Abstract
Specialization in cell function and morphology is influenced by the differential expression of mRNAs, many of which are expressed at low abundance and restricted to certain cell types. Detecting such transcripts in cDNA libraries may require sequencing millions of clones. Massively parallel signature sequencing (MPSS) is well-suited for identifying transcripts that are expressed in discrete cell types and in low abundance. We have made MPSS libraries from microdissections of three inner ear tissues. By comparing these MPSS libraries to those of 87 other tissues included in the Mouse Reference Transcriptome (MRT) online resource, we have identified genes that are highly enriched in, or specific to, the inner ear. We show by RT-PCR and in situ hybridization that signatures unique to the inner ear libraries identify transcripts with highly specific cell-type localizations. These transcripts serve to illustrate the utility of a resource that is available to the research community. Utilization of these resources will increase the number of known transcription units and expand our knowledge of the tissue-specific regulation of the transcriptome.
Keywords: Ear, Inner, MPSS, Transcription, Genetic
Abbreviations: ESTs, expressed sequence tags; GEO, gene expression omnibus; MoCR, mouse organ of Corti MPSS library; MoSV, mouse stria vascularis MPSS library; MoVB, mouse vestibular sensory epithelia MPSS library; MPSS, massively parallel signature sequencing; MRT, mouse reference transriptome; SAGE, serial analysis of gene expression; tpm, transcripts per million
Introduction
Specialized cell functions and structures are the product of differential expression of mRNAs, many of which are restricted to certain cell types and may be represented by as little as 10 copies per cell [1]. The problem of detecting rare mRNAs is especially acute in the mammalian inner ear. The inner ear is a complex tissue of multiple morphologically discrete cell types with functions that are not yet completely known. A profile of gene expression would provide insight into the physiology and function of these various cell types. Many of the special features of inner ear cell morphology and function are conferred by low-abundance mRNAs, which may have thus far eluded detection due to the relative inaccessability and small mass of this tissue. For example, the sensory cells of the organ of Corti, the inner and outer hair cells, comprise no more than ~5% of this tissue (Fig. 1). With perhaps 250,000 mRNAs expressed by an average cell, a typical low-abundance message unique to the hair cells would be expected to be present at a rate of 1 in one million transcripts. The identification of such mRNAs by sequencing cochlear cDNA libraries at the conventional depth of 4- to 6-fold coverage would be a daunting task. Enriching the cDNA population for rare transcripts by subtractive hybridization against cDNA from another tissue source [2] potentially introduces biases that could result in the loss of certain species of cDNA – for example, members of gene families whose homologs are abundantly expressed in the normalizing library. In addition, information regarding relative abundance is lost or obscured by subtraction or normalization.
Figure 1.
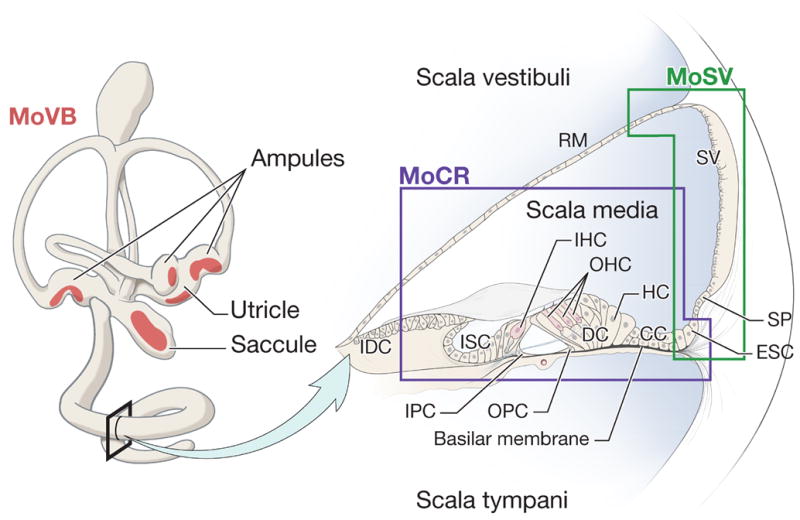
Schematic representation of tissues dissected for RNA isolation in the production of the inner ear MPSS libraries. The MoVB library was made from microdissected sensory epithelia of the vestibular receptor organs. Red shading indicates the approximate locations of sensory epithelia patches comprising utricular and saccular maculae and three cristae ampulares. The right panel shows a cross section through one turn of the cochlea. The approximate boundaries of the dissected material used to generate the organ of Corti (MoCR) and stria vascularis (MoSV) libraries are indicated. CC Claudius cells; DC, Deiter’s cells; ESC, external sulcus cells; HC, Hensen cells; IDC, interdental cells; IHC, inner hair cell; IPC, inner pillar cell; ISC, inner sulcus cells; OHC, outer hair cells; OPC, outer pillar cells; RM, Reissner’s membrane; SP, spiral prominence; SV, stria vascularis. The cross-section of the cochlea is redrawn from the Hereditary Hearing Loss Homepage (http://webhost.ua.ac.be/hhh/), with the permission of Guy Van Camp.
Alternatively, transcripts may be identified by generation of short sequence tags in a non-normalized, non-subtracted library, as in SAGE (Serial Analysis of Gene Expression) [3,4] or MPSS (Massively Parallel Signature Sequencing) [5]. In the MPSS technique, a 20-nucleotide signature, encompassing a DpnII site nearest to the poly-A tail, is generated from each mRNA. Greater than 90% of the signatures can be mapped to a known transcript, or to a genomic location in the case where no transcript is known [5,6]. The genomic signatures might be artifactual, or might originate from genes that have previously escaped detection due to low abundance and/or restricted spatial/temporal expression patterns. MPSS typically generates greater than 2 million signatures from less than 100 μg total RNA, therefore it is well-suited for identifying low abundance messages restricted to specific cell types in a complex tissue. Because of the depth of the coverage, it is also not necessary to normalize or subtract the starting cDNA material. Thus the rate at which a signature is identified in a library, measured in transcripts per million (tpm), approximates the relative transcript abundance in the original tissue. MPSS technology can reliably detect signatures down to a tpm of 3, which corresponds to approximately 1 transcript per cell of a homogeneous sample [7].
We generated MPSS libraries from microdissections of three tissues from mouse inner ear: the organ of Corti, stria vascularis and the vestibular sensory epithelia (Fig. 1). The organ of Corti is a complex multicellular epithelium separating the scala media and scala tympani chambers of the coiled cochlea. It includes the sensory cells of sound perception (the inner and outer hair cells), and their supporting cells. The stria vascularis is a pigmented tissue that forms part of the lateral wall of the scala media and is primarily responsible for maintaining the ionic composition of the endolymph. The vestibular epithelia are discrete patches of sensory hair cells and their supporting cells located in the utricle, saccule, and three ampules, and are responsible for detecting gravity, linear and angular acceleration.
Here we describe the general characteristics of these three inner ear libraries, and give examples of several genes identified through MPSS signatures. By comparing the signatures in these libraries to those derived from other mouse tissues in the Mouse Reference Transcriptome (MRT) Project, we are able to identify signatures that represent transcripts that are preferentially or exclusively expressed in the inner ear. These include signatures that appear to originate from novel or putative transcripts. We confirmed the presence and structures for several putative transcripts by sequencing PCR products from inner ear cDNA, and we demonstrate highly restricted cellular origins for several transcripts by in situ hybridization.
Results
Dissections of three tissues from male and female mouse postnatal day 7 inner ears (Fig. 1) were the source of RNA for the production of three MPSS libraries. The libraries are designated MoCR (mouse organ of Corti), MoSV (mouse stria vascularis) and MoVB (mouse vestibular epithelium). For each library, greater than 2 million signature clones were sequenced using the method described by Brenner et al. [5] In total, there are 29,672 discrete signatures that are significant (greater than 3 transcripts per million, or tpm) in at least one of the NIDCD inner ear libraries (Table 1). In addition to the signature sequences and their corresponding tpm statistics, Solexa provides annotation extracted from a table of automatically generated “virtual signatures”. The table of annotations corresponding to the signatures found in the inner ear libraries is available at the Gene Expression Omnibus (GEO) [8,9] website under the accession GPL1010. The unigene cluster number and transcript description are provided, or the chromosome and nucleotide location are provided if the signature matches genomic sequence only. Of the 29,672 signatures in the three inner ear libraries, 27,623 (93.1%) can be assigned to at least one unigene cluster or genomic location, and 24,392 (82.2%) can be assigned unambiguously to a single unigene cluster or genomic location. Although signatures that have ambiguous localization may be potentially interesting and provide real expression level data, only signatures with unambiguous localizations will be discussed as examples in this paper.
Table 1.
Abundance of discrete MPSS signatures and their classification in three MPSS libraries derived from inner ear tissues.
| All Signatures | Unique Signatures | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Signature Class | Summarized Class Description | MoSV | MoCR | MoVB | Combined Inner Ear | MoSV | MoCR | MoVB | Combined Inner Ear |
| 0 | Repetitive | 271 | 281 | 177 | 409 | 7 | 10 | 5 | 27 |
| 1–6 | Known Gene, Sense Orientation | 9,466 | 10,302 | 8,286 | 15,897 | 622 | 711 | 243 | 1,742 |
| 11–16 | Known Gene, Antisense Orientation | 1,644 | 1,902 | 653 | 3,460 | 544 | 716 | 99 | 1,459 |
| 22–26 | Known Gene, Orientation Ambiguous | 977 | 1,134 | 720 | 2,145 | 179 | 220 | 93 | 549 |
| 1000 | Genome Only | 1,985 | 2,643 | 1,787 | 5,712 | 773 | 1,085 | 783 | 2,748 |
| -- | Unclassifiable | 816 | 935 | 658 | 2,049 | 416 | 496 | 318 | 1,323 |
| Total | 15,159 | 17,197 | 12,281 | 29,672 | 2,541 | 3,238 | 1,541 | 7,848 | |
Numbers are signatures with tpm > 3. Unique signatures are those that have a maximum tpm < 4 in any mouse library in the MRT, excluding testis (MoTe.Male) or day 18 embryo (MoEm.MF18). Numbers in the “combined inner ear” column are not the sum of the three inner ear library totals because some signatures occur in multiple libraries.
Solexa classifies the signatures primarily on the basis of orientation relative to the best-hit unigene cluster, and then by proximity to a poly-A tail or signal. Briefly, classes 1–6 are signatures that match the sense strand of a unigene cluster, and are ranked corresponding to their proximity to a known or predicted poly-A signal or poly-A tail; classes 11–16 match the antisense strand of a unigene cluster; classes 22–26 match a unigene cluster whose orientation is ambiguous; and class 1000 matches genomic sequence only. The classification system is available at the Solexa website (http://sgbpub.lynxgen.com/). For simplicity, we have collapsed the classification scheme and summarized the results in Table 1.
There is a higher percentage of signatures that are classified 1–6 (sense orientation to a known gene) in the MoVB library compared to the MoSV and MoCR libraries (67.5% versus 62.4% and 59.9%, respectively). This difference is even more pronounced when considering the percentages that are classes 1–3 (i.e. those that are in the sense orientation and clearly in close proximity to a defined 3′ end of the transcript), which are 56.6%, 46.3% and 44.3%, respectively. These differences are probably due to a higher percentage of artifactual signatures in the MoCR and MoSV libraries. Artifactual signatures can be generated in the antisense orientation, or from intronic sequence, or from DpnII sites at the 5′ ends of transcripts. We generated the MoCR and MoSV libraries in 2003 before technical improvements in the MPSS process were implemented by Solexa (Irina Khrebtukova and Daixing Zhou, personal communication). The MoVB library was produced in 2004 using the improved protocol. Thus, when selecting individual signatures that appear to be unique to the inner ear, we are more confident in those from the MoVB library.
By comparing MPSS data from 32 human tissues, Jongeneel et al. [10] estimated that one-fifth of all expressed genes are tissue-specific. In order to distinguish between signatures that represent likely housekeeping genes and those that are specific to inner ear tissues, we compared our data to that of the Mouse Reference Transcriptome Project (MRT) (http://www.ncbi.nlm.nih.gov/projects/geo/info/mouse-trans.html). The MRT is an NIH initiative comprising 87 MPSS libraries generated from various mouse tissues (personal communication Chris Austin, NHGRI, 2005). For most tissues, separate male and female libraries were made. The protocol used for generating the MRT libraries is the same as that employed for the MoVB library (personal communication Daixing Zhou, Solexa, 2005). We imported the classification and tpm data for each signature of the inner ear libraries into a FilemakerPro v7 database (Filemaker Inc. Santa Clara, CA) and then imported the corresponding library and tpm data for each matching signature in the 87 MRT libraries (series GSE1581) to facilitate comparison of transcript abundance between libraries.
There are 7,848 signatures that are unique to one or more of the three combined inner ear libraries: 1,541 in the MoVB (12% of the MoVB total); 3,238 (18%) in the MoCR; 2,541 (16.7%) in the MoSV; 528 occur in more than one inner ear library, but do not occur in any MRT library (Table 1). The increase in percentage of unique signatures in the MoCR and MoSV libraries over that of the MoVB library may be a reflection of greater cell-type heterogeneity in those tissues, or it may simply be the result of increased numbers of artifactual signatures as mentioned above. Of the 7,848 signatures unique to one or more inner ear library, 3,750 can be mapped to a unigene cluster. Of these unigene cluster matches, 639 appear once. In other words, there are 639 unigene clusters that are represented by a single signature unique to the inner ear libraries. Overall, 344 (54%) of these single-unigene unique signatures map to their unigene cluster in the sense orientation (i.e. Solexa class 1–6), while 185 (29%) map in the reverse orientation (classes 11–16). Antisense transcription is a well-documented phenomenon in mammalian genomes [11–15]. However, the improvement in ratio of sense to antisense signatures present only in the MoVB library (83/138 or 60% sense, 27/138 or 19% antisense) is probably additional evidence of the better quality of the MoVB library.
Table 2 lists the most abundant signatures that are unique to the inner ear libraries, present in the MoVB library, and have unambiguous localization (single genome or unigene hit). The signatures are sorted by their maximum tpm in any of the three inner ear libraries. Almost all of the most abundant signatures originate from genes well-known to be unique to the inner ear and important for inner ear function, including otospiralin, alpha-tectorin, beta-tectorin, otogelin, col9a1, col9a3, and otoancorin. The most abundant signatures are, of course, likely to be associated with genes already known by virtue of mRNAs or ESTs submitted to Genbank. The surprising exception is the second-most abundant signature on this list which identifies a gene not previously described, but annotated originally as an in-silico predicted gene, GM741 (GeneID 327956; Refseq XM_282996). The locus name has been changed to Vmo1. The predicted protein has a single membrane spanning region and a vitelline membrane outer layer protein I (VMO-I) domain, which was first described in a protein isolated from chicken eggs. It is hypothesized to be involved in carbohydrate binding and synthesis. There are only 3 ESTs from two clones supporting this hypothetical gene (one of them from an inner ear library), despite the high abundance predicted by our MPSS libraries. We confirm the presence and the structure of Vmo1 by PCR from inner ear cDNA (Fig. 2) and demonstrate the tissue localization of messenger RNA by in situ hybridization (Fig. 3A and B). This transcript localizes specifically to Reissner’s membrane, which is a two-cell thick epithelial sheet dividing the scala media from the scala vestibuli. We believe that this is the first example of a transcript expressed exclusively in Reissner’s membrane.
Table 2.
Loci corresponding to the ten most abundant MPSS signatures unique to one or more of the NIDCD mouse inner ear libraries.
| signature abundance in tpm (transcripts per million) | |||||
|---|---|---|---|---|---|
| Signature | Locus | Description | MoVB | MoCR | MoSV |
| GATCGAGGACATGGCCAGAA | Otos | Otospiralin, mouse mutant is deaf [41] | 592 | 9,212 | 2,229 |
| GATCCGGCTACACTGCACTC | Gm741 | novel, Vmo1 (gene model 741) | 218 | 4,650 | 171 |
| GATCCTCAAACCCAGCCCTT | Tecta | alpha-tectorin, major component of tectorial membrane; mutations in human TECTA cause hearing loss (DFNA8, DFNA12, DFNB21) [42,43] | 489 | 2,737 | 0 |
| GATCCCCTCCTTTCTGCACT | Otog | Otogelin, inner ear specific glycoprotein, mouse mutant (twister) is deaf [44] | 1,832 | 1,105 | 4 |
| GATCTTCTAACCCTTCAAAA | Col9a3 | Mutations in human COL9A3 cause progressive hearing loss [45] | 1,649 | 1,194 | 1,431 |
| GATCTCAGATTATTCATTCA | Col9a1 | Mouse KO causes progressive deafness [46] | 476 | 183 | 1,398 |
| GATCTGCAGAGATTATTGCC | Oc90 | Otoconin, 90% of otoconia mass [47] | 1216 | 572 | 123 |
| GATCTGTTTGCGGGAGTAGA | Tectb | Beta-tectorin, major component of the tectorial membrane, [48] | 4 | 819 | 15 |
| GATCCTGCGAGCTCCAGACT | Cib3 | Calcium and integrin binding family member 3 (predicted), function unknown | 675 | 0 | 0 |
| GATCACGGGGCCTTGGAAAA | Otoa | Otoancorin, mutations in human OTOA cause autosomal recessive deafness, DFNB22 [49] | 379 | 296 | 0 |
Signatures >3 tpm in at least one inner ear library, <4 tpm in any MRT library, and are class 1–6. Signatures identifying alternate 3′ ends of genes already listed in the table are omitted, i.e. there are two signatures each for Otos and Col9a1 that would rank in the top ten for abundance.
Figure 2.
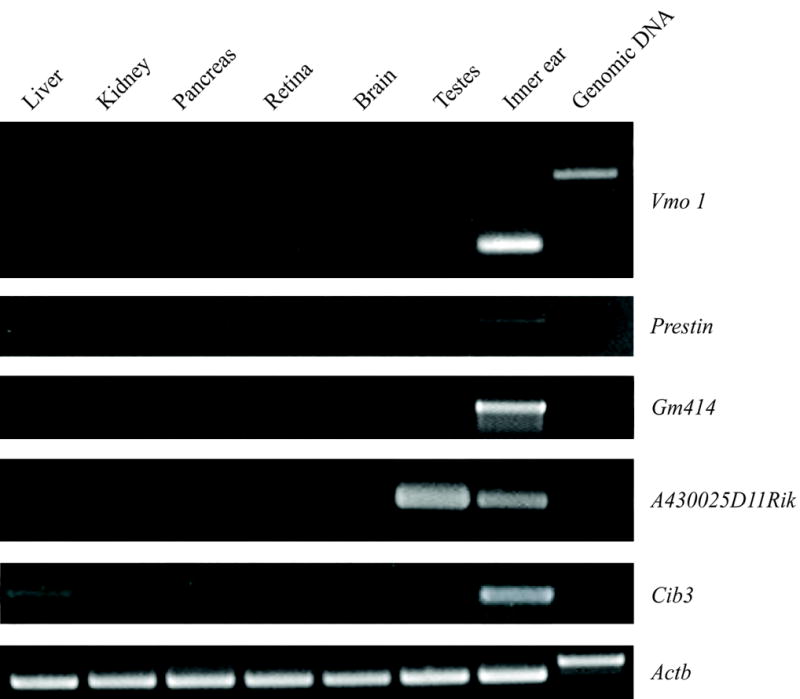
RT-PCR experiments confirming expression of predicted transcripts in the inner ear. Templates were mRNA derived from liver, kidney, pancreas, retina, brain, testes, inner ear and genomic DNA. Primers were designed to span at least one intron at the 3′ end of a transcript predicted by an MPSS signature. Control primers for the mouse beta-actin gene, Actb, amplify different size products from cDNA and genomic DNA templates, and are used in RT-PCR as a loading control. The RT-PCR products depicted here were cloned and sequenced, and were the templates for in situ RNA probes.
Figure 3.
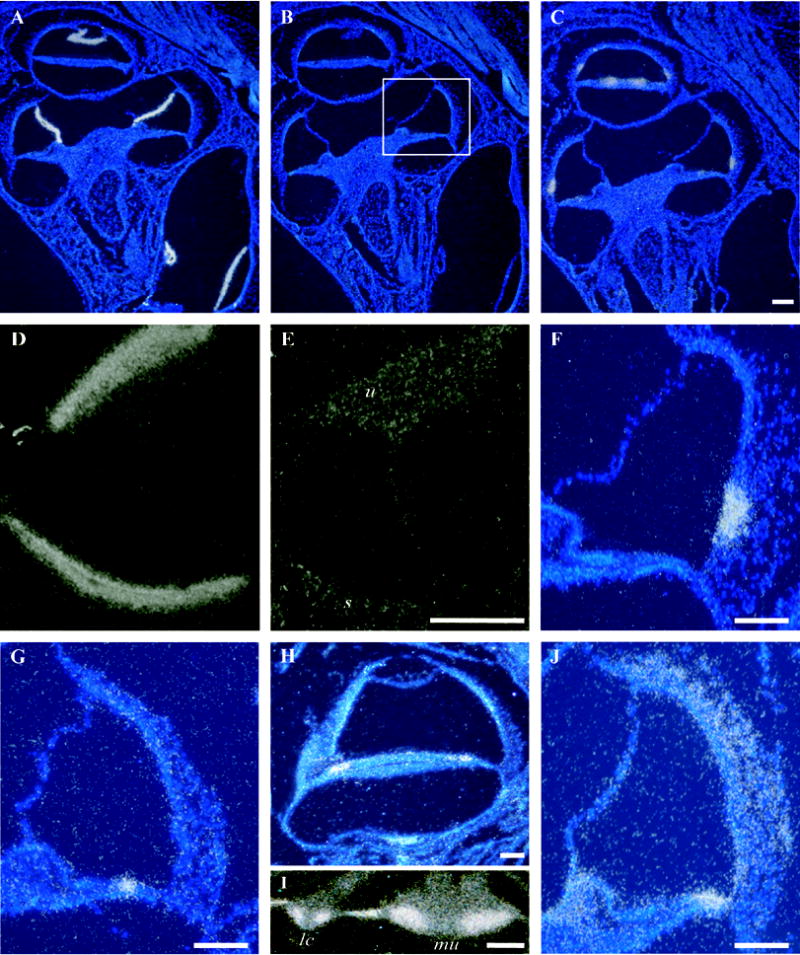
In situ hybridization in cross sections of the mouse inner ear. A) Antisense probe for Vmo1 shows discrete localization of the signal to Reissner’s membrane in cross sections through several cochlear turns. B) Control (sense) probe for Vmo1. C) A similar section to A and B shows hybridization signals obtained using an antisense probe for GM414, a mouse homolog of the zebrafish gene Otolin-1. D) The same probe as in C shows dense signal for GM414 in the maculae of the utricle and saccule. E) Control (sense) probe results for GM414 in the utricle (u) and saccule (s) in a section adjacent to the one depicted in panel D. F) A cross section through one turn of the cochlea at approximately the same level as the boxed region in panel B. Discrete localization of the Scg2 message to cells of the spiral prominence. G) An antisense probe for Umodl1 (olfactorin) reveals discrete localization of the message restricted to the middle of the organ of Corti, possibly in the hair cells or their supporting cells. H) An antisense probe for A430025D11Rik shows hybridization signal in the organ of Corti despite the lack of any signature in the MoCR library. A signature for this gene is found at moderate levels (160 tpm) in the MoVB library. In situ hybridization confirms expression in the vestibular sensory epithelia (data not shown). I) Cib3 in situ hybridization signal in a lateral crista ampularis (lc) and the macula of the utricle (mu). The intense signal on the slopes of the crista and faint signal at the apex is consistent with either supporting cells or hair cells. J) widespread in situ hybridization signal for antisense probe to the novel gene 1110017I16Rik. Scale bars = 100 μm.
There are currently six EST libraries in the mouse unigene database (build #151) derived from the ear, comprising approximately 43,000 sequences. The largest of these, dbEST Library ID.9974 (22,576 sequences), was enriched for inner ear specific clones by subtraction and inspected by Beisel et al. [16] for the presence of four genes known to be specific to hair cells and, therefore, are expressed in low-abundance in the inner ear: two acetylcholine receptor genes, Chrna10 and Chrna9; Myo15a, an unconventional myosin which localizes to the tips of stereocilia [17]; and Pres which encodes Prestin, a motor protein that is unique to outer hair cells [18]. They identified three clones for Chrna9 in their cDNA library, but none for the remaining three inner ear benchmark genes. In our inner ear MPSS libraries, there are no signatures for Chrna10, nor are there in any of the libraries of the MRT. However, there are several signatures in the inner ear libraries for the other three benchmark genes. A signature unique to the MoCR library may identify an alternate 3′ end of Chrna9 (Table 3). A signature for Myo15a is abundant in the MoVB library (445 tpm) and in pituitary libraries (284 tpm male, 182 tpm female) from the MRT (Table 3). This is consistent with the previously published localization data for Myo15a [19,20].
Table 3.
Selected MPSS signatures described in this paper.
| Signature abundance in transcripts per millin (tpm) | |||||||
|---|---|---|---|---|---|---|---|
| Locus | Signature | Class | Notes | MoVB | MoCR | MoSV | MRT* |
| Chrna9 | GATCTGGTGTGGAGGCCGGA | 4 | exon 3 | 0 | 8 | 0 | 0 |
| GATCCCCCAGAATACTGACA | 4 | exon 5 | 0 | 0 | 0 | 27 (Thymus.F) | |
| Myo15a | GATCTGCTCCGGCGTCACAC | 2 | 445 | 3 | 0 | 284 (Pituitary.M) | |
| Pres | GATCTGACAGGCAAGGCAGG | 1000 | ~2.5 kb 3′ to Pres | 0 | 232 | 0 | 42 (Brain.Amygdala.F) |
| GATCTCCTTTTTCTTCTAAC | 1000 | ~6 kb 3′ to Pres | 0 | 6 | 0 | 0 | |
| GM414 (Otolin-1) | GATCAAGTGTGGCTGGAAGT | 1000 | coding sequence | 482 | 13 | 54 | 6 (EmbryoE18.M/F) |
| GATCCTCTGCTAGTTGGTTC | 1000 | reverse | 60 | 20 | 0 | 0 | |
| GATCCATGTTTGTGTCTGTT | 1000 | reverse | 53 | 0 | 0 | 0 | |
| GATCGCCTTCTATCCTAAGG | 1000 | ~3 kb 3′ to GM414 | 51 | 1 | 19 | 0 | |
| Scg2 | GATCTGCTTCACTTATTCTG | 1 | 7 | 0 | 228 | 1211 (Pituitary.F) | |
| GATCAGGGCTTTGGAGTACA | 4 | 0 | 0 | 830 | 2466 (Pituitary.M) | ||
| Umodl1 (Olfactorin) | GATCATCTGGGGGAAACGAG | last coding exon | 0 | 120 | 0 | 0 | |
| Otof | GATCTACCGAGGCAGTGCCA | 4 | short isoform of Otoferlin | 41 | 6 | 0 | 0 |
| A430025D11Rik | GATCCTAAAGCTCAGAGAGG | 1 | 160 | 0 | 0 | 0 | |
| Cib3 | GATCCTGCGAGCTCCAGACT | 1 | 675 | 0 | 0 | 0 | |
| 1110017I16Rik LOC68527 | GATCGAAGCATAAAGGTTTT | 1 | 1037 | 59 | 259 | 17 (Cartilage.xiphoid.M) | |
Maximum tpm in the MRT, name of library, if any, given in parentheses.
There are no signatures in any library that match the Pres unigene cluster. However, two signatures are present in the MoCR library that matches genomic sequence near Pres (Table 3). Inspection of the genomic location of these signatures using a web page tool provided by Solexa shows that they are approximately 2.5 kb and 6 kb from the 3′ end of the Pres gene annotation and in the sense orientation. Sequencing of PCR products of inner ear cDNA confirms that these signatures are derived from Pres transcripts (Fig. 2 and data not shown) and demonstrates that the 3′ UTR of Pres is at least 2 kb larger than previously reported [21]. This extension of the 3′ UTR now encompasses four mouse ESTs, including one (Genbank Acc. No. BQ568786) from the inner ear library dbEST ID.9974. Thus, a signature for a gene represented by only 1 clone in 22,000 in an enriched and subtracted cochlear library has an abundance of 232 tpm in one of our non-subtracted, non-normalized MPSS libraries.
Overall, the presence of signatures for three of the four “benchmark” genes suggests that our inner ear MPSS libraries may be useful in detecting novel and/or rare transcripts. For example, there are four signatures unique to the inner ear libraries that localize to mouse genomic region 3qE2, within 3 kb of the 3′ end of locus GM414 (GeneID 229389; Refseq XM_143327) (Table 3). Two of the signatures are in the sense orientation relative to GM414, two are antisense. GM414 is a putative gene supported by mRNA and ESTs from mouse inner ear. The predicted encoded protein is homologous to the zebrafish gene otolin-1, which is a matrix protein necessary for proper otolith formation [22,23]. We confirmed the presence and structure of this gene by sequencing RT-PCR products of mouse inner ear mRNA (Fig. 2). In situ hybridization reveals that GM414 is expressed in the maculae of the vestibular receptor organs as well as in discrete locations in the organ of Corti (Fig. 3C, D and E). In the organ of Corti it is most evident in the cells of the spiral prominence and in supporting cells surrounding the hair cells in the apical-most turn of the cochlea. The presence of this transcript in the organ of Corti is somewhat surprising given its putative role in the formation of the otolith in the vestibular system.
Transcripts with limited spatial patterns, such as the ones described above, potentially will be helpful in the identification of regulatory elements that determine cell-specific expression. Differential spatial expression of genes is evident by comparing signatures not only between the inner ear and the MRT libraries, but among the three inner ear libraries as well. For example, although signatures corresponding to the Scg2 gene (secretogranin II; GeneID 20254; Refseq NM_009129) are abundant in many MRT libraries from neural and endocrine tissues, among the inner ear libraries they are preferentially found in the MoSV library (Table 3). In situ hybridization confirms that, in the inner ear, Scg2 is expressed exclusively by cells of the spiral prominence, adjacent to the stria vascularis (Figure 3F).
Of particular interest are those transcripts whose expression is limited to hair cells. One signature found exclusively in the MoCR library corresponds to Umodl1 (olfactorin; GeneID 52020; Refseq NM_177465), a gene which was originally identified in silico by virtue of its potential extracellular protein motifs, including a zona pellucida domain, and later amplified from human kidney, testis and mouse olfactory epithelium by RT-PCR [24,25]. Its expression is reported to be limited to the olfactory epithelium and vomeronasal organ in mouse (tissues not represented in the MRT), but here we demonstrate that olfactorin message is present in the organ of Corti (Fig. 3G), as predicted by the MPSS signature data (Table 3). The compact in situ signal is consistent with either hair cells or their supporting cells (pillar and/or Deiter’s cells) – the limits of resolution using 35S-labelled riboprobes do not allow us to distinguish among these cell types.
Because of the higher proportion of hair cells in the vestibular sensory epithelia compared to hair cells in the organ of Corti, signatures that are present in the MoVB library but absent or in low-abundance in the MoCR library, might indicate hair cell specific transcription. For example, a signature for Otof (otoferlin; GeneID 83762; Refseq NM_031875), which was previously shown to be expressed exclusively in inner hair cells in the organ of Corti [26], is present at only 6 tpm in the MoCR library, but at 41 tpm in the MoVB and absent from any MRT library. Similarly, a novel gene, A430025D11Rik (GeneID 214191; Refseq NM_172526), which is predicted to encode hypothetical protein LOC214191, has a corresponding signature found only in the MoVB library (160 tpm). A possible second signature located approximately 600 bp upstream is found only in the testis library in the MRT (132 tpm). RT-PCR confirms transcription of this hypothetical gene in the inner ear and testes (Fig. 2). In situ hybridization demonstrates modest expression of A430025D11Rik in the vestibular sensory epithelium. In situ analysis of the organ of Corti demonstrates a signal approximately in the region of the hair cells despite the absence of this signature from the MoCR (Fig. 3H).
Of course, signatures from the MoVB also identify genes that are expressed exclusively in the vestibular sensory epithelia. One of the most abundant signatures unique to the MoVB (675 tpm) corresponds to Cib3 (GeneID 234421; Refseq XM_356089), a gene predicted to encode a protein with calcium and integrin binding motifs (Table 3). Figure 3I shows the in situ hybridization signal from the cristae of two vestibular ampules. No signal was detected from sections of the organ of Corti.
Exclusivity to one of the inner ear MPSS libraries may be an unnecessarily stringent criterion for selecting transcripts that contribute to unique cell type function/morphology. For example, the criterion for tissue-specificity used by Jongeneel et al. [10] in their comparison of 32 human MPSS libraries was 512-fold higher expression in one library compared to all other tissues combined. If we consider signatures that are not unique to the inner ear, but show many-fold higher tpm than the greatest tpm in the MRT, or than the average tpm in the MRT, additional interesting genes are revealed. For example, a signature corresponding to 1110017I16Rik (GeneID 68527; Refseq NM_026754; hypothetical protein LOC68527) occurs with significant tpm in only one MRT library (male xiphoid cartilage; 17 tpm), but is present in all three inner ear libraries at highly significant levels, including 1037 tpm in the MoVB (61 times greater than the maximum value and 4,378 times greater than the average value in the MRT) (Table 3). In situ hybridization reveals this gene is highly expressed in the maculae of the utricle and saccule (data not shown). In the cochlea, a more diffuse pattern of expression is seen in the spiral ligament of the lateral wall and the spiral limbus, with some concentration of signal in the external sulcus (Figure 3J). The predicted protein LOC68527 has no known domains and no homology to any proteins with known or predicted function.
Discussion
We have generated MPSS libraries from microdissected inner ear tissues in order to identify transcripts that are relevant to inner ear physiology and development. We chose to collect mRNA from postnatal day 7 (P7) mouse pups because of the combination of ease of dissection and developmental changes that occur approximately at this stage. The length of the cochlea reaches adult size by P7, while the inner and outer hair cells attain adult numbers by P3, and remain constant through adulthood [27]. Although hearing function in mice apparently begins between P9 and P11, the mechanoelectrical transduction current can be recorded as early as P1 [28].
By comparing the signature abundance in the inner ear libraries with that of libraries derived from 87 other mouse tissues (MRT project) we focused on those genes that are preferentially expressed in the inner ear. Signatures with a unique genomic localization, but no unigene database hit (class 1000), often can be shown to originate from nearby known or predicted genes, and represent previously unknown transcribed sequences. When evaluating MPSS libraries from two human cell lines, Jongeneel et al. [29] estimated that greater than 70% of class 1000 signatures are derived from uncharacterized regions of known genes. In this paper we give illustrations of signatures that correspond to predicted genes and to previously unknown 3′UTRs of known genes.
The identification of the 3′UTR of genes, and especially the alternate 3′UTR specific to particular tissues, is one of the major benefits of analyzing MPSS data. The 3′UTR of transcripts can be the location of regulatory elements that help determine temporal and spatial expression [30]. Many of these regulatory sequences are identified by comparison of 3′UTRs within and between species, thus the full elucidation of the 3′UTRs or expressed sequences will accelerate the discovery of 3′UTR regulatory sequences and the mechanisms of gene regulation [31–34]. The alternate 3′UTRs discovered by MPSS signatures in the above examples illustrate both the utility of MPSS libraries for identifying 3′ ends of transcripts, and the extent of alternate 3′UTR variation. It has been suggested that half of all human genes encode alternate transcripts with differing 3′UTRs [35].
While signatures that are unique to the inner ear may represent novel transcribed regions, some of them, especially those with low abundance, are artifacts. However, the occurrence of the same signature in another library provides corroborative evidence that they originate from transcripts, and are not due to errors in the decoding process. Overlap between inner ear libraries and certain libraries in the MRT is especially interesting given the syndromic nature of many hereditary deafnesses in humans [36]. For example, there are over 60 deaf-blindness syndromes that have been described [37]. There are 48 signatures that are present in the inner ear libraries and in the eye libraries of the MRT, but in no other library. The most abundant of these signatures appear to identify alternate transcripts of Trpm3 and Trpm1 (data not shown), and two correspond to genes that cause vision defects in the mouse: Slc45a2 and Grk1tm1Citb which are mouse models of Oculocutaneous albinism type IV (MIM 606574) and Oguchi disease (MIM 258100), respectively [38,39]. It may be interesting to fully characterize the alternate transcripts associated with this set of signatures, and to test the hearing status of mice mutant for Slc45a2 and Grk1.
The MoCR, MoSV and MoVB MPSS libraries will be a valuable resource for researchers studying the molecular components of the inner ear. Here we show that exploitation of the comparative data provided by the libraries in the Mouse Reference Transcriptome (MRT) leads to the identification of transcripts that are spatially restricted to discrete cell types within the inner ear. A more general application of comparative MPSS analyses utilizing the MRT data will lead to transcript discovery in a variety of tissues, and ultimately will greatly facilitate the discovery of regulatory elements, especially those located in the 3′UTRs of transcripts. The signature data has been deposited in GEO under the accession numbers GPL3835 (for the signature annotation), GSM112130 (MoVB), GSM112131 (MoCR), GSM112132 (MoSV), with the latter three comprising series GSE4965.
Materials and methods
Isolation of total RNA
The organ of Corti, stria vascularis and vestibular sensory epithelia were dissected from the cochleae of C57B16 mice (Charles River Laboratories) at postnatal days 7 and 8. The dissected material from male and female mice were pooled together in 1X PBS, 4°C, pH 7.4. and then the total RNA was extracted using TRIzol (Invitrogen) following the manufacturer’s protocol. The dissected portion from the organ of Corti included the sensory hair cells and their adjacent supporting cells, such as inner sulcus border, pillar, inner phalangeal, Hensen’s, Böttcher’s, and Claudius cells. Reissner’s membrane and the spiral limbus were also included. The dissected tissue of the lateral wall of the cochlea consisted of stria vascularis and spiral ligament includes marginal, intermediate, and basal cells, spiral prominence cells, external sulcus cells, and fibroblasts. A small part of Reissner’s membrane was also present in the dissected tissue. The vestibular dissection included the sensory epithelium and the surrounding non-sensory cells of the utricle, saccule, ampullae, and three semi-circular ducts. A total of 59 μg of total RNA was isolated from 175 dissections of the organ of Corti; 149 μg of total RNA was isolated from 179 lateral wall tissues; and 89 μg of total RNA was isolated from 412 vestibular labyrinths. All RNA samples had OD 260:280 ratios greater than 1.8.
Generation of MPSS Libraries
Total RNA was sent to Lynx Therapeutics, Inc. (now Solexa, Inc.; http://www.solexa.com/wt/page/index) for polyA RNA isolation. The RNA was converted to cDNA, immobilized on microbeads, and the 16 nucleotides adjacent to the 3′prime-most DpnII site were decoded using the method detailed in Brenner et al. [5] For each library, a minimum of 2 million signatures were generated. The sequence and abundance (expressed in ‘transcripts per million’ or ‘tpm’) were determined for each distinct 17 and 20 nucleotide signature in the MoCR (mouse organ of Corti), MoSV (mouse stria vascularis), and MoVB (mouse vestibular epithelium) libraries.
The MoCR and MoSV libraries were produced first, and frequently contained numerous signatures per locus, most of which are presumably artifactual signatures that did not correspond to the true 3′ ends of the transcripts from which they originated. These “shadow” signatures could be distributed throughout the gene, in either orientation. Prior to the generation of the MoVB (and MRT) libraries, Lynx modified their decoding process to suppress production of these, and other, artifacts.
MPSS signature classification and annotation
Signatures for MoCR, MoSVand MoVB libraries, and their corresponding tpm, were imported into a Filemaker Pro database. Each signature occurs as a separate record. Signature annotations in the mouse genome and/or mouse unigene database are provided by Solexa. A Microsoft Excel spreadsheet containing all unique MoVB, MoCR and MoSV signatures, their classfication, and annotation, is available as a supplemental file. Solexa automatically classifies the signatures according to their orientation relative to known genes/transcripts. The annotations corresponding to signatures in the three NIDCD libraries were imported into our Filemaker Pro v7 database. In addition, the tpm data for all NIDCD signatures also present in the ~ 87 libraries comprising the Mouse Reference Transcriptome Project (MRT; http://www.ncbi.nlm.nih.gov/projects/geo/info/mouse-trans.html) were imported. Signatures were then sorted by their abundance, annotation, or occurrence in other libraries. Only signatures that had a unique localization, either in the genome or in unigene database, were considered for analyses. Signatures that were present only in the NIDCD libraries, or showed limited expression levels in other libraries, were selected for further study. The genomic locations of the signatures were inspected using a web browser tool supplied by Solexa. When signatures appeared to be in the correct orientation relative to predicted genes or unidentified transcripts, primers were designed to amplify the 3′ ends from mouse inner ear cDNA. All RT-PCR fragments were cloned and sequenced.
In situ Hybridization
For the gene of interest, a probe spanning 300–700 nucleotides of the mouse 3′UTR was amplified by RT-PCR from oligo-dT primed inner ear mRNA and the PCR products were cloned into PCR-Script Amp vector (Stratagene). Primer sequences are A430025D11Rik (5′-ccttgtccataccggctaaa-3′ and 5′-gacctcagccagggtacaaa-3′); Cib3 (5′-caggtctggatcaagctggt-3′ and 5′-ccaggtcttgtcctctgagc-3′); GM414 (Otolin-1)(5′-caggaaaaccaggagagcaa-3′ and 5′-ctcctcggtagcctttctcc-3′); Pres (5′-tacgcaggtcaattttgtgg-3′ and 5′-ccttgcctgtcagatctgtgt-3′); Scg2 (5′-ccctatgccttgaatctgga-3′ and 5′-ctgcccacagcattcactaa-3′); Umodl1 (Olfactorin)(5′-acccaactgaccaggagatg-3′ and 5′-gggaaacatgcaagaggtgt-3′); Vmo1(5′-ggcctgagatgtgtcctgat-3′ and 5′-ggtaaaagacagtactggcagagc-3′); 1110017I16Rik (5′-ttggcggttgtagaggtagg-3′ and 5′-tgcggagggagtattacgag-3′). Inserts were sequenced using T7 and T3 primers and Big Dye Terminatior v3.1 sequencing kits (Applied Biosystems). Probes were prepared by in vitro transcription of linearized plasmid using T7 polymerase (Stratagene) for the generation of antisense and sense RNA probes in the presence of [α-35S]UTP (NEN LifeScientific Products). P5 mouse heads were hemi-dissected, fixed, embedded in paraffin, and 10 μm serial cross-sections were mounted on microscope slides for in situ hybridization. Hybridization was performed at 52°C for 16 hr using 50,000 c.p.m./μl [α-35S]UTP-labeled RNA as previously described [40]. Slides were washed in 5X SSC, 0.1 M DTT at 50°C and then followed by 50% formamide, 2X SSC, 0.1 M DTT at 65°C. The slides were treated with RNaseA (Roche), rinsed, dehydrated and processed for standard autoradiography using NTB-2 Kodak emulsion (Eastman Kodak) and then exposed for 17 days at 4°C. Histological staining of sections was performed with nuclear dye Hoechst stain solution 3325B (Sigma) and analyzed using darkfield optics on a Nikon Eclipse 80i microscope.
Web site references
Mouse Reference Transcriptome (MRT): http://www.ncbi.nlm.nih.gov/projects/geo/info/mouse-trans.html
Gene Expression Omnibus (GEO): http://www.ncbi.nlm.nih.gov/geo/
Solexa, Inc.: http://www.solexa.com/wt/page/index
Supplementary Material
Acknowledgments
Our thanks to Daixing Zhou and Irina Khrebtukova (Solexa, Inc.) for their assistance in analyzing and annotating the MPSS signature data. We thank Chris Austin (NHGRI), and other members of the Mouse Transcriptome Project consortium. We thank Susan Sullivan (NIDCD), Doris Wu (NIDCD), and Dennis Drayna (NIDCD) for their suggestions in improving the manuscript. This work was supported by intramural funds from the NIDCD Z01 DC000035-07 and Z01 DC000039-07 to TBF and by a professional services contract from the NIDCD to Lynx Therapeutics (now Solexa, Inc.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–72. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 2.Bonaldo MF, Lennon G, Soares MB. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- 3.Harbers M, Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nat Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
- 4.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–7. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 5.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, Roth R, George D, Eletr S, Albrecht G, Vermaas E, Williams SR, Moon K, Burcham T, Pallas M, DuBridge RB, Kirchner J, Fearon K, Mao J, Corcoran K. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–4. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 6.Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 2004;14:1641–53. doi: 10.1101/gr.2275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandenberger R, Khrebtukova I, Thies RS, Miura T, Jingli C, Puri R, Vasicek T, Lebkowski J, Rao M. MPSS profiling of human embryonic stem cells. BMC Dev Biol. 2004;4:10. doi: 10.1186/1471-213X-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles--database and tools. Nucl Acids Res. 2005;33:D562–566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jongeneel CV, Delorenzi M, Iseli C, Zhou D, Haudenschild CD, Khrebtukova I, Kuznetsov D, Stevenson BJ, Strausberg RL, Simpson AJG, Vasicek TJ. An atlas of human gene expression from massively parallel signature sequencing (MPSS) Genome Res. 2005;15:1007–1014. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 12.Lehner B, Williams G, Campbell RD, Sanderson CM. Antisense transcripts in the human genome. Trends Genet. 2002;18:63–5. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- 13.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 14.Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–65. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 15.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, Nemzer S, Pinner E, Walach S, Bernstein J, Savitsky K, Rotman G. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–86. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 16.Beisel KW, Shiraki T, Morris KA, Pompeia C, Kachar B, Arakawa T, Bono H, Kawai J, Hayashizaki Y, Carninci P. Identification of unique transcripts from a mouse full-length, subtracted inner ear cDNA library. Genomics. 2004;83:1012–23. doi: 10.1016/j.ygeno.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Belyantseva IA, Boger ET, Friedman TB. Myosin XVa localizes to the tips of inner ear sensory cell stereocilia and is essential for staircase formation of the hair bundle. PNAS. 2003;100:13958–13963. doi: 10.1073/pnas.2334417100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng J, Madison LD, Oliver D, Fakler B, Dallos P. Prestin, the motor protein of outer hair cells. Audiol Neurootol. 2002;7:9–12. doi: 10.1159/000046855. [DOI] [PubMed] [Google Scholar]
- 19.Liang Y, Wang A, Belyantseva IA, Anderson DW, Probst FJ, Barber TD, Miller W, Touchman JW, Jin L, Sullivan SL, Sellers JR, Camper SA, Lloyd RV, Kachar B, Friedman TB, Fridell RA. Characterization of the human and mouse unconventional myosin XV genes responsible for hereditary deafness DFNB3 and shaker 2. Genomics. 1999;61:243–58. doi: 10.1006/geno.1999.5976. [DOI] [PubMed] [Google Scholar]
- 20.Lloyd RV, Vidal S, Jin L, Zhang S, Kovacs K, Horvath E, Scheithauer BW, Boger ETA, Fridell RA, Friedman TB. Myosin XVA Expression in the Pituitary and in Other Neuroendocrine Tissues and Tumors. Am J Pathol. 2001;159:1375–1382. doi: 10.1016/S0002-9440(10)62524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng J, Shen W, He DZ, Long KB, Madison LD, Dallos P. Prestin is the motor protein of cochlear outer hair cells. Nature. 2000;405:149–55. doi: 10.1038/35012009. [DOI] [PubMed] [Google Scholar]
- 22.Murayama E, Herbomel P, Kawakami A, Takeda H, Nagasawa H. Otolith matrix proteins OMP-1 and Otolin-1 are necessary for normal otolith growth and their correct anchoring onto the sensory maculae. Mechanisms of Development. 2005;122:791–803. doi: 10.1016/j.mod.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Murayama E, Takagi Y, Ohira T, Davis JG, Greene MI, Nagasawa H. Fish otolith contains a unique structural protein, otolin-1. Eur J Biochem. 2002;269:688–696. doi: 10.1046/j.0014-2956.2001.02701.x. [DOI] [PubMed] [Google Scholar]
- 24.Di Schiavi E, Riano E, Heye B, Bazzicalupo P, Rugarli EI. UMODL1/Olfactorin is an extracellular membrane-bound molecule with a restricted spatial expression in olfactory and vomeronasal neurons. European Journal of Neuroscience. 2005;21:3291–3300. doi: 10.1111/j.1460-9568.2005.04164.x. [DOI] [PubMed] [Google Scholar]
- 25.Shibuya K, Nagamine K, Okui M, Ohsawa Y, Asakawa S, Minoshima S, Hase T, Kudoh J, Shimizu N. Initial characterization of an uromodulin-like 1 gene on human chromosome 21q22.3. Biochemical and Biophysical Research Communications. 2004;319:1181–1189. doi: 10.1016/j.bbrc.2004.05.094. [DOI] [PubMed] [Google Scholar]
- 26.Yasunaga S, Grati M, Chardenoux S, Smith TN, Friedman TB, Lalwani AK, Wilcox ER, Petit C. OTOF encodes multiple long and short isoforms: genetic evidence that the long ones underlie recessive deafness DFNB9. Am J Hum Genet. 2000;67:591–600. doi: 10.1086/303049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zine A, Romand R. Development of the auditory receptors of the rat: a SEM study. Brain Res. 1996;721:49–58. doi: 10.1016/0006-8993(96)00147-3. [DOI] [PubMed] [Google Scholar]
- 28.Geleoc GS, Holt JR. Developmental acquisition of sensory transduction in hair cells of the mouse inner ear. Nat Neurosci. 2003;6:1019–20. doi: 10.1038/nn1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jongeneel CV, Iseli C, Stevenson BJ, Riggins GJ, Lal A, Mackay A, Harris RA, O'Hare MJ, Neville AM, Simpson AJG, Strausberg RL. Comprehensive sampling of gene expression in human cell lines with massively parallel signature sequencing. PNAS. 2003;100:4702–4705. doi: 10.1073/pnas.0831040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuersten S, Goodwin EB. The Power of the 3′ UTR: Translational Control and Development. Nature Reviews Genetics. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 31.Elnitski L, Hardison RC, Li J, Yang S, Kolbe D, Eswara P, O'Connor MJ, Schwartz S, Miller W, Chiaromonte F. Distinguishing regulatory DNA from neutral sites. Genome Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shalgi R, Lapidot M, Shamir R, Pilpel Y. A catalog of stability-associated sequence elements in 3′ UTRs of yeast mRNAs. Genome Biol. 2005;6:R86. doi: 10.1186/gb-2005-6-10-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs Confer Robustness to Gene Expression and Have a Significant Impact on 3′UTR Evolution. Cell. 2005;123:1133–46. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 34.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iseli C, Stevenson BJ, de Souza SJ, Samaia HB, Camargo AA, Buetow KH, Strausberg RL, Simpson AJ, Bucher P, Jongeneel CV. Long-range heterogeneity at the 3′ ends of human mRNAs. Genome Res. 2002;12:1068–74. doi: 10.1101/gr.62002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toriello H, Reardon W, Gorlin R, editors. Hereditary Hearing Loss and its Syndromes. Oxford University Press; Oxford: 2004. [Google Scholar]
- 37.Kimberling W. Genetic Hearing Loss Associated with Eye Disorders. In: Toriello H, Reardon W, Gorlin R, editors. Hereditary Hearing Loss and its Syndromes. Oxford University Press; Oxford: 2004. pp. 126–165. [Google Scholar]
- 38.Chen CK, Burns ME, Spencer M, Niemi GA, Chen J, Hurley JB, Baylor DA, Simon MI. Abnormal photoresponses and light-induced apoptosis in rods lacking rhodopsin kinase. PNAS. 1999;96:3718–3722. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweet HO, Brilliant MH, Cook SA, Johnson KR, Davisson MT. Brief communication. A new allelic series for the underwhite gene on mouse chromosome 15. J Hered. 1998;89:546–551. doi: 10.1093/jhered/89.6.546. [DOI] [PubMed] [Google Scholar]
- 40.Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–55. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 41.Delprat B, Ruel J, Guitton MJ, Hamard G, Lenoir M, Pujol R, Puel JL, Brabet P, Hamel CP. Deafness and Cochlear Fibrocyte Alterations in Mice Deficient for the Inner Ear Protein Otospiralin. Mol Cell Biol. 2005;25:847–853. doi: 10.1128/MCB.25.2.847-853.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhoeven K, Van Laer L, Kirschhofer K, Legan PK, Hughes DC, Schatteman I, Verstreken M, Van Hauwe P, Coucke P, Chen A, Smith RJ, Somers T, Offeciers FE, Van de Heyning P, Richardson GP, Wachtler F, Kimberling WJ, Willems PJ, Govaerts PJ, Van Camp G. Mutations in the human alpha-tectorin gene cause autosomal dominant non-syndromic hearing impairment. Nat Genet. 1998;19:60–2. doi: 10.1038/ng0598-60. [DOI] [PubMed] [Google Scholar]
- 43.Mustapha M, Weil D, Chardenoux S, Elias S, El-Zir E, Beckmann JS, Loiselet J, Petit C. An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum Mol Genet. 1999;8:409–12. doi: 10.1093/hmg/8.3.409. [DOI] [PubMed] [Google Scholar]
- 44.Simmler MC, Cohen-Salmon M, El-Amraoui A, Guillaud L, Benichou JC, Petit C, Panthier JJ. Targeted disruption of otog results in deafness and severe imbalance. Nat Genet. 2000;24:139–43. doi: 10.1038/72793. [DOI] [PubMed] [Google Scholar]
- 45.Asamura K, Abe S, Fukuoka H, Nakamura Y, Usami S-i. Mutation analysis of COL9A3, a gene highly expressed in the cochlea, in hearing loss patients. Auris Nasus Larynx. 2005;32:113. doi: 10.1016/j.anl.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki N, Asamura K, Kikuchi Y, Takumi Y, Abe S, Imamura Y, Hayashi T, Aszodi A, Fassler R, Usami S-i. Type IX collagen knock-out mouse shows progressive hearing loss. Neuroscience Research. 2005;51:293–298. doi: 10.1016/j.neures.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Kowalski PE, Thalmann I, Ornitz DM, Mager DL, Thalmann R. Otoconin-90, the mammalian otoconial matrix protein, contains two domains of homology to secretory phospholipase A2. PNAS. 1998;95:15345–15350. doi: 10.1073/pnas.95.26.15345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Legan PK, Rau A, Keen JN, Richardson GP. The Mouse Tectorins. Modular Matrix Proteins of the Inner Ear Homologous to Components of the Sperm-Egg Adhesion System. J Biol Chem. 1997;272:8791–8801. doi: 10.1074/jbc.272.13.8791. [DOI] [PubMed] [Google Scholar]
- 49.Zwaenepoel I, Mustapha M, Leibovici M, Verpy E, Goodyear R, Liu XZ, Nouaille S, Nance WE, Kanaan M, Avraham KB, Tekaia F, Loiselet J, Lathrop M, Richardson G, Petit C. Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. PNAS. 2002;99:6240–6245. doi: 10.1073/pnas.082515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


