Abstract
The specific binding of HIV-1 nucleocapsid protein (NC) to the different forms assumed in vitro by the stemloop 1 (Lai variant) of the genome’s packaging signal has been investigated using electrospray ionization-Fourier transform mass spectrometry (ESI-FTMS). The simultaneous observation of protein-RNA and RNA-RNA interactions in solution has provided direct information about the role of NC in the two-step model of RNA dimerization and isomerization. In particular, two distinct binding sites have been identified on the monomeric stemloop structure, corresponding to the apical loop and stem-bulge motifs. These sites share similar binding affinities that are intermediate between those of stemloop 3 (SL3) and the putative stemloop 4 (SL4) of the packaging signal. Binding to the apical loop, which contains the dimerization initiation site (DIS), competes directly with the annealing of self-complementary sequences to form a metastable kissing-loop (KL) dimer. In contrast, binding to the stem-bulge affects indirectly the monomer-dimer equilibrium by promoting the rearrangement of KL into the more stable extended duplex (ED) conformer. This process is mediated by the duplex-melting activity of NC, which destabilizes the intramolecular base pairs surrounding the KL stem-bulges and enables their exchange to form the inter-strand pairs that define the ED structure. In this conformer, high-affinity binding takes place at stem-bulge sites that are identical to those present in the monomeric and KL forms. In this case, however, the NC-induced ‘breathing’ does not result in dissociation of the double-stranded structure because of the large number of intermolecular base pairs. The different binding modes manifested by conformer-specific mutants have shown that NC can also provide low affinity interactions with the bulged-out adenines flanking the DIS region of the ED conformer, thus supporting the hypothesis that these exposed nucleotides may constitute ‘base-grips’ for protein contacts during the late stages of the viral lifecycle.
Keywords: RNA dimerization, RNA isomerization, NC binding, NC chaperone, mass spectrometry
Introduction
Human immunodeficiency virus type-1 (HIV-1) selectively packages two copies of genomic RNA that are non-covalently linked by the dimer linkage structure (DLS) of the 5’ leader.1–5 The formation of DLS involves the annealing of a self-complementary sequence located in the loop region of the highly conserved stemloop 1 (SL1) called the dimerization initiation site (DIS).2; 3; 6; 7 Deletions and mutations of the DIS sequence affect not only dimerization, but also packaging of genomic RNA in vivo,8–10 thus suggesting a close functional relationship between these crucial steps of the viral lifecycle.11 This hypothesis is further supported by the fact that both processes are mediated by the nucleocapsid (NC) domain of the viral Gag polyprotein,1; 12; 13 which exercises broad nucleic acid binding and chaperone activities.14; 15 Unfortunately, the specific roles played by RNA-RNA and protein-RNA interactions in dimerization and packaging are still not completely understood in spite of extensive study.16; 17
A two-step model has been proposed for genome dimerization, which involves the initial hybridization of the DIS palindromes of homologous strands to form a transient kissing-loop (KL) dimer (Scheme 1).2; 3; 7; 18–20 Experiments performed in vitro have shown that the metastable KL complex can subsequently isomerize into a more stable extended duplex (ED) that involves extensive intermolecular base-pairing along the entire SL1 sequence (Scheme 1).21–25 The transition between KL and ED forms is catalyzed in vitro by NC,23; 26–29 in a process that could explain the increased thermal stability observed for genomic RNA of HIV-1 and Moloney murine leukemia virus upon protease-dependent viral maturation.30–32 The 3D structures of SL1 constructs in monomeric,33–35 KL,36–40 and ED form38; 41–43 support the two-step dimerization model. However, further progress in elucidating the mechanism of dimer isomerization has been hampered by the lack of equivalent structural information for the individual complexes formed by NC with the different forms assumed by SL1.
Scheme 1.
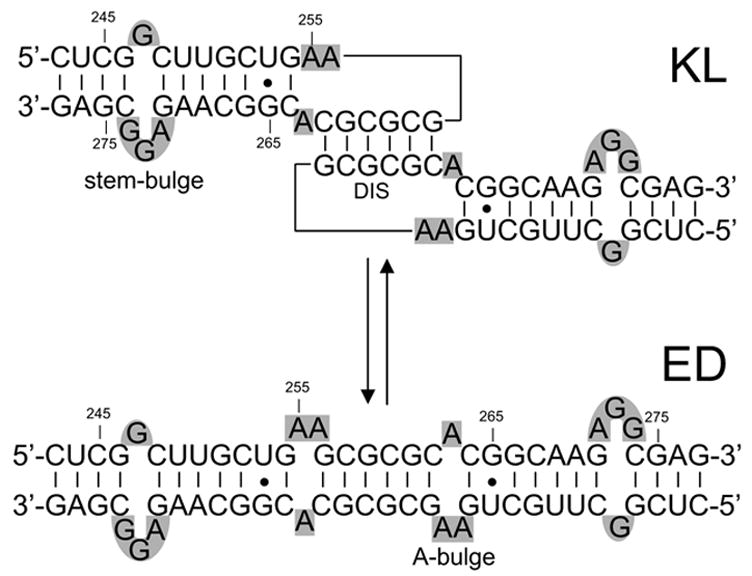
Sequence and secondary structures of the kissing loop (KL) and extended duplex (ED) dimers formed by wild-type SL1. Nucleotides are numbered according to the Lai variant genome of HIV-1. Stem- and A-bulges are highlighted in gray.
The specific interactions between NC and monomeric SL1 have been studied in vitro using fluorescence quenching techniques that suggested the presence of two distinct binding sites located on the loop and stem-bulge of the RNA stemloop (Scheme 2).35; 44 However, a construct with mutated loop and wild-type bulge was found incapable of binding NC by native gel electrophoresis and NMR,34 thus casting doubts on the actual number and location of the binding sites present on the stemloop structure. The interactions with dimeric SL1 have been investigated with the goal of understanding their effects on RNA conformation, while the common practice of removing the protein prior to RNA analysis has precluded the concomitant acquisition of direct information about the nature of the protein-RNA interactions driving the isomerization process. In the absence of high-resolution data, important questions regarding the activities performed by NC in the processes of RNA dimerization and structural rearrangement remain unanswered, including: a) the location of the NC binding sites on the wild-type SL1 structure, b) which oligomeric form of SL1 is recognized by NC, and c) the number of NC molecules participating in the process.
Scheme 2.
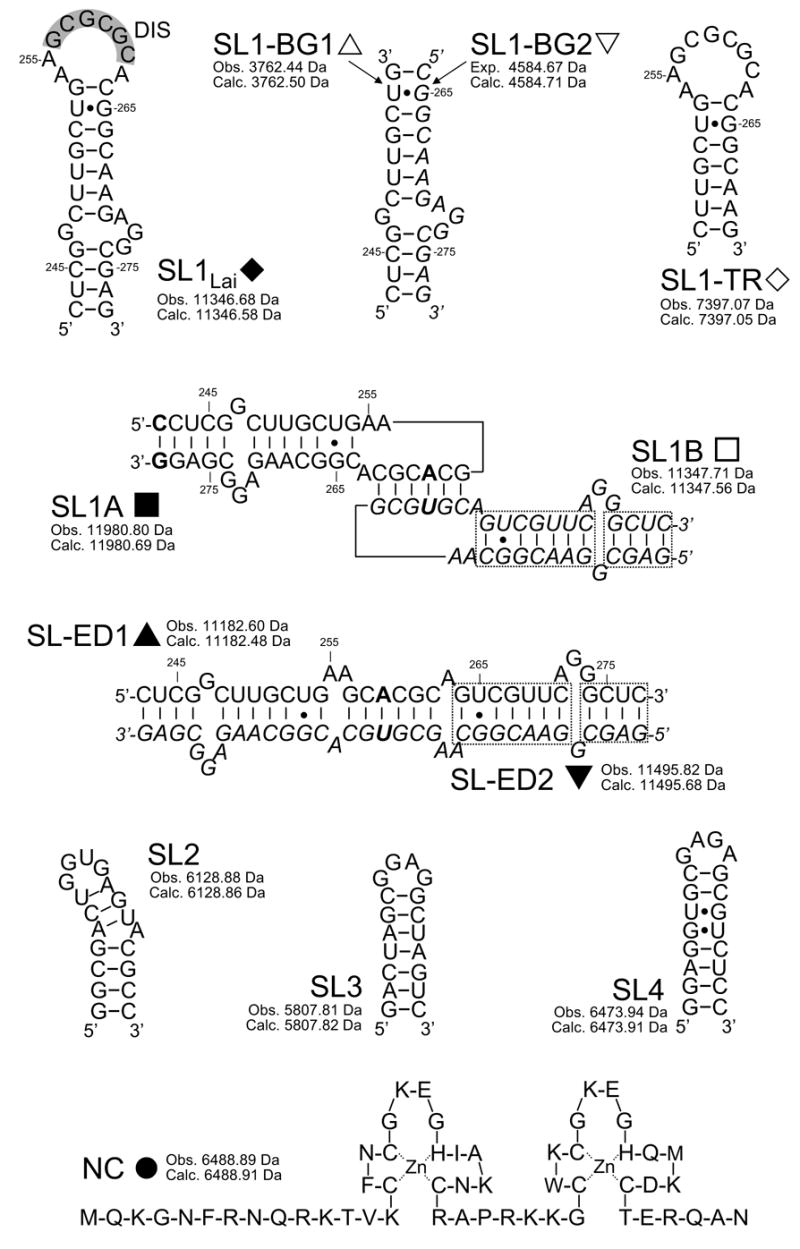
Sequences and secondary structures of the species used in the study. Nucleotides are numbered according to the Lai variant of the HIV-1 genome. The self-complementary sequence of wild-type SL1Lai is highlighted in gray. In SL1A, the G to A substitution in position 259 and a GC pair added at the end of the stem are marked in bold (see text for complete explanations). In SL1B, the G to U substitution in position 260 is marked in bold. The dashed-boxes highlight stem sequences that were swapped to obtain conformer-specific constructs. In all dimeric/duplex structures, one of the individual constructs was italicized for clarity. For each species, the monoisotopic masses observed experimentally and calculated from sequence are included.
These questions were addressed using electrospray ionization Fourier transform mass spectrometry (ESI-FTMS),45–47 which allowed us to overcome the intrinsic experimental hurdles associated with monitoring simultaneously the RNA dimerization and the protein binding events. This soft ionization technique can transfer noncovalent assemblies into the gas-phase while preserving their solution-phase association state.48–50 For this reason, it can be employed to determine the composition, stoichiometry, and binding affinity of large macromolecular complexes consisting of proteins, DNA, RNA, small molecule ligands, and other components (reviewed in ref.s 51–53). Taking advantage of this characteristic, we have recently employed ESI-FTMS to determine the NC binding modes with short single-stranded DNA oligonucleotides54 and with the hairpins SL2, SL3, and SL4,55 which are present with SL1 in the packaging signal of viral RNA.56–58 We have now extended this approach to investigate the specific interactions between NC and full-length SL1 (Lai variant) and the effects of protein binding on dimerization and isomerization. The direct observation of RNA-RNA and protein-RNA interactions has provided valuable information on the role of NC in the two-step model. The long term goal of these studies is to elucidate the molecular basis of the interactions governing genome dimerization and packaging, which is necessary for the development of new antiviral strategies aimed at interfering with these crucial processes of the HIV-1lifecycle.
Results
NC interactions with monomeric SL1Lai
The investigation of the binding modes of NC with SL1Lai can be confounded by the concomitant presence of monomer-dimer equilibria in solution. For this reason, a mutant was initially employed to decouple protein binding from RNA dimerization and facilitate the elucidation of the determinants of NC binding. This construct was obtained by replacing a central loop guanine with adenine (SL1A, Scheme 2), which has been shown to inhibit dimerization by disrupting the self-complementary nature of the DIS sequence.19 In control experiments performed on this construct, only monomeric SL1A was observed by ESI-FTMS under conditions optimized for noncovalent complexes (see Materials and Methods), thus confirming the desired effects of the DIS mutation on RNA dimerization (Figure 1a). Addition of NC to the dimerization-deficient mutant provided stable assemblies with a 1:1 and a 2:1 protein to RNA stoichiometry (Figure 1b). The observed masses were in excellent agreement with those calculated from the sequence of the protein and RNA components (provided in Scheme 2), including two zinc(II) ions per NC unit.55 Further increases of NC in solution did not produce higher order complexes, thus confirming the presence of two distinct sites on the SL1A structure (Figure 1c). The detection of a 2:1 complex in the presence of free SL1A in solution (Figure 1b) indicates that binding to the second site was initiated before saturation of the first was complete, which is consistent with sites sharing similar affinities and negligible cooperativity.
Fig. 1.
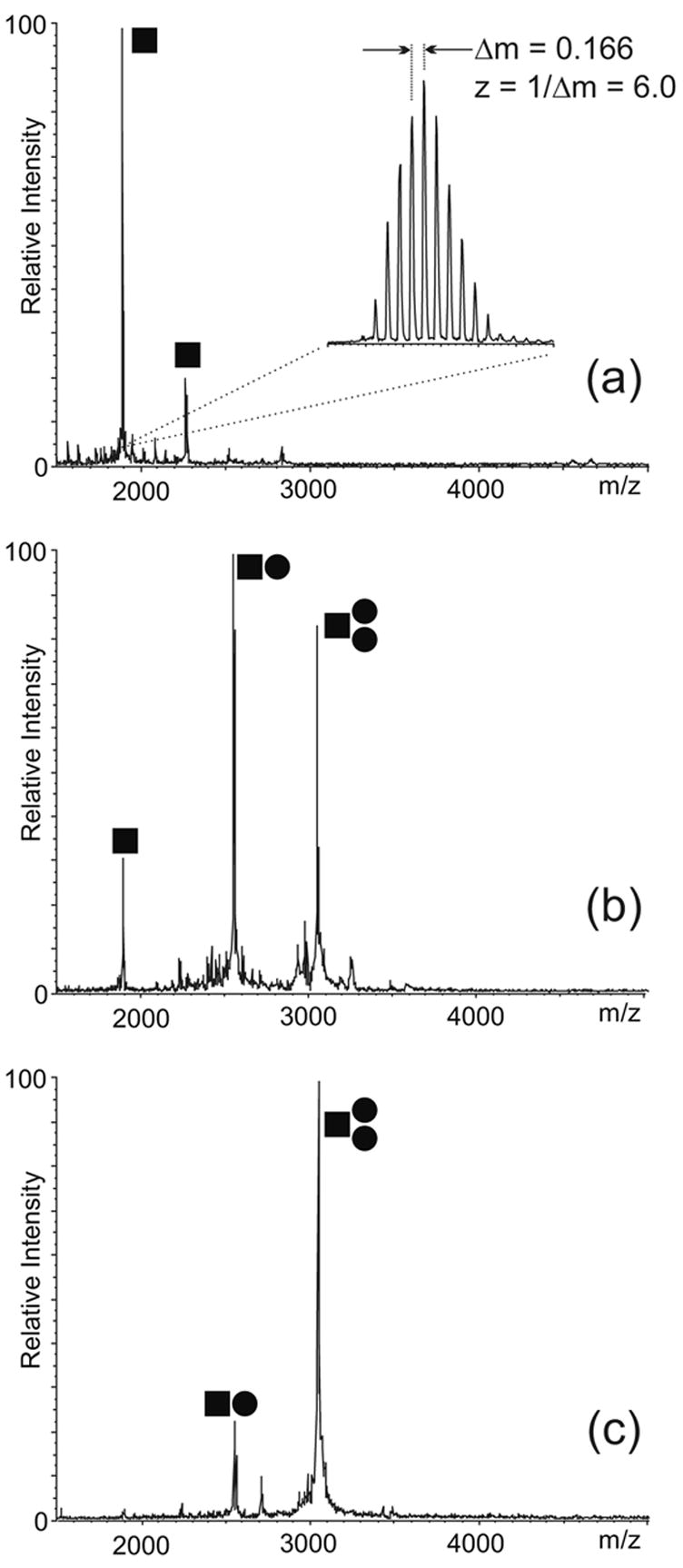
ESI-FTMS of NC binding to 4 μM SL1A in 100 mM ammonium acetate (pH 7.0): (a) control; (b) 4 μM NC; (c) 12 μM NC. The ● symbol corresponds to NC, while ■ corresponds to SL1A. The signal corresponding to the 6- ion is enlarged in the inset to show how the charge state of each species is unambiguously determined from the respective isotopic spacing. The mass is then readily obtained from the charge state and 23 the position of the peak on the m/z scale. This sample provided a monoisotopic mass of 11330.74 Da, which matches very closely the theoretical mass of 11330.59 Da calculated from sequence. In this direction, Scheme 2 provides experimental and calculated masses for the constructs used in the study, which enable to assess the typical mass accuracy achieved by these determinations. (Note that the SL1A construct used here lacks the non-native GC base pair present in the mutant in Scheme 2, which is reflected by the different masses).
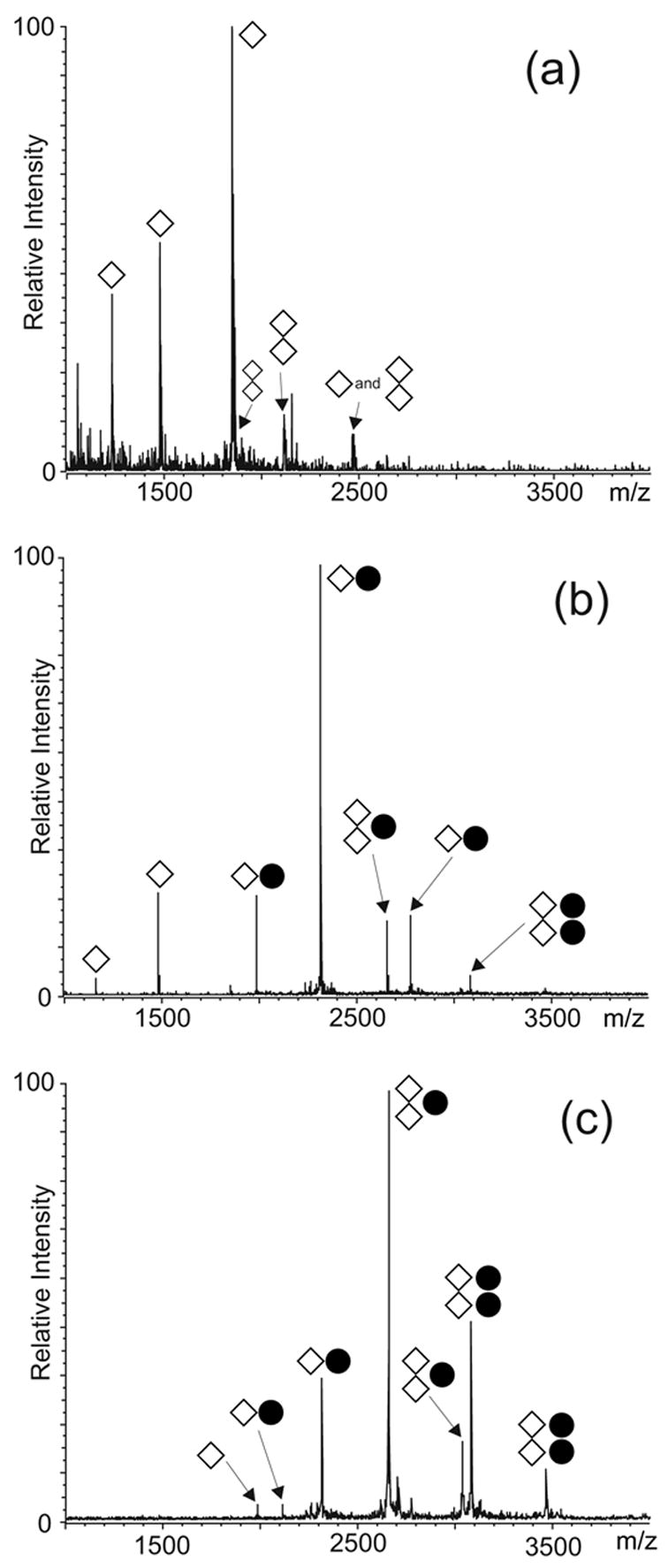
An unambiguous evaluation of the affinity of NC for monomeric SL1Lai was provided by competitive binding experiments in which SL1A was directly compared with isolated SL2, SL3, and SL4 stemloops of the packaging signal.56–58 This approach relies on the intrinsic ability of ESI-FTMS to resolve multiple complexes in solution according to their unique molecular masses and without resorting to labeling or separation techniques. As demonstrated in earlier work, the comparable size and chemical composition of these protein-RNA assemblies translate into very similar ionization characteristics.55 In this way, the simultaneous detection of the complexes in a single analysis provides an unambiguous and unbiased scale of relative binding affinities. According to this approach, NC was titrated into a 150 mM ammonium acetate solution (pH 7.0) containing 5 bM of each RNA hairpin (Figure 2a). At low protein concentrations, the NC·SL2 and NC·SL3 assemblies were readily observed with relatively low free/bound ratios in solution (Figure 2b), which were consistent with the high binding affinities manifested by these stemloops under the selected experimental conditions.55 A stable NC·SL1A complex including one protein unit was also detected with a free/bound ratio consistent with a weaker binding affinity, while the NC·SL4 assembly exhibited a ratio corresponding to the weakest affinity of the series (Figure 2b). Further protein additions induced a significant increase of both NC·SL2 and NC·SL3, the binding of a second unit of NC to SL1A, and a moderate increase of the NC·SL4 complex (Figure 2c). Overall, these experiments provided the following relative scale of binding affinities in 150 mM ammonium acetate: SL2 > SL3 > SL1’ ≈ SL1” > SL4.
Fig. 2.
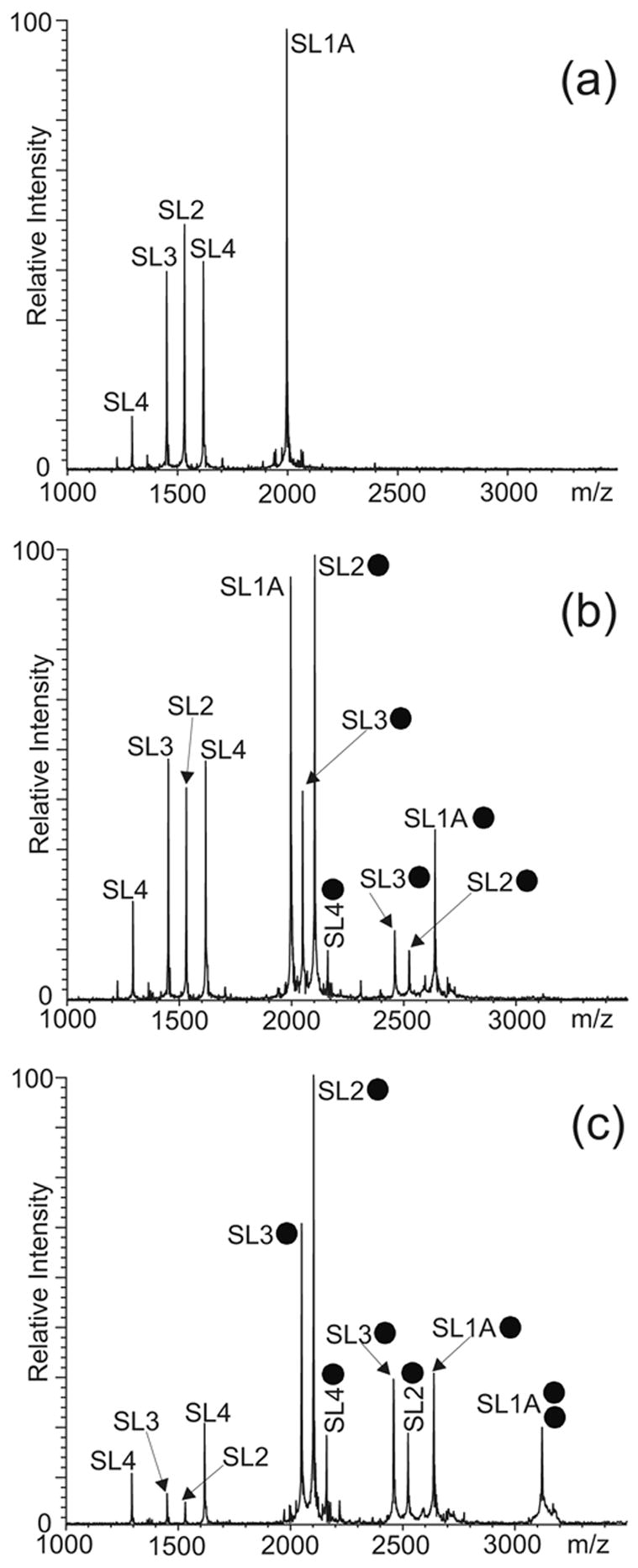
ESI-FTMS of competitive NC binding between 5 μM each of SL1A, SL2, SL3, and SL4 hairpins of HIV-1 packaging signal in 150 mM ammonium acetate (pH 7.0): (a) control; (b) 5 μM NC; (c) 20 μM NC. The ● symbol corresponds to NC. The charge states of the different anionic species are not indicated.
The possibility that the G to A mutation introduced in SL1A might have unexpected consequences on the NC binding mode was tested in competition experiments comparing the dimerization-deficient and wild-type SL1Lai constructs (data not shown). In this case, a lower ionic strength (i.e., 10 mM ammonium acetate, pH 7.0) was employed to enable a fair competition between monomeric substrates by weakening the RNA-RNA interactions that promote the dimerization of wild-type SL1Lai. The fact that the competing monomers manifested comparable affinities indicated that the single base mutation did not cause adverse effects on the specific NC interactions, thus validating the use of the dimerization-deficient SL1A as the model monomeric form for binding experiments within the selected range of ionic strengths (i.e., 10–150 mM ammonium acetate).
In light of the preference of NC for single-stranded nucleic acids,54; 59–61 the structural motifs comprising distinctive unpaired regions were reproduced in separate constructs to investigate their individual ability to sustain protein binding. The first was a mutant SL1 truncated just above the internal bulge, which provided a structure spanning the DIS loop and the upper stem region (SL1-TR, Scheme 2). The second was obtained by annealing two individual RNA strands that recreated the stem-bulge, but eliminated the apical loop (SL1-BG1/2, Scheme 2). When titrated with NC, the monomeric SL1-TR and intact duplex SL1-BG1/2 formed stable protein-RNA complexes (vide infra), thus confirming that the apical loop and the stem-bulge can function as individual binding sites. The site locations on the SL1 structure were further confirmed by footprinting and crosslinking experiments in a separate study (E. Yu and D. Fabris, manuscript in preparation).
Effects of NC binding on SL1 dimerization
The interactions of NC with the two sites have distinctive effects on the dimerization process in vitro. In fact, in addition to possessing ideal features for NC binding, the apical loop contains the palindromic DIS sequence that promotes the initial intermolecular base-pairing essential to dimer formation (Scheme 1). Therefore, the ability of NC to interact with DIS is expected to directly affect the oligomeric state of SL1Lai and vice versa, in a possible competition between protein binding and RNA dimerization. In contrast, the stem-bulge motif is conserved in the secondary structures of monomeric as well as dimeric SL1Lai (Scheme 1). For this reason, specific binding to this structure is expected to play only an indirect role in the process of RNA dimerization. The possible interplay between these concomitant but independent effects was explored by using preformed dimers in either KL or ED conformation.
The intrinsic difficulty of differentiating the two conformers on the basis of their identical molecular mass was overcome by designing SL1 mutants of different base composition, which presented unique masses and could assume only one of the dimeric forms without inter-conversion. An obligated KL dimer was obtained by combining the SL1A mutant with a construct that included a C to U mutation in position 260 and a reversed base-pairing pattern in the stem structure (SL1B, Scheme 2).20; 23; 28 The base substitution was introduced to enable exclusive loop-loop interactions with the complementary SL1A sequence and induce the formation of an immediately recognizable KL heterodimer (Scheme 2). The remaining modifications eliminated any intermolecular complementarity between the corresponding stem regions, thus ensuring that no ED structure could be produced by isomerization of the initial KL conformer. In analogous way, an obligated ED dimer was obtained by swapping the stem strands to provide two individual constructs, called SL1-ED1 and SL1-ED2 (Scheme 2), which lacked the ability to form the intramolecular base pairs necessary to fold into individual stemloops, but maintained the extensive inter-strand complementarity necessary to produce a bulged hetero-duplex analogous to the wild-type ED structure.23 As shown by the ESI-FTMS spectrum obtained from a solution including both conformer-specific samples (Figure 3a), only the expected SL1A/B and SL1-ED1/2 heterodimers were obtained by appropriately mixing and heat-refolding the constructs in 150 mM ammonium acetate (see Materials and Methods). The absence of homodimeric species in solution (e.g., SL1A/A, SL1B/B, SL1-ED1/1, or SL1-ED2/2) proved the validity of the mutants design by showing that the observed RNA-RNA interactions followed the base-pairing rules. Furthermore, the absence of mismatched dimers or higher order RNA complexes confirmed the ability of this analytical platform to provide a valid representation of the solution equilibria without nonspecific strand aggregation.62 These experiments served also to demonstrate that the KL dimer could be observed intact despite the relatively weak intermolecular stabilization offered by the presence of only 6 inter-strand base pairs in its structure.
Fig. 3.
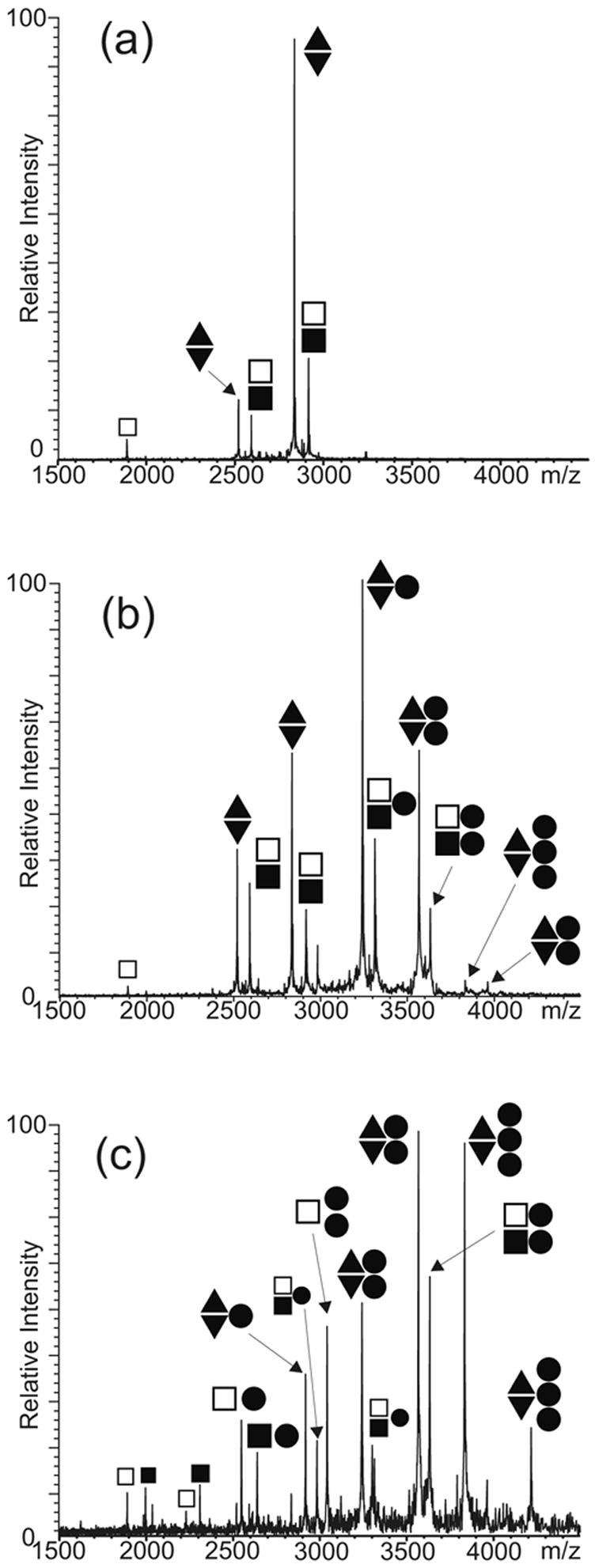
ESI-FTMS of competitive NC binding between 5 μM each of preformed SL1A/B and SL1-ED1/2 dimers in 150 mM ammonium acetate (pH 7.0): (a) control; (b) 10 μM NC; (c) 30 μM NC. The ● symbol corresponds to NC; ■ to SL1A; □ to SL1B; ▲ to SL1-ED1; and ▼ to SL1-ED2. The charge states of the different anionic species are not indicated.
The binding of NC to the dimeric forms was investigated by titrating preformed SL1A/B and SL1-ED1/2 complexes in competition experiments. Initial addition of 10 bM NC to a solution containing 5 bM each of the KL- and ED-obligated constructs produced predominantly 1:2 and 2:2 protein to RNA assemblies for both dimeric forms (Figure 3b). No significant differences in relative affinities could be detected between substrates, thus suggesting that NC bound to structural features present in both conformers. Increasing protein concentrations, however, induced very distinctive effects on the oligomeric state of the two substrates. In the case of the KL-obligated dimer, the addition of 30 μM NC induced a noticeable decrease of the overall proportion of dimeric assemblies in solution, which was accompanied with an increase of the corresponding stemloop components bound to one or two NC units (Figure 3c). In contrast, no dissociation of the ED-obligated dimer into individual strands was observed at this protein concentration. Furthermore, the maximum stoichiometry detected for the KL-dimer remained 2:2 protein to RNA, while abundant 3:2 assemblies were obtained for the ED conformer in the same sample solution (Figure 3c). A 4:2 assembly was also observed for the SL1-ED1/2 construct at higher protein concentrations. The binding of two NC units to both KL and ED structures could be accounted for by the presence of the conserved stem-bulge motifs in both conformers. However, additional binding to the ED-obligated dimer can be explained only by the presence of alternative sites that are unique to this conformer’s structure and become active only after the primary sites have been occupied.
Competitive binding experiments between the SL1-TR mutant and the SL1-ED1/2 dimer were carried out to test whether the unpaired adenines flanking the DIS sequence (A-bulges, Scheme 1) could constitute suitable sites for protein interactions (Figure 4). In this case, the truncated construct was appropriately heat-refolded in 150 mM ammonium acetate to obtain an extended duplex lacking the stem-bulges characteristic of full-length constructs (see Materials and Methods).42; 43 At low concentrations, NC was shown to bind exclusively to the SL1-ED1/2 dimer with formation of 1:2 and 2:2 protein to RNA assemblies (Figure 4b). Only at higher concentrations did NC bind to the SL1-TR duplex, while a 3:2 complex was obtained from the SL1-ED1/2 substrate in the same solution (Figure 4c). Considering that the A-bulges constitute the only single-stranded regions present in the truncated duplex, the detection of this NC-complex is consistent with the ability of the DIS-flanking structures to sustain effective protein interactions. The fact that the full-length dimer titrated at lower NC concentrations than the truncated duplex indicated that the G-rich stem-bulges possess a higher affinity than the A-bulges, which agrees with the known preference of NC for unpaired Gs.54; 59–61 Therefore, these results suggest that the stem-bulges constitute the primary binding sites on the ED conformer, while the A-bulges provide viable but weaker alternatives.
Fig. 4.
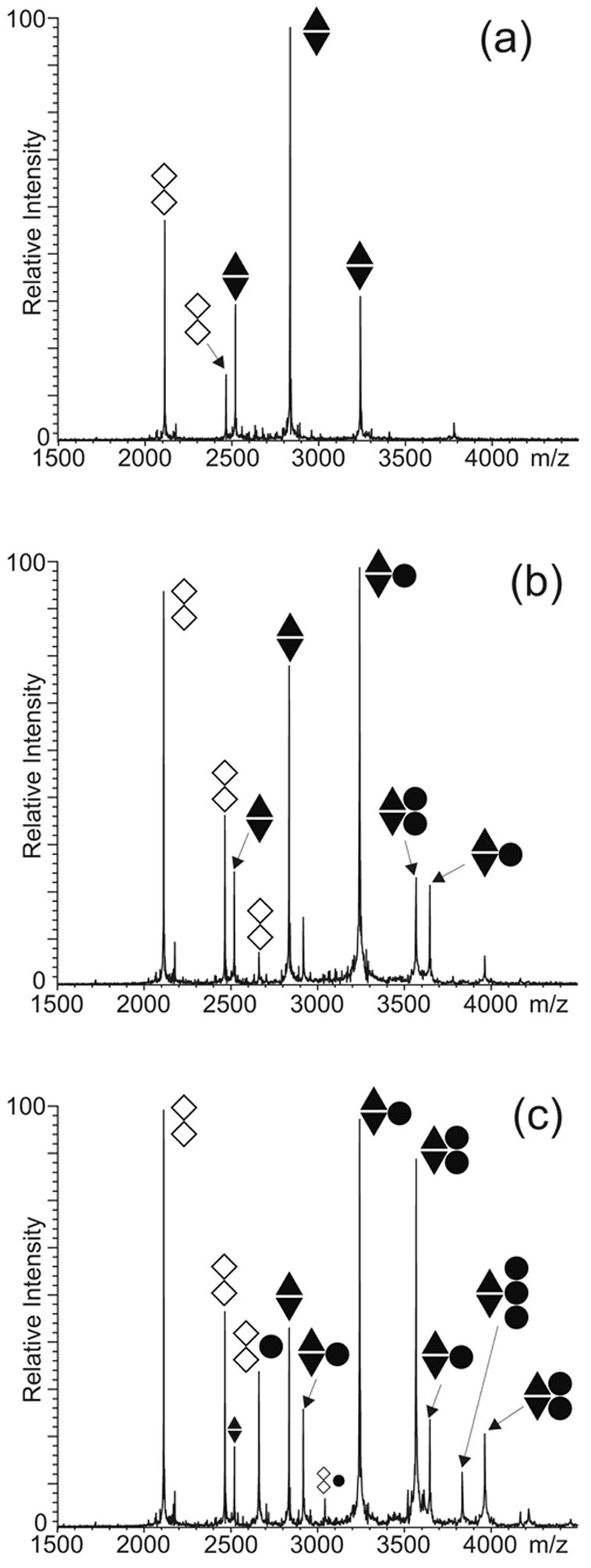
ESI-FTMS of competitive NC binding between 5 μM each of preformed SL1-ED1/2 and SL1-TR folded in the ED conformation (see text) in 150 mM ammonium acetate (pH 7.0): (a) control; (b) 5 μM NC; (c) 10 μM NC. The ● symbol corresponds to NC; ▲ to SL1-ED1; ▼ to SL1-ED2; and ◇ to SL1-TR. The charge states of the different anionic species are not indicated.
Effects of NC on dimer isomerization
The characteristic stoichiometries observed for the conformer-specific substrates provided the key to interpret the results obtained from the wild-type construct capable of assuming both KL and ED conformations. When a 5μM SL1Lai sample (Scheme 2) was heat-denatured, snap-cooled on ice, and then incubated at 37°C for 30 min. in 150 mM ammonium acetate (see Materials and Methods), both monomeric and dimeric species were readily observed in solution, as expected from a monomer-dimer equilibrium in solution (Figure 5a). Consistent with the fast annealing procedure and with the results of parallel crosslinking experiments (E. Yu and D. Fabris, manuscript in preparation), the detected dimer represented predominantly the kinetically trapped KL product. Incubating this sample in the presence of 5μM NC did not significantly increase the proportion of dimer versus monomer in solution, while protein binding was observed for both monomeric and dimeric substrates. In particular, abundant 1:2 and 2:2 protein to RNA assemblies were detected together with a weaker but recognizable 3:2 complex (Figure 5b). Based on the inability of the KL-obligated dimer to bind more than two protein units (Figure 3b-c), the 3:2 species was ascribed to the presence of small amounts of ED conformer in solution, which could be either formed with very low yields during the initial heat-refolding, or result from the NC-induced isomerization of the initial kinetic product KL. However, the dramatic increase of higher order complexes (i.e., 3:2 and 4:2) upon addition of 10 μM NC in solution could be attributed only to the transition of initial KL dimer to the thermodynamic ED product, which is a direct result of the chaperone activity of NC (Figure 5c). When added in large excess, NC induced complete dimer dissociation and allowed for the detection of 2:1 and 3:1 complexes of monomeric SL1Lai, as expected from its duplex melting capabilities (Figure 5d).14; 15
Fig. 5.
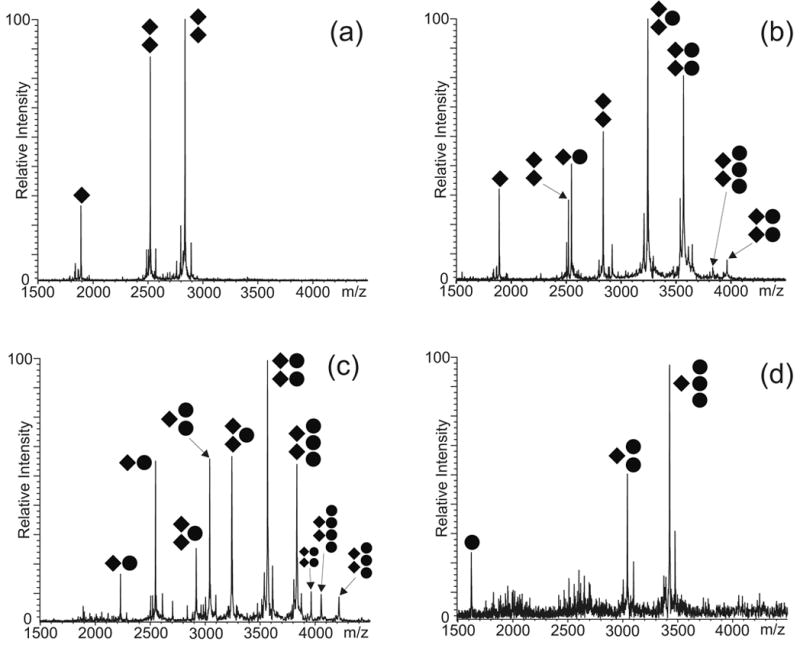
ESI-FTMS resulting from the NC titration of 5 μM of wild-type SL1Lai in 150 mM ammonium acetate (pH 7.0): (a) control showing approximately 82% dimer; (b) 5 μM NC showing ~86% dimer; (c) 10 μM NC, ~60% dimer; (d) 20 μM NC, ~0% dimer. The ● symbol corresponds to NC; ◆ to SL1Lai. The charge states of the different anionic species are not indicated. Percent dimer was calculated as described in Material and Methods.
The previously described SL1-TR and SL1-BG1/2 substrates (Scheme 2) were employed to isolate the possible roles of the individual binding sites in the isomerization process. After preliminary experiments had shown that only dimeric species could be exclusively detected in SL1-TR samples prepared in 150 mM ammonium acetate (see for example Figure 4a), the ionic strength was significantly decreased to weaken the RNA-RNA interactions and facilitate the observation of the monomer-dimer equilibrium (e.g., 10 mM ammonium acetate, Figure 6a). As observed for full-length SL1Lai, addition of protein provided assemblies involving RNA monomer and dimers (Figure 6b and c). Unlike in the wild-type’s case, however, NC induced also a very significant increase of the proportion of dimeric versus monomeric species in solution, in spite of the rather unfavorable ionic strength. In contrast, when the SL1-BG1/2 construct was titrated with NC, progressive dissociation of the duplex structure was found to accompany the formation of complexes with double- and single-stranded substrates in solution (Figure 7a-c). The dissociating effects were observed even in environments of higher ionic strength (i.e., in the 100 mM ammonium acetate solution used here), which are expected to strengthen the interactions between complementary RNA strands. These experiments clearly highlighted how seemingly contrasting functions, such as strand annealing and duplex melting, can be performed by NC depending on the substrate structural context. As discussed below, the combination of the different facets of the chaperone activity of NC is critical for the isomerization process of full-length SL1Lai.
Fig. 6.
ESI-FTMS resulting from the NC titration of 4 μM of SL1-TR in 10 mM ammonium acetate (pH 7.0): (a) control showing approximately ~5% dimer; (b) 4 μM NC, ~8% dimer; (c) 8 μM NC, ~71% dimer. The ● symbol corresponds to NC; ⋄ to SL1-TR. The charge states of the different anionic species are not indicated. Percent dimer was calculated as described in Material and Methods.
Fig. 7.
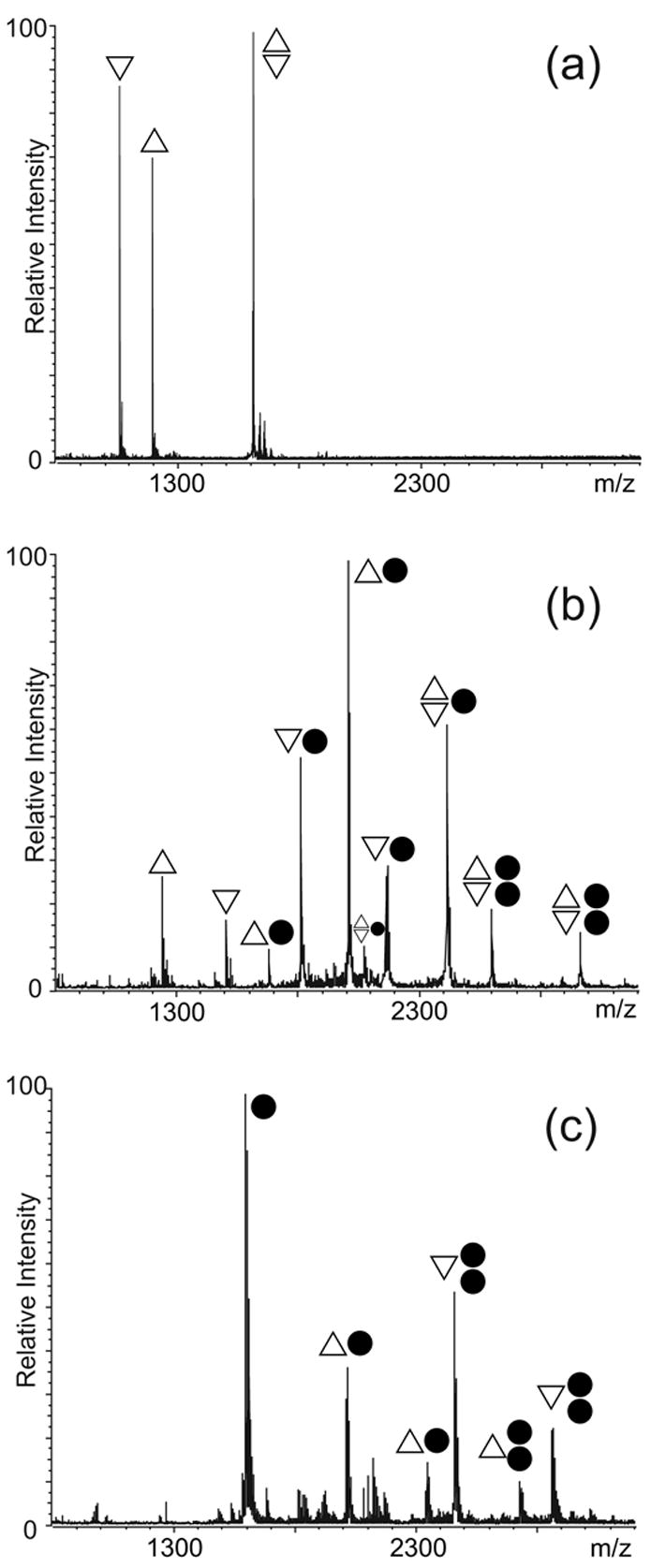
ESI-FTMS resulting from the NC titration of 5 μM of SL1-BG in 100 mM ammonium acetate (pH 7.0): (a) control showing approximately 42% dimer; (b) 5 μM NC, ~17% dimer; (c) 20 μM NC, 0% dimer. The ● symbol corresponds to NC; △ to SL1-BG1; ▽ to SL1-BG2. The charge states of the different anionic species are not indicated. Percent dimer was calculated as described in Material and Methods.
Discussion
The NC binding modes with the different forms assumed by SL1Lai in vitro have been dissected by a concerted strategy based on ad hoc RNA mutants and ESI-FTMS detection. In this way, protein-RNA complexes representing salient intermediates in the NC-mediated processes of dimerization and isomerization were effectively trapped and characterized using titrations and competitive binding experiments. In the absence of RNA-RNA interactions, experiments involving either the dimerization-deficient SL1A, or the wild-type SL1Lai under conditions unfavorable to RNA dimerization (i.e., low ionic strength), have unambiguously demonstrated the presence of two distinct binding sites on the monomeric stemloop structure. The apical loop and the stem-bulge were identified as the regions involved in protein-RNA interactions using substrates that alternatively eliminated these characteristic motifs (e.g., SL-TR and SL1-BG1/2). The presence of several unpaired Gs and the preference of NC for single-stranded nucleic acids54; 59–61 account for the stability of the observed complexes. The results provided by our approach corroborate earlier fluorescence studies pointing toward these regions as the putative sites for high-affinity interactions on the SL1 structure.35; 44
The binding patterns observed during the titration experiments indicate that the two sites share comparable affinities toward NC. In particular, the data obtained near the titration midpoint clearly show that occupancy of the second site initiates before the first is completely saturated (Figure 1b), which can be explained only on the basis of a fair competition between sites. In addition, the observation that a complex with one occupied site was accumulated in solution is consistent with the absence of significant cooperativity between sites, which would have enabled the second to be more readily saturated once the first was bound. The same considerations are valid also in regard of possible cooperativity involving protein-protein interactions, rather than communication between RNA sites.
The affinity of either SL1Lai site towards NC is intermediate between those exhibited by SL3 and SL4, as demonstrated by competition experiments including all four hairpins of the packaging signal (Figure 2). The simultaneous titration of the individual substrates combined in solution leaves no doubts about their relative affinities for NC, which follow the SL2 > SL3 > SL1’ ≈ SL1” > SL4 scale in 150 mM ammonium acetate. Although no dissociation constant (Kd) was explicitly determined for either SL1Lai site, close estimates could be inferred by considering that Kd values of 178 nM and 1.3 μM were respectively obtained for the NC·SL3 and NC·SL4 complexes using the same experimental approach.55 The observed scale of relative affinities is in excellent agreement with fluorescence studies of similar stemloop structures,44 but contrasts with earlier filter binding experiments that ranked SL1 at the top of the isolated hairpins.57 It should be noted, however, that direct comparisons between binding data provided by different techniques are made very difficult by the widely heterogeneous experimental conditions and by the significant effects of concentrations and ionic strength on binding, as shown earlier for the NC complexes with SL2, SL3, and SL4.55
Dimeric SL1 substrates provide different structures that are capable of sustaining specific NC interactions, as demonstrated by conformer-obligated mutants that could not undergo isomerization. In the KL dimer, a maximum of two binding sites were active under the selected experimental conditions (Figure 3), which were identified with the stem-bulges of the two stemloop components. In the ED structure, this motif is formed by nucleotides contributed by the cognate strands, but its structural features are identical to those observed in the monomeric and KL forms.38; 63 For this reason, it is not surprising that the stem-bulges provided similar binding affinities regardless of the oligomeric state of the actual substrate (Figure 3).
Additional sites on the ED structure are offered by the A-bulges flanking the annealed DIS sequences, which become competitive at higher NC concentrations (Figure 4). According to the high-resolution structures available for this dimeric conformer, A255 can alternatively participate in a zipper-like motif with the adenines on the opposite strand 42; 43, or assume a bulged-out conformation that could promote protein-RNA or RNA-RNA interactions.41 In this context, our results provide strong experimental support to the hypothesis that the A-bulges may indeed act as ‘base-grips’ for NC recognition. Furthermore, considering that the zipper-like motif increases the stability of strand association, protein binding could induce local destabilization by locking unpaired nucleobases in less favorable, bulged-out conformations. An analogous situation takes place in the KL dimer, in which the hinges between the stems and the loops are defined by a rather dynamic situation of the DIS-flanking bases.36; 37; 40; 64 In this case, NC interactions are also expected to favor destabilizing bulged-out conformations, which may induce the dissociation of the weaker KL dimer, as shown in Figure 3b and c.
In light of the very distinctive effects induced by the binding of NC to the different sites of monomeric and dimeric SL1Lai, a putative mechanism can be proposed for the conformer transition in vitro, which takes into account the intervening protein-RNA and RNA-RNA interactions (Scheme 3.) In agreement with the two-step model for RNA dimerization and isomerization, the initial SL1Lai substrate can readily establish an equilibrium between monomer and KL dimer in solution (process 1a), but the latter is unable to spontaneously transition to ED (process 1b) because of the large energy barrier to dissociating the intramolecular base pairs that stabilize the KL stemloops.65; 66 The addition of NC, shown in Scheme 3 with increasing concentrations from left to right, has the potential to result in many complex binding events. Considering first the monomeric substrate, high-affinity protein interactions can occur either at the apical loop or at the stem-bulge (process 2a). The former compete directly with the formation of intermolecular base pairs between self-complementary DIS sequences, which constitutes the initial step of RNA dimerization (process 1a). Given the ability of NC to occupy an average of seven (6.8 ± 0.3) nucleotides upon binding to single-stranded nucleic acids,61; 67; 68 its interactions with the G-rich loop are expected to completely obstruct the palindrome and prevent stemloop annealing.
Scheme 3.
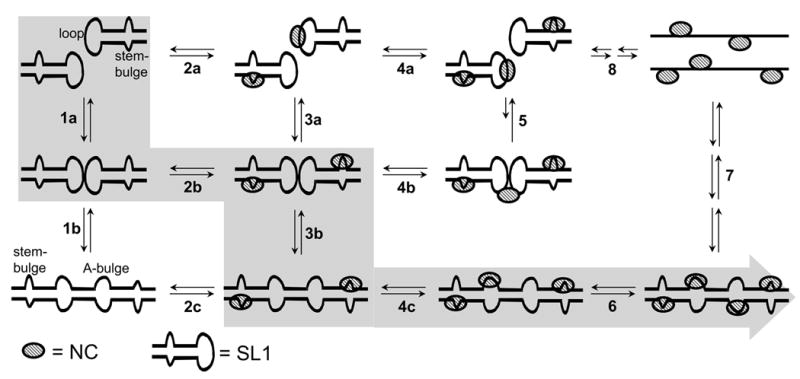
Putative mechanism of dimerization and isomerization of SL1Lai assisted in vitro by NC. The main sequence of events observed at increasing NC concentrations is highlighted. Double-arrows are employed to suggest the reversible nature of a certain process, rather than to represent a fully balanced equilibrium (see text for complete explanations).
In the context of the KL structure, the stem-bulge constitutes the only site available for high-affinity protein interactions (process 2b). As clearly demonstrated by SL1-BG1/2, NC weakens the base pairs surrounding this motif, thus leading to melting of the double-stranded structure (Figure 7). In the KL dimer, the destabilized stems are in close proximity to each other and available for exchange to form the intermolecular base pairs that stabilize the ED conformer (Scheme 3, process 3b). The intervention of NC is essential for structural rearrangement, as proved by the fact that wild-type SL1 could undergo isomerization in 150 mM ammonium acetate only in the presence of protein (Figure 5). In contrast, the SL1-TR mutant was readily isomerized under the same conditions even in the absence of NC, because of the lower stabilization offered by the 4 fewer base pairs in the stem region.
In the ED conformer, as well, the stem-bulges represent the high-affinity binding sites. In this case, however, the greater number of inter-strand base pairs (i.e., 28 in ED versus 6 in KL) provides greater stability to the dimer association. Consequently, this double-stranded structure is only marginally affected by the localized strand ‘breathing’ induced by NC, even at the higher protein concentrations that induce binding to the low affinity A-bulges (process 4c). In this direction, the detection of higher order protein-RNA complexes with the wild-type and ED-obligated constructs (i.e., 3:2 and 4:2, Figures 5c and 3c) has clearly demonstrated that complete occupancy of all the stem- and A-bulge sites in this conformer is still compatible with a stable dimeric state (process 6). In contrast, NC binding to the low affinity hinges of the KL structure (process 5) appears to be utterly unfavorable to dimer association, as demonstrated by the described inability to obtain higher order complexes from the KL-obligated construct under the same experimental conditions (Figure 3c). The fact that an overall decrease of the percentage of dimeric species was also observed for the wild-type construct (Figure 5c) is a direct result of the destabilizing effects of hinge binding on the KL dimer, which could compete with the stem-bulge-meditated isomerization at the higher NC concentrations.
Eventually, greater amounts of NC in solution can cause the dissociation of the dimer into its corresponding NC-bound strands (process 7). The 3:1 stoichiometry observed in the presence of a large excess of protein over RNA (Fig. 5c) can be explained with binding to newly exposed single-stranded regions. This effect is consistent with the unfolding of the stem-loop structures and non-specific binding to the accessible nucleotides (process 8), which was observed also for the other hairpins of the packaging signal in the presence of excess NC.55 We could speculate that if NC were removed from solution by protease digestion or phenol extraction before analysis of the SL1 oligomeric state, as performed in other studies, this would allow the reversible binding equilibria to shift back to the left, thus providing only the RNA species involved in the two-step model shown on the left side of Scheme 3.
Conclusions
The simultaneous observation of protein-RNA and RNA-RNA interactions has provided new insights on the roles played by NC and the different RNA motifs in the mechanism of dimerization and isomerization of SL1 (Lai variant) in vitro. In agreement with the two-step model, our results indicate that the apical loop needs to be unobstructed by protein binding to enable the initial annealing of self-complementary DIS palindromes. The comparable binding affinities manifested by the loop and stem-bulge motifs ensure that this condition is met in the presence of sub-saturating concentrations of NC, which leave unoccupied a sufficient proportion of sites in solution. The initial formation of the KL structure constitutes an obligated step in the dimerization process, since isolated SL1A or SL1B mutants are unable to form homodimers even in the presence of NC, consistent with other studies.24; 28; 65 On the other hand, this observation is not compatible with the possibility that dimerization might involve the direct annealing of the stem regions to form a cruciform-type complex, which would altogether dispense with the need for a transient KL structure.25
The NC-induced destabilization of the KL stems is an essential condition for structural rearrangement, which our data show is achieved through specific binding to the stem-bulge of the stemloop components. This duplex-melting effect is due to the ability of NC to decrease the cooperativity of the helix-coil transition and increase base pair ‘breathing’ in double-stranded structures.61; 69–71 In the context of the KL-ED transition, this activity enables the gradual dissociation of the stems’ intramolecular base pairs, which can then reform in a concerted fashion as intermolecular pairs with the complementary strands, possibly following a zippering mechanism analogous to that described for the tRNA primer annealing to the complementary sequence of the primer binding site (PBS).15; 72 The possibility that strand exchange might occur through a trans-esterification mechanism37 was not corroborated by the ESI-FTMS determinations, which under no circumstances showed either the formation of cleaved RNA products, or the formal addition of water (18 Da mass shift) expected from the hydrolysis of phosphodiester backbones.
The importance of the stem-bulge motif in viral replication is upheld by the observation that its sequence is highly conserved across all HIV-1 isolates.73 In addition, mutations or deletions in the bulge region have been shown to dramatically reduce both dimerization and packaging.8; 10; 74 Our data clearly indicate that this structure constitutes an active site of protein interactions regardless of the overall conformation assumed in vitro by SL1Lai. Therefore, beyond its specific role in targeting the chaperone activity of NC to promote isomerization, this site could serve in vivo as a general point of contact between NC/Gag and genomic RNA during different stages of the processes of genome recognition, dimerization, packaging, and maturation. Our data show that also the DIS-flanking A-bulges could constitute weaker but viable sites for NC interactions in vitro. The fact that this binding mode was observed only for the ED conformer suggests that these motifs may constitute ‘base-grips’ for NC/Gag contacts during the late stages of the viral lifecycle, when the SL1 domain may assume the more stable, mature conformation.30–32
While varying the protein concentration in vitro has proven very helpful to highlight the binding properties of the different RNA structures, the significance of analogous variations in vivo remains to be established. According to a recent model that involves the localization of the Gag-genome complex at the host membrane during the packaging process,17 the effective concentration of Gag in this region is likely to differ significantly from that of the cytosol bulk. In the same way, the effective concentration of NC in the vicinity of the viral genome is expected to be yet different in mature virions, after compaction of the nucleic acid component and rearrangement of the viral structures.31; 75 Future work will be directed toward investigating possible correlations between the concentration-dependent effects observed in vitro and the actual processes taking place in the different cellular and viral compartments in vivo.
Materials and Methods
NC and RNA constructs
NC was expressed in E. coli BL21 (DE3)-pLysE and purified under non-denaturing conditions as previously described.76 The protein was then extensively desalted using Centricon YM-3 ultrafiltration devices (Millipore, Billerica, MA) against ammonium acetate buffer of the desired concentration with pH adjusted to 7.0. The purity and integrity of the protein, including the presence of the coordinated zinc ions, were confirmed by ESI-FTMS.55
RNA constructs corresponding to the wild-type and mutant sequences shown in Scheme 2 were purchased from IDT (Coralville, IA) and desalted using Centricon YM-3 ultrafiltration devices. Hairpins SL2, SL3, and SL4 of the packaging signal were prepared as previously described.55 The purity of each RNA sample was confirmed by ESI-FTMS prior to use, while concentrations were determined by UV absorbance using molar extinction coefficients calculated from their sequences.77 Unless otherwise specified, the constructs were individually heated to 95°C for 5 minutes and then quickly cooled on ice immediately before use to allow for proper folding of the RNA hairpins. For experiments with the duplexes SL1-BG1/2, SL1-ED1/2, and the ED form of SL1-TR, the RNA was also heated to 95°C for 5 minutes, but it was then allowed to slowly cool to room temperature over one hour to allow for the formation of the ED fold.
Binding and competition experiments
All binding experiments were performed in buffered solutions of ammonium acetate (pH 7.0). The concentration of ammonium acetate present was adjusted to appropriately vary the strength of the protein-RNA and RNA-RNA interactions. Because of the divergent effects of ionic strength on these two types of interactions, a moderate concentration of 100–150 mM was used for most experiments, as indicated in the text. Appropriate volumes of each RNA stock were diluted in ammonium acetate buffer to a 5–10 bM final concentration, as indicated in the text. The RNA samples were incubated with NC at room temperature or 37°C for 15–30 minutes prior to analysis to ensure that a binding equilibrium had been established. Alterations to these experimental conditions, as dictated by the specific interactions under investigation, are noted in the text.
Mass spectrometry
Analyses were performed on a Bruker Daltonics (Billerica, MA) Apex III FTMS equipped with a 7T actively-shielded superconducting magnet and a nano-ESI source build in house. This nano-ESI apparatus consisted of a quartz nano-ESI needle (New Objective, Woburn, MA) mounted on an x-y-z stage approximately 1 mm in front of a stainless steel, resistively-heated desolvation capillary. Desolvation temperature, skimmer voltage, and other source parameters known to control the ionization process were optimized to allow for the observation of the RNA-RNA and protein-RNA noncovalent complexes, as previously described.55
Analyte solutions were mixed with iso-propanol immediately before analysis to a final concentration of 10% in volume to assist desolvation. 5 bl samples were loaded into the needle and the spray voltage (< 1 kV) was applied to the solution through a stainless steel wire inserted into back of the needle. No solvent pumps were necessary, as the solution flow-rate was dictated by the applied voltage and the size of the nano-ESI needle tip (~1–2 bm). Spectra were acquired in negative ionization mode and processed with XMASS 7.0.2 (Bruker Daltonics, Billerica, MA). Scans were performed in broadband mode that allowed for a typical 150,000 resolving power at m/z 2000. The spectra were externally calibrated using a 1 mg/ml solution of CsI, which produced a series of peaks throughout the mass range of 1000–6000 m/z and enabled to achieve a typical mass accuracy of 20 ppm or better across this range. Each analysis was performed at least in triplicate and only representative spectra were shown.
Data analysis
The identity and stoichiometry of the noncovalent protein-RNA and RNA-RNA complexes were obtained directly from the detected masses, rather than from the observation of a bulk spectroscopic property, or from relative migrations in a native gel. The average masses determined from the mass spectra were in agreement with the theoretical values calculated from the individual RNA and protein components (Scheme 2). The high-resolution and mass accuracy that are a hallmark of FTMS enabled us to resolve any ambiguities that might have resulted from overlapping peaks in the spectra.55
In some experiments, the proportion of dimeric species in solution was determined to evaluate the effects of NC of the dimerization state. This semi-quantitative estimation was based on the signal intensity of each species of interest divided by its respective charge state. The normalized intensity of all dimeric species, both free and NC-bound, were added together and divided by the sum of all the RNA species detected in solution (monomeric, dimeric, free, and NC-bound). The use of normalized intensities is legitimated by the fact that NC binding was never found to induce complete neutralization of the nucleic acid component, but rather a mere shift in charge distribution (see 55 and ref.s therein). The percentages provided were only used to compare multiple spectra within a single titration experiment and not to determine actual equilibrium binding constants.
Acknowledgments
This research was funded by the National Institutes of Health (R01-GM643208) and the National Science Foundation (CHE-0439067). N.A.H. was also supported by an NIH Chemistry Biology Interface Training Fellowship (T32-GM066706). The authors would like to thank R. Karpel and M. Summers for helpful discussions, and A. Hawkins, D. Smith, K. Turner, and K. Williams for technical assistance at different stages of the project.
References
- 1.Darlix JL, Gabus C, Nugeyre MT, Clavel F, Barré-Sinussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 2.Laughrea M, Jetté L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 3.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci U S A. 1994;91:4945–9. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Höglund S, Öhagen Å, Goncalves J, Panganiban A, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 5.Coffin JM, Hughes SH, Varmus H. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. [PubMed] [Google Scholar]
- 6.Berkhout B, van Wamel JL. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–32. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paillart JC, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie. 1996;78:639–53. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 8.Clever J, Parslow TG. Mutant Human Immunodeficiency Virus Type I Genomes with Defects in RNA Dimerization or Encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paillart JC, Berthoux L, Ottmann M, Darlix JL, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–54. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen N, Jetté L, Liang C, Wainberg MA, Laughrea M. Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J Virol. 2000;74:5729–35. doi: 10.1128/jvi.74.12.5729-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell RS, Liang C, Wainberg MA. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology. 2004;1:23. doi: 10.1186/1742-4690-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickson C, Eisenman R, Fan H, Hunter E, Reich N. Protein biosynthesis and assembly. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA Tumor Viruses. 2. Cold Spring Harbor Laboratory Press; Plainview, NY: 1985. pp. 513–648. part 2. [Google Scholar]
- 13.Darlix JL, Lapadat-Tapolsky M, de Roquigny H, Roques BP. First glimpse at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 14.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for retroviral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 15.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–86. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 16.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat Rev Microbiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 17.D’Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 18.Muriaux D, Girard PM, Bonnet-Mathonière B, Paoletti J. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270:8209–16. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 19.Clever J, Wong ML, Parslow TG. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci U S A. 1996;93:5572–7. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muriaux D, Fossé P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35:5075–82. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 22.Laughrea M, Jetté L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–98. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi KI, Baba S, Chattopadhyay P, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. Structural requirement for the two-step dimerization of human immunodeficiency virus type 1 genome. RNA. 2000;6:96–102. doi: 10.1017/s1355838200991635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Windbichler N, Werner M, Schroeder R. Kissing complex-mediated dimerisation of HIV-1 RNA: coupling extended duplex formation to ribozyme cleavage. Nucleic Acids Res. 2003;31:6419–27. doi: 10.1093/nar/gkg873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernacchi S, Ennifar E, Toth K, Walter P, Langowski J, Dumas P. Mechanism of hairpin-duplex conversion for the HIV-1 dimerization initiation site. J Biol Chem. 2005;280:40112–21. doi: 10.1074/jbc.M503230200. [DOI] [PubMed] [Google Scholar]
- 26.Feng YX, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci U S A. 1996;93:7577–81. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muriaux D, De Rocquigny H, Roques BP, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J Biol Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 28.Rist MJ, Marino JP. Mechanism of nucleocapsid protein catalyzed structural isomerization of the dimerization initiation site of HIV-1. Biochemistry. 2002;41:14762–70. doi: 10.1021/bi0267240. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi K, Baba S, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. Two basic regions of NCp7 are sufficient for conformational conversion of HIV-1 dimerization initiation site from kissing-loop dimer to extended-duplex dimer. J Biol Chem. 2001;276:31274–8. doi: 10.1074/jbc.M104577200. [DOI] [PubMed] [Google Scholar]
- 30.Prats AC, Roy C, Wang PA, Erard M, Housset V, Gabus C, Paoletti C, Darlix JL. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990;64:774–83. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–9. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu W, Gorelick RJ, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–8. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greatorex J, Gallego J, Varani G, Lever A. Structure and stability of wild-type and mutant RNA internal loops from the SL-1 domain of the HIV-1 packaging signal. J Mol Biol. 2002;322:543–57. doi: 10.1016/s0022-2836(02)00776-3. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence DC, Stover CC, Noznitsky J, Wu Z, Summers MF. Structure of the intact stem and bulge of HIV-1 Psi-RNA stem-loop SL1. J Mol Biol. 2003;326:529–42. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 35.Yuan Y, Kerwood DJ, Paoletti AC, Shubsda MF, Borer PN. Stem of SL1 RNA in HIV-1: structure and nucleocapsid protein binding for a 1 x 3 internal loop. Biochemistry. 2003;42:5259–69. doi: 10.1021/bi034084a. [DOI] [PubMed] [Google Scholar]
- 36.Mujeeb A, Clever JL, Billeci TM, James TL, Parslow TG. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat Struct Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 37.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat Struct Biol. 2001;8:1064–8. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 38.Baba S, Takahashi K, Noguchi S, Takaku H, Koyanagi Y, Yamamoto N, Kawai G. Solution RNA Structures of the HIV-1 Dimerization Initiation Site in the Kissing-Loop and Extended-Duplex Dimers. J Biochem (Tokyo) 2005;138:583–92. doi: 10.1093/jb/mvi158. [DOI] [PubMed] [Google Scholar]
- 39.Ennifar E, Dumas P. Polymorphism of bulged-out residues in HIV-1 RNA DIS kissing complex and structure comparison with solution studies. Journal of Molecular Biology. 2006 doi: 10.1016/j.jmb.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 40.Kieken F, Paquet F, Brulé F, Paoletti J, Lancelot G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006;34:343–52. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ennifar E, Yusupov M, Walter P, Marquet R, Ehresmann B, Ehresmann C, Dumas P. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveal an extended duplex with two adenine bulges. Struct Fold Des. 1999;7:1439–1449. doi: 10.1016/s0969-2126(00)80033-7. [DOI] [PubMed] [Google Scholar]
- 42.Girard F, Barbault F, Gouyette C, Huynh-Dinh T, Paoletti J, Lancelot G. Dimer initiation sequence of HIV-1Lai genomic RNA: NMR solution structure of the extended duplex. J Biomol Struct Dyn. 1999;16:1145–57. doi: 10.1080/07391102.1999.10508323. [DOI] [PubMed] [Google Scholar]
- 43.Mujeeb A, Parslow TG, Zarrinpar A, Das C, James TL. NMR structure of the mature dimer complex of HIV-1 genomic RNA. FEBS Lett. 1999;458:387–392. doi: 10.1016/s0014-5793(99)01183-7. [DOI] [PubMed] [Google Scholar]
- 44.Shubsda MF, Paoletti AC, Hudson BS, Borer PN. Affinities of packaging domain loops in HIV-1 RNA for the nucleocapsid protein. Biochemistry. 2002;41:5276–5282. doi: 10.1021/bi016045+. [DOI] [PubMed] [Google Scholar]
- 45.Yamashita M, Fenn JB. Electrospray ion source. Another variation on the free-jet theme. J Phys Chem. 1984;88:4671–4675. [Google Scholar]
- 46.Comisarow MB, Marshall AG. Fourier transform ion cyclotron resonance. Chem Phys Lett. 1974;25:282–283. [Google Scholar]
- 47.Hendrickson CL, Emmett MR, Marshall AG. Electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Annu Rev Phys Chem. 1999;50:517–536. doi: 10.1146/annurev.physchem.50.1.517. [DOI] [PubMed] [Google Scholar]
- 48.Ganem B, Li YT, Henion JD. Detection of non-covalent receptor-ligand complexes by mass spectrometry. J Am Chem Soc. 1991;113:6294–6296. [Google Scholar]
- 49.Ganguly AK, Pramanik BN, Tsarbopoulos A, Covey TR, Huang E, Fuhrman SA. Mass spectrometric detection of the noncovalent GDP-bound conformational state of the human H-ras protein. J Am Chem Soc. 1992;114:6559–6560. [Google Scholar]
- 50.Loo JA, Holler TP, Foltin SK, McConnell P, Banotal CA, Horne NM, Mueller WT, Stevenson TI, Mack DP. Application of electrospray ionization mass spectrometry for studying human immunodeficiency virus protein complexes. Proteins Suppl. 1998;2:28–37. doi: 10.1002/(sici)1097-0134(1998)33:2+<28::aid-prot5>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 51.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 52.Daniel JM, Friess SD, Rajagopalan S, Wendt S, Zenobi R. Quantitative determination of noncovalent binding interactions using soft ionization mass spectrometry. Int J Mass Spectrom Ion Proc. 2002;216:1–27. [Google Scholar]
- 53.Hofstadler SA, Sannes-Lowery KA, Hannis JC. Analysis of nucleic acids by FTICR MS. Mass Spectrom Rev. 2005;24:265–285. doi: 10.1002/mas.20016. [DOI] [PubMed] [Google Scholar]
- 54.Fisher RJ, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, Shoemaker R, McNitt KL, Johnson DG, Hixson CV, Gorelick RJ, Fabris D, Henderson LE, Rein A. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Research. 2006;34:472–484. doi: 10.1093/nar/gkj442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hagan N, Fabris D. A direct mass spectrometric determination of the stoichiometry and binding affinity of the complexes between HIV-1 nucleocapsid protein and RNA stem-loops hairpins of the HIV-1 Ψ-recognition element. Biochemistry. 2003;42:10736–10745. doi: 10.1021/bi0348922. [DOI] [PubMed] [Google Scholar]
- 56.Aldovini A, Young RA. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clever JL, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McBride MS, Panganiban AT. The human immunodeficiency virus type-1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casa-Finet JR, Medaglia N, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vuilleumier C, Bombarda E, Morellet N, Gérard D, Roques BP, Mély Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- 61.Urbaneja MA, Wu M, Casas-Finet JR, Karpel RL. HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity. J Mol Biol. 2002;318:749–764. doi: 10.1016/S0022-2836(02)00043-8. [DOI] [PubMed] [Google Scholar]
- 62.Ding J, Anderegg RJ. Specific and nonspecific dimer formation in the electrospray ionization mass spectrometry of oligonucleotides. J Am Soc Mass Spectrom. 1995;6:159–164. doi: 10.1016/1044-0305(94)00102-6. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi K, Baba S, Hayashi Y, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. NMR analysis of intra- and inter-molecular stems in the dimerization initiation site of the HIV-1 genome. J Biochem (Tokyo) 2000;127:681–6. doi: 10.1093/oxfordjournals.jbchem.a022657. [DOI] [PubMed] [Google Scholar]
- 64.Mihailescu MR, Marino JP. A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex. Proc Natl Acad Sci U S A. 2004;101:1189–94. doi: 10.1073/pnas.0307966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Theilleux-Delalande V, Girard F, Huynh-Dinh T, Lancelot G, Paoletti J. The HIV-1(Lai) RNA dimerization. Thermodynamic parameters associated with the transition from the kissing complex to the extended dimer. Eur J Biochem. 2000;267:2711–9. doi: 10.1046/j.1432-1327.2000.01292.x. [DOI] [PubMed] [Google Scholar]
- 66.Aci S, Mazier S, Genest D. Conformational pathway for the kissing complex-->extended dimer transition of the SL1 stem-loop from genomic HIV-1 RNA as monitored by targeted molecular dynamics techniques. J Mol Biol. 2005;351:520–30. doi: 10.1016/j.jmb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 67.Mély Y, de Rocquigny H, Sorinas-Jimeno M, Keith G, Roques B, Marquet R, Gerard D. Binding of the HIV-1 nucleocapsid protein to the primer tRNA3Lys, in vitro, is essentially not specific. J Biol Chem. 1995;270:1650–56. doi: 10.1074/jbc.270.4.1650. [DOI] [PubMed] [Google Scholar]
- 68.Urbaneja MA, Kane BP, Johnson DG, Gorelick RJ, Henderson LE, Casas-Finet JR. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J Mol Biol. 1999;287:59–75. doi: 10.1006/jmbi.1998.2521. [DOI] [PubMed] [Google Scholar]
- 69.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc Nat Acad Sci USA. 2001;98:6121–6. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams MC, Rouzina I, Bloomfield VA. Thermodynamics of DNA interactions from single molecule stretching experiments. Acc Chem Res. 2002;35:159–66. doi: 10.1021/ar010045k. [DOI] [PubMed] [Google Scholar]
- 71.Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, Rein A, Rouzina I, Williams MC. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hargittai MR, Gorelick RJ, Rouzina I, Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J Mol Biol. 2004;337:951–68. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 73.Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B, editors. HIV Sequence Compendium 2005. Theoretical Biology and Biophysics Group; Los Alamos National Laboratory: 2005. [Google Scholar]
- 74.Shen N, Jetté L, Wainberg MA, Laughrea M. Role of stem B, loop B, and nucleotides next to the primer binding site and the kissing-loop domain in human immunodeficiency virus type 1 replication and genomic-RNA dimerization. J Virol. 2001;75:10543–9. doi: 10.1128/JVI.75.21.10543-10549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci U S A. 2001;98:5246–51. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Guzman RN, Turner RB, Summers MF. Protein-RNA recognition. Biopolymers. 1998;48:181–95. doi: 10.1002/(SICI)1097-0282(1998)48:2<181::AID-BIP7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 77.Gray DM, Hung SH, Johnson KH. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]


