to 200 bp) than bacterial ISL3 family members. This group includes ISM1 itself, from Methanobrevibacter smithii; ISMst1 (Methanosphaera stadtmanae); ISMbu2 (M. burtonii); ISMma11 (M. mazei and related MITEs); ISMac19 (M. acetivorans; full and partial copies and one copy split into three segments by independent insertions of ISMac5 and ISMac9); and ISMba4 (M. barkeri; one complete copy containing many in-phase stop codons [but see “Lost in translation,” above], partial copies, and related MITEs). ISM1 has long IRs of 34 bp beginning with 5′-G and also generates DRs of 8 or 9 bp, as do the canonical ISL3 elements.
IS1595 group.
The second, and more distant, group (IS1595) is represented by six archaeal ISs. Four are complete while two are present as partial copies. Two closely related elements are present in Halobacterium: ISH4 (one complete chromosomal copy and one in pNRC100, 1,004 bp, IR of 23/29 bp) and ISH50 (one complete copy in H. salinarium). ISHma4 is present in one copy in H. marismortui chromosome I, one complete copy in pNG400, and a partial copy in chromosome II. ISNph2 is present as one copy in the N. pharaonis chromosome and one in pL131.
These elements are approximately 1,000 bp long, encode a unique polypeptide bordered by IRs of 25 to 29 bp, and are flanked by DRs of 8 bp with no clear similarities. The terminal IRs do not appear to be related to those of the bacterial ISL3 family. These two elements have some similarities with a very short ORF present in a single copy in the genomes of P. horikoshii (PH1854) and P. abyssi (PAB2064). No IRs and DRs could be found associated with these two Pyrococcus elements, and they could therefore represent degenerate truncated copies of ISs belonging to this family. This family contains many bacterial relatives, notably IS1595 from Xanthomonas oryzae (L. Gagnevin and P. Siguier, unpublished data).
IS66-Related Elements: the New Subgroup ISBst12
The IS66 family comprises elements found mainly in Proteobacteria (34). They are characterized by a large size (>2.5 kb), carry multiple ORFs (generally three: TnpA, TnpB, and TnpC), and are flanked by very similar IRs of between 15 to 27 bp, with a conserved terminal 5′-GTA. Insertion results in an 8-bp DR (34). Simpler but closely related elements carrying TnpC but not TnpA or TnpB can be found in both Archaea and Bacteria. These are shorter than IS66 (1,529 to 1,839 bp) and are bordered by IRs of 12 to 30 bp, which are closely related and include the conserved IS66 family terminal 5′-GTA trinucleotide. Insertion generates an 8-bp DR (34). The Tpases align well with TnpC and carry a DDE catalytic motif (34). The function of the products of the other reading frames is unclear.
These elements include ISH10 and ISH10B in Halobacterium sp.; ISMac8 (M. acetivorans); ISMbu5 (Methanococcoides burtonii); ISMhu3 (M. hungatei); a second but partial copy (Mhun1220) in M. hungatei; ISMma13, ISMma14, and ISMma15 (M. mazei); ISArch7 (from an uncultured archaeon); and a partial copy from T. volcanium (TVN0684). All ISMma13, ISMma14, and ISMma15 copies include a TAG termination codon within the Tpase gene (20) (see “Lost in translation,” above). ISMhu3 and ISArch7 also carry the small ORF found in the ISA1214 family (see “ISA1214-related elements,” above). In these cases it is in the same orientation upstream of, and partially overlapping, the Tpase gene. These constitute a new IS66 group whose members are significantly shorter and which carry a single Tpase ORF.
IS1182
Members of the recently identified IS1182 family are present in both Archaea and Bacteria (P. Siguier, unpublished data). Most encode a single long ORF of more than 450 aa. Several appear to encode two ORFs, and expression may involve translational or transcriptional frameshifting. All include a clear DDE motif. Members of this group carry inverted terminal repeats of 14 to 16 bp.
We have identified eight distinct archaeal members, at present restricted to the methanogens: ISMac1, ISMac2, and ISMac20 (M. acetivorans); ISMma2 (M. mazei); ISMhu1 and ISMhu2 (M. hungatei); and ISArch1 and ISArch2 (uncultured archaeon). All except ISMac20 generate a DR of 4 to 5 bp, while ISMac20 inserts into a palindromic target sequence.
ISH6
The ISH6 group is very distantly related to the IS256 family. It is at present restricted to halophilic archaea, except for a partial copy in A. fulgidus (AF0828). Members include ISH6 (H. salinarium and pNRC200 of Halobacterium sp.), ISHs1 (H. salinarium), ISHma5 (pNG100 and pNG500 of H. marismortui), and ISNph3 (N. pharaonis plasmid pL131). No bacterial members have yet been identified. Members are 1,450 bp in length and carry a single ORF generating a Tpase of about 440 aa, and, while the overall alignment with IS256 Tpases is not good, the conserved DDE motif clearly aligns with that of IS256 family members. They are flanked by imperfect IRs of 27 bp and generate 8-bp AT-rich DR.
ISC1217
The ISC1217 group contains three members: ISC1217 (S. solfataricus), ISC1205 (Sulfolobus sp.), and ISSto10 (S. tokodaii). ISSto10-derived MITEs have also been identified. This group is very distantly related to the IS4 superfamily.
MITEs, MICs, AND SOLO IRs
MITEs
MITEs (for “miniature inverted-repeat transposable elements”) have been known for some time to be present in the Eukarya and Bacteria (27). They are extremely numerous in certain plant genomes, where they were first discovered (38), but have been identified in various other eukaryotic genomes and in several bacteria, including Neisseria gonorrhoea, N. meningitides (10), and Streptococcus pneumoniae (55).
MITEs are generally thought to derive from ISs that specify DDE Tpases and are composed of flanking terminal IRs but no interstitial Tpase gene. They range in size from less than 100 bp to more than 300 bp and do not carry other ORFs. MICs (mobile insertion cassettes) are similar to MITEs but carry passenger genes unrelated to transposition (19). MITEs and MICs are considered to be nonautonomous transposable elements, mobilizable in trans by Tpases of full-length genomic copies of the parent transposon.
In eukaryotes, many MITEs are related to the Tc/mariner family elements, as judged both by the similarity between their IRs and by their target site duplication. The Tc/mariner family is distantly related to the bacterial IS630 family, and MITEs derived from IS630 were the first bacterial examples to be described (10, 55). Other eukaryotic MITEs are related to other DNA transposons, such as PIF/Harbinger (itself related to bacterial IS5 elements), and probably to members of the hAT, CACTA, and Mutator elements (27). MITEs showing similarities to one of several of the IS families have now been identified in bacteria and in several archaeal genomes, including A. pernix, S. solfataricus and S. tokodaii, M. jannaschii, M. mazei, M. acetivorans, and M. barkeri (8, 9, 71; also this work). However, their transposition activities have yet to be analyzed in any detail. Transposition activity is implied from observations showing that they may be present in some but not all copies of a given multicopy IS, and sometimes a given element has been identified in several different ISs. Moreover, the insertions within an IS have permitted determination of the length of target repeat generated by comparing empty and occupied insertion sites (71). A single transposition event involving an S. solfataricus MITE has recently been observed (6).
The most numerous archaeal MITEs (Table 3) are observed in the Sulfolobales and Methanosarcinales. A systematic study of the Sulfolobales revealed relatively high numbers in both S. solfataricus and S. tokodaii but not in S. acidocaldarius (8, 9, 71). They were divided into several classes (called SM1 to SM6). SM5 and SM6 appeared to be limited to S. tokodaii, and SM4 appeared to be limited to S. solfataricus. For the Methanosarcinales, significant numbers are found in M. acetivorans and M. barkeri but M. mazei contains many fewer full copies.
The MITE families are described below according to their probable “parental” IS family. Where not stated, they were identified by simple BLAST analysis using the supposed parental IS as a seed sequence. Other groups were not identifiable in this way either because no entire parental IS copy was present in the database or because the IRs are degenerate. In these cases the sequences were initially identified as repeated sequences within the genome (8). The relationship of these with the parental IS is therefore more tenuous. We have not attempted to identify others of this type.
IS1.
Two larger elements (315 and 317 bp) with IR ends identical to those of ISSto7 were identified in S. tokodaii. In addition, two groups of MITEs, SM4 and SM7 in S. solfataricus, are related to IS1. SM4 (147 to 168 bp) has 27-bp IRs, similar in length to IS1 IRs, which are 49% identical to those of ISC1173 and are flanked by 8-bp target repeats (8). The second group, SM7 (330 bp) has only 13-bp IRs, significantly shorter than those of ISC1173, with >90% identity but with no obvious DRs (M. F. White, P. Redder, and R. A. Garrett, personal communication). A third group, SM6 (127 bp), related to ISSto9 (ISC794) is present in S. tokodaii with a high degree of identity (>95%).
IS4.
The methanogens carry MITEs related to two ISs of the IS4 family: ISMba11 and ISMba12. Those related to ISMba11 are 241 bp in length and are found as complete and partial copies (with the left or right IR deleted) in M. barkeri, M. mazei, and M. acetivorans. Those related to ISMba12, found in M. barkeri, are 278 bp in length.
Among the Halobiales, we have identified IS4-related MITEs only in H. marismortui. These are related to ISH20 and are present in single copies of 277 and 287 bp.
IS5.
Five groups of MITEs related to ISMac11 are found in M. acetivorans. These all have similar lengths (130 to 131 bp), but their sequences are clearly different and form five distinguishable clusters. A limited number of MITEs related to ISMba5 in M. barkeri (two copies of 131 bp and one of 152 bp) and to ISMbu10 in M. burtonii (185 bp) have also been found.
The S. solfataricus genome (71) carries 40 copies of SM3. These are between 127 and 139 bp long and are divided into two subgroups (SM3A and SM3B) based on sequence identities (75 to 97% within each group and only 60% between the groups). These appear to be distantly related to ISC1058 and include a DR of 9 bp, although they are not revealed by a simple BLAST analysis. The S. tokodaii genome also carries four copies of a MITE of 329 to 332 bp related to ISSto3.
Halobacterium sp. also carries MITE derivatives of 180 bp related to ISH9.
IS6.
Eight MITE-like sequences with lengths between 358 and 360 bp were identified in M. jannaschii. These represent internal deletions of ISMja1 (703 bp) between positions 263 and 604. IS6-related MITEs, SM5, have also been described in the S. tokodaii genome with a high level of conservation (92 to 100%). They show limited identity to ISC774, which is too low to be detected by BLAST analysis (8).
IS200/IS605.
Note that IS200/IS605 are the only MITEs yet identified from non-DDE-type ISs. All archaeal ISs carrying only orfA1 have MITE derivatives. These are ISMma21 (248 bp; M. mazei) ISMba16 (three types: 272 bp, 248 bp with 87% identity to ISMma21 and 195 bp in M. barkeri), ISMba18 (four types: 209 bp in M. barkeri, M. acetivorans, and M. mazei; 55 bp in M. barkeri and M. acetivorans; 174 bp in M. barkeri; and 178 bp in M. barkeri).
Some MITEs derived from IS605-related ISs are also observed in S. tokodaii, related to ISSto1 (356 to 357 bp, equivalent to SMA [8]), and in M. mazei, related to ISMma19 (206 bp).
IS607-related elements also give rise to MITEs. These are derived from ISSto12 (274 bp) and ISSto13 (266 to 271 bp), both in S. tokodaii.
IS630.
Fifteen complete copies of a 172-bp MITE related to ISMac13 were identified in the M. acetivorans genome. In addition, a group of MITEs in S. solfataricus and S. tokodaii, SM1, whose members are >95% conserved and between 79 and 80 bp long, are distantly related to ISC1048.
ISM1.
Several MITEs belonging to the ISM1 subgroup and related to ISMma11 (199 bp) and ISMba4 (193 to 302 bp) are at present limited to the methanogens.
ISC1217.
An additional class of MITEs found in both S. solfataricus and S. tokodaii is related to ISC1217 and ISSto10. Members of this group, SM2, are between 183 and 186 bp and present in 25 and 36 copies, respectively, with >95% conservation. A single longer MITE of 295 bp, related to ISSto10, is also present in S. tokodaii.
Nonclassified MITEs.
Multiple short sequences resembling MITEs (called MJREs) have been found in M. jannaschii (85). These were divided into three groups based on sequence similarities. There are a total 141 copies: MJRE1 has 59 copies and is 108 ± 7 bp long with IRs of about 17 bp, MJRE2 has 69 copies and is 96 ± 3 bp long with IRs of about 16 bp, and MJRE3 has 13 copies and is 101 ± 4 bp long with IRs of 15 bp. No particular regional bias was observed in the M. jannaschii genome. These sequences were not observed in other Archaea, and no corresponding complete IS elements with similar ends were found.
Solo IRs
Another type of IS fragment found in many genomes are isolated solo IRs. We have observed significant numbers in certain archaeal and bacterial genomes (Tables 2 and 3). They may represent deletion derivatives of MITEs or of full-length elements, but their impact on the host genome has not been investigated. It is possible that they attract insertions of homologous elements by homologous recombination or, as is the case for bacterial elements IS911 and IS30, act as transposition insertion hotspots under certain conditions (48, 58).
COMPOUND TRANSPOSONS, BITS, AND PIECES
Several more-complicated IS-related structures were identified. The most obvious were those resembling compound transposons.
Compound Transposons
Two copies of ISMba2 (IS1 family) form a potential composite transposon: they flank a 6,912-bp DNA segment (carrying an Fe-S oxidoreductase, a tRNA nucleotidyltransferase and three uncharacterized genes) in direct repetition, and the entire structure is flanked by a DR of 8 bp. Interestingly, the interior IR (IRR) of the first copy is mutated in the terminal dinucleotide, presumably rendering the IS inactive for transposition while retaining a wild-type Tpase. Moreover, the termination codon of the orfA gene of the second copy is mutated. This would place OrfB out of phase and thus produce an inactive Tpase. Thus, transposition of the whole would presumably be dependent on expression of an active Tpase from the flanking IS copy with the inactive inner end. The Tpase would be capable of acting both on the IS from which it was expressed (in cis) and on the other flanking IS, which does not encode an entire Tpases gene (in trans). These mutations would thus serve to increase the “coherence” of the composite transposon (as in the case of Tn10 and Tn5 [72]).
Another potential compound transposon was identified in P. furiosus. This is composed of a 16-kb region carrying an actively transcribed maltose and trehalose ABC transport system flanked by two insertion sequences from the IS6 family (now called ISPfu1) (21). It is absent from both P. abyssi and P. horikoshii. Interestingly, this region was also identified in Thermococcus litoralis, where it was flanked by very short (12- and 18-bp) sequences that might be IS remnants.
Uncharacterizable IS-Like Sequences
While analyzing M. acetivorans ISMac13 (IS630), we observed an insertion of 1,695 bp. A further six copies of this are present in the M. acetivorans genome but are not associated with other ISs. They include IRs of 17/21 bp, do not exhibit DRs, and encode a potential protein of 397 aa, which has no apparent relation to any known Tpase or any other gene in the microbial database.
Concatenated ISs
A “Russian doll”-like structure observed in S. solfataricus is consistent with an insertion of ISC1190 into an ISC1217 element, followed by insertion of ISC1058 and ISC1048 into the ISC1190 copy. The structure also includes two copies of an IS630-derived MITE, SM1 (8). In another structure of this type, two ISC1385 IRs (IS630 family) with a duplicated flanking TA flank two truncated copies of ISC1491 (IS110 family), which in turn flank an entire copy of ISC1048 (IS630 family). In a third example, two identical copies of ISC1225 (IS4) are both interrupted by an IS5 family member, suggesting that the entire structure may have transposed. Additional comparable structures can also be identified not only in Sulfolobus but in other Archaea, in particular in the Methanosarcina.
GENOME COMPARISONS: IS DISTRIBUTION, ABUNDANCE, AND GEOGRAPHICAL VARIATIONS
Table 1 shows the relative abundance of IS material in the complete genomes of Archaea (plasmids and chromosomes).
Intergenome Distribution and Abundance
As in Bacteria, plasmids generally carry a higher density of IS material than do chromosomes, particularly the megaplasmids of halophiles. In pNRC100 from H. salinarum, this reaches more than 20% (Table 1). In most cases, the presence of ISs in the plasmids is correlated with the presence of the corresponding element (complete or fragmented) in the host chromosome, suggesting a dynamic exchange of ISs between plasmid and chromosome.
Again, as in the case of Bacteria, the number of IS copies can vary greatly between chromosomes of different species. They are particularly numerous in Methanosarcinales (2.6 to 4.4%), some halophiles (3.5% for chromosome II of H. marismortui), and some Sulfolobus sp. (S. solfataricus). It is striking to observe the large variation in IS numbers in closely related genomes. This has been previously underlined for Sulfolobus (many ISs are present in S. solfataricus but no entire copies at all in S. acidocaldarius [8]). A survey of the ISs in S. solfataricus also gives the overall impression that they have undergone a high rate of mutation by nested insertions and deletions. Large differences in IS load are also seen in Pyrococcus (from 0.16% to 1.65%) and in Thermoplasma (from 0.48% to 1.72%).
Intragenome Distribution
Heterogeneity in the localization of the ISs around the genome has been reported for S. solfataricus, where two large regions with few elements were identified. The larger contains many genes required for translation. Heterogeneity between the two replicores around the supposed single origin of replication was noted. The authors suggested that ISs spread independently in each replicore. However, recent data demonstrate the existence of three origins of replication in S. solfataricus (50), and this observation must now be integrated into the previous hypothesis. Other IS-rich archaeal genomes (e.g., M. burtonii) can also show nonhomogenous IS distribution (see the section “Genomes” at www-is.biotoul.fr), as do those of many bacteria.
IS-rich regions may result from horizontal acquisition of blocks of DNA or reflect IS exclusion from other regions, differential IS extinction (90), or IS insertion specificity. For example, In Bacteria, Tn7 shows regional specificity in both of its parallel transposition pathways (61), and IS903 shows distinct regional preferences that are dependent on the nucleoid protein H-NS (86). Thus, the interpretation of regional specificity is likely to reflect a complex interaction of different influences.
Large Genomic Rearrangements
Brugger et al. (9) have performed a genome comparison of the three Sulfolobus genomes and present evidence that in at least two regions of 100 kbp (region I) and 70 kbp (region II), insertion sequence activity has resulted in significant rearrangements. The contiguous sequences of each region carried by S. acidocaldarius, in which we have been able to identify only partial IS copies (Tables 1 and 2), are found to be rearranged, with bordering ISs, into four segments (region I) and six segments (region II) in S. sulfolobus and as two segments each in S. tokodaii.
Genome analysis of P. furiosus, P. abyssi, and P. horikoshii showed that P. furiosus carries a set of 29 complete ISs and 9 truncated copies. It has been reported that P. abyssi and P. horikoshii lack complete IS elements (21); however, we have now been able to identify at least three different elements belonging to the relatively newly defined IS607 and IS1595 families. Additional studies with Pyrococcus (33) have shown that the major differences between the P. furiosus and P. woesei genomes appear to be due to gene clusters present in P. furiosus and lacking in P. woesei. These clusters include one (PF1737 to PF1751) involved in maltose/trehalose metabolism and flanked by ISPfu1. The authors suggest that the MalI cluster in P. furiosus is a composite transposon that undergoes replicative transposition.
Geographical Variations
A limited number of studies have addressed geographical variations in the IS content of archaeal strains. Such studies can provide important information on IS distribution and activities in shaping genomes.
Analysis of diverse Sulfolobus species from Siberia and the western United States, with 5-fluoroorate resistance used as a screen, led to the identification of seven additional ISs distinct from those identified in the sequenced Sulfolobus genomes (6). This indicates the existence of large regional disparity in IS content and suggests, moreover, that a large fraction of the true diversity of ISs in Archaea remains to be discovered.
Another study analyzed IS6 elements in a collection of 36 isolates of Pyrococcus from the Pacific Ocean and the Mediterranean Sea. It revealed that IS6 derivatives are present in almost all of the analyzed strains. These either are isoforms or are very closely related to IS-pfu1 (= ISPfu2 in ISfinder standard nomenclature), IS-pfu2 (= ISPfu1), and IS-pfu3 (= ISPfu5) (33). The authors suggest that such ISs could play an important role in genetic drift, leading to geographic diversification of hyperthermophilic archaea (24).
EVOLUTIONARY HISTORY OF ISs IN ARCHAEA: A POSSIBLE SCENARIO
Using the most recent sequence-based consensual phylogeny of archaea derived from comparative genomics (C. Brochier, personal communication; also reference 7), together with phylogenetic analysis and the distribution of each IS family, we propose a possible scenario for the evolutionary history of ISs in archaeal genomes. For each family, we indicate the most parsimonious scenario of IS gain by mapping acquisition of elements at each node (Fig. 14). Detection of IS loss, on the other hand, is a more difficult task and obviously becomes impossible if no IS scars (partial copies, MITEs, or solo IRs) are present. Two families, IS6 and IS605, could be considered ancestral in Archaea. They exhibit a large distribution in archaeal genomes and form a phylogenetic cluster distinct from most of their bacterial counterparts (Fig. 6 and 13). This scenario would also imply several independent events of subsequent gene loss occurring in almost all lineages of the Crenarchaeota, except for Sulfolobus species, and in most of the basal taxa of the Euryarchaeota (Thermococcales, Methanopyrus, and Nanoarchaeum, etc.).
FIG. 14.
Possible evolutionary scenario of ISs in archaea. A phylogeny of the Archaea is represented, and for each IS family we indicate the most parsimonious scenario of IS gain by mapping acquisition of elements at each node. The distribution by IS families is also indicated for each taxon; complete and partial elements are indicated in black and in gray, respectively. The various Archaea have been color-coded as follows for clarity: Sulfolobales, red; Thermoplasmatales, magenta; halophiles, green; methanogens, blue; “other,” orange.
Five additional families have a broad distribution, suggesting that they arose in ancient lineages. IS4, IS5, IS110/IS1111, IS481, and IS630 have a large distribution in the Crenarchaeota: with the exception of Picrophilus torridus and Haloferax volcanii, almost all lineages of the Crenarchaeota contain numerous elements belonging to these four families.
Horizontal transfer between Bacteria and Archaea has almost certainly occurred. For example, the incongruencies in the phylogenies of orfA and orfB of the IS605 family (Fig. 13) suggest a mosaic evolution of these elements (multiple events of replacement of one gene by another). In addition, several bacterial ISs belonging to the IS6 and IS605 families are more similar to archaeal ISs than to other bacterial ISs. For example, in the IS605 family, the TnpB proteins of ISBce3 (Bacillus cereus) and ISTma1 (Thermotoga maritima) are related to archaeal elements. This observation suggests that archaeal genomes may occasionally be a source of new ISs in bacterial genomes (and vice versa).
Due to the “erratic” phylogenetic distribution of the remaining families, it is likely that the large majority of IS families arose after the last common ancestor of the Archaea. Many ISs have a sporadic distribution in archaea, with only one or few members. These elements are more likely to have been recently acquired by archaea and include IS3 and ISL3 in Thermoplasma; IS256 in Thermoplasmatales; IS91, IS1, ISL3, IS256, and IS21 in Methanosarcinales; and IS982 in Pyrococcus. For the mesophilic archaea, such as the Methanosarcinales, this is consistent with the presence of numerous Bacteria-related genes in their genomes (20). At present, there are too few sequenced genomes of hyperthermophilic bacteria available to draw such inferences for the hyperthermophilic archaea.
An example of a possible massive invasion of ISs in the Archaea comes from the genome of the Sulfolobus species. The ancestors of Sulfolobus could have been invaded by ISs from the IS1, IS4, IS5, IS30, IS110, IS256, and IS630 families (Fig. 14). S. solfataricus has remained largely colonized (82), whereas S. tokodaii may have lost several of these elements (IS5, IS256, and IS630), as it retains a small IS subset, and S. acidocaldarius has lost all complete elements and retains only short and inactive copies. This is reminiscent of the IS distribution in bacteria, where IS expansion is often observed and for which it has been suggested that periodic extinction of transposable elements occurs (90).
Taken together, these results suggest that a small set of ISs existed in the last common ancestor of the Archaea and that subsequent lateral acquisition of new bacterial IS families has enriched archaeal phyla. It is interesting to note that Eukarya-type transposons were not detected either in Archaea or in Bacteria. This may indicate that lateral gene transfers of ISs between prokaryotes and eukaryotes did not occur before the emergence of the three kingdoms. However, Eukarya contains numerous mobile genetic elements that transpose by using DDE enzymes or tyrosine recombinases. It is tempting to speculate that these archaeal, bacterial, and eukaryal enzymes shared common ancestors but that eukaryotic enzymes have diverged to such an extent that homology is now undetectable from the sole primary sequences. This result suggests that the last universal common ancestor most likely had DNA transposons that transposed by using DDE or tyrosine enzymes. Alternatively, eukaryotic DNA transposons may have originated from mitochondria or chloroplasts and subsequently invaded the eukaryotic nucleus. Such a scenario has also been proposed for the origin of spliceosomal introns (35) in Eukarya by capture of mitochondrial group II introns (11, 51). If Eukarya are the result of the fusion of an archaeon and a bacterium (49, 51, 75), eukaryotic transposons could derive from either bacterial or archaeal ISs initially present in the two partners. An alternative hypothesis is that chemically there are a limited number of ways in which DNA can undergo the cleavages and strand transfers required for transposition and that at least some of the similarities observed result from convergent evolution. We note, however, that eukaryotic viruses infecting algae and amoebae surprisingly carry Bacteria- and Archaea-type ISs belonging to the IS4 and IS605/IS607/IS608 families (27a). These ISs have most likely been acquired by these viruses along with other prokaryotic genes. Similar types of virus, providing a genetic interface between prokaryotes and eukaryotes, may have been responsible for introducing prokaryote-type transposons during the early steps of eukaryotic evolution. Such transposons may then have diverged so extensively during the evolutionary course of the Eukarya that only the catalytic sites remain conserved. Finally, we should also mention that the archaeal Acidianus two-tailed virus carries four ISs belonging to the IS605/IS607/IS608 family (69). This observation further suggests that viruses would be an efficient system for laterally transmitting ISs between species.
CONCLUSIONS
We have attempted to provide an integrated overview of the diversity and distribution of ISs within archaeal genomes. We also review the limited understanding of various aspects of the regulation of IS activity. This snapshot covers the entire collection of available genomes (chromosomes and plasmids) whose nucleotide sequences have been determined (as of June 2006), together with several ISs from unfinished genomes or from metagenomic sequencing studies. The data are posted at www-is.biotoul.fr, the ISfinder website. During this study we performed an extensive annotation of the ISs, which, in most cases, includes the noncoding DNA regions in addition to their detectable ORFs. We have also compared the archaeal ISs with those from Bacteria and find that there is a common core of representatives of “classical” IS families in both of these domains of life. In addition, we have identified certain Archaea-specific subgroups in some of these families while for others both archaeal and bacterial members are clearly intermingled. We have tried to underline the many similarities and the few differences between ISs of the Archaea and the Bacteria. In doing this, we hope to have provided a solid base for future analysis of the impact of ISs in shaping the archaeal genome. Very few studies have addressed the mechanism and regulation of transposition in the Archaea. The possibility that some of these processes are Archaea-specific (e.g., readthrough of termination codons in Methanosarcina) requires further examination. The role of noncoding RNA in these regulations also requires experimental examination, as does the intervention of the archaeal replication apparatus. It also seems clear to us that very little is known concerning IS targeting. In the light of the limited understanding of global genome targeting in the Bacteria (factors affecting regional insertion preferences), it is clear that the nature of host factors in target choice needs to be addressed.
ADDENDUM IN PROOF
The IS4 family has recently been redefined (D. De Palmenaer, P. Siguier, and J. Mahillon, submitted for publication). The ISH8 subgroup remains a subgroup of the IS4 family. However, subgroups IS1634, ISH3, and IS701 are now defined as autonomous families.
Acknowledgments
We thank members of the Chandler laboratory—B. Tong-Hoang, P. Rouseau, G. Duval-Valentin, C. Guynet, N. Pouget, and E. Gueguen—for fruitful discussion, and we thank Jocelyne Perochon and Laurent Lestrade for crucial informatics support. Celine Brochier, Olivier Fayet, Roger Garrett, and Daniel De Palmenaer kindly provided advice and unpublished information.
Intramural funding was provided by the Centre National de la Recherche Scientifique (CNRS) (France), and extramural funding was provided by European contract LSHM-CT-2005-019023. J.F. was supported by the CNRS and by the European contract.
REFERENCES
- 1.Ammendola, S., L. Politi, and R. Scandurra. 1998. Cloning and sequencing of ISC1041 from the archaeon Sulfolobus solfataricus MT-4, a new member of the IS30 family of insertion elements. FEBS Lett. 428:217-223. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., D. D. Leipe, and E. V. Koonin. 1998. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26:4205-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baranov, P. V., O. Fayet, R. W. Hendrix, and J. F. Atkins. 2006. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 22:174-181. [DOI] [PubMed] [Google Scholar]
- 4.Bini, E., V. Dikshit, K. Dirksen, M. Drozda, and P. Blum. 2002. Stability of mRNA in the hyperthermophilic archaeon Sulfolobus solfataricus. RNA 8:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blinkowa, A. L., and J. R. Walker. 1990. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 18:1725-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blount, Z. D., and D. W. Grogan. 2005. New insertion sequences of Sulfolobus: functional properties and implications for genome evolution in hyperthermophilic archaea. Mol. Microbiol. 55:312-325. [DOI] [PubMed] [Google Scholar]
- 7.Brochier, C., S. Gribaldo, Y. Zivanovic, F. Confalonieri, and P. Forterre. 2005. Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales? Genome Biol. 6:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brugger, K., P. Redder, Q. She, F. Confalonieri, Y. Zivanovic, and R. A. Garrett. 2002. Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 206:131-141. [DOI] [PubMed] [Google Scholar]
- 9.Brugger, K., E. Torarinsson, P. Redder, L. Chen, and R. A. Garrett. 2004. Shuffling of Sulfolobus genomes by autonomous and non-autonomous mobile elements. Biochem. Soc. Trans. 32:179-183. [DOI] [PubMed] [Google Scholar]
- 10.Buisine, N., C. M. Tang, and R. Chalmers. 2002. Transposon-like Correia elements: structure, distribution and genetic exchange between pathogenic Neisseria sp. FEBS Lett. 522:52-58. [DOI] [PubMed] [Google Scholar]
- 11.Cavalier-Smith, T. 1991. Intron phylogeny: a new hypothesis. Trends Genet. 7:145-148. [PubMed] [Google Scholar]
- 12.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 13.Cornet, F., and M. Chandler. 2004. Non-homologous recombination, p. 36-66. In C. J. Miller and M. Day (ed.), Evolution: gene establishment, survival, and exchange. ASM Press, Washington, DC.
- 14.Craig, N. L., R. Craigie, M. Gellert, and A. Lambowitz (ed.). 2002. Mobile DNA, vol. 2. ASM Press, Washington, DC.
- 15.Curcio, M. J., and K. M. Derbyshire. 2003. The outs and ins of transposition: from mu to kangaroo. Nat. Rev. Mol. Cell. Biol. 4:865-877. [DOI] [PubMed] [Google Scholar]
- 16.DasSarma, S. 1989. Mechanisms of genetic variability in Halobacterium halobium: the purple membrane and gas vesicle mutations. Can. J. Microbiol. 35:65-72. [DOI] [PubMed] [Google Scholar]
- 17.DasSarma, S., U. L. RajBhandary, and H. G. Khorana. 1983. High-frequency spontaneous mutation in the bacterio-opsin gene in Halobacterium halobium is mediated by transposable elements. Proc. Natl. Acad. Sci. USA 80:2201-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLong, E. F. 1998. Everything in moderation: archaea as “non-extremophiles”. Curr. Opin. Genet. Dev. 8:649-654. [DOI] [PubMed] [Google Scholar]
- 19.De Palmenaer, D., C. Vermeiren, and J. Mahillon. 2004. IS231-MIC231 elements from Bacillus cereus sensu lato are modular. Mol. Microbiol. 53:457-467. [DOI] [PubMed] [Google Scholar]
- 20.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 21.Diruggiero, J., D. Dunn, D. L. Maeder, R. Holley-Shanks, J. Chatard, R. Horlacher, F. T. Robb, W. Boos, and R. B. Weiss. 2000. Evidence of recent lateral gene transfer among hyperthermophilic archaea. Mol. Microbiol. 38:684-693. [DOI] [PubMed] [Google Scholar]
- 22.Duval-Valentin, G., B. Marty-Cointin, and M. Chandler. 2004. Requirement of IS911 replication before integration defines a new bacterial transposition pathway. EMBO J. 23:3897-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enright, A. J., S. Van Dongen, and C. A. Ouzounis. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30:1575-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Escobar-Paramo, P., S. Ghosh, and J. DiRuggiero. 2005. Evidence for genetic drift in the diversification of a geographically isolated population of the hyperthermophilic archaeon Pyrococcus. Mol. Biol. Evol. 22:2297-2303. [DOI] [PubMed] [Google Scholar]
- 25.Evguenieva-Hackenberg, E., P. Walter, E. Hochleitner, F. Lottspeich, and G. Klug. 2003. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 4:889-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felsenstein, J. 1989. Phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 27.Feschotte, C., X. Zhang, and S. Wessler. 2002. Miniature inverted repeat transposable elements and their relationship to established DNA transposons, p. 1147-1158. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 27a.Filée, J., P. Siguier, and M. Chandler. 2007. I am what I eat and I eat what I am: acquisition of bacterial genes by giant viruses. Trends Genet. 23:10-15. [DOI] [PubMed]
- 28.Forterre, P., C. Brochier, and H. Philippe. 2002. Evolution of the Archaea. Theor. Popul. Biol. 61:409-422. [DOI] [PubMed] [Google Scholar]
- 29.Grindley, N. D. F. 2002. The movement of Tn3-like elements: transposition and cointegrate resolution, p. 230-271. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 30.Haas, D., B. Berger, S. Schmid, T. Seitz, and C. Reimmann. 1996. Insertion sequence IS21: related insertion sequence elements, transpositional mechanisms, and application to linker insertion mutagenesis, p. 238-249. In T. Nakazawa (ed.), Molecular biology of pseudomonads. ASM Press, Washington, DC.
- 31.Halladay, J. T., J. G. Jones, F. Lin, A. B. MacDonald, and S. DasSarma. 1993. The rightward gas vesicle operon in Halobacterium plasmid pNRC100: identification of the gvpA and gvpC gene products by use of antibody probes and genetic analysis of the region downstream of gvpC. J. Bacteriol. 175:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamilton, P. T., and J. N. Reeve. 1985. Structure of genes and an insertion element in the methane producing archaebacterium Methanobrevibacter smithii. Mol. Gen. Genet. 200:47-59. [DOI] [PubMed] [Google Scholar]
- 33.Hamilton-Brehm, S. D., G. J. Schut, and M. W. W. Adams. 2005. Metabolic and evolutionary relationships among Pyrococcus species: genetic exchange within a hydrothermal vent environment. J. Bacteriol. 187:7492-7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han, C. G., Y. Shiga, T. Tobe, C. Sasakawa, and E. Ohtsubo. 2001. Structural and functional characterization of IS679 and IS66 family elements. J. Bacteriol. 183:4296-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haugen, P., D. M. Simon, and D. Bhattacharya. 2005. The natural history of group I introns. Trends Genet. 21:111-119. [DOI] [PubMed] [Google Scholar]
- 36.Hofman, J. D., L. C. Schalkwyk, and W. F. Doolittle. 1986. ISH51: a large, degenerate family of insertion sequence-like elements in the genome of the archaebacterium Halobacterium volcanii. Nucleic Acids Res. 14:6983-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber, H., M. J. Hohn, R. Rachel, T. Fuchs, V. C. Wimmer, and K. O. Stetter. 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417:63-67. [DOI] [PubMed] [Google Scholar]
- 38.Jiang, N., Z. Bao, X. Zhang, H. Hirochika, S. R. Eddy, S. R. McCouch, and S. R. Wessler. 2003. An active DNA transposon family in rice. Nature 421:163-167. [DOI] [PubMed] [Google Scholar]
- 39.Kanoksilapatham, W., J. M. Gonzalez, D. L. Maeder, J. DiRuggiero, and F. T. Robb. 2004. A proposal to rename the hyperthermophile Pyrococcus woesei as Pyrococcus furiosus subsp. woesei. Archaea 1:277-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapitonov, V. V., and J. Jurka. 2001. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 98:8714-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kersulyte, D., N. S. Akopyants, S. W. Clifton, B. A. Roe, and D. E. Berg. 1998. Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene 223:175-186. [DOI] [PubMed] [Google Scholar]
- 42.Koonin, E. V., and T. V. Ilyina. 1993. Computer-assisted dissection of rolling circle DNA replication. Biosystems 30:241-268. [DOI] [PubMed] [Google Scholar]
- 43.Krebs, M. P., U. L. RajBhandary, and H. G. Khorana. 1990. Nucleotide sequence of ISH11, a new Halobacterium halobium insertion element isolated from the plasmid pGRB1. Nucleic Acids Res. 18:6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lao-Sirieix, S. H., L. Pellegrini, and S. D. Bell. 2005. The promiscuous primase. Trends Genet. 21:568-572. [DOI] [PubMed] [Google Scholar]
- 45.Lecompte, O., R. Ripp, V. Puzos-Barbe, S. Duprat, R. Heilig, J. Dietrich, J. C. Thierry, and O. Poch. 2001. Genome evolution at the genus level: comparison of three complete genomes of hyperthermophilic archaea. Genome Res. 11:981-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee, H. H., J. Y. Yoon, H. S. Kim, J. Y. Kang, K. H. Kim, J. Kim Do, J. Y. Ha, B. Mikami, H. J. Yoon, and S. W. Suh. 2006. Crystal structure of a metal ion-bound IS200 transposase. J. Biol. Chem. 281:4261-4266. [DOI] [PubMed] [Google Scholar]
- 47.Leipe, D. D., L. Aravind, and E. V. Koonin. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27:3389-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loot, C., C. Turlan, P. Rousseau, B. Ton-Hoang, and M. Chandler. 2002. A target specificity switch in IS911 transposition: the role of the OrfA protein. EMBO J. 21:4172-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez-Garcia, P., and D. Moreira. 2006. Selective forces for the origin of the eukaryotic nucleus. Bioessays 28:525-533. [DOI] [PubMed] [Google Scholar]
- 50.Lundgren, M., A. Andersson, L. Chen, P. Nilsson, and R. Bernander. 2004. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl. Acad. Sci. USA 101:7046-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin, W., and E. V. Koonin. 2006. A positive definition of prokaryotes. Nature 442:868. [DOI] [PubMed] [Google Scholar]
- 52.Martusewitsch, E., C. W. Sensen, and C. Schleper. 2000. High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J. Bacteriol. 182:2574-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagy, Z., and M. Chandler. 2004. Regulation of transposition in bacteria. Res. Microbiol. 155:387-398. [DOI] [PubMed] [Google Scholar]
- 54.Ng, W. V., S. A. Ciufo, T. M. Smith, R. E. Bumgarner, D. Baskin, J. Faust, B. Hall, C. Loretz, J. Seto, J. Slagel, L. Hood, and S. DasSarma. 1998. Snapshot of a large dynamic replicon in a halophilic archaeon: megaplasmid or minichromosome? Genome Res. 8:1131-1141. [DOI] [PubMed] [Google Scholar]
- 55.Oggioni, M. R., and J. P. Claverys. 1999. Repeated extragenic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology 145:2647-2653. [DOI] [PubMed] [Google Scholar]
- 56.Ohta, S., K. Tsuchida, S. Choi, Y. Sekine, Y. Shiga, and E. Ohtsubo. 2002. Presence of a characteristic D-D-E motif in IS1 transposase. J. Bacteriol. 184:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohtsubo, H., K. Nyman, W. Doroszkiewicz, and E. Ohtsubo. 1981. Multiple copies of iso-insertion sequences of IS1 in Shigella dysenteriae chromosome. Nature 292:640-643. [DOI] [PubMed] [Google Scholar]
- 58.Olasz, F., T. Fischer, M. Szabo, Z. Nagy, and J. Kiss. 2003. Gene conversion in transposition of Escherichia coli element IS30. J. Mol. Biol. 334:967-978. [DOI] [PubMed] [Google Scholar]
- 59.Partridge, S. R., and R. M. Hall. 2003. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J. Bacteriol. 185:6371-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paul, L., D. J. Ferguson, Jr., and J. A. Krzycki. 2000. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J. Bacteriol. 182:2520-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peters, J. E., and N. L. Craig. 2001. Tn7: smarter than we thought. Nat. Rev. Mol. Cell. Biol. 2:806-814. [DOI] [PubMed] [Google Scholar]
- 62.Pfeifer, F., and M. Betlach. 1985. Genome organization in Halobacterium halobium: a 70 kb island of more (AT) rich DNA in the chromosome. Mol. Gen. Genet. 198:449-455. [DOI] [PubMed] [Google Scholar]
- 63.Pfeifer, F., and U. Blaseio. 1990. Transposition burst of the ISH27 insertion element family in Halobacterium halobium. Nucleic Acids Res. 18:6921-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfeifer, F., J. Friedman, H. W. Boyer, and M. Betlach. 1984. Characterization of insertions affecting the expression of the bacterio-opsin gene in Halobacterium halobium. Nucleic Acids Res. 12:2489-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pfeifer, F., and P. Ghahraman. 1993. Plasmid pHH1 of Halobacterium salinarium: characterization of the replicon region, the gas vesicle gene cluster and insertion elements. Mol. Gen. Genet. 238:193-200. [DOI] [PubMed] [Google Scholar]
- 66.Pfeifer, F., G. Weidinger, and W. Goebel. 1981. Characterization of plasmids in halobacteria. J. Bacteriol. 145:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfeifer, F., G. Weidinger, and W. Goebel. 1981. Genetic variability in Halobacterium halobium. J. Bacteriol. 145:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Philippe, H. 1993. MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Res. 21:5264-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prangishvili, D., G. Vestergaard, M. Haring, R. Aramayo, T. Basta, R. Rachel, and R. A. Garrett. 2006. Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J. Mol. Biol. 359:1203-1216. [DOI] [PubMed] [Google Scholar]
- 70.Pritham, E. J., C. Feschotte, and S. R. Wessler. 2005. Unexpected diversity and differential success of DNA transposons in four species of entamoeba protozoans. Mol. Biol. Evol. 22:1751-1763. [DOI] [PubMed] [Google Scholar]
- 71.Redder, P., Q. She, and R. A. Garrett. 2001. Non-autonomous mobile elements in the crenarchaeon Sulfolobus solfataricus. J. Mol. Biol. 306:1-6. [DOI] [PubMed] [Google Scholar]
- 72.Reznikoff, W. S. 2002. Tn5 transposition, p. 403-422. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 73.Reznikoff, W. S., S. R. Bordenstein, and J. Apodaca. 2004. Comparative sequence analysis of IS50/Tn5 transposase. J. Bacteriol. 186:8240-8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rezsohazy, R., B. Hallet, J. Delcour, and J. Mahillon. 1993. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol. Microbiol. 9:1283-1295. [DOI] [PubMed] [Google Scholar]
- 75.Rivera, M. C., and J. A. Lake. 2004. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431:152-155. [DOI] [PubMed] [Google Scholar]
- 76.Robertson, C. E., J. K. Harris, J. R. Spear, and N. R. Pace. 2005. Phylogenetic diversity and ecology of environmental Archaea. Curr. Opin. Microbiol. 8:638-642. [DOI] [PubMed] [Google Scholar]
- 77.Ronning, D. R., C. Guynet, B. Ton-Hoang, Z. N. Perez, R. Ghirlando, M. Chandler, and F. Dyda. 2005. Active site sharing and subterminal hairpin recognition in a new class of DNA transposases. Mol. Cell 20:143-154. [DOI] [PubMed] [Google Scholar]
- 78.Rousseau, P., C. Normand, C. Loot, C. Turlan, R. Alazard, G. Duval-Valentin, and M. Chandler. 2002. Transposition of IS911, p. 366-383. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 79.Sapienza, C., M. R. Rose, and W. F. Doolittle. 1982. High-frequency genomic rearrangements involving archaebacterial repeat sequence elements. Nature 299:182-185. [DOI] [PubMed] [Google Scholar]
- 80.Schleper, C., R. Roder, T. Singer, and W. Zillig. 1994. An insertion element of the extremely thermophilic archaeon Sulfolobus solfataricus transposes into the endogenous beta-galactosidase gene. Mol. Gen. Genet. 243:91-96. [DOI] [PubMed] [Google Scholar]
- 81.Schmidt, F. J., R. A. Jorgensen, M. de Wilde, and J. E. Davies. 1981. A specific tetracycline-induced, low-molecular-weight RNA encoded by the inverted repeat of Tn10 (IS10). Plasmid 6:148-150. [DOI] [PubMed] [Google Scholar]
- 82.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simons, R. W., and N. Kleckner. 1983. Translational control of IS10 transposition. Cell 34:683-691. [DOI] [PubMed] [Google Scholar]
- 84.Simsek, M., S. DasSarma, U. L. RajBhandary, and H. G. Khorana. 1982. A transposable element from Halobacterium halobium which inactivates the bacteriorhodopsin gene. Proc. Natl. Acad. Sci. USA 79:7268-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suyama, M., W. C. Lathe III, and P. Bork. 2005. Palindromic repetitive DNA elements with coding potential in Methanocaldococcus jannaschii. FEBS Lett. 579:5281-5286. [DOI] [PubMed] [Google Scholar]
- 86.Swingle, B., M. O'Carroll, D. Haniford, and K. M. Derbyshire. 2004. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol. Microbiol. 52:1055-1067. [DOI] [PubMed] [Google Scholar]
- 87.Tang, T. H., N. Polacek, M. Zywicki, H. Huber, K. Brugger, R. Garrett, J. P. Bachellerie, and A. Huttenhofer. 2005. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 55:469-481. [DOI] [PubMed] [Google Scholar]
- 88.Ton-Hoang, B., C. Guynet, D. R. Ronning, B. Cointin-Marty, F. Dyda, and M. Chandler. 2005. Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. EMBO J. 24:3325-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ton-Hoang, B., C. Turlan, and M. Chandler. 2004. Functional domains of the IS1 transposase: analysis in vivo and in vitro. Mol. Microbiol. 53:1529-1543. [DOI] [PubMed] [Google Scholar]
- 90.Wagner, A. 2006. Periodic extinctions of transposable elements in bacterial lineages: evidence from intragenomic variation in multiple genomes. Mol. Biol. Evol. 23:723-733. [DOI] [PubMed] [Google Scholar]
- 91.Woods, W., and M. Dyall-Smith. 1996. Revised nucleotide sequence of an archaeal insertion element (ISH28) reveals a putative transposase gene. Gene 182:219-220. [DOI] [PubMed] [Google Scholar]
- 92.Woods, W. G., K. Ngui, and M. L. Dyall-Smith. 1999. An improved transposon for the halophilic archaeon Haloarcula hispanica. J. Bacteriol. 181:7140-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu, W. L., and W. F. Doolittle. 1983. Structure of the archaebacterial transposable element ISH50. Nucleic Acids Res. 11:4195-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu, W., I. Mierau, A. Mars, E. Johnson, G. Dunny, and L. L. McKay. 1995. Novel insertion sequence-like element IS982 in lactococci. Plasmid 33:218-225. [DOI] [PubMed] [Google Scholar]
- 95.Zivanovic, Y., P. Lopez, H. Philippe, and P. Forterre. 2002. Pyrococcus genome comparison evidences chromosome shuffling-driven evolution. Nucleic Acids Res. 30:1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]