Abstract
The long interspersed element-1 (LINE-1 or L1) is a highly successful retrotransposon in mammals. L1 elements have continued to actively propagate subsequent to the human–chimpanzee divergence, ~6 million years ago, resulting in species-specific inserts. Here, we report a detailed characterization of chimpanzee-specific L1 subfamily diversity and a comparison with their human-specific counterparts. Our results indicate that L1 elements have experienced different evolutionary fates in humans and chimpanzees within the past ~6 million years. Although the species-specific L1 copy numbers are on the same order in both species (1200–2000 copies), the number of retrotransposition-competent elements appears to be much higher in the human genome than in the chimpanzee genome. Also, while human L1 subfamilies belong to the same lineage, we identified two lineages of recently integrated L1 subfamilies in the chimpanzee genome. The two lineages seem to have coexisted for several million years, but only one shows evidence of expansion within the past three million years. These differential evolutionary paths may be the result of random variation, or the product of competition between L1 subfamily lineages. Our results suggest that the coexistence of several L1 subfamily lineages within a species may be resolved in a very short evolutionary period of time, perhaps in just a few million years. Therefore, the chimpanzee genome constitutes an excellent model in which to analyze the evolutionary dynamics of L1 retrotransposons.
Keywords: L1 elements, Retrotransposons, Human, Chimpanzee, Species-specific, Polymorphism
1. Introduction
Long interspersed elements-1 (LINE-1 or L1) are the most successful autonomous retrotransposons in mammals. A full-length functional L1 element is about 6 kb in length and contains a 5′ untranslated region (UTR) bearing an internal RNA polymerase II promoter, two non-overlapping open reading frames (ORF1 and ORF2), which are separated by an ~60-bp-long intergenic spacer, and a 3′ UTR ending in a poly(A) tail (Kazazian and Moran, 1998). ORF1 encodes an RNA-binding protein that has nucleic acid chaperone activity in vitro, and ORF2 encodes both reverse transcriptase and endonuclease activities (Mathias et al., 1991; Feng et al., 1996; Kolosha and Martin, 1997). L1 elements propagate through an RNA intermediate in a process known as retrotransposition, which is thought to occur by a mechanism termed target primed reverse transcription; the insertion process typically results in 7–20-bp-long target site duplications flanking each side of the L1 element (Fanning and Singer, 1987; Luan et al., 1993).
With >500,000 copies, L1 elements account for ~17% of the human genome (Lander et al., 2001). The L1 family emerged around 120 million years (myrs) ago (Smit et al., 1995; Khan et al., 2006) and is still actively expanding in humans, as demonstrated by the existence of highly polymorphic L1 elements in human populations (Sheen et al., 2000; Myers et al., 2002; Badge et al., 2003; Boissinot et al., 2004; Seleme et al., 2006; Wang et al., 2006) and de novo L1 insertions responsible for genetic disorders (Chen et al., 2005). The detection of several hundred species-specific L1 insertions in both the human and chimpanzee genomes further supports the recent mobilization of this family of retrotransposons (Mathews et al., 2003; CSAC, 2005; Mills et al., 2006). Contrary to the non-autonomous Alu retrotransposons in which different subfamilies are capable of concomitant expansions (Batzer and Deininger, 2002; Xing et al., 2004; Hedges et al., 2005), a single line of successive L1 subfamilies has amplified within the past 40 myrs in the primate lineage leading to humans (Khan et al., 2006). L1 subfamilies can be distinguished by diagnostic substitutions that are shared by all members of any given subfamily. For example, five subfamilies are thought to have amplified in hominoid primates (i.e. humans and apes) within the past 25 myrs, named L1PA1 to L1PA5 (Smit et al., 1995; Boissinot et al., 2000; Lander et al., 2001; Khan et al., 2006). The most recently evolved, Homo sapiens-specific (Hs) L1 subfamilies have been well characterized (Boissinot et al., 2000; Myers et al., 2002; Ovchinnikov et al., 2002; Salem et al., 2003a; Boissinot et al., 2004) and the recent completion of the chimpanzee genome sequence (CSAC, 2005) facilitates comparisons of the recent patterns of diversity and evolution of L1 subfamilies since the divergence of human and chimpanzee, ~6 million years ago (Goodman et al., 1998). Global overviews of Hs and Pan troglodytes-specific (Pt) L1 elements have previously been published (CSAC, 2005; Mills et al., 2006). Here, we report a detailed characterization of Pt L1 subfamily diversity and a comparison with their Hs counterparts. Our results indicate that L1 elements have experienced drastically different evolutionary fates in humans and chimpanzees within the past ~6 myrs.
2. Materials and methods
2.1. Computational identification of L1 elements
We identified all L1 elements with complete 3′ end sequences in the human genome (hg16, UCSC July 2003 freeze) by Basic Local Alignment Search Tool (BLAST) querying the genome with the 3′-most 50 bp preceding the poly-A tail of the L1 consensus sequence. This strategy yielded ~110,000 candidate elements, corresponding to the most recent fraction of all L1 elements inserted in the human genome. Next, 300-bp-long sequences covering each L1 3′-end and 100 bp of flanking sequence immediately downstream the poly-A tail were extracted. The exact terminus of the poly-A tails in these L1 sequences was determined by a BLASTsearch with the 50-bp L1 consensus sequence to which a tract of 100 adenosines was added. The sequences were used as queries for BLAST searches against the chimpanzee genome sequence (UCSC Nov. 2003 freeze). Queries with matches limited to the 100-bp L1 3′ end flanking regions in human were collected as candidates representing the orthologous pre-integration sites of the human L1 insertions. Then, we extracted the 800-bp region centered at the chimpanzee pre-integration site, along with the human L1 insertion and 400-bp upstream and downstream flanking sequence. To reduce false positives, pairs of chimpanzee and human non-L1 genomic sequences were required to exhibit >95% identity over their entire length. This resulted in 1989 candidate Hs L1 insertions. The procedure was repeated by reversing the order of the human and chimpanzee genome sequences to identify candidate Pt L1 insertions, resulting in the recovery of 1207 loci. All candidate loci were subsequently subjected to manual verification, yielding a total of 1835 Hs and 1190 Pt L1 elements.
2.2. PCR amplification and DNA sequencing
Cell lines used to isolate DNA samples were as follows: human (H. sapiens) HeLa (American Type Culture Collection [ATCC] number CCL2), common chimpanzee Clint (P. troglodytes; cell line NS06006B), gorilla (Gorilla gorilla; cell line AG05251) and orangutan (Pongo pygmaeus; cell line ATCC CR6301). DNA samples from 20 European, 20 African American and 20 Asian human individuals isolated from peripheral blood lymphocytes were available from previous studies in our lab, and DNA samples from 20 South American individuals were obtained from the Coriell Institute for Medical Research. A common chimpanzee (P. troglodytes) population panel composed of 12 unrelated individuals of unknown geographic origin was obtained from the Southwest Foundation for Biomedical Research.
Oligonucleotide primers for the PCR amplification of L1 elements were designed using the software Primer3 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). PCR amplification of each locus was performed in 25 μl reactions using 10–50 ng DNA, 200 nM of each oligonucleotide primer, 200 μM dNTPs in 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris–HCl (pH 8.4) and 2.5 U Taq DNA polymerase. Each sample was subjected to an initial denaturation step of 5 min at 95 °C, followed by 35 cycles of PCR at 1 min of denaturation at 95 °C, 1 min at the annealing temperature, 1 min of extension at 72 °C, followed by a final extension step of 10 min at 72 °C. The resulting products were loaded on 2% agarose gels, stained with ethidium bromide, and visualized using UV fluorescence. Detailed conditions for all PCR assays designed in this study are available in Supplemental Table 1.
Individual PCR products were purified from the gels using the Wizard® gel purification kit (Promega) and cloned into vectors using the TOPO-TA Cloning® kit (Invitrogen), according to the manufacturer's instructions. DNA sequencing was performed using chain termination sequencing on an Applied Biosystems 3100 automated DNA sequencer. The DNA sequences from this study have been deposited in GenBank under accession numbers DQ375560–DQ375750.
PCR amplification of 5 full-length L1 loci was performed in 50μl reactions using 200 ng DNA, 300 nM of each oligonucleotide primer, 200 μM dNTPs, 1 mM MgSO4, 2% DMSO, and 2 U KOD Hifi DNA polymerase (Novagen). Each sample was subjected to heating for 2 min at 94 °C to activate the polymerase, followed by 35 cycles of PCR at 15 s of denaturation at 94 °C, 30 s of annealing at 60 °C, and 5 min of extension at 72 °C. The PCR products were purified using the Wizard® PCR clean-up system (Promega). DNA sequencing was completed using 26 L1 internal primers (Supplemental Table 2, (Seleme et al., 2006)). These DNA sequences have been deposited in GenBank under accession numbers DQ456 866–DQ456870.
2.3. Data analyses
We aligned 864 bp corresponding to ORF2 3′ end and entire 3′ UTR (excluding the G4TG6AG6AG3 repeat exhibiting variable length among sequences) of 1000 Hs and 207 Pt L1 elements, using the software BioEdit v.7.0 (Hall, 1999). L1 subfamily consensus sequences were generated based on putative diagnostic substitutions using the module MegAlign available in the package DNAStar. The relationships among the subfamilies were reconstructed using a median-joining network (Bandelt et al., 1999; Cordaux et al., 2004), as implemented in the software NETWORK 4.111 (http://www.fluxus-engineering.com/sharenet.htm). The age of the subfamilies was calculated with NETWORK, based on the divergence among all the copies of each subfamily. We used a nucleotide mutation rate of 0.15% per site per myr (Miyamoto et al., 1987), assuming that L1 elements accumulate mutations at the neutral rate after their insertion (Voliva et al., 1984; Pascale et al., 1993). The software MEGA 3.1 (Kumar et al., 2004) was used to build neighbor-joining trees of the 5′ UTR consensus sequences of two Pt L1 subfamily lineages and other L1 subfamilies (L1Hs and L1PA2-13; (Khan et al., 2006)), based on the observed number of nucleotide differences and Kimura 2-parameters distance. Support for the branching patterns was evaluated based on 1000 bootstrap replicates.
For flanking sequence GC content analysis, we used the BLAST-Like Alignment Tool (BLAT) server (http://genome.ucsc.edu/cgi-bin/hgBlat) to extract 20 kb of flanking sequence in either direction of each L1 element examined, after excluding 100 bp downstream of the polyadenylation signal to prevent bias towards excessive adenosine residues. The percentage of GC nucleotides in the flanking sequence of each L1 element was calculated using the EMBOSS GeeCee server (http://bioweb.pasteur.fr/seqanal/interfaces/geecee.html). For the gene density analysis, we counted the number of genes within 2 Mb sequences surrounding the 5′ and 3′ ends of each L1 element examined.
3. Results and discussion
3.1. L1 elements and nomenclature used in this study
Our comparison of the human and chimpanzee genome sequences resulted in the identification of 1835 Hs and 1190 Pt L1 elements. These figures compare favorably with previous estimates, considering the differences in the computational methodologies and requirements for validation of candidate loci used in the different studies (Mathews et al., 2003; CSAC, 2005; Mills et al., 2006). Because L1 elements are often truncated or rearranged (Smit et al., 1995; Szak et al., 2002), we based our analyses of L1 subfamily diversity and relationships on 864 bp-long sequences encompassing the last 665 bp of ORF2 and the entire 3′ UTR, to maximize the number of elements included in the analyses. This approach resulted in the inclusion of 1000 Hs and 207 Pt L1 elements. While this represents more than half of all Hs L1 elements identified, it barely accounts for one fifth of all Pt elements, suggesting that Pt L1 elements tend to be more severely truncated than Hs L1 elements (see below).
In the following text, we refer to species-specific L1 subfamilies as Hs and Pt for human and chimpanzee, respectively, and we use the RepeatMasker subfamily assignment for shared L1 subfamilies (Table 1 and Fig. 1). Each subfamily name is further identified by an Arabic numeral indicating the L1 subfamily lineage to which it belongs, followed by an upper-case letter identifying the subfamily within the sequential lineage (lower-case letters are also added for isolated subfamilies outside of the sequential lineage). Upper- and lower-case letters follow the Latin alphabet, starting from the oldest subfamily in the lineage. For example, subfamily L1Pt-2A is the oldest (A) L1 subfamily belonging to P. troglodytes-specific (Pt) subfamily lineage 2. Subfamily L1PA2-1D is the fourth oldest (D) L1PA2 subfamily belonging to the subfamily lineage 1 shared between human and chimpanzee. Subfamily L1PA2-1Da is the oldest isolated subfamily (a) stemming from L1PA2-1D. Throughout the manuscript we use the designations commonly employed in the literature for the previously characterized Hs subfamilies PreTa, Ta0 and Ta1 (Skowronski et al., 1988), which could also be referred to as L1Hs-1C, L1Hs-1D and L1Hs-1E, respectively, according to the terminology applied to the other L1 subfamilies.
Table 1.
Subfamily classification and age estimates for species-specific L1 elements
| Classification in present study | RepeatMasker classification | Age±SD (myrs) | Polymorphism level | Proportion of species-specific L1 elementsa |
|
|---|---|---|---|---|---|
| Chimp (%) | Human (%) | ||||
| L1 subfamilies shared by human, chimpanzee and gorilla, but not orangutan | |||||
| L1PA3-1A | L1PA3 | 12.7±0.8b | 0% (0/49) | 26.6 | 25.6 |
| L1PA3-1Aa | L1PA3 | 12.2±0.8 | |||
| L1PA3-1B | L1PA3/L1PA2 | 12.6±0.6b | |||
| L1PA3-1Ba | L1PA3/L1PA2 | 10.3±0.7 | |||
| L1PA3-1Bb | L1PA3/L1PA2 | 10.2±0.5b | |||
| L1PA2-1A | L1PA2 | 9.0±0.4b | |||
| L1 subfamilies shared by human and chimpanzee, but not gorilla | |||||
| L1PA2-1B | L1PA2 | 7.6±0.5b | 7% (5/67) | 27.1 | 34.8 |
| L1PA2-1C | L1PA2 | 8.0±0.5b | |||
| L1PA2-1D | L1PA2 | 7.9±0.5b | |||
| L1PA2-1Da | L1PA2 | 7.8±0.5 | |||
| L1PA2-1Db | L1PA2 | 6.0±0.5 | |||
| L1PA2-1E | L1PA2 | 6.5±0.4b | |||
| Human-specific L1 subfamilies | |||||
| L1Hs-1A | L1PA2 | 5.7±0.8 | 9% (1/11) | 0 | 38.0 |
| L1Hs-1B | L1PA2 | 4.4±0.4 | |||
| L1Hs-preTa | L1Hs-preTa | 3.1±0.3 | 14%c | ||
| L1Hs-Ta0 | L1Hs-Ta0 | 2.7±0.2 | 45%c | ||
| L1Hs-Ta1 | L1Hs-Ta1 | 1.9±0.2 | |||
| Chimpanzee-specific L1 subfamilies | |||||
| L1Pt-1A | L1PA2 | 6.2±0.8 | 30% (3/10) | 15.0 | 0 |
| L1Pt-1B | L1PA2 | 3.9±0.5 | |||
| L1Pt-2A | L1PA2 | 4.7±0.9 | 80% (8/10) | 27.5 | |
| L1Pt-2B | L1PA2 | 2.9±0.3 | |||
| L1Pt-2C | L1PA2 | 2.9±0.4 | |||
| L1Pt-2D | L1PA2 | 2.4±0.5 | |||
| Others | 3.8 | 1.6 | |||
Based on 1000 Hs and 207 Pt L1 elements.
Estimated from both Hs and Pt L1 elements.
Data from Salem et al. (2003a) and Myers et al. (2002).
Fig. 1.
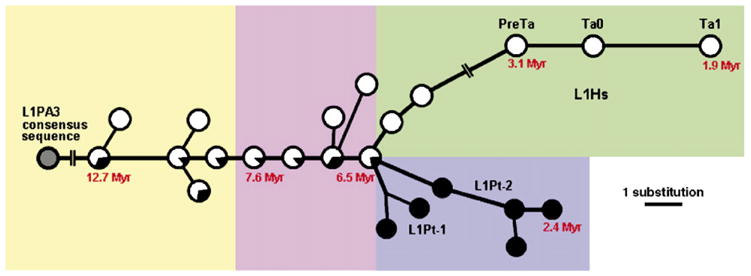
Median-joining network of L1 subfamilies. The network was reconstructed using Hs and Pt L1 elements. Empty circles represent Hs L1 subfamilies. Filled circles represent Pt L1 subfamilies. Lineage names and ages of some nodes are shown. The lines represent substitution steps, with a one-step distance indicated in the bottom-right corner. The network is also classified as yellow (L1PA3 and L1PA2-1A), pink (L1PA2), green (L1Hs) and blue (L1Pt) shadow boxes.
3.2. L1 subfamily diversity
We arbitrarily set the minimum number of elements to form a subfamily as 1% of all species-specific elements examined, or 10 Hs and 2 Pt L1 elements. Using this criterion, we could assign greater than 98% of all species-specific L1 elements to 17 human subfamilies containing 10–131 copies and 14 chimpanzee subfamilies containing 5–27 copies (Table 1). By extrapolation to total genome size, these figures imply that at least 20–30 copies of each subfamily are present in their respective genomes.
With respect to human subfamilies, we recovered the previously identified preTa, Ta0 and Ta1 Hs subfamilies (Skowronski et al., 1988), that account for 31.5% of all Hs L1 elements. All other Hs L1 elements were assigned to the older L1PA2 or L1PA3 subfamilies by RepeatMasker. Interestingly, although we analyzed species-specific L1 elements, eight subfamilies were shared between the human and chimpanzee genomes, all of which were estimated to be older than 6 myrs (Table 1), an age consistent with the human–chimpanzee divergence time (Goodman et al., 1998). These results underscore the important distinction that needs to be made about the species-specific nature of L1 individual copies versus subfamilies. Four additional human L1 subfamilies have ages estimated to be greater than 6 myrs, but are apparently not shared with chimpanzee (Table 1). However, since only about one fifth of all Pt L1 elements could be examined, it is conceivable that these four apparently Hs L1 subfamilies are actually present in the chimpanzee genome but are truncated to such an extent that they were not recognized or included in our analyses. By contrast, the two remaining human subfamilies also absent from chimpanzee (i.e. L1Hs-1A and L1Hs-1B) have estimated ages of 4–6 myrs; they are therefore likely true Hs subfamilies.
With respect to the 14 L1 subfamilies identified in chimpanzee, beyond the eight subfamilies shared with human, the six other subfamilies that account for 42.5% of all Pt elements are not shared with human (Table 1). Given that our human sample includes 1000 L1 copies, it is very unlikely that these subfamilies would appear to be Pt as a consequence of not having been sampled from the entire set of Hs L1 elements. Moreover, these six subfamilies are estimated to be 2–6 myrs-old, therefore postdating the human–chimpanzee divergence time (Goodman et al., 1998). Therefore we believe they are true Pt L1 subfamilies.
3.3. Phylogenetic relationships of L1 subfamilies
To reconstruct the relationships among the different L1 subfamilies identified in human and chimpanzee, we applied the median-joining network method (Bandelt et al., 1999; Cordaux et al., 2004) using the consensus sequences of each L1 subfamily (Fig. 1 and Supplemental Fig. 1). This network, rooted with the older L1PA3 consensus sequence, shows the global sequential order in which the successive L1 subfamilies arose (Fig. 1). Moreover, the ages estimated independently for individual subfamilies based on within-subfamily sequence diversity are in complete agreement with this phylogenetic structure (Fig. 1 and Table 1). In particular, the sequential order observed for the subfamilies shared between human and chimpanzee, and Hs subfamilies is in perfect agreement with previous studies (Boissinot et al., 2000; Khan et al., 2006). In sharp contrast with the human L1 subfamily single-lineage structure, the 6 Pt subfamilies belong to two independent L1 lineages, termed L1Pt-1 and L1Pt-2 (Fig. 1 and Table 1), which encompass two and four subfamilies, respectively.
3.4. Comparison of 5′ UTR sequences
It has recently been proposed that the number of retrotransposition-active L1 lineages at a given period of primate evolution is correlated with the extent of 5′ UTR sequence variation among subfamilies (Khan et al., 2006). Therefore, we analyzed the 5′ UTR sequences of the two L1Pt lineages we identified (i.e. L1Pt-1 and L1Pt-2) in conjunction with the 5′ UTR of other L1 subfamilies (i.e. L1Hs and L1PA2-13). Our results indicate that the 5′ UTRs of both L1Pt subfamily lineages are highly similar to each other (Fig. 2) and to the L1Hs and L1PA2 5′ UTRs. More generally, both L1Pt subfamily lineages fall within the cluster of L1 subfamilies which have been sharing a common 5′ UTR presumably recruited ~40 myrs ago (Khan et al., 2006). The presence of two L1 subfamily lineages with similar 5′ UTRs in the chimpanzee genome suggests that they might be (or might have been recently) competing with each other for the same transcription factors (Khan et al., 2006). If so, two lines of evidence suggest that the L1Pt-2 lineage may have had an advantage over the L1Pt-1 lineage. Indeed, not only is the L1Pt-2 lineage represented by twice as many copies as the L1Pt-1 lineage, but three of the four L1Pt-2 subfamilies are 2–3 myrs-old, whereas the youngest L1Pt-1 subfamily is ~4 myrs-old (Table 1). Interestingly, we identified two full-length L1Pt-2 copies with intact ORF1 and ORF2, while L1Pt-1 does not possess any detectable full-length copy with intact ORFs (i.e. putatively retrotransposition-competent) in the chimpanzee genome reference sequence (see below). Because L1 retrotransposition molecules exhibit strong cis-preference (Wei et al., 2001; Dewannieux et al., 2003), the differential number of retrotransposition-competent L1 copies among lineages may provide an advantage in the putative competition among L1 lineages. However, it is currently unknown whether the preservation of ORFs in some L1 copies is only the result of chance (i.e. because of the stochastic occurrence of ORF-disrupting mutations, all but two full-length L1Pt copies have been inactivated so far and they both happen to belong to the L1Pt-2 lineage) or because a selective process is acting to specifically preserve the integrity of the ORFs of these two particular L1Pt-2 copies. It is worthy to note here that although competition is a plausible explanation for the differential evolutionary successes of the L1Pt-1 and L1Pt-2 lineages, random chance alone could have led to the same evolutionary outcome.
Fig. 2.
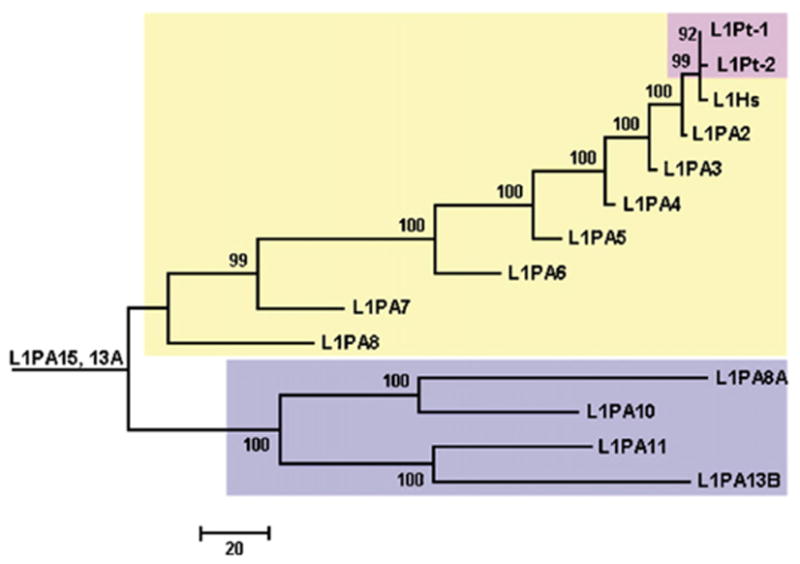
Phylogenetic tree of 5′ UTR consensus sequence of L1Pt, L1Hs and L1PA subfamilies. This neighbor-joining tree is built by using 5′ UTR consensus sequences, based on the observed number of nucleotide differences. The 5′ UTR consensus sequences of L1Hs and L1PA2-13 families in light yellow and blue shadow boxes were from Khan et al. (2006) and the sequences of two L1Pt lineages in a pink shadow box were generated in this study. Bootstrap values (%) are shown above each branch.
3.5. Insertion polymorphism levels of L1 subfamilies
To estimate the polymorphism levels (i.e. the proportion of polymorphic elements for insertion presence/absence) associated with the different L1 subfamilies, we analyzed a total of 147 L1 elements from the different subfamilies using locus-specific PCR reactions. Eighty two Hs elements were genotyped in 80 humans and 65 Pt elements were genotyped in 12 chimpanzees. As expected (Hedges et al., 2005), polymorphism levels decreased with subfamily ages (Table 1). For example, 45–80% of L1 elements belonging to subfamilies younger than ~3 myrs are polymorphic, and 9–30% of L1 elements are polymorphic in subfamilies that are estimated to be ~3–6 myrs-old. By contrast, in ~6–8 myrs-old subfamilies, only 7% of the L1 elements are polymorphic, and in subfamilies older than ~9 myrs, no elements are polymorphic. This result is consistent with the polymorphism levels observed for Alu subfamilies of similar ages, in which Alu subfamilies older than ~10 myrs, for example, virtually lack polymorphic elements (Xing et al., 2003; Salem et al., 2005).
The comparison between Pt and Hs L1 subfamilies of similar ages indicates that the polymorphism levels of Pt subfamilies is about twice as high as that of Hs subfamilies, e.g. 80% vs. 45% for <3 myrs-old L1 subfamilies and 30% vs. 9–14% for 3–6 myrs-old L1 subfamilies (Table 1). These results are consistent with those observed for Hs and Pt Alu elements, that also showed that the polymorphism levels of Pt Alu subfamilies are about twice as high as those of Hs Alu subfamilies (Hedges et al., 2004).
3.6. Comparisons with gorilla and orangutan
As shown in Table 1, several L1 subfamilies exhibit ages predating the human–chimpanzee divergence ~6 myrs ago (Goodman et al., 1998), based on subfamily sequence diversity. In fact, the oldest L1 subfamilies containing species-specific elements are estimated to be about twice as old as the human–chimpanzee divergence time (Table 1). To investigate whether these represent L1 subfamilies that have been producing new copies over extended periods of time or if the L1 elements have inserted prior to the human–chimpanzee divergence but were lost in either species (for example as a result of lineage sorting events), we genotyped the 147 L1 elements described in the previous section in gorilla and orangutan. None of the 147 elements were present in the orangutan genome. This result is consistent with the fact that the oldest L1 subfamilies examined are ~12 myrs-old (Table 1) and thus they postdate the divergence of orangutans and the ancestor of gorillas, chimpanzees and humans, estimated to have taken place ~14 myrs ago (Goodman et al., 1998). By contrast, 16 out of 49 L1 elements belonging to the 6 oldest subfamilies examined (i.e. ~9–12 myrs-old, Table 1) were present in gorilla but absent from either humans or chimpanzees in our panel (Fig. 3). DNA sequence analysis of the PCR products derived from these L1 elements showed that they are shared between gorilla and either human or chimpanzee and are identical-by-descent rather than derived from parallel, independent insertion events.
Fig. 3.
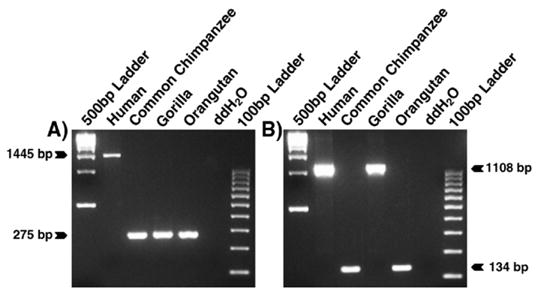
Species-specific L1 insertions. Agarose gel chromatographs derived from the analysis of two loci are shown. The DNA template used in each lane is shown at top. The product sizes for filled and empty alleles are displayed at the left or right. (A) An Hs L1 insertion. (B) Lineage sorting of an L1 insertion.
Because these elements belong to L1 subfamilies which have presumably expanded before the divergence of gorillas and the ancestor of humans and chimpanzees, it is not unexpected that some elements are shared with gorilla. One explanation for this phylogenetic distribution is that the L1 elements inserted prior to the divergence of the three species and were still polymorphic at the time of speciation. As a result, some elements have become fixed in some species while being lost in others; many examples illustrating this process of lineage sorting of mobile element insertion polymorphisms involving closely related species exist in the literature (Salem et al., 2003b; Hedges et al., 2004; Ray et al., in press). It is likely that most individual copies of the shared L1 subfamilies are also shared by the different primate species, but since our analyses were designed to detect L1 elements differentially inserted between human and chimpanzee, shared L1 elements would not be recovered.
By contrast, none of the 98 L1 elements belonging to 8 myrs-old or younger L1 subfamilies was present in the gorilla genome. Therefore, our data suggest that the divergence of gorillas and the ancestor of humans and chimpanzees occurred ~8–9 myrs ago, corresponding to the time window between the oldest L1 subfamilies shared by human and chimpanzee to the exclusion of gorilla (L1PA2-1B/C/D) and the youngest L1 subfamily shared by human, chimpanzee and gorilla (L1PA2-1A) (Table 1). Our results therefore suggest that the successive speciation events leading to the human, chimpanzee and gorilla lineages occurred within a restricted period of time, consistent with previous studies (Goodman et al., 1998). Such limited time periods between speciation events are particularly prone to lineage sorting of genetic variants because polymorphic L1 loci at the time of speciation can be independently fixed or lost in each species, as exemplified by the analysis of retrotransposon insertions among African cichlid fish species which are thought to have experienced radiation several myrs ago (Takahashi et al., 2001; Terai et al., 2003).
3.7. Structural comparison of human and chimpanzee L1 insertions
To investigate structural differences between L1 insertions that are differentially inserted in human and chimpanzee, we focused on the comparison of the genomic sequences of human and chimpanzee chromosomes 1 and 21 (using the new chimpanzee chromosome designation). We identified 138 Hs and 103 Pt L1 elements on these chromosomes. On average, Hs L1 elements were about fourfold longer than Pt L1 elements (i.e. 2533 vs. 641 bp; Fig. 4). This sharp difference is explained by the fact that ~30% (41/138) of Hs L1 elements were full-length vs. only ~2% (2/103) of Pt L1 elements (Boissinot et al., 2000; Myers et al., 2002; Boissinot et al., 2004; Mills et al., 2006) (Fig. 4). By contrast, ~86% (89/103) of Pt L1 elements are shorter than 1 kb vs. only ~48% (66/138) of Hs L1 elements (Fig. 4). Therefore, Pt L1 elements appear to be more severely truncated than their Hs counterparts. The reason for such structural differences between Hs and Pt L1 elements is currently unknown. We cannot presently exclude the possibility that this observation is the result of lower genome coverage or sequence quality available for the chimpanzee genome as compared to the highly refined human genome draft sequence. It is also possible that one or several biological processes are responsible for these differences. For example, assuming that full-length or relatively long L1 elements are more deleterious than severely truncated elements (Boissinot et al., 2001), the size differences observed between chimpanzee and human L1 elements could be explained by a higher efficiency of selection in chimpanzees than in humans, given that the chimpanzee effective population size is higher than that of humans (Graur and Li, 2000; Fischer et al., 2004) and that the efficiency of selection theoretically increases with effective population size (Graur and Li, 2000). An alternative explanation might be that, due to innovations in the host or L1 biology, L1 elements have become less adept at integrating themselves into the chimpanzee genome.
Fig. 4.
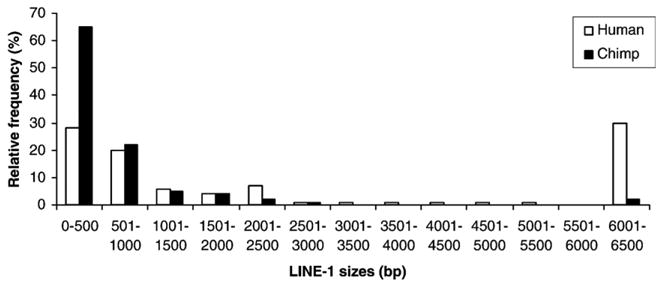
Size distribution of species-specific L1 elements. A comparison of the sizes of species-specific L1 insertions from chromosomes 1 and 21 are shown. The Hs and Pt L1 elements on chromosomes 1 and 21 are grouped in 500-bp bins.
Among the truncated L1 elements inserted on chromosomes 1 and 21, 29% (28/97) and 21% (21/101) of the Hs and Pt L1 elements, respectively, showed 5′ inversions. The inverted L1 elements were grouped into three classes, according to the structure of the junctions between the two inverted segments: deletion, overlap and precise join, as previously described (Szak et al., 2002; Martin et al., 2005). Examination of the junctions showed that 57% (16/28) and 43% (12/28) of truncated Hs L1 elements belonged to the deletion and overlap class, respectively. By comparison, 81% (17/21), 14% (3/21) and 5% (1/21) of the truncated Pt elements belonged to the deletion, overlap and precise join classes. Hence, the deletion class of inverted L1 elements was the most frequent in chimpanzee, similar to what has been reported in human and mouse (Gilbert et al., 2002, 2005; Martin et al., 2005).
Next, we examined the coding sequence of full-length L1 elements to investigate whether they are intact and thus encode putatively functional proteins required for retrotransposition. We found that 32 out of 41 full-length Hs L1 elements inserted on chromosomes 1 and 21 contained substitutions introducing premature stop codons within ORF1 or ORF2, while 9 elements encoded putatively functional proteins. Given that chromosomes 1 and 21 represent ~9% of the entire human genome, we would predict that ~100 (9/9%) intact L1 elements exist in the human genome. This figure is very close to the ~90 human retrotransposition-competent L1 elements previously identified in a genome-wide analysis (Brouha et al., 2003). The similarity between the two values suggests that the features of L1 elements inserted on chromosomes 1 and 21 constitute a good approximation of genome-wide patterns of L1 diversity. By contrast with humans, none of the full-length Pt L1 elements located on chromosome 1 and 21 possessed intact ORFs. Given this result, we extended our investigation of full-length Pt L1 elements to the whole chimpanzee genome. We identified a total of 19 full-length Pt L1 elements genome-wide, one of which contained an Alu element inserted in ORF1. However, again, none of the L1 elements was apparently intact. Strikingly, the chimpanzee L1 elements showed a frequent occurrence of 1- or 2-bp insertions responsible for frameshifts and the introduction of premature stop codons (Table 2). In most cases, those insertions were located in homopolymeric tracts (e.g. presence of four T nucleotides in a row in one copy with a frameshift, whereas the consensus of all other L1 sequences examined would possess only three T nucleotides preserving the ORF). These results suggest that at least some of these insertions may not be authentic, for example resulting from sequencing errors in the draft sequence of the chimpanzee sequence used in this study (Mills et al., 2006). To test this hypothesis, we selected 5 full-length Pt L1 elements and resequenced them using DNA from the chimpanzee individual analyzed in the chimpanzee genome project, known as Clint (CSAC, 2005). None of the 64 insertions of 1 or 2 bp present in the chimpanzee genome reference sequence (Nov. 2003 freeze) were found in our sequence analysis (Table 2). By contrast, the single 3-bp insertion detected in the reference sequence was confirmed as an authentic event. It turns out that this insertion introduced a codon that did not disrupt the ORF of the L1 element. In addition, all but one deletion sequenced (7/8) were confirmed as authentic events. These results suggest that small insertions are likely to be artifacts whereas most small deletions appear to be authentic. Therefore, we reanalyzed the 19 full-length Pt L1 elements computationally after removing all 1- or 2-bp insertions. Using this approach, we identified five intact L1 elements in the chimpanzee genome, that is considerably lower than the ~90 retrotransposition-competent L1 elements identified in the human genome (Brouha et al., 2003). Two of the intact chimpanzee L1 elements belong to the subfamily lineage L1Pt-2B and three are L1PA2 members. As discussed above (see Section 3.4 “Comparison of 5′ UTR sequences”), this may contribute to explain why the L1Pt-2 subfamily lineage seems to have been more successful than the L1Pt-1 lineage in recent chimpanzee evolution.
Table 2.
Insertions and deletions in the coding region of 5 full-length chimpanzee-specific L1 elements
| Size | Insertions
|
Deletions
|
||||||
|---|---|---|---|---|---|---|---|---|
| 1 bp | 2 bp | 3 bp | 1 bp | 2 bp | 3 bp | 4 bp | 6 bp | |
| Number in chimpanzee genome sequence (Nov. 2003 freeze) | 56 | 8 | 1 | 2 | 1 | 4 | 1 | 1 |
| Number confirmed by DNA sequencing in this study | 0 | 0 | 1 | 1 | 1 | 4 | 1 | ? |
3.8. Genomic distribution of human and chimpanzee L1 insertions
To test whether Hs and Pt L1 elements inserted in genomic regions with similar properties, we analyzed the GC content and gene density of genomic regions flanking the L1 elements inserted on chromosomes 1 and 21. We examined the GC content of 20 kb flanking genomic sequence each side of the L1 elements. The results showed that Hs and Pt L1 elements had very similar GC content distributions, both being skewed towards AT-rich regions of the genome (Fig. 5A). Indeed, 74% (102/138) and 83% (86/103) of Hs and Pt L1 elements, respectively are found in AT-rich regions (defined as regions with GC content less than the 41% genome-wide average), whereas, in comparison, 58% of the human genome consists of AT-rich regions (Lander et al., 2001). We also compared the gene density of 1 Mb flanking genomic sequence each side of L1 elements. Again, we found that Hs and Pt L1 elements had similar gene density distributions, skewed towards gene-poor regions of the genomes (Fig. 5B). These results are not unexpected, however, since there is a positive correlation between GC content and gene density (Lander et al., 2001; Versteeg et al., 2003).
Fig. 5.
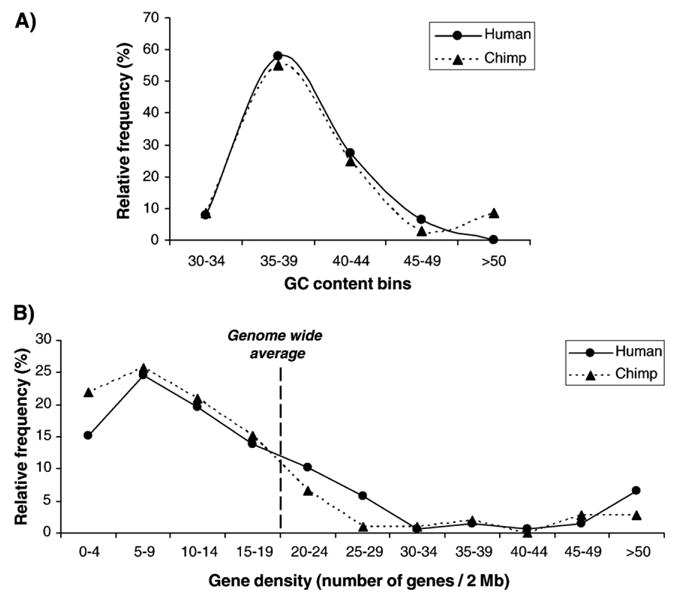
Analysis of genomic environment adjacent to species-specific L1 elements. (A) Analysis of GC content. The vertical axis represents the number of L1 loci within each GC bin. The highest frequency of species-specific L1 loci is shown in 35–39% GC bin from both the human and chimpanzee genomes. (B) Number of genes flanking L1 elements. The distribution of the number of genes flanking species-specific L1 elements is similar in both the human and chimpanzee genomes.
To investigate global polymorphism levels of Hs and Pt L1 elements regardless of subfamily affiliation, we randomly selected 31 Hs and 31 Pt L1 elements located on chromosomes 1 and 21 and genotyped them in our relevant human or chimpanzee population panels. We found that 10% (3/31) and 23% (7/31) of the Hs and Pt L1 elements, respectively, were polymorphic. Hence, consistent with the L1 subfamily-specific polymorphism results (see above) and previously reported Alu element results (Hedges et al., 2004), the global L1 insertion polymorphism level is about twice as high in chimpanzees as in humans.
4. Conclusions
Our analyses indicate that L1 elements have had very different evolutionary dynamics in the chimpanzee and human genomes, within the past ~6 myrs. Although the species-specific L1 copy numbers are on the same order in both species (1200–2000 copies; this study, (CSAC, 2005)), the number of retrotransposition-competent elements appears to be much higher in the human genome than in the chimpanzee genome. Nevertheless, in the human genome, only a subset of all retrotransposition-competent L1 elements may be responsible for most L1 insertions (Brouha et al., 2003; Seleme et al., 2006), indicating that the total number of apparently intact L1 elements in a genome is not necessarily predictive of the overall L1 activity. Interestingly, we identified two recent lineages of L1 subfamilies in the chimpanzee genome. The two lineages seem to have coexisted for several myrs, but only one shows evidence of expansion within the past three myrs. This lineage contains twice as many copies as the other lineage and we identified two retrotransposition-competent L1 elements belonging to this most recently active lineage in the chimpanzee genome, whereas no retrotransposition-competent L1 element can be identified in the other, apparently less active lineage. If the differential evolutionary dynamics of these two L1 subfamily lineages is not the result of chance, our results suggest that the coexistence of several L1 lineages might be unstable (Khan et al., 2006), and that a situation of competition between two L1 subfamily lineages may be resolved in a very short evolutionary period of time, perhaps on the order of just a few myrs. Our data suggest that speciation events and associated host demographic changes (Hedges et al., 2004; Cordaux and Batzer, 2006) may facilitate the coexistence of multiple L1 subfamily lineages within species. Therefore, cases of coexistence of multiple L1 subfamily lineages may have been quite common during evolution. However, if this situation is evolutionarily unstable and quickly leads to the loss of activity of one of the lineages, then it would appear on a large evolutionary time scale as though all or most L1 subfamilies in one species belong to one major lineage of subfamilies, as previously reported (Khan et al., 2006). Within the chimpanzee genome, two Pt L1 subfamily lineages can be unambiguously detected, presumably because of the short evolutionary time-depth involved. Therefore, the chimpanzee genome constitutes an excellent model in which to further analyze the evolutionary dynamics of L1 retrotransposons.
Supplementary Material
Acknowledgments
This research was supported by the National Science Foundation BCS-0218338 and EPS-0346411 (MAB), Louisiana Board of Regents Millennium Trust Health Excellence Fund HEF (2000-05)-05, (2000-05)-01 and (2001-06)-02 (MAB), National Institutes of Health RO1 GM59290 (MAB), R03 CA101515 (PL), P30 CA16056 (Roswell Park Cancer Institute), and the State of Louisiana Board of Regents Support Fund (MAB).
Abbreviations
- LINE-1 or L1
long interspersed element-1
- UTR
untranslated region
- ORF
open reading frame
- Pt
Pan troglodytes-specific
- Hs
Homo sapiens-specific
- myrs
million years
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.gene.2006.08.029.
References
- Badge RM, Alisch RS, Moran JV. ATLAS: a system to selectively identify human-specific L1 insertions. Am J Hum Genet. 2003;72:823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev, Genet. 2002;3:70–79. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Young L, Munson PJ, Furano AV. The insertional history of an active family of L1 retrotransposons in humans. Genome Res. 2004;14:1221–1231. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet. 2005;117:411–427. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing, Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. Teaching an old dog new tricks: SINEs of canine genomic diversity. Proc Natl Acad Sci U S A. 2006;103:1157–1158. doi: 10.1073/pnas.0510714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Hedges DJ, Batzer MA. Retrotransposition of Alu elements: how many sources? Trends Genet. 2004;20:464–467. doi: 10.1016/j.tig.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Fanning T, Singer M. The LINE-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Res. 1987;15:2251–2260. doi: 10.1093/nar/15.5.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Fischer A, Wiebe V, Paabo S, Przeworski M. Evidence for a complex demographic history of chimpanzees. Mol Biol Evol. 2004;21:799–808. doi: 10.1093/molbev/msh083. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M, et al. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Graur D, Li WH. Fundamentals of Molecular Evolution. 2. Sinauer Associates; Sunderland: 2000. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Series. 1999;41:95–98. [Google Scholar]
- Hedges DJ, Callinan PA, Cordaux R, Xing J, Barnes E, Batzer MA. Differential alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges DJ, et al. Modeling the amplification dynamics of human alu retrotransposons. PLoS Comput Biol. 2005;1:333–340. doi: 10.1371/journal.pcbi.0010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci U S A. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bio-inform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Martin SL, Li WL, Furano AV, Boissinot S. The structures of mouse and human L1 elements reflect their insertion mechanism. Cytogenet Genome Res. 2005;110:223–228. doi: 10.1159/000084956. [DOI] [PubMed] [Google Scholar]
- Mathews LM, Chi SY, Greenberg N, Ovchinnikov I, Swergold GD. Large differences between LINE-1 amplification rates in the human and chimpanzee lineages. Am J Hum Genet. 2003;72:739–748. doi: 10.1086/368275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Mills RE, et al. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet. 2006;78:671–679. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto MM, Slightom JL, Goodman M. Phylogenetic relations of humans and African apes from DNA sequences in the psi eta-globin region. Science. 1987;238:369–373. doi: 10.1126/science.3116671. [DOI] [PubMed] [Google Scholar]
- Myers JS, et al. A comprehensive analysis of recently integrated human Ta L1 elements. Am J Hum Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov I, Rubin A, Swergold GD. Tracing the LINEs of human evolution. Proc Natl Acad Sci U S A. 2002;99:10522–10527. doi: 10.1073/pnas.152346799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale E, Liu C, Valle E, Usdin K, Furano AV. The evolution of long interspersed repeated DNA (L1, LINE 1) as revealed by the analysis of an ancient rodent L1 DNA family. J Mol Evol. 1993;36:9–20. doi: 10.1007/BF02407302. [DOI] [PubMed] [Google Scholar]
- Ray DA, Xing J, Salem AH, Batzer MA. SINEs of a nearly perfect character. Syst Biol. doi: 10.1080/10635150600865419. in press. [DOI] [PubMed] [Google Scholar]
- Salem AH, Myers JS, Otieno AC, Watkins WS, Jorde LB, Batzer MA. LINE-1 preTa elements in the human genome. J Mol Biol. 2003a;326:1127–1146. doi: 10.1016/s0022-2836(03)00032-9. [DOI] [PubMed] [Google Scholar]
- Salem AH, et al. Alu elements and hominid phylogenetics. Proc Natl Acad Sci U S A. 2003b;100:12787–12791. doi: 10.1073/pnas.2133766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem AH, Ray DA, Hedges DJ, Jurka J, Batzer MA. Analysis of the human Alu Ye lineage. BMC Evol Biol. 2005;5:18. doi: 10.1186/1471-2148-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleme MC, Vetter MR, Cordaux R, Bastone L, Batzer MA, Kazazian HH., Jr Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc Natl Acad Sci U S A. 2006;103:6611–6616. doi: 10.1073/pnas.0601324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen FM, et al. Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF, Toth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Genome Biol. Vol. 3. 2002. Molecular archeology of L1 insertions in the human genome; p. research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Terai Y, Nishida M, Okada N. Phylogenetic relationships and ancient incomplete lineage sorting among cichlid fishes in Lake Tanganyika as revealed by analysis of the insertion of retroposons. Mol Biol Evol. 2001;18:2057–2066. doi: 10.1093/oxfordjournals.molbev.a003747. [DOI] [PubMed] [Google Scholar]
- Terai Y, Takahashi K, Nishida M, Sato T, Okada N. Using SINEs to probe ancient explosive speciation: “hidden” radiation of African cichlids? Mol Biol Evol. 2003;20:924–930. doi: 10.1093/molbev/msg104. [DOI] [PubMed] [Google Scholar]
- Versteeg R, et al. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13:1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voliva CF, Martin SL, Hutchison CA, III, Edgell MH. Dispersal process associated with the L1 family of interspersed repetitive DNA sequences. J Mol Biol. 1984;178:795–813. doi: 10.1016/0022-2836(84)90312-7. [DOI] [PubMed] [Google Scholar]
- Wang J, Song L, Grover D, Azrak S, Batzer MA, Liang P. dbRIP: a highly integrated database of retrotransposon insertion polymorphisms in humans. Hum Mutat. 2006;27:323–329. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing JC, et al. Comprehensive analysis of two Alu Yd subfamilies. J Mol Evol. 2003;57:S76–S89. doi: 10.1007/s00239-003-0009-0. [DOI] [PubMed] [Google Scholar]
- Xing J, Hedges DJ, Han K, Wang H, Cordaux R, Batzer MA. Alu element mutation spectra: molecular clocks and the effect of DNA methylation. J Mol Biol. 2004;344:675–682. doi: 10.1016/j.jmb.2004.09.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


