Abstract
Human APOBEC3G (hA3G) is a member of the APOBEC-1 Related Protein (ARP) family of cytidine deaminases. hA3G functions as a natural defense against endogenous retrotransposons and a multitude of retroviruses, most notably human immunodeficiency virus type 1 (HIV-1). Nothing is known about the cellular function of hA3G, however, upon HIV-1 infection hA3G functions as an antiviral factor by mutating viral single stranded DNA during reverse transcription. Whereas homologous deaminases such as APOBEC-1 and AID act on RNA and DNA, respectively, in the cell nucleus, hA3G mutagenic activity appears to be restricted to the cytoplasm. We demonstrate that hA3G is not a nucleo-cytoplasmic shuttling protein like APOBEC-1 and AID, but is strongly retained in the cytoplasm through a mechanism that involves both the N and C-terminal regions of the protein.
Keywords: APOBEC3G, AID, APOBEC-1, trafficking, NLS, NES
Introduction
Human APOBEC3G (hA3G) belongs to the APOBEC-1 related protein (ARP) family of proteins that carry out deamination of cytidine/deoxycytidine in the context of nucleic acids [1]. The family is characterized by one or two zinc-dependent deaminase domains (ZDD, Fig. 1) that has a conserved amino acid sequence of (C/H)xExnPCxxC distinct from other zinc-binding motifs [2].
Fig. 1.
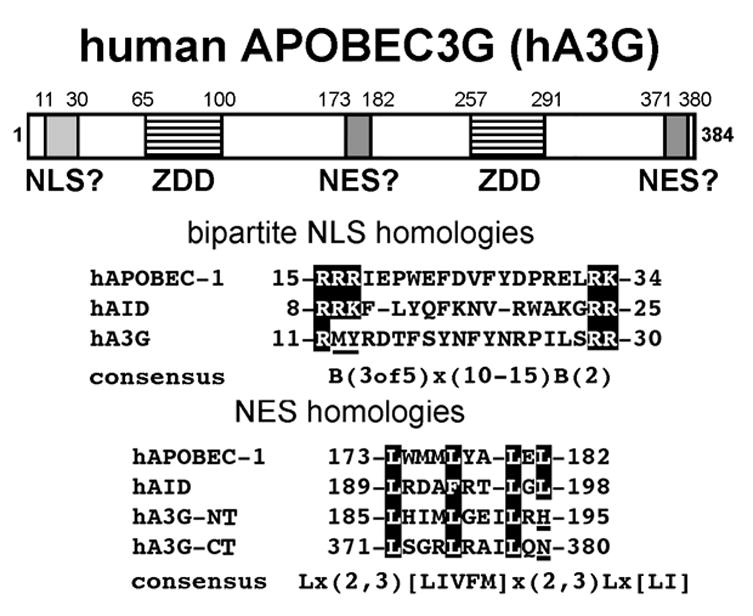
A diagram depicting the two zinc-dependent deaminase (ZDD) domains of human APOBEC3G (hA3G) and the regions homologous to the bipartite NLS and NES regions of hAPOBEC-1 and hAID. The sequence homologies of the bipartite NLS are shown in the boxed regions in hAPOBEC-1 and hAID. The underlined region in hA3G, demonstrates that a bipartite NLS is not conserved in hA3G. The NES homologies show three conserved leucines (boxed) and one leucine not conserved (underlined) in both the N-terminal (hA3G-NT) and C-terminal (hA3G-CT) regions of hA3G corresponding to critical leucines that make up the NES regions of hAPOBEC-1 and hAID, respectively (the human sequences are representative of the respective regions of APOBEC-1, AID and A3G in a multitude of mammalian species). The consensus NLS and NES are indicated below the respective homologies where x means any amino acid and B means basic amino acids lysine and arginine.
hA3G is an antiretroviral factor that hypermutates single stranded viral RNA during reverse transcription [3, 4]. HIV-1 accessory protein Vif is able to shuttle hA3G to the proteasomal degradation pathway in order to counteract hA3G antiviral activity [5, 6]. Aside from its antiviral activity not much is known about the cellular function of hA3G. Therefore, we analyzed hA3G’s homologies to other ARPs to gain insight into hA3G’s cellular function. Apolipoprotein B editing complex-1 (APOBEC-1) and activation-induced cytidine deaminase (AID) are ARP family members with one ZDD that have well characterized cellular functions. APOBEC-1 is the catalytic subunit of an editosome that edits apolipoprotein B (apoB) mRNA at C6666 in liver and intestine of most mammals [7, 8, 9 ]. The editing event introduces a premature stop codon resulting in the translation of a shortened apoB 48 protein while unedited transcripts make apoB 100 [10, 11]. AID’s deaminase activity is required for gene conversion, somatic hypermutation and class switch recombination which are all key mechanisms involved in diversification of immunoglobulins in B lymphocytes [12, 13]. hA3G contains two ZDD domains, each homologous to APOBEC-1 and AID and we have analyzed the individual halves of hA3G, along with regions homologous to the nuclear localization signal (NLS) and nuclear export signal (NES) domains of the other ARP family members [1] to determine whether nucleo-cytoplasmic trafficking is also a part of hA3G’s function.
There are three main reasons why we analyzed the trafficking capabilities of hA3G. First, nucleo-cytoplasmic trafficking is crucial for both APOBEC-1 and AID cellular functions, respectively [14–19]. Trafficking of both deaminases requires a bipartite NLS in the N-terminus, a C-terminal NES and interaction with chaperone proteins [15–17, 20]. Second, hA3G has an unknown cellular function but it is known to specifically target single stranded DNA [4]. Therefore, trafficking to the nucleus would suggest hA3G has a specific genomic DNA target for deaminase activity whereas cytoplasmic restriction suggests that hA3G is kept in the cytoplasm to prevent genotoxicity. Lastly, hA3G antiviral activity is not entirely dependent on deaminase activity. Deaminase inactive mutants retain antiviral activity [21, 22], and Vif independent hA3G antiviral activity in resting CD4+ T cells does not cause hypermutations in the HIV genome [23]. If hA3G does traffic, multiple subcellular localizations could contribute to deaminase-dependent and deaminase-independent activities.
Materials and Methods
Plasmid constructions
hA3G and hAID cDNAs were produced from oligo(dT)-primed total cellular RNA using avian myoblastosis virus reverse transcriptase (Promega) from H9 cells, and Raji cells, respectively. hA3G was subsequently subcloned into a pIRES-P [24] vector with N-terminal 6 His and HA tags, respectively. hAID, was subcloned into pIRES-P with a C-terminal V5 tag. The N-terminal and C-terminal deletion clones of hA3G were subcloned from full length hA3G described above and were separated in the predicted linker region between the two regions of shared homology [1, 2] and a V5 tag was added to the N-terminus of the N-terminal deletion clone. All NES hA3G mutations were made with site-directed mutagenesis using the Quikchange system (Stratagene) and sequence verified. The SV40 NLS was inserted into a unique EcoRV site between the HA tag and first codon of hA3G for all full length hA3G clones and the C-terminal deletion or between the V5 tag and the 209th codon of hA3G in the N-terminal deletion, all clones were sequence verified for proper sequence and directionality. Design and construction of CMPK and NLS-CMPK have been described previously [17].
Cell culture and transfections
HeLa cells obtained from ATCC (Manassas, VA) were maintained in DMEM containing 10% (v/v) fetal bovine serum plus penicillin/streptomycin/fungizone (Cellgro), and non-essential amino acids (Invitrogen) and were transfected using Lipofectamine 2000 according to the manufacture’s protocol (Invitrogen). Leptomycin B (Calbiochem) was added for 4 h at 20 ng/mL 20 h after transfections.
Immunocytochemistry
24 h after HeLa cells were transiently transfected in 6 well clusters they were fixed for 15 min at room temperature with 2% paraformaldehyde in PBS. Fixed cells were permeabilized with 40 μg/mL digitonin (Sigma) in PBS and immunolabeled for 1 h at room temperature with anti-HA (Convance) 1:500 or anti-V5 (Invitrogen) 1:500 in PBS containing 1% BSA, followed by incubation for 1 h at room temperature with FITC conjugated goat anti-mouse IgG antibody (Organon Teknika-Cappel) 1:500 in PBS containing 1% BSA. Cells were maintained at 4 °C in Vectashield mounting medium with DAPI (Vector) and were imaged by a QICIM-IR fast 12 bit mono chrome camera and Q capture software (Q-Imaging) through a 40X Olympus objective with an Olympus IX 70 inverted fluorescence microscope and label specific chrome filters.
Results
hA3G does not traffic between the nucleus and cytoplasm
Both APOBEC-1 and AID require a bipartite NLS within the first 35 amino acids (a.a.) and cytoplasmic chaperones to traffic to the nucleus [14, 17, 20]. BLAST analysis of both APOBEC-1 and AID revealed that a bipartite NLS is conserved among nine and eight mammalian species for APOBEC-1 and AID, respectively. However, inspection of hA3G showed it lacks key basic amino acids associated with an NLS (Fig. 1). Immunocytochemistry (ICC) suggested that hA3G was localized only in the cytoplasm (Fig. 2B). This distribution has been described previously [25, 26], but could arise from a trafficking equilibrium that favors the cytoplasm. The nuclear export inhibitor leptomycin B (LMB) is useful for trapping CRM1-dependent trafficking proteins in the cell nucleus and has been used to demonstrate nuclear accumulation of AID in B cells [15, 20] and APOBEC-1 in heterokaryons [16]. Therefore, we used AID as a positive control for LMB treatment in our cell system. Although LMB treatment resulted in nuclear accumulation of AID (Fig. 2A) LMB was unable to trap hA3G in the nucleus (Fig. 2B). These data suggest that hA3G does not undergo CRM1-dependent nuclear pore export. Since sequence homolog analysis revealed that hA3G does not contain a canonical bipartite NLS as seen in AID, we added a strong NLS (PKKKRKV) from the SV40 large T antigen [27] to hA3G’s N-terminus to determine whether hA3G could be imported to the nucleus. NLS-hA3G remained unable to localize to the nucleus with or without LMB (Fig. 2C). CMPK is frequently used as a nonspecific 'cargo' protein in demonstrating NLS function as it is an established cytoplasmic resident control (Fig. 3A). Consistent with prior reports [17, 27, 28], CMPK trafficked to the nucleus when coupled to the SV40 NLS (Fig. 3B). These findings demonstrate that hA3G does not, and cannot be made to import to the nucleus and suggested that it may be actively retained in the cytoplasm.
Fig. 2.

hA3G does not traffic to the nucleus. (A) Cellular distribution of AID. C-terminal V5 tagged hAID transfected into HeLa cells localized to the cytoplasm (−LMB). Upon treatment with 20 ng/mL of LMB for 4 h (+LMB) hAID localized to the nucleus. (B) N-terminal HA tagged hA3G transfected into HeLa cells localized in the cytoplasm with and without LMB treatment (+/− LMB) and (C) NLS-hA3G with a SV40 NLS added to the N-terminus remained cytoplasmic with and without LMB treatment (+/− LMB). The upper and lower panels for A–C are two representative fields of the same treatment group. DAPI staining indicates the position of the cell nuclei in each panel.
Fig. 3.

The two NES-like leucine-rich domains in hA3G are nonfunctional. (A) Chicken muscle pyruvate kinase (CMPK) transfected into HeLa cells localized to the cytoplasm. (B) SV40 NLS added to the N-terminus of CMPK enables trafficking to the nucleus. hA3G with three leucine to alanine mutations (boxed in hA3G-NT and hA3G-CT from Fig. 1) in either the N or C-termini (C and E, respectively) or in both regions (G) localized in the cytoplasm when transfected into HeLa cells. An N-terminal SV40 NLS added to the hA3G mutants in C, E, and G, (D, F and H, respectively) were also localized in the cytoplasm when transfected into HeLa cells. The side-by-side panels for A–H are two representative fields of the same treatment group. DAPI staining indicates the position of the cell nuclei in each panel.
hA3G’s leucine-rich regions are not functional nuclear export signals
Along with an N-terminal NLS, both APOBEC-1 and AID have C-terminal leucine-rich, nuclear export signal (NES) domains (Fig. 1). In fact, for APOBEC-1 the NES is able to override an N-terminal SV40 NLS, resulting in a cytoplasmic localization of the protein [16, 17]. BLAST analysis of both APOBEC-1 and AID revealed that the C-terminal NES is conserved among nine and eight mammalian species for APOBEC-1 and AID, respectively. Analysis of hA3G sequence shows two leucine-rich regions located within the N- and C-terminal halves of hA3G conserved among ten mammalian species. These sequences are homologous to those of APOBEC-1 and AID with the important difference that sequences of hA3G both are lacking the last leucine in the consensus NES (Fig. 1). Given that only 36% of the experimentally verified leucine-rich NES sequences contain the consensus sequence [29], we set out to demonstrate whether the NES-like regions of hA3G were functional. Three leucines were mutated to alanines in both halves of hA3G or in each half separately and evaluated for their effect on the cellular distribution of hA3G with or without the SV40 NLS. ICC revealed that regardless of the point mutations, hA3G mutants localized to the cytoplasm (Fig. 3C–H). Thus, despite the need for a functional leucine-rich NES for cytoplasmic localization of APOBEC-1 and AID [15–17, 19, 20], the leucine-rich motifs were not determining factors in retaining hA3G in the cytoplasm.
hA3G half molecules are retained in the cytoplasm
The nuclear pore restricts simple diffusion of proteins into the nucleus to those less than 60 kDa. hA3G (384 a.a.) APOBEC-1 (229 a.a.) and AID (198 a.a.) are each predicted to form higher order multimers [2, 23, 30–32] whose aggregate size may prohibit passive entry into the nucleus. We investigated whether size contributed to hA3G cytoplasmic retention. To this end, hA3G half molecules were designed based on the structure of the yeast cytidine deaminase and RNA editing enzyme, CDD1 [2] and APOBEC3 secondary structure predictions [1]. N- and C-terminal halves of hA3G were separated at the predicted linker region between the two homologous zinc deaminase domains to produce a C-terminal deletion of hA3G containing a.a. 1-208 and a N-terminal deletion of hA3G containing a.a. 209-384. ICC revealed that each of the hA3G halves localized to the cytoplasm irrespective of an N-terminal SV40 NLS or the nuclear export inhibitor LMB (Fig. 4). The data suggested that there is sufficient information in either half of hA3G to confer cytoplasmic retention.
Fig. 4.

Both halves of hA3G are responsible for cytoplasmic retention. Constructs were designed that split hA3G in the predicted linker region between the two homologous halves of the protein. The C-terminal deletion hA3G mutant, a.a. 1-208 (A), N-terminal deletion hA3G mutant, a.a. 209-384 (C) and each mutant with an N-terminal SV40 NLS (B and D, respectively) localized to the cytoplasm when transfected into HeLa cells with or without (+/−) LMB treatment. The upper and lower panels for A–D are two representative fields of the same treatment group. DAPI staining indicates the position of the cell nuclei in each panel. The dashed lines indicate the missing portions of the deletion mutants.
Discussion
We have shown that hA3G is strongly retained in the cytoplasm and does not undergo nucleo-cytoplasmic trafficking like its paralogs APOBEC-1 and AID. Moreover, hA3G’s cytoplasmic retention was not mediated by the NES-like, leucine-rich regions homologous to those identified in APOBEC-1 and AID [15–17, 19, 20]. hA3G contains an N-terminal and C-terminal cytidine deaminase domain and both halves of hA3G contain sufficient information to confer cytoplasmic retention. Consistent with the predicted evolutionary origin of hA3G as a tandem duplication of a primordial ARP [1, 2, 33], perhaps like APOBEC-1 and AID, which each harbor a single ZDD motif per polypeptide chain [30, 31], each half of hA3G is capable of multimerization thereby excluding it from the nucleus.
However, cytoplasmic retention in the presence of an SV40 NLS suggests more than multimerization kept hA3G in the cytoplasm. In fact, ARP family members APOBEC3A (containing one ZDD) and APOBEC3B (containing two ZDDs) have a predominantly nuclear localization [34, 35], while the most comparable paralog to hA3G, APOBEC3F (containing two ZDDs), is strictly cytoplasmic [35]. Inspection of the protein sequences of APOBEC3A, 3B, and 3F reveal no obvious NLS, although APOBEC3B, unlike hA3G, does contain the last leucine in its N-terminal leucine-rich region giving it a consensus NES, suggesting that APOBEC3B might be a trafficking protein like APOBEC-1 and AID. APOBEC3 paralogs therefore are distributed in the cell through yet-to-be discovered domains and/or through interactions with RNA or protein chaperones.
The focus of our future research will be to determine whether novel domains, protein-protein/RNA interactions, or post-translational modifications in both halves of the molecule are responsible for the strong cytoplasmic retention of hA3G. Recent reports show hA3G in mRNA processing bodies, polysomes and stress granules where hA3G has only been shown to interact with proteins through an RNA intermediate [35, 36]. Moreover, hA3G forms a RNA-dependent higher order complex with reduced antiviral activity in activated T cells [23] and a more active low molecular mass complex in macrophages and resting T cells [37, 38] suggesting that the interaction between hA3G and RNA is closely linked to its antiviral potency [23, 37, 38] and could be involved in the cytoplasmic retention of hA3G.
Both APOBEC-1 and AID are tightly regulated in the cell but overexpression in cells that do not express appropriate amounts of regulatory cofactors leads to a loss of substrate specificity that can result in tumorigenesis [39–43]. A recent report showed that retroviral infection of pre-B cells in bone marrow triggers expression of AID leading to DNA damage that marks the virus infected cell for destruction by NK cells [44]. Since hA3G has been specifically shown to deaminate single stranded DNA but not RNA [4], hA3G localization to the cytoplasm restricts the deaminase from entering the nucleus keeping it away from genomic DNA. APOBEC-1 and AID both function in the nucleus and must traffic to be active in their respective cell types. hA3G has an as-of-yet undiscovered cellular role but its antiviral activity is clearly restricted to the cytoplasm where its high level of expression can be achieved without the risk of genotoxicity.
Acknowledgments
We would like to thank J. Reeder for guidance in the use of the fluorescence microscope and imaging software. This work was supported by an NIH grant R21 AI58789 awarded to HCS. The authors also acknowledge NIH training grant T32 AI49815 for support of RPB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wedekind JE, Dance GS, Sowden MP, Smith HC. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 2003;19:207–216. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- 2.Xie K, Sowden MP, Dance GS, Torelli AT, Smith HC, Wedekind JE. The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1. Proc Natl Acad Sci U S A. 2004;101:8114–8119. doi: 10.1073/pnas.0400493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 4.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 5.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 6.Stopak K, De Noronha C, Yonemoto W, Greene WC. HIV-1 Vif Blocks the Antiviral Activity of APOBEC3G by Impairing both Its Translation and Intracellular Stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 7.Chen SH, Habib G, Yang CY, Gu ZW, Lee BR, Weng SA, Silberman SR, Cai SJ, Deslypere JP, Rosseneu M, et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987;238:363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- 8.Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 9.Smith HC. Apolipoprotein B mRNA editing: the sequence to the event. Semin Cell Biol. 1993;4:267–278. doi: 10.1006/scel.1993.1032. [DOI] [PubMed] [Google Scholar]
- 10.Chan L, Chang BH, Nakamuta M, Li WH, Smith LC. Apobec-1 and apolipoprotein B mRNA editing. Biochim Biophys Acta. 1997;1345:11–26. doi: 10.1016/s0005-2760(96)00156-7. [DOI] [PubMed] [Google Scholar]
- 11.Davidson NO. Apolipoprotein B mRNA editing: a key controlling element targeting fats to proper tissue. Ann Med. 1993;25:539–543. [PubMed] [Google Scholar]
- 12.Smith HC, Bottaro A, Sowden MP, Wedekind JE. Activation induced deaminase: the importance of being specific. Trends Genet. 2004;20:224–227. doi: 10.1016/j.tig.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 14.Blanc V, Kennedy S, Davidson NO. A novel nuclear localization signal in the auxiliary domain of apobec-1 complementation factor regulates nucleocytoplasmic import and shuttling. J Biol Chem. 2003;278:41198–41204. doi: 10.1074/jbc.M302951200. [DOI] [PubMed] [Google Scholar]
- 15.Ichikawa HT, Sowden MP, Torelli AT, Bachl J, Huang P, Dance GS, Marr SH, Robert J, Wedekind JE, Smith HC, Bottaro A. Structural Phylogenetic Analysis of Activation-Induced Deaminase Function. J Immun. 2006 doi: 10.4049/jimmunol.177.1.355. in press. [DOI] [PubMed] [Google Scholar]
- 16.Chester A, Somasekaram A, Tzimina M, Jarmuz A, Gisbourne J, O'Keefe R, Scott J, Navaratnam N. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. Embo J. 2003;22:3971–3982. doi: 10.1093/emboj/cdg369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Smith HC. Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc Natl Acad Sci U S A. 1997;94:13075–13080. doi: 10.1073/pnas.94.24.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sowden MP, Ballatori N, Jensen KL, Reed LH, Smith HC. The editosome for cytidine to uridine mRNA editing has a native complexity of 27S: identification of intracellular domains containing active and inactive editing factors. J Cell Sci. 2002;115:1027–1039. doi: 10.1242/jcs.115.5.1027. [DOI] [PubMed] [Google Scholar]
- 19.Brar SS, Watson M, Diaz M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem. 2004;279:26395–26401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- 20.Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, Nakata M, Honjo T. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bishop KN, Holmes RK, Malim MH. Antiviral Potency of APOBEC Proteins Does Not Correlate with Cytidine Deamination. J Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 23.Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 24.Hobbs S, Jitrapakdee S, Wallace JC. Development of a bicistronic vector driven by the human polypeptide chain elongation factor 1alpha promoter for creation of stable mammalian cell lines that express very high levels of recombinant proteins. Biochem Biophys Res Commun. 1998;252:368–372. doi: 10.1006/bbrc.1998.9646. [DOI] [PubMed] [Google Scholar]
- 25.Wichroski MJ, Ichiyama K, Rana TM. Analysis of HIV-1 viral infectivity factor-mediated proteasome-dependent depletion of APOBEC3G: correlating function and subcellular localization. J Biol Chem. 2005;280:8387–8396. doi: 10.1074/jbc.M408048200. [DOI] [PubMed] [Google Scholar]
- 26.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 27.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 28.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 2003;31:393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Shinkura R, Muramatsu M, Nagaoka H, Kinoshita K, Honjo T. Identification of a specific domain required for dimerization of activation-induced cytidine deaminase. J Biol Chem. 2006 doi: 10.1074/jbc.M601645200. [DOI] [PubMed] [Google Scholar]
- 31.Oka K, Kobayashi K, Sullivan M, Martinez J, Teng BB, Ishimura-Oka K, Chan L. Tissue-specific inhibition of apolipoprotein B mRNA editing in the liver by adenovirus-mediated transfer of a dominant negative mutant APOBEC-1 leads to increased low density lipoprotein in mice. J Biol Chem. 1997;272:1456–1460. doi: 10.1074/jbc.272.3.1456. [DOI] [PubMed] [Google Scholar]
- 32.Opi S, Takeuchi H, Kao S, Khan MA, Miyagi E, Goila-Gaur R, Iwatani Y, Levin JG, Strebel K. Monomeric APOBEC3G is catalytically active and has antiviral activity. J Virol. 2006;80:4673–4682. doi: 10.1128/JVI.80.10.4673-4682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 34.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 35.Wichroski MJ, Robb GB, Rana TM. Human Retroviral Host Restriction Factors APOBEC3G and APOBEC3F Localize to mRNA Processing Bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The Anti-HIV-1 Editing Enzyme APOBEC3G Binds HIV-1 RNA and Messenger RNAs that Shuttle Between Polysomes and Stress Granules. J Biol Chem. 2006 doi: 10.1074/jbc.M601901200. in press. [DOI] [PubMed] [Google Scholar]
- 37.Chen K, Huang J, Zhang C, Huang S, Nunnari G, Wang FX, Tong X, Gao L, Nikisher K, Zhang H. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J Virol. 2006;80:7645–7657. doi: 10.1128/JVI.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng G, Lei KJ, Jin W, Greenwell-Wild T, Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med. 2006;203:41–46. doi: 10.1084/jem.20051512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oppezzo P, Vuillier F, Vasconcelos Y, Dumas G, Magnac C, Payelle-Brogard B, Pritsch O, Dighiero G. Chronic lymphocytic leukemia B cells expressing AID display dissociation between class switch recombination and somatic hypermutation. Blood. 2003;101:4029–4032. doi: 10.1182/blood-2002-10-3175. [DOI] [PubMed] [Google Scholar]
- 40.Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamanaka S, Balestra ME, Ferrell LD, Fan J, Arnold KS, Taylor S, Taylor JM, Innerarity TL. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sowden M, Hamm JK, Smith HC. Overexpression of APOBEC-1 results in mooring sequence-dependent promiscuous RNA editing. J Biol Chem. 1996;271:3011–3017. doi: 10.1074/jbc.271.6.3011. [DOI] [PubMed] [Google Scholar]
- 43.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 44.Gourzi P, Leonova T, Papavasiliou FN. A role for activation-induced cytidine deaminase in the host response against a transforming retrovirus. Immunity. 2006;24:779–786. doi: 10.1016/j.immuni.2006.03.021. [DOI] [PubMed] [Google Scholar]


