Abstract
Background
The bacterial biothreat agents Burkholderia mallei and Burkholderia pseudomallei are the cause of glanders and melioidosis, respectively. Genomic and epidemiological studies have shown that B. mallei is a recently emerged, host restricted clone of B. pseudomallei.
Results
Using bacteriophage-mediated immunoscreening we identified genes expressed in vivo during experimental equine glanders infection. A family of immunodominant antigens were identified that share protein domain architectures with hemagglutinins and invasins. These have been designated Burkholderia Hep_Hag autotransporter (BuHA) proteins. A total of 110/207 positive clones (53%) of a B. mallei expression library screened with sera from two infected horses belonged to this family. This contrasted with 6/189 positive clones (3%) of a B. pseudomallei expression library screened with serum from 21 patients with culture-proven melioidosis.
Conclusion
Members of the BuHA proteins are found in other Gram-negative bacteria and have been shown to have important roles related to virulence. Compared with other bacterial species, the genomes of both B. mallei and B. pseudomallei contain a relative abundance of this family of proteins. The domain structures of these proteins suggest that they function as multimeric surface proteins that modulate interactions of the cell with the host and environment. Their effect on the cellular immune response to B. mallei and their potential as diagnostics for glanders requires further study.
Background
Burkholderia mallei is the causative agent of glanders, a serious Gram-negative infection that predominantly affects horses and other equines [1]. Natural B. mallei infection has largely been eradicated and human infection is extremely rare, but renewed interest in this organism parallels its classification as a category B biothreat agent. There is a need to develop an effective vaccine for individuals at risk of exposure from deliberate release, and understanding the immune response elicited during infection with B. mallei is central to this process. The relative importance of cellular versus humoral responses in the development of protective immunity against B. mallei is under investigation. Experimental evidence indicates that both play a role, but that stimulation of a Th1-like immune response may be important for protection [2-5]. One study of murine monoclonal antibodies (mAbs) against B. mallei administered to mice prior to a lethal aerosol challenge reported non-sterilizing protection [6]. Passive protection in the mouse model has also been described for the highly related B. pseudomallei using murine mAbs specific for B. pseudomallei polysaccharide [7].
The development of improved diagnostic tests for the diagnosis of glanders is also a necessity. PCR has been described for the detection of B. mallei during acute presentation [8-10], but the antibody response to infection is poorly understood and there are no serological tests currently recommended for the diagnosis of glanders in patients who have been partially or fully treated and in whom B. mallei is not detectable. Humoral immune response profiling to infection is a logical approach to the evaluation of immunogenic antigens, and has the ability to define novel targets for both diagnostics and vaccines. Profiling using proteome microarrays has been described for a vaccinia virus proteome consisting of 185 individual viral proteins and used to determine Ab profiles in serum from vaccinia virus-immunized humans, primates, and mice [11]. This technology has not been described in the literature to date for antibody profiling in response to infection with B. mallei or B. pseudomallei, although polysaccharide microarray technology has been described for the detection of B. pseudomallei and B. mallei antibodies to capsular antibodies [12]. An alternative strategy for immunoscreening is the use of a bacteriophage expression library. This has recently been used to detect in vivo expressed antigens of Staphylococcus aureus and to utilize this information to develop a vaccine to protect against experimental nasal carriage [13]. Although complex molecules such as LPS will not be represented, this technique offers an immediately available, rapid and simple means of identifying and testing putative candidate vaccines and targets for serological diagnostic tests. We have applied bacteriophage-mediated immunoscreening to identify antibody responses to in vivo expressed genes during experimental equine glanders infection, and have compared this with those elicited during human melioidosis.
Results
B. mallei expression library screening with glanders serum
The B. mallei expression library was individually probed with sera from two experimentally infected horses taken seven days after intra-tracheal bacterial inoculation with B. mallei ATCC 23344. This equine glanders model has been described previously [14]. Positive plaques were picked and purified by additional rounds of screening. A total of 207 positive clones were identified and partially sequenced. These corresponded to 71 different loci (comprising a contiguous DNA region from single or overlapping clones) containing 228 known or putative protein coding sequences (CDSs) of B. mallei. Additional file 1 provides full details of the B. mallei antigens recognized by one or both of the infected horse sera. The distribution of loci was not equal between the two chromosomes; 50 loci were present on chromosome 1, and 21 loci were present on chromosome 2 in B. mallei ATCC 23344.
The specificity of these results for experimental glanders infection as opposed to the presence of pre-existing cross reactive antibodies to other bacterial species was tested by screening serum taken from one healthy experimental horse prior to infection with B. mallei. A total of 20 screenings failed to reveal any positive plaques (data not shown). This strongly suggests that the antibodies detected in sera from the two infected horses are specific to the experimental B. mallei infection.
Four loci were highly over-represented amongst the positive clones; these are shown in Table 1. These four loci together represented 110 (53%) of all positive clones. The most frequent was locus number 14, which was represented in 40 clones, followed by locus 10 (24 clones), locus 54 (24 clones) and locus 61 (22 clones). This compares with the fifth most frequent locus, which occurred only 6 times (Additional file 1). Both horse sera recognized each of the four common loci. The specificity of this result was confirmed by Western blot of one representative clone of each of the four loci (data not shown).
Table 1.
The four most frequent loci identified on immunoscreening of the B. mallei library.
| Locus | Locus co-ordinates | Clone co-ordinates | Clone occurrence | Genes within locus (with co-ordinates) | Name, putative function of homology |
| 10 | 875663–881753 | 875663–880118 | 1 | BMA0838 (875959–876654) | DNA-binding response regulator |
| 876370–881176 | 1 | BMA0839 (876702–876944) | Hypothetical protein | ||
| 876765–880560 | 2 | BMA0840 (877030–880302) | Haemagglutinin family protein | ||
| 876928–880933 | 1 | BMA0841 (880422–881093) | OmpA family protein | ||
| 876928–880678 | 3 | BMA0842 (881095–882624) | TPR Domain domain protein | ||
| 877235–881753 | 1 | ||||
| 877402–880118 | 1 | ||||
| 877579–880118 | 2 | ||||
| 877579–879740 | 3 | ||||
| 877235–880659 | 3 | ||||
| 877575–880933 | 1 | ||||
| 877581–880663 | 2 | ||||
| 878122–880678 | 1 | ||||
| 879170–880659 | 1 | ||||
| 879216–880873 | 1 | ||||
| 14 | 1072256–1078767 | 1072256–1076403 | 1 | BMA1024 (1072412–1073026) | Type-1 fimbrial protein, A subunit |
| 1072709–1077699 | 1 | BMA1025 (1073192–1073653) | Pseudogene | ||
| 1072731–1076799 | 2 | BMA1026 (1073869–1074054) | Hypothetical protein | ||
| 1073129–1078023 | 2 | BMA1027 (1074198–1077236) | Outer membrane protein, putative | ||
| 1073358–1076799 | 3 | BMA1028 (1077244–1077492) | Hypothetical protein | ||
| 1073358–1078533 | 1 | BMA1029 (1077515–1077694) | Pseudogene | ||
| 1073625–1076884 | 3 | BMA1030 (1077715–1077978) | Pseudogene | ||
| 1074020–1076884 | 2 | BMA1031 (1077953–1078135) | Pseudogene | ||
| 1074281–1076800 | 1 | BMA1032 (1078196–1078453) | Pseudogene | ||
| 1074570–1076800 | 2 | BMA1033 (1078422–1079098) | Pseudogene | ||
| 1074570–1076992 | 8 | ||||
| 1074570–1077699 | 1 | ||||
| 1074570–1077970 | 2 | ||||
| 1074598–1076992 | 1 | ||||
| 1074858–1076799 | 1 | ||||
| 1075270–1078767 | 1 | ||||
| 1075270–1076799 | 1 | ||||
| 1075414–1077699 | 1 | ||||
| 1075414–1078023 | 1 | ||||
| 1075414–1078078 | 2 | ||||
| 1075414–1078309 | 1 | ||||
| 1075421–1077970 | 1 | ||||
| 1075438–1077970 | 1 | ||||
| 54 | 657988–664905 | 657988–661537 | 2 | BMAA0649 (658368–662975) | Hep_Hag family protein/haemagglutinin motif family |
| 657991–661537 | 3 | protein/YadA-like domain protein | |||
| 658070–660535 | 1 | BMAA0650 (658368–662975) | Hypothetical protein | ||
| 658080–661537 | 1 | BMAA0651 (663846–664187) | H-NS histone family protein | ||
| 658345–660890 | 1 | BMAA0652 (664378–665118) | Transcriptional regulator, IclR family | ||
| 658485–663240 | 1 | ||||
| 658485–660037 | 1 | ||||
| 659301–663317 | 1 | ||||
| 659301–663508 | 2 | ||||
| 659440–661453 | 1 | ||||
| 659519–661453 | 1 | ||||
| 659675–663508 | 1 | ||||
| 659947–663508 | 1 | ||||
| 660568–662869 | 1 | ||||
| 660906–663317 | 1 | ||||
| 660906–663909 | 2 | ||||
| 661040–663303 | 2 | ||||
| 661040–664905 | 1 | ||||
| 61 | 1422362–1427764 | 1422362–1425667 | 1 | BMAA1320 (1421605–1423098) | Amidase family protein |
| 1422767–1427459 | 1 | BMAA1321 (1423230–1423490) | Hypothetical protein | ||
| 1424023–1426561 | 1 | BMAA1322 (1423632–1424066) | UspA family protein | ||
| 1424255–1427209 | 3 | BMAA1323 (1424227–1425042) | Outer membrane protein OmpA/SmpA/OmlA family | ||
| 1424255–1427211 | 1 | BMAA1324 (1425116–1427611) | Haemagglutinin family protein | ||
| 1424263–1427203 | 1 | ||||
| 1424846–1426767 | 2 | ||||
| 1424886–1426776 | 1 | ||||
| 1424846–1426977 | 1 | ||||
| 1424846–1427211 | 1 | ||||
| 1424846–1427764 | 2 | ||||
| 1425017–1427764 | 2 | ||||
| 1425067–1427764 | 1 | ||||
| 1425768–1426773 | 2 | ||||
| 1425774–1426766 | 2 | ||||
The CDSs in each of the four common loci were examined to identify the putative antigen responsible for immune stimulation. This was achieved by mapping overlapping clones at each locus. A complete or partial copy of BMA0840 was present in all clones of locus 10; the same was true for BMA1027 in locus 14, BMAA0649 for locus 54 and BMAA1324 for locus 61. Furthermore, BMA0840 was the only gene present in 6 independent clones, BMA1027 was the only gene present in 14 clones, BMAA0649 was present alone in 12 clones, and BMAA1324 was present alone in 8 clones. Based on these observations, we propose that these CDSs encode immuno-stimulatory proteins during experimental equine infection. We cannot infer from this that all four genes were expressed during equine infection; the presence of highly homologous regions between the four proteins could lead to antibody cross-reactivity. Furthermore, our results do not exclude the possibility that additional CDSs encoding surface-expressed proteins present in the four commonly represented loci also contributed towards the antibody response. For example, BMA0841 (locus 10) encodes a putative OmpA family protein, and BMAA1323 (locus 61) encodes a putative outer membrane protein OmpA/SmpA/OmlA family.
B. mallei Burkholderia Hep_Hag autotransporter (BuHA) proteins
Three of the four putatively immunodominant antigens were annotated as hemagglutinin family proteins, and the fourth was annotated as a putative outer membrane protein (Table 1). The domain organization of these proteins was defined and compared using the Pfam protein family database [15]. All four proteins contain a C-terminal YadA domain (Pfam domain PF03895), together with several HIM (Pfam domain PF05662) and Hep_Hag (Pfam domain PF05658) domains (Figure 1). The Hep_Hag domain is a repetitive region comprising a 7 amino acid repeat that is found in haemagglutinins and invasins. The HIM domain is another short motif (~20 amino acids) that is often found in conjunction with the Hep_Hag motif. The YadA domain is composed of approximately 120 amino acids, and is found in the C-terminal regions of surface-exposed Gram-negative bacterial proteins associated with autotransporters [16]. Proteins containing the YadA domain often have a characteristic functional organization, with an extended conserved N-terminal signal sequence (data not shown) and a central region that often includes Hep_Hag and HIM domains. In addition to the conserved domains, these proteins contain variable domains, which are often composed of low complexity sequence that are not conserved between family members. The domain conservation of these proteins has led us to designate them as Burkholderia Hep_Hag autotransporter (BuHA) proteins.
Figure 1.

Domain architecture of Burkholderia mallei BuHA proteins. Pfam [36] domain organization of 4 immunodominant BuHA proteins of B. mallei, and 2 additional CDSs from B. mallei that contain Hep_Hag domains. Also included is the YadA protein from Yersinia enterocolitica (accession number YADA1_YEREN). Keys are shown for the domains in the bead diagrams.
To define further the immunodominant region of these proteins, mapping was carried out to define the minimum region present in all positive clones at each of the four loci. For BMA0840, BMA1027 and BMAA1324, all of the clones contained Hep_Hag domains. For BMAA0649 all but one of the clones contained Hep_Hag domains, the errant clone contained the first 48 amino acids at the N-terminus. The Hep_Hag domain may therefore represent the minimum epitope required to stimulate an antibody response.
In silico analysis was conducted to define the presence of B. mallei BuHA proteins using the whole genome sequence of B. mallei ATCC 23344 [17]. Six genes were identified, four of which were the CDSs identified above (BMA0840, BMA1027, BMAA0649, and BMAA1324; Figure 1). Genes encoding the two non-immunogenic B. mallei BuHA proteins were examined. BMAA0810 contained a C-terminal YadA domain and a single HIM domain, but no Hep_Hag domains. The CDS encoding this protein had an IS element (IS407A) located immediately upstream and lacked an N-terminal signal sequence. IS element mediated recombination, a feature of the evolution of the B. mallei genome, appears to have resulted in the truncation of this CDS; it is likely that this CDS is a pseudogene. The other non-immunogenic member of this family is BMAA0749. This CDS contains Hep_Hag and HIM domains (Figure 1), but lacks a strong Pfam YadA domain match in the C-terminus; a weaker match to the YadA domain with a score below the Pfam gathering threshold (score 21.10, e-value 7.4e-06) was detected. This CDS also lacked an N-terminal signal sequence, and it is likely that the product is not processed to the external surface of the cell. These observations could explain the failure to detect these two loci by immuno-screening.
Six B. mallei isolates are currently undergoing whole genome sequencing by TIGR [18], and are publicly available via the Pathema database [19]. Alleles of all four immunodominant proteins were identified in strains 10399, FMN, JHU, and GB8; the number of residues in the encoded proteins was also comparable. Strain GB8 is a horse passaged derivative of ATCC 23344, and strains FMN and JHU are human passaged derivatives of ATCC 23344 isolated during the same infective episode. Two of the strains, NCTC 10229 and NCTC 10247, did not contain alleles of BMA1027. Additionally, both of these strains contained identical alleles of BMAA0649, which encoded proteins with additional internal residues (172 amino acids) in comparison to the ATCC 23344 protein. NCTC 10229 was originally isolated in Hungary in 1961, and NCTC 10247 was isolated in Turkey in 1960. Given their different origins, it is surprising that they both lack BMA1027 and have identical variants of BMAA049.
The distribution of the four immunogenic B. mallei BuHA proteins was investigated in collections of B. mallei strains (n = 21) using PCR. All isolates were positive for BMA0840 and BMAA1324, whilst BMA1027 and BMAA0649 showed some variability: 12/21 (57%) of strains were positive for BMA1027 and 18/21 (86%) of strains were positive for BMAA0649. The screening was expanded to include a collection of B. pseudomallei strains (n = 100). All isolates were positive for the B. pseudomallei homologs of BMA0840 and BMAA1324, while homologs of BMA1027 and BMAA0649 were variable but more widely distributed; 96/100 (96%) and 92/100 (92%) of the strains were positive for BMA1027 and BMAA0649, respectively.
Distribution of BuHA proteins
We hypothesized that the BuHA proteins are important virulence determinants in B. mallei and B. pseudomallei, and that these would not be present in the highly related but non-pathogenic B. thailandensis genome [20]. In silico analysis using the whole genome sequences of B. mallei ATCC 23344 [17], B. pseudomallei K96243 [21] and B. thailandensis strain E264 [20] was performed to identify orthologs of the B. mallei BuHA proteins.
The genome of B. pseudomallei K96243 contained 9 genes encoding BuHA proteins (Figure 2). This included orthologs of all four B. mallei immunodominant proteins and the other two non-immunogenic proteins (Table 2). With the exception of the B. pseudomallei proteins BPSL1631 and BPSS0796, all of the orthologs contained a similar number of amino acid residues and were of a similar size. In the case of BPSL1631 and BPSS0796, there were increased residues in the repeat regions of these CDSs; BPSL1631 contained an additional 113 amino acid residues that corresponded to 4 Hep_Hag domains (Figures 1 and 2); and BPSS0796 contained an additional 118 amino acid residues in a low complexity region (Figures 1 and 2). The B. pseudomallei ortholog of BMAA0810, BPSS1439, is not truncated and contains an N-terminal signal sequence, HIM domains, and a C-terminal YadA domain, but does not contain Hep_Hag domains.
Figure 2.
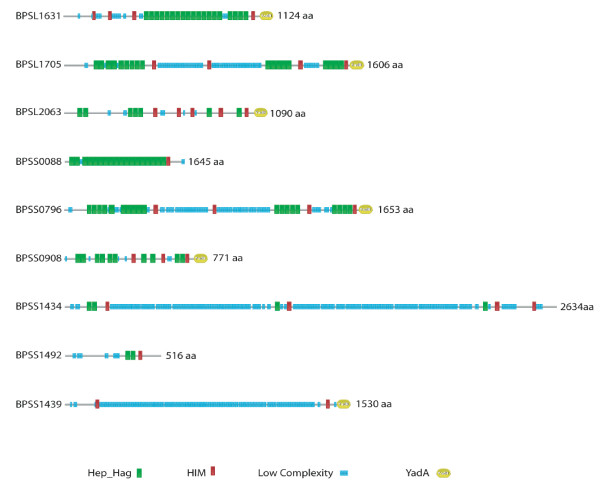
Domain architecture of Burkholderia pseudomallei BuHA proteins. Pfam [36] domain organization of BuHA proteins from B. pseudomallei. Keys are shown for the domains in the bead diagrams.
Table 2.
Orthologs of BuHA proteins in Burkholderia pseudomallei, Burkholderia mallei and Burkholderia thailandensis.
| B. mallei | B. pseudomallei | B. thailandensis |
| BMA0840 | BPSL2063 | - |
| BMA1027 | BPSL1631 | - |
| - | BPSL1705 | - |
| BMAA0649 | BPSS0796 | - |
| BMAA1324 | BPSS0908 | BTH_II1489 |
| BMAA0810*§ | BPSS1439§ | BTH_II0954*§+ |
| BMAA0749+ | BPSS1492+ | BTH_II0875+ |
| - | BPSS1434+ | BTH_II0957 |
| - | BPSS0088+ | BTH_II0112 |
| - | - | BTH_II0878+ |
Orthologs of BuHA proteins identified in the complete genomes of B. pseudomallei K96243, B. mallei ATCC 23344 and B. thailandensis E264. The presence of a BuHA protein is indicated by the systematic identifier for that protein in the genome, – indicates absence of an orthologue. Orthology was determined by viewing pairwise BLASTN [41] comparisons of the genomes in ACT [47] to identify loci displayed similarity and conserved surrounding gene order. * Pseudogene, §Below Pfam trusted threshold Hep_Hag domain match, + Below Pfam trusted threshold or no YadA domain match.
In addition to the orthologs of the B. mallei BuHA proteins, the B. pseudomallei genome contained three other related CDSs: BPSL1705, BPSS0088 and BPSS1434. All of these proteins contain HIM, Hep_Hag and YadA domains; the YadA domain matches for BPSS0088 and BPSS1434 are below the Pfam gathering threshold (score 15.2, e-value 2.1e-05 and score 21.8, e-value 4.8e-06 respectively).
The genome of B. thailandensis strain E264 contained 6 BuHA proteins (Table 2): 4 with Hep-Hag and YadA domains (BTH_II0112, BTH_II0878, BTH_II0957 and BTH_II1489); 1 protein with Hep-Hag domains but lacking a C-terminal domain (BTH_II0875); and 1 gene remnant (BTH_II0954) of the B. mallei BMAA0810 and B. pseudomallei BPSS1439 orthologs. Only one of these proteins was orthologous to the B. mallei immunodominant proteins (Table 2); two were orthologous to the non-immunogenic proteins, two have B. pseudomallei orthologs, but not B. mallei orthologs, and one is unique to B. thailandensis.
The identification of numerous hemagglutinin family proteins in the three Burkholderia genomes is in marked contrast to other bacterial species, including some other members of the Burkholderiacae which have lower numbers. The phylogenetic relationship of this family of proteins was investigated by aligning the C-terminal YadA domain regions. The repetitive distribution of Hep_Hag domains within proteins, and the variability and low complexity composition of other regions, renders whole protein comparisons unsuitable for phylogenetic analysis. The BuHA proteins from B. mallei, B. pseudomallei, B. thailandensis and 2 other sequenced Burkholderia species (Burkholderia xenovorans and Burkholderia sp. 383), were compared with BuHA proteins from other bacteria (Figure 3). The genomes of other Burkholderiaceae contained varying number of BuHA proteins (Burkholderia sp. 383 contains 6; Burkholderia xenovorans LB400 contains 4; and Ralstonia solanacearum GMI1000 contains 1). The BuHA proteins from B. mallei and B. pseudomallei were represented in two lineages. The B. mallei protein BMAA1324 clustered with orthologs from B. pseudomallei and B. thailandensis (Table 2) and a protein from Burkholderia sp. 383, and formed part of a larger cluster that was made up predominantly of proteins from outside the Burkholderiaceae. The other BuHA proteins from B. mallei, B. pseudomallei and B. thailandensis resided within a second cluster.
Figure 3.
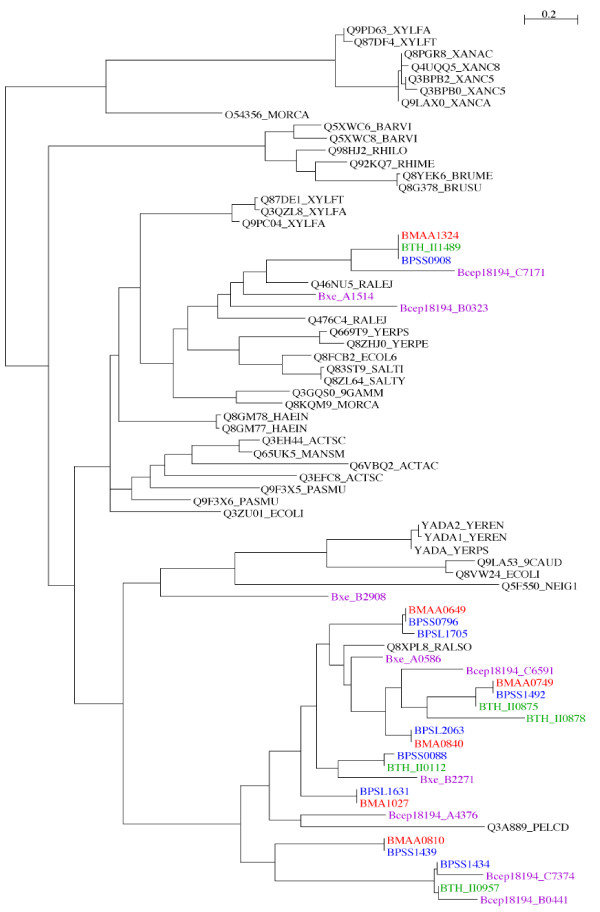
Phylogenetic relationships of BuHA proteins based on C-terminal YadA domains. Unrooted Maximum likelihood tree built from an alignment of YadA domain sequences from Hep_Hag family proteins. YadA domain sequences were extracted from B. mallei ATCC 23344, B. pseudomallei K96243, B. thailandensis E264, B. xenovorans LB400, and Burkholderia sp. 383 genome annotations; additional sequences were downloaded from the Pfam website [36,46]. B. mallei proteins are coloured red, B. pseudomallei proteins are coloured blue, B. thailandensis proteins are coloured green, other Burkholderia species proteins are coloured purple.
The orthogous relationships identified for the B. mallei and B. pseudomallei proteins using comparative genomic analysis were also evident in the phylogenetic tree (Figure 3). In addition, the tree topology suggested paralogous relationships for some of the BuHA proteins. For example, the orthologous pair B. pseudomallei protein BPSS0796 and B. mallei protein BMAA0649 clustered with B. pseudomallei protein BPSL1705. BPSL1705 resides in a genomic island (GI 8) [21] on chromosome I and BPSS0796 resides on chromosome II, but phylogenetic analysis of the YadA domains suggests that the two loci arose from a duplication event. The conservation of Hep_Hag and HIM domains organization in these two proteins also suggests a common ancestry. On this basis, it is possible that mobile genetic elements may have had a role in the expansion of BuHA proteins in B. pseudomallei.
B. pseudomallei expression library screening with melioidosis patient serum
The finding that B. pseudomallei K96243 contains 9 genes encoding BuHA proteins raised the possibility that these may also be immunogenic during human melioidosis. A B. pseudomallei K96243 expression library was probed with twenty-one separate serum samples from patients presenting to Sappasithiprasong Hospital in northeast Thailand with culture-proven melioidosis. Patient age ranged from 10–71 years (median 49 years, IQR 31–56 years), and 15 were male. Diabetes mellitus was present in 12 patients, a proportion that reflects the presence of this condition in the larger melioidosis population. Manifestations of disease were highly variable, as follows: 7 patients had disseminated melioidosis (blood culture positive plus >1 non-contiguous foci of infection), 4 patients had bacteremia with a single or no identifiable focus of infection, 2 patients had multifocal infection (>1 non-contiguous foci of infection and blood culture negative), and 8 patients had localized disease (single focus of infection and blood culture negative).
A total of 189 clones, corresponding to 131 different loci (comprising a contiguous DNA region from single or overlapping clones) containing 428 known or putative open reading frames of B. pseudomallei were isolated. The BuHA proteins were uncommon; 5 of the putative B. pseudomallei BuHA proteins were present in either one (BPSL2063, BPSS0908, BPSS0796 and BPSS1439), or two clones (BPSS1492) (total 6/189 (3%) of clones overall); the other members of the BuHA protein group were not observed. Three of the four B. mallei homologs were each represented once.
Discussions and conclusion
The findings of this study indicate that the BuHA proteins generate a strong antibody response in the experimental equine model of glanders. Bacteriophage-mediated immunoscreening demonstrated that four BuHA proteins were highly over-represented. Comparative sequence analysis of the cloned inserts of these four loci identified the minimum regions present in all clones. For each protein the mapped regions contained Hep_Hag motifs. We propose that this domain represents the immunostimulatory region of these proteins; the presence of multiple Hep_Hag protein domains may indicate that there are multiple sites for immune recognition. In silico analysis of the B. mallei ATCC 23344 genome identified two further members of this protein family (BMAA0749 and BMAA0810); their absence in the library can be explained by the fact that these were predicted not to be expressed or processed correctly.
In addition to the Hep_Hag domain, the four B. mallei immunostimulatory BuHA proteins contain C-terminal YadA trimeric autotransporter domains. This domain is found in a large family of surface proteins, and has been shown to be important in protein processing and transport. The YadA domain inserts into the outer membrane forming a trimeric structure, which then translocates the passenger domains to the cell surface side of the membrane [16]. Unlike other autotransporter proteins (for example, proteins containing the Pfam domain PF03797), the passenger domains of YadA domain proteins are not cleaved and remain covalently linked to the translocator domain. Stable trimerization has been shown to be essential for native folding and stability of the functional passenger domain [22]. Several of the YadA family proteins have been functionally characterized and shown to be adhesins that mediate bacterial interactions with host cells or extracellular matrix proteins [23-28]. The prototypical protein of this family is the YadA adhesin from Yersinia enterocolitica [29]. In addition to the C-terminal YadA domain, the protein also contains 4 Hep_Hag domains and 1 HIM domain. YadA has a trimeric head-stalk-anchor architecture [30], with the Hep_Hag and HIM domains forming part of the head domain [31] (Figure 1). This region of the protein has a left-handed parallel β-roll (LPBR) structure, which has been shown to bind collagen [31]. In particular, residues within the Hep_Hag domain regions of YadA have been shown to be essential for YadA-mediated collagen binding [32]. Epitope mapping of YadA identified 7 epitopes that were uniquely recognized by an anti-YadA antiserum that was able to inhibit collagen binding [32]. Four of these epitopes shared a motif NSVAIG-S that is repeated eight times within the N-terminal half of YadA and forms part of the Hep_Hag motif.
The role of B. mallei immunostimulatory BuHA proteins is unknown, but the similarity in domain organisation to adhesins such as YadA suggests they may function as cell surface binding proteins that potentially modulate host-cell interactions. The redundancy of proteins in the genomes, and the diversity of the proteins, suggests that they may have functionally distinct roles. Although the Hep_Hag domain of the YadA protein of Y. enterocolitica has been shown to bind collagen, it is not known if any of the B. mallei proteins are able to bind this host molecule. Further work is needed to characterize the functions and specificities of these proteins, and the role they play in the pathogenesis of B. mallei.
Orthologues of all the B. mallei BuHA proteins were identified in the B. pseudomallei K96243 genome, together with three further CDSs (BPSL1705, BPSS0088 and BPSS1434). These additional CDSs were in regions of the B. mallei ATCC23344 genome that appear to have been deleted in comparison to the B. pseudomalllei K96243 genome [21]. However, this family of proteins was not strongly immunostimulatory during human melioidosis, despite evidence for expression in vivo as indicated by a small number of positive clones in the library. Possible explanations for the difference in immunostimulatory capacity of B. mallei in horses and B. pseudomallei in humans includes variation in the nature of the immune response or other aspects of disease pathogenesis in the horse versus human host, and different rates of gene regulation and expression between the two bacterial species. It is also possible that protein folding and associated immunogenicity differs between the two bacterial species, although this seems less tenable given their degree of relatedness [33].
The implications of these findings for vaccine development and diagnostics are currently uncertain. It is unclear whether the BuHA proteins are immunogenic during human B. mallei infection. Given the relative lack of immune response to the BuHA proteins during human melioidosis, it seems unlikely that detection of antibodies to BuHA proteins will have diagnostic utility for this infection. Although there are apparent differences in the immunostimulatory profiles of the hemaglutinin family proteins of B. mallei and B. pseudomallei in their respective disease model, both pathogens share an expanded number of these proteins in comparison to the non-pathogenic B. thailandensis. Both B. pseudomallei and B. thailandensis can be isolated from the soil. We speculate on the basis of a reduced number of hemaglutinin family proteins othologues in B. thailandensis that they may have a role in modulating host-cell interactions in B. pseudomallei. Further studies will be required to investigate the in vivo expression of these proteins in the host, and the role they play in the pathology of melioidosis.
Methods
Bacterial strains
E. coli strains were grown in Luria-Bertani medium, using selection with kanamycin (50 μg/ml), where appropriate. B. pseudomallei and B. mallei were grown in TSB or TSB plus 4% glycerol, respectively.
Serum
Sera taken from two separate horses obtained 7 days after intra-tracheal inoculation of a suspension containing B. mallei ATCC 23344 together with a control serum from a horse pre-inoculation were kindly provided by Donald E Woods, Faculty of Medicine, University of Calgary. Sera from 21 patients with culture-confirmed melioidosis were collected on admission to Sappasithiprasong Hospital, Ubon Ratchathani, northeast Thailand between 1998 and 2003. Attributable days of symptoms prior to presentation ranged from 3 to 150 days (median 14 days, inter-quartile range (IQR) 7 to 28 days).
Screening of B. mallei and B. pseudomallei expression libraries with equine or human sera
Bacteriophage were propagated on E. coli XL1-Blue MRF' and plaque lifted onto Immobilon-NC nitrocellulose membranes (Millipore) pre-soaked in 10 mM IPTG, according to the manufacturer's instructions. After overnight blocking at 4°C in TBST (10 mM Tris-HCl [pH 7.4], 0.15 M NaCl, 0.05% [vol/vol] Tween 20) containing 6% (wt/vol) skimmed milk powder, hybridization was carried out with equine or human sera diluted 1:1,000 (in TBST-milk) for 90 min at room temperature. Blots were then incubated with alkaline phosphatase-conjugated anti-human or anti-equine IgG gamma chain specific monoclonal antibodies (Sigma) diluted 1:50,000 (in TBST) for 30 min at room temperature. Bound antibodies were detected using nitroblue tetrazolium (NBT)-BCIP (5-Bromo-4-chloro-3-indolylphosphate) solution (Roche). After a second purification round of screening, phagemids containing cloned B. mallei or B. pseudomallei DNA were excised into E. coli XLOLR according to the manufacturer's instructions.
Excised phagemid DNA was purified using QIAprep spin miniprep columns (QIAGEN) and the B. pseudomallei or B. mallei insert DNA sequenced using a DYEnamic ET Dye Terminator Kit (Amersham) and MegaBACE 500 sequencer. BLASTN searches using sequences from either end of the insert were carried out against the B. mallei ATCC 23344 genome [17,18], or the B. pseudomallei K96243 genome [21,34]. Mapping of positive clones were performed using Artemis software [35]. Domain organization of proteins was defined and compared using the Pfam [15] website [36].
PCR detection of genes encoding BuHA proteins in B. mallei and B. pseudomallei
Two bacterial strain collections were examined; 21 B. mallei isolates have been described previously [37], and 100 B. pseudomallei isolates were from patients with melioidosis presenting to Sappasithiprasong Hospital during 2001 (n = 50) or recovered from the environment in northeast Thailand (n = 50). A single colony picked from solid agar was inoculated into TSB and incubated overnight in air at 37°C, after which genomic DNA was extracted using the Wizard Genomic DNA purification kit (Promega).
The presence or absence of four BuHA proteins in B. mallei and their homologs in B. pseudomallei was defined by PCR. The primers used were as follows: BMA0840-f (5'-AGCGGCGCGGGGTCCTATTC), BMA0840-r (5'-CCGCCGCCACGTTGATGAG); BMA1027-f (5'-AGCGCGGGCAGCATTTACTTCC), BMA1027-r (5'-GATTCGCGTTGACCGTGCTGAGG); BMAA0649-f (5'-CGGTGAACGGCTCGCAGATGAATG), BMAA0649-r (5'-GGCCCCGCGAACCGAACGACAC); and BMAA1324-f (5'-GCCGCGGCGCAGGTCAG), BMAA1324-r (5'-CCGTCGCCCGCCGCTTCC).
The final concentrations of the PCR mixtures were 1X reaction buffer, 1.5 mM MgCl2, 0.35 μM of each primer, 5 μl of 1:20 dilution template DNA, 200 μM dNTPs and 2.5 unit Taq polymerase (Promega). Samples were held at 95°C for 2 min and then subjected to 40 cycles of 95°C for 30 s, 68°C (BMA0840 & BMA1027) or 70°C (BMAA0649 & BMAA1324) for 30 s, and 72°C for 60 s, followed by a final extension step of 72°C for 5 min. Reactions for BMA 0840 and BMA 1027 were multiplexed. PCR amplifications were performed using a PTC-0200 DNA engine (MJ Research, Cambridge, MA), and aliquots of reaction mixtures were analyzed by 2% agarose gel electrophoresis.
In silico identification of orthologs of BuHA proteins
Orthologs of BuHA proteins in the genomes of B. pseudomallei K96243 (accession numbers BX571965 and BX571966) [21], B. mallei ATCC 23344 (accession numbers CP000010 and CP000011) [17] and B. thailandensis E264 (accession numbers CP000086 and CP000085) [20] was carried out by reciprocal FASTA [38] analysis as previously described [39]. Coding sequences containing Hep_Hag (PF05658) and YadA (PF03895) domains were identified using HMMER [40] and the respective Pfam profile HMMs downloaded from the Pfam website [36]. The identification of alleles of BuHA proteins in 6 B. mallei isolates (10399, FMN, JHU, GB8, 10229 and NCTC10247) currently undergoing whole genome sequencing by The Institute of Genome Research (TIGR) [18] was carried out using BLASTN [41]. Additional Burkholderiaceae genomes sequences used in this study were from: Burkholderia sp. 383 (accession numbers CP000150, CP000151 and CP000152), Burkholderia xenovorans LB400 (accession numbers CP000270, CP000271 and CP000272) and Ralstonia solanacearum GMI1000 (accession numbers AL646052 and AL646053) [42].
Phylogenetic analysis
An unrooted maximum likelihood tree built using Phylip (Version 3.6) [43] was drawn using NJplot [44]. Sequences were aligned using ClustalX (Version 1.82) [45].
Authors' contributions
RT carried out the in vivo gene expression component, MTGH carried out the bioinformatics component and drafted the manuscript, ST carried out the in vivo gene expression component, SR contributed critical review, SRC contributed to the experimental concept and design and provided technical support, SJF contributed to the experimental concept and design and provided technical support, WCN assisted with the bioinformatics component and assisted with drafting the manuscript, NPJD contributed towards study design and undertook critical review of the manuscript, and SJP conceived of the study, participated in the study design and coordination, and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Loci identified on immunoscreening of the B. mallei library.
Acknowledgments
Acknowledgements
We are grateful for the support of staff at the Sappasithiprasong Hospital, in particular to Professor Wipada Chaowagul, and to the staff of the Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand. SJP is supported by a Wellcome Trust Career Development Award in Clinical Tropical Medicine (Grant ref.066735). This study was funded by the Wellcome Trust.
Contributor Information
Rachaneeporn Tiyawisutsri, Email: rachaneeporn_t@hotmail.com.
Matthew TG Holden, Email: mh3@sanger.ac.uk.
Sarinna Tumapa, Email: jay@tropmedres.ac.
Sirirat Rengpipat, Email: rsirirat05@yahoo.com.
Simon R Clarke, Email: s.clarke@sheffield.ac.uk.
Simon J Foster, Email: s.foster@sheffield.ac.uk.
William C Nierman, Email: wnierman@tigr.org.
Nicholas PJ Day, Email: nickd@tropmedres.ac.
Sharon J Peacock, Email: sharon@tropmedres.ac.
References
- DeShazer D, Waag D. Glanders: new insights into an old disease. In: Lindler L, Lebeda F, Korch GW, editor. Biological weapons defense: infectious diseases and counterbioterrorism. Totowa, N.J. , The Humana Press, Inc.; 2004. pp. 209–237. [Google Scholar]
- Amemiya K, Bush GV, DeShazer D, Waag DM. Nonviable Burkholderia mallei induces a mixed Th1- and Th2-like cytokine response in BALB/c mice. Infect Immun. 2002;70:2319–2325. doi: 10.1128/IAI.70.5.2319-2325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya K, Meyers JL, Trevino SR, Chanh TC, Norris SL, Waag DM. Interleukin-12 induces a Th1-like response to Burkholderia mallei and limited protection in BALB/c mice. Vaccine. 2006;24:1413–1420. doi: 10.1016/j.vaccine.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Ulrich RL, Amemiya K, Waag DM, Roy CJ, DeShazer D. Aerogenic vaccination with a Burkholderia mallei auxotroph protects against aerosol-initiated glanders in mice. Vaccine. 2005;23:1986–1992. doi: 10.1016/j.vaccine.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Waag DM, McCluskie MJ, Zhang N, Krieg AM. A CpG oligonucleotide can protect mice from a low aerosol challenge dose of Burkholderia mallei. Infect Immun. 2006;74:1944–1948. doi: 10.1128/IAI.74.3.1944-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino SR, Permenter AR, England MJ, Parthasarathy N, Gibbs PH, Waag DM, Chanh TC. Monoclonal antibodies passively protect BALB/c mice against Burkholderia mallei aerosol challenge. Infect Immun. 2006;74:1958–1961. doi: 10.1128/IAI.74.3.1958-1961.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SM, Ellis JF, Russell P, Griffin KF, Oyston PC. Passive protection against Burkholderia pseudomallei infection in mice by monoclonal antibodies against capsular polysaccharide, lipopolysaccharide or proteins. J Med Microbiol. 2002;51:1055–1062. doi: 10.1099/0022-1317-51-12-1055. [DOI] [PubMed] [Google Scholar]
- Scholz HC, Joeseph M, Tomaso H, Al Dabouk S, Witte A, Kinne J, Hagen RM, Wernery R, Wernery U, Neubauer H. Detection of the reemerging agent Burkholderia mallei in a recent outbreak of glanders in the United Arab Emirates by a newly developed fliP-based polymerase chain reaction assay. Diagn Microbiol Infect Dis. 2006;54:241–247. doi: 10.1016/j.diagmicrobio.2005.09.018. [DOI] [PubMed] [Google Scholar]
- U'Ren JM, Van Ert MN, Schupp JM, Easterday WR, Simonson TS, Okinaka RT, Pearson T, Keim P. Use of a real-time PCR TaqMan assay for rapid identification and differentiation of Burkholderia pseudomallei and Burkholderia mallei. Journal of Clinical Microbiology. 2005;43:5771–5774. doi: 10.1128/JCM.43.11.5771-5774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich MP, Norwood DL, Christensen DR, Ulrich RL. Using real-time PCR to specifically detect Burkholderia mallei. J Med Microbiol. 2006;55:551–559. doi: 10.1099/jmm.0.46350-0. [DOI] [PubMed] [Google Scholar]
- Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, Nguyen TT, Kalantari-Dehaghi M, Crotty S, Baldi P, Villarreal LP, Felgner PL. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy N, Deshazer D, England M, Waag DM. Polysaccharide microarray technology for the detection of Burkholderia pseudomallei and Burkholderia mallei antibodies. Diagn Microbiol Infect Dis. 2006 doi: 10.1016/j.diagmicrobio.2006.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SR, Brummell KJ, Horsburgh MJ, McDowell PW, Mohamad SA, Stapleton MR, Acevedo J, Read RC, Day NP, Peacock SJ, Mond JJ, Kokai-Kun JF, Foster SJ. Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J Infect Dis. 2006;193:1098–1108. doi: 10.1086/501471. [DOI] [PubMed] [Google Scholar]
- Lopez J, Copps J, Wilhelmsen C, Moore R, Kubay J, St-Jacques M, Halayko S, Kranendonk C, Toback S, DeShazer D, Fritz DL, Tom M, Woods DE. Characterization of experimental equine glanders. Microbes Infect. 2003;5:1125–1131. doi: 10.1016/j.micinf.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. The Pfam Protein Families Database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter SE, Surana NK, St Geme JW., 3rd Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 2005;13:199–205. doi: 10.1016/j.tim.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Nierman WC, DeShazer D, Kim H, Tettelin H, Nelson KE, Feldblyum TV, Ulrich RL, Kolonay JF, Brinkac LM, Daugherty SC, Deboy RT, Dodson RJ, Durkin AS, Gwinn ML, Madupu R, Nelson WC, Sullivan SA, Davidsen TD, Zafar N, Haft DH, Selengut J, Zhou LW, Yu Y, Shamblin C, Sarria S, Khouri H, Radune D, Mohammoud Y, Dimitrov G, Russell D, Fraser CM. Structural flexiblility in the Burkholderia mallei genome. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14246–14251. doi: 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Institute for Genomic Research (TIGR) http://www.tigr.org/
- Pathema Bioinformatics Resource Center http://pathema.tigr.org/
- Yu Y, Kim HS, Chua HH, Lin CH, Sim SH, Lin D, Derr A, Engels R, DeShazer D, Birren B, Nierman WC, Tan P. Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol. 2006;6:46. doi: 10.1186/1471-2180-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter SE, Surana NK, Grass S, St Geme JW., 3rd Trimeric autotransporters require trimerization of the passenger domain for stability and adhesive activity. J Bacteriol. 2006;188:5400–5407. doi: 10.1128/JB.00164-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenkamp SJ, St Geme JW., 3rd Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol Microbiol. 1996;19:1215–1223. doi: 10.1111/j.1365-2958.1996.tb02467.x. [DOI] [PubMed] [Google Scholar]
- Cole LE, Kawula TH, Toffer KL, Elkins C. The Haemophilus ducreyi serum resistance antigen DsrA confers attachment to human keratinocytes. Infect Immun. 2002;70:6158–6165. doi: 10.1128/IAI.70.11.6158-6165.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comanducci M, Bambini S, Brunelli B, Adu-Bobie J, Arico B, Capecchi B, Giuliani MM, Masignani V, Santini L, Savino S, Granoff DM, Caugant DA, Pizza M, Rappuoli R, Mora M. NadA, a novel vaccine candidate of Neisseria meningitidis. J Exp Med. 2002;195:1445–1454. doi: 10.1084/jem.20020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope LD, Lafontaine ER, Slaughter CA, Hasemann CA, Jr., Aebi C, Henderson FW, McCracken GH, Jr., Hansen EJ. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J Bacteriol. 1999;181:4026–4034. doi: 10.1128/jb.181.13.4026-4034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggenkamp A, Neuberger HR, Flugel A, Schmoll T, Heesemann J. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence and mouse virulence. Mol Microbiol. 1995;16:1207–1219. doi: 10.1111/j.1365-2958.1995.tb02343.x. [DOI] [PubMed] [Google Scholar]
- St Geme JW, 3rd, Cutter D, Barenkamp SJ. Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J Bacteriol. 1996;178:6281–6287. doi: 10.1128/jb.178.21.6281-6287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiczyk E, Roggenkamp A, Reichenbecher M, Lupas A, Heesemann J. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. Embo J. 2000;19:5989–5999. doi: 10.1093/emboj/19.22.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koretke KK, Szczesny P, Gruber M, Lupas AN. Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica. J Struct Biol. 2006;155:154–161. doi: 10.1016/j.jsb.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Nummelin H, Merckel MC, Leo JC, Lankinen H, Skurnik M, Goldman A. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. Embo J. 2004;23:701–711. doi: 10.1038/sj.emboj.7600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir YE, Kuusela P, Skurnik M. Functional mapping of the Yersinia enterocolitica adhesin YadA. Identification Of eight NSVAIG - S motifs in the amino-terminal half of the protein involved in collagen binding. Mol Microbiol. 2000;37:192–206. doi: 10.1046/j.1365-2958.2000.01992.x. [DOI] [PubMed] [Google Scholar]
- Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, Spratt BG. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol. 2003;41:2068–2079. doi: 10.1128/JCM.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Sanger Institute http://www.sanger.ac.uk/ [DOI] [PubMed]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- The Wellcome Trust Sanger Institute Pfam website http://www.sanger.ac.uk/Software/Pfam/ [DOI] [PubMed]
- Chantratita N, Vesaratchavest M, Wuthiekanun V, Tiyawisutsri R, Ulziitogtokh T, Akcay E, Day NP, Peacock SJ. Pulsed-field gel electrophoresis as a discriminatory typing technique for the biothreat agent Burkholderia mallei. Am J Trop Med Hyg. 2006;74:345–347. [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved Tools for Biological Sequence Comparison. Proc Natl Acad Sci U S A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell KS, Sebaihia M, Pritchard L, Holden MTG, Hyman LJ, Holeva MC, Thomson NR, Bentley SD, Churcher LJC, Mungall K, Atkin R, Bason N, Brooks K, Chillingworth T, Clark K, Doggett J, Fraser A, Hance Z, Hauser H, Jagels K, Moule S, Norbertczak H, Ormond D, Price C, Quail MA, Sanders M, Walker D, Whitehead S, Salmond GPC, Birch PRJ, Parkhill J, Toth IK. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp atroseptica and characterization of virulence factors. Proc Natl Acad Sci U S A. 2004;101:11105–11110. doi: 10.1073/pnas.0402424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlat M, Billault A, Brottier P, Camus JC, Cattolico L, Chandler M, Choisne N, Claudel-Renard C, Cunnac S, Demange N, Gaspin C, Lavie M, Moisan A, Robert C, Saurin W, Schiex T, Siguier P, Thebault P, Whalen M, Wincker P, Levy M, Weissenbach J, Boucher CA. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, Rutherford K, Berriman M, Rajandream MA, Barrell B, Parkhill J. ACT: the Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Loci identified on immunoscreening of the B. mallei library.


