Abstract
There is widespread interest in defining factors and mechanisms that stimulate proliferation of pancreatic islet cells. Wnt signaling is an important regulator of organ growth and cell fates, and genes encoding Wnt-signaling factors are expressed in the pancreas. However, it is unclear whether Wnt signaling regulates pancreatic islet proliferation and differentiation. Here we provide evidence that Wnt signaling stimulates islet β cell proliferation. The addition of purified Wnt3a protein to cultured β cells or islets promoted expression of Pitx2, a direct target of Wnt signaling, and Cyclin D2, an essential regulator of β cell cycle progression, and led to increased β cell proliferation in vitro. Conditional pancreatic β cell expression of activated β-catenin, a crucial Wnt signal transduction protein, produced similar phenotypes in vivo, leading to β cell expansion, increased insulin production and serum levels, and enhanced glucose handling. Conditional β cell expression of Axin, a potent negative regulator of Wnt signaling, led to reduced Pitx2 and Cyclin D2 expression by β cells, resulting in reduced neonatal β cell expansion and mass and impaired glucose tolerance. Thus, Wnt signaling is both necessary and sufficient for islet β cell proliferation, and our study provides previously unrecognized evidence of a mechanism governing endocrine pancreas growth and function.
Keywords: Cyclin D2, diabetes mellitus, islets of Langerhans, pancreas, Pitx2
Relative or absolute deficiency of pancreatic β cell mass underlies the pathogenesis of type 1 and type 2 diabetes (1, 2). Thus, there is considerable interest in understanding the mechanisms that stimulate pancreatic islet cell growth and differentiation. Recent studies identified several cell-autonomous factors, including Cyclin D2, c-Myc, cyclin-dependent kinase 4 (CDK4), menin, and other nuclear proteins that are essential for regulating growth of pancreatic islet β cells, the sole source of insulin in postnatal animals (3, 4). However, little is known about the signaling factors that control expression or activity of these β cell-autonomous growth regulators.
Wnt proteins are a family of highly conserved secreted proteins that regulate multiple developmental processes, including proliferation of organ-specific stem/progenitor cell populations, growth control and cell fate determination in diverse organs, and tissue patterning (5). In the canonical Wnt signaling pathway, binding of Wnt ligand to cognate membrane-spanning receptor proteins called Frizzleds (Fz), triggers a signaling cascade that results in the stabilization and nuclear localization of β-catenin, whose interactions with T cell-specific factor/lymphoid enhancer-binding factor (TCF/LEF) transcription factors control transcription of target genes like Pitx2 (6, 7). In the absence of Wnt signaling, cytoplasmic β-catenin in Wnt-responsive cells is targeted for proteosome-mediated degradation by a heteromeric protein complex that includes Axin, glycogen synthase kinase 3β (GSK-3β), and other proteins (8–10).
Recent studies document expression of several Wnt ligands and Fz receptors in the embryonic and postnatal pancreas. In one study, conditional disruption of pancreatic β-catenin expression produced pancreatitis and reduced islet mass, but the mechanism for impaired islet cell proliferation was not identified (11). In other studies, Wnt signaling disruption led to clear changes in exocrine pancreas growth without detectable changes in endocrine cell proliferation (12–14). Thus, it remains unclear whether Wnt signaling controls islet cell growth. Here we use gain- and loss-of-function approaches to show that Wnt signaling controls islet β cell proliferation and to unveil a mechanism underlying this regulation.
Results
Wnt Signaling Stimulates Expression of Essential β Cell Cycle Regulators.
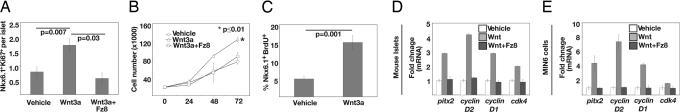
To test whether Wnt signaling controls islet β cell growth, we exposed isolated pancreatic islets to purified Wnt3a, a Wnt family member that is expressed in the developing pancreas and in adult human islets (15). In prior studies we showed that purified Wnt3a led to β-catenin stabilization and induction of established Wnt target genes (16). To quantify β cell proliferation, we measured expression of Ki67, a marker of late G1, S, G2, and M phases previously used to assess β cell cycle progression. In islets treated with Wnt3a, we found a 2-fold increase of Ki67+ β cells (Fig. 1A), which characteristically coexpressed insulin and the transcription factor Nkx6.1, markers not expressed by other mature islet cells [supporting information (SI) Fig. 6; ref. 17]. The addition of soluble Fz 8-cysteine-rich domain (Fz8-CRD), a potent specific Wnt inhibitor (D.t.B. and R.N., unpublished results) eliminated this stimulatory effect of Wnt3a, indicating that active Wnt signaling was assessed (Fig. 1A). Consistent with these data, we found that growth of MIN6 cells, a transformed mouse β cell line responsive to normal growth cues (18, 19), was significantly increased by purified Wnt3a, compared with treatment with vehicle alone (Fig. 1B). To test whether Wnt3a promoted MIN6 cell proliferation, we measured incorporation of BrdU, which proved more consistent than Ki67 as a proliferation marker in our MIN6 cultures. Compared with vehicle-treated controls, Wnt3a-treated cells had a 3-fold increase in BrdU incorporation (Fig. 1C). Collectively, these results show that Wnt3a exposure in vitro was sufficient to increase β cells proliferation.
Fig. 1.
Purified Wnt3a induces expression of Pitx2, cyclin D2, and other cell cycle regulators in β cells and promotes cell expansion. (A) Expression of Ki67 in Nkx6.1+ cells of purified WT P8 islets stimulated with Wnt3a, vehicle, or Wnt3a + Fz8-CRD. (B) Growth of MIN6 cells stimulated with vehicle only, Wnt3a, or Wnt3a + Fz8-CRD and counted at 24, 48, and 72 h. Each condition was performed in triplicate. (C) MIN6 cells stimulated with Wnt3a or vehicle for 8 h were assayed for BrdU incorporation by immunofluorescence analysis. (A–C) Data are presented as the average ± SEM. All RT-PCR results were normalized to β-actin and are the average value from triplicate experiments. P values are indicated in each graph. (D) Real-time RT-PCR analysis of Pitx2, cyclin D2, cyclin D1, and cdk4 cDNA from purified P8 islets treated with vehicle only, purified Wnt3a, or Wnt3a + Fz8-CRD for 24 h. (E) Real-time RT-PCR analysis of Pitx2, cyclin D2, cyclin D1, and cdk4 cDNA from MIN6 cells treated with vehicle only, purified Wnt3a, or Wnt3a + Fz8-CRD for 24 h.
Several cell cycle regulators, including Cyclin D1, Cyclin D2, CDK4, and c-myc, have been shown to control β cell proliferation in vivo (20–23). To identify the mechanism of Wnt-stimulated β cell proliferation, we tested whether expression of these established β cell cycle regulators was controlled by Wnt signaling. Cyclin D2 expression in islet β cells peaks between postnatal day (P) 4 and P8, corresponding with a period of increased β cell proliferation and expansion (21), suggesting competence for endogenous signals that regulate proliferation. Thus, we tested whether expression of cyclin D2 and other cell cycle regulators was stimulated by Wnt3a protein in islets from mice at P8. Wnt3a treatment of islets for 24 h led to significantly increased mRNA levels of cyclin D1, cyclin D2, and CDK4 compared with controls, a response that was completely abolished by Fz8-CRD (Fig. 1D). To test whether Wnt3a affected β cells, we then exposed MIN6 cells to Wnt3a. Similar to our finding in islets, mRNA levels of cyclin D1, cyclin D2, and CDK4 were clearly increased by Wnt3a exposure (Fig. 1E). The addition of soluble Fz8-CRD eliminated the stimulatory effects of Wnt3a on each of these targets in MIN6 cells (Fig. 1E). Thus, Wnt3a was sufficient to stimulate expression of cell cycle regulators in purified islets and cultured β cells, which was matched by increased β cell proliferation. To test whether Wnt signaling regulation of cell cycle regulators was conserved, we assessed the effects of Wnt3a exposure on cultured human islets. Real-time RT-PCR studies revealed that Wnt3a increased mRNA levels of cyclinD2 and Pitx2, a direct transcriptional regulator of cyclinD2 (see below): These effects were blocked by Fz8-CRD (SI Fig. 7). Additional studies are needed to assess whether Wnt signaling is sufficient to stimulate expansion of cultured human islet cells.
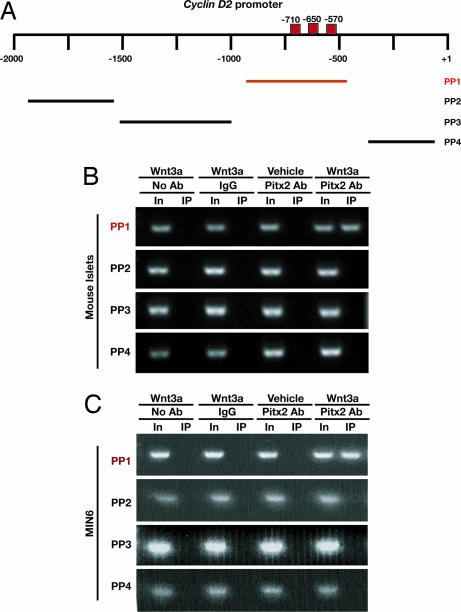
Wnt3a Promotes Direct Association of Pitx2 with Regions of the cyclin D2 Gene in β Cells.
In neuroendocrine cells, Wnt signaling stimulates expression of the transcription factor Pitx2, which in turn can activate transcription of cyclin D2 and c-myc (7). We found that exposure of islets or MIN6 cells to purified Wnt3a led to a 3- to 4-fold increase in Pitx2 mRNA levels, an effect abolished by Fz8-CRD (Fig. 1 D and E and SI Fig. 7). To determine whether Wnt signaling promotes the direct association of Pitx2 with target genes, we performed ChIP studies in MIN6 cells and postnatal pancreatic islets. Before Wnt3a exposure, Pitx2 was not detectably associated with genomic DNA −980 to −525 bp immediately 5′ of the cyclin D2 transcriptional start site, a region that contains consensus bicoid-like binding sites (Fig. 2A, red boxes) known to bind Pitx2 (7). After exposure of islets (Fig. 2B) and MIN6 cells (Fig. 2C) to purified Wnt3a protein, ChIP analysis revealed Pitx2 association with elements within −980 bp of the transcription start site of cyclin D2 (Fig. 2 A and B). By contrast, we did not detect Pitx2 association with regions in the cyclinD2 locus distal to −980 bp that lack these consensus sites (Fig. 2B) or in regions proximal to −525 bp. Although Wnt3a stimulation results in increased transcription of cyclin D1 and Cdk4 in MIN6 cells and purified islets (Figs. 1 D and E), we did not observe direct association of Pitx2 with these genes by ChIP analysis (data not shown). Thus, Wnt3a stimulated Pitx2 association with cyclin D2 in islets. Collectively, our results provide previously unrecognized evidence that Wnt signaling is sufficient to induce Pitx2 binding to elements of cyclin D2 in β-cells, promoting increased Cyclin D2 expression and proliferation.
Fig. 2.
ChIP demonstrates that Pitx2 associates with specific elements in the cyclin D2 promoter upon Wnt3a stimulation. (A) Position of DNA regions flanked by primer pairs (PP) used for PCR to assess Pitx2 association with elements within the cyclin D2 promoter. Consensus bicoid binding sequences are marked by red boxes. (B) Anti-Pitx2 ChIP performed on 8-h Wnt3a-stimulated islet cultures derived from WT P8 mice reveals Pitx2 binding to the cyclin D2 promoter at sites −980 to −525 upstream of the transcriptional start site. (C) Anti-Pitx2 ChIP performed on MIN6 cells treated with Wnt3a for 3 h and subsequently analyzed with PCR shows a similar binding pattern to the cyclin D2 promoter.
Expression of Activated β-Catenin Is Sufficient to Stimulate Islet β Cell Expansion.
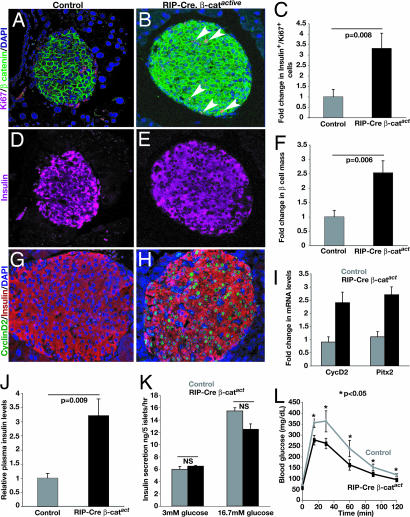
To test whether Wnt signaling activation is sufficient to promote β cell proliferation in vivo, we intercrossed mice to create strains expressing Cre-recombinase in β cells from the rat insulin promoter (RIP-Cre) (24, 25) and harboring the β-catactive allele, which encodes a loxP-flanked constitutively active form of β-catenin (see Methods; refs. 14 and 26). Previously, we used mice expressing Cre-recombinase under the control of the Pdx1 promoter (24) to induce expression of β-catactive in adult islets, but expression of Cre-recombinase was mosaic in islet β cells and did not result in significant changes in β cell mass (14). In 3-month-old bitransgenic RIP-Cre, β-catactive mice, immunohistology revealed increased levels of β-catenin in the cytoplasm of β cells, with some cells also exhibiting nuclear localization of β-catenin (Fig. 3A and B and SI Fig. 8). By contrast, β-catenin in β cells from control mice appeared to be localized exclusively to the plasma membrane (SI Fig. 8). Expression of Ki67 by β cells in adult RIP-Cre, β-catactive mice was increased 3-fold, corresponding well with a 2.5-fold increase in β cell mass (Fig. 3 C–F). Consistent with our in vitro islet studies, the in vivo expression of Cyclin D2 was increased in β cells of RIP-Cre, β-catactive mice (Fig. 3 G and H). Real-time RT-PCR measures revealed a 2-fold increase of cyclinD2 and Pitx2 mRNA levels in RIP-Cre, β-catactive islets compared with controls (Fig. 3I). In 3-month-old RIP-Cre, β-catactive mice, cyclinD2+ β cells maintained expression of insulin (Fig. 3 G and H). Moreover, expression of adult β cell markers, including Pdx1, Glut2, and Nkx6.1 were also maintained in the islet cells of these mice (data not shown). Together, these findings suggest that activation of β-catenin was sufficient to stimulate expansion of β cells that maintained their differentiated fate. Consistent with our finding of increased β cell mass in RIP-Cre, β-catactive mice, serum insulin levels in these mice were increased ≈3-fold (Fig. 3J). However, insulin content per islet cell was not detectably changed in RIP-Cre, β-catactive mice (SI Fig. 8). Moreover, compared with islets from control mice, insulin secretion by islets from RIP-Cre, β-catactive mice after stimulation with glucose (Fig. 3K) or other secretogogues such as arginine (SI Fig. 8) was indistinguishable. Consistent with their hyperinsulinemia, the blood glucose concentration in fasted RIP-Cre, β-catactive mice was significantly reduced, and i.p. glucose challenge revealed that glucose disposal was improved in RIP-Cre, β-catactive mice compared with controls (Fig. 3L). Collectively, these results support the conclusion that hyperinsulinemia in RIP-Cre, β-catactive mice principally reflects increased β cell mass. Thus, induction of β-catenin in vivo was sufficient to promote expansion of functional pancreatic β cells.
Fig. 3.
Conditional expression of activated β-catenin in RIP-Cre, β-catactive islets. (A and B) Elevated β-catenin expression and an increased number of proliferating Ki67+ cells in RIP-Cre, β-catactive islets compared with controls. (C) Quantification of insulin+, Ki67+ cells in control and RIP-Cre, β-catactive islets. (D and E) Immunostaining reveals insulin expression is maintained in proliferating Ki67+ cells in RIP-Cre, β-catactive islets. (F) Quantification of pancreatic β cell mass in control and RIP-Cre, β-catactive mice. (G and H) Immunostaining reveals increased numbers of cyclin D2-expressing insulin+ cells in RIP-Cre, β-catactive islets. (I) Real-time PCR quantification of cyclin D2 mRNA levels in isolated control and RIP-Cre, β-catactive islets. All data are statistically significant. P < 0.005. (J) Serum insulin levels in fed control and RIP-Cre, β-catactive mice. (K) Assessment of insulin secretion in response to increased glucose as performed on cultured islets. P values were not significant (NS). (L) i.p. glucose tolerance tests of control and RIP-Cre, β-catactive mice after overnight fasting. (J–L) Values represent mean ± SEM. P values are indicated where appropriate.
Conditional Axin Expression Impairs Pitx2 Expression and β Cell Expansion.
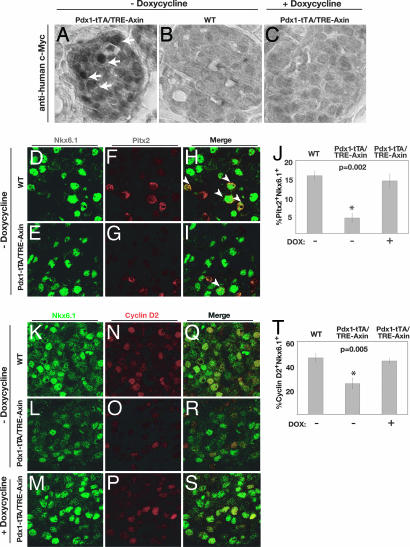
To address whether Wnt signaling was required for normal β cell expansion in vivo, we generated transgenic mice that permitted conditional pancreatic expression of Axin, a potent inhibitor of Wnt signaling. Intercrosses produced mice that harbor a transgene encoding Axin adjacent to a tetracycline response element (TRE) promoter (TRE-Axin) (27) and the Pdx1-tetracycline-regulated transactivator (tTA) transgene (28, 29) in which the tTA replaces the coding region of Pdx1, a gene expressed in pancreatic progenitor cells and mature β cells (30–32). Bitransgenic Pdx1-tTA/TRE-Axin progeny and mice of other genotypes from this intercross were obtained at Mendelian frequency from >20 litters. Transgenic Axin expression in Pdx1-tTA/TRE-Axin islets was clearly detectable by immunohistochemistry (Fig. 4) and by Western blotting (SI Fig. 9). Axin was detected in the islet core, where Pdx1-expressing β cells reside, in pancreatic islets of bitransgenic mice, but not littermate controls on P4 (Fig. 4 A and B), a neonatal stage when β cells are actively proliferating (21). As expected, continuous exposure of Pdx1-tTA/TRE-Axin mice to doxycycline (Dox), a tetracycline analogue, from the time of conception prevented expression of TRE-Axin in Pdx1+ cells (Fig. 4C; see Methods). Thus, in Pdx1-tTA/TRE-Axin mice, TRE-Axin was expressed in Pdx1+ cells and conditionally repressed by Dox exposure.
Fig. 4.
Dox-dependent expression of Axin, Pitx2, and Cyclin D2 in Pdx1-tTA/TRE-Axin islets. (A–C) Immunohistochemical detection of Myc-tagged Axin expression in islets. Pdx1-tTA/TRE-Axin P4 mice (A) express Myc-tagged Axin (arrows), which is absent in WT (B) and Pdx1-tTA/TRE-Axin (+ Doxycycline) (C) control mice at P4. (Original magnification: ×63.) (D–I) Immunofluorescent detection of Nkx6.1 (green), Pitx2 (red), and merge (yellow) in the islets of P4 WT and Pdx1-tTA/TRE-Axin mice and Pdx1-tTA/TRE-Axin mice on Dox from the time of conception. (H and I) White arrowheads indicate Nkx6.1+ /Pitx2+ nuclei. (J) Percentage of Nkx6.1+ that are Pitx2+ in Pdx1-tTA/TRE-Axin and control mice. (K–S) Immunofluorescent detection of Nkx6.1 (green) and Cyclin D2 (red) in the islets of P4 WT and Pdx1-tTA/TRE-Axin mice and Pdx1-tTA/TRE-Axin mice on Dox. (T) Percentage of Nkx6.1+ that are Cyclin D2+ in Pdx1-tTA/TRE-Axin and control mice. Data are presented as the average ± SEM. P values are indicated in the figures. Data are from at least five mice per genotype. (Original magnification: ×100.)
To assess Axin effects on Wnt signaling in Pdx1-tTA/TRE-Axin mice at P4, we analyzed the expression of Pitx2, a direct Wnt signaling target (7). Immunohistology showed that Pitx2 was localized to β cell nuclei (Fig. 4 D–I). Compared with WT controls or to Pdx1-tTA/TRE-Axin mice administered Dox, the percentage of Pitx2+ β cells per pancreas was reduced by 75% in Pdx1-tTA/TRE-Axin mice (Fig. 4 D–J). Thus, conditional Axin induction reduced Pitx2 expression, providing evidence of Wnt-signaling disruption in the endocrine pancreas of Pdx1-tTA/TRE-Axin mice.
Prior studies showed that Cyclin D2-deficient mice have a 70% reduction in β cell mass and glucose intolerance without changes in total pancreatic mass (21), phenotypes that are reminiscent of the changes we noted in Pdx1-tTA/TRE-Axin mice. On the basis of these findings and our in vitro studies, we postulated that cyclin D2 expression by nascent β cells in Pdx1-tTA/TRE-Axin mice might be impaired. Immunohistology showed that Cyclin D2 protein colocalized with Nkx6.1+ cells in WT and Pdx1-tTA/TRE-Axin mice, consistent with our studies of MIN6 cells and cultured islets (Fig. 4 K–S). However, analysis of Pdx1-tTA/TRE-Axin mice revealed a 60% reduction of Cyclin D2 expression in Nkx6.1+ cells, an effect reversed by Dox (Fig. 4 K–T). These changes correlated with a significant reduction of BrdU incorporation by Nkx6.1+ cells (SI Fig. 9). In contrast to these proliferation phenotypes, we did not detect changes in pancreatic apoptosis by using terminal deoxynucleotidyltransferase (TdT)-mediated 2′-deoxyuridine 5′-triphosphate (dUTP) nick-end labeling (data not shown). Collectively, these studies suggest that Wnt signaling regulates in vivo expression of Cyclin D2, an essential regulator of β cell proliferation.
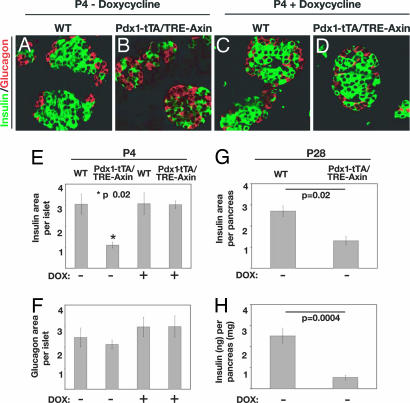
To identify the effects of Axin expression on pancreas islets, we analyzed pancreata from Pdx1-tTA/TRE-Axin and control mice. The morphology of the pancreas from Pdx1-tTA/TRE-Axin mice appeared grossly normal, and changes in acinar or ductal cell morphology and number were not detected (data not shown). However, immunohistology and morphometry revealed several defects in postnatal pancreatic islet composition and morphology in Pdx1-tTA/TRE-Axin pancreata (Fig. 5). Compared with islets from WT mice or bitransgenic mice exposed to Dox, islets from bitransgenic Pdx1-tTA/TRE-Axin mice had a significant reduction of β cells (Fig. 5 B–E) and lacked the characteristic architecture of insulin+ cells surrounded by glucagon+ cells. The number of non-β islet cells, including glucagon+, somatostatin+, and PP+ cells and the total number of islets, was not significantly altered in bitransgenic mice (Fig. 5 B, E, and F and data not shown). At weaning, Pdx1-tTA/TRE-Axin mice had persistent β cell hypoplasia, with a 50–60% reduction of β cells compared with littermate controls (Fig. 5G). Compared with controls, including Pdx1-tTA mice, total pancreatic insulin content in Pdx1-tTA/TRE-Axin mice was reduced (Fig. 5H). Although weight gain, blood glucose, and serum insulin levels during random feeding appeared normal in Pdx1-tTA/TRE-Axin mice (data not shown), i.p. glucose challenge revealed impaired glucose tolerance that was more severe than in Pdx1-tTA mice, which have mild glucose intolerance (SI Fig. 10; ref. 28) or in other littermate controls. Thus, glucose tolerance was significantly worsened by Axin expression in Pdx1-tTA/TRE-Axin mice.
Fig. 5.
Conditional Axin expression in Pdx1+ progenitors impairs β cell development. (A–D) Immunofluorescent detection of insulin+ (green) and glucagon+ (red) cells in WT (A) and Pdx1-tTA/TRE-Axin (B) islets (no Dox) and in WT (C) and Pdx1-tTA/TRE-Axin (D) islets administered Dox from the time of conception. (Original magnification: ×100.) (E and F) The relative insulin+ (E) and glucagon (F) area per islet compared in WT and Pdx1-tTA/TRE-Axin mice and in WT and Pdx1-tTA/TRE-Axin mice administered Dox from the time of conception. (E) The P value for insulin+ area between Pdx1-tTA/TRE-Axin islets and control islets is ≤0.02. (F) The P value for glucagon+ area among all genotypes is not significant. (G) Morphometric analysis of insulin+ area as a percentage of total pancreas area from P28 WT and Pdx1-tTA/TRE-Axin mice. (H) Total insulin (nanograms) per pancreas (milligrams) in P28 Pdx1-tTA/TRE-Axin and WT mice. All data are from at least three litters, with four to eight mice per genotype. Data are presented as the average ± SEM.
Discussion
These studies reveal that pancreatic Wnt signaling controls islet β cell proliferation. Prior studies provided evidence that pancreatic growth and differentiation are regulated by Wnt signaling (11–15, 33, 34). However, these reports did not present a mechanism for the Wnt-mediated action nor did they test whether Wnt signaling was sufficient to stimulate β cell proliferation in pancreatic islets. Here, treatment of β cells with purified Wnt protein or stimulation with activated β-catenin permitted us to test whether Wnt pathway activation might promote islet β cell proliferation and allowed elucidation of mechanisms underlying Wnt-mediated β cell growth. Here we show that Wnt signaling stimulated expression of multiple β cell cycle regulators, including Cyclins D1 and D2, resulting in enhanced islet proliferation. Previous studies showed that these D-type cyclins, CDK4, c-myc, and other cell-autonomous factors control β cell proliferation in pancreas development (21–23, 35) but did not reveal how expression of these factors was controlled. Our studies reveal that Wnt signals promote expression of Pitx2, a transcriptional activator that directly associates with cis-regulatory elements in the cyclin D2 gene. On the basis of these results, we hypothesize that Wnt-signaling induction of Pitx2 promotes cyclin D2 expression to drive proliferation of nascent β cells. Pitx2 has also been shown to regulate expression of c-Myc and cyclin D1 (6, 36), but in isolated islets we did not detect direct association of Pitx2 with promoter-proximal sequences in these genes. Thus, the basis of enhanced expression of c-Myc, cyclin D1, and CDK4 in Wnt-treated β cells and the contribution of these factors to Wnt-stimulated β cell proliferation requires further investigation. After Wnt stimulation, β cell proliferation was not accompanied by a loss of characteristic β cell markers, such as insulin or Nkx6.1 (37). On the contrary, our studies of RIP-Cre, β-catactive mice show that Wnt signaling had a clear physiologic impact by enhancing serum insulin levels and glucose disposal. Thus, exposure to Wnt signals in some contexts can promote islet growth and hallmark β cell features, although prolonged activation of Wnt signaling may affect the differentiation state of islet β cells (14). Exposure to purified mouse Wnt3a protein can stimulate expression of Pitx2 and cyclinD2 in cultured human cadaveric islets, but we have not yet found that Wnt signaling is sufficient to induce β cell expansion in human islets. Thus, further studies are needed to test the possibility that Wnt signaling may be useful for expanding functional islets for therapeutic goals.
We also showed that conditional Axin expression impaired proliferation of neonatal β cells, demonstrating a requirement for Wnt signaling during β cell expansion in vivo. Consistent with our findings, a prior study (11) reported comparable β cell hypoplasia after conditional inactivation of β-catenin, but no mechanism for reduced islet cell proliferation was described, nor was impaired glucose control detected in that study. Here, we show that Axin expression impaired normal expression of islet Pitx2 and cyclinD2. Together with in vitro studies of Wnt-stimulated islets, our results collectively suggest that Wnt signaling is required for β cell Pitx2 and Cyclin D2 expression to promote in vivo β cell growth. Mutations in Pitx2 are associated with Rieger syndrome in humans, an autosomal-dominant condition that includes impaired glucose tolerance, reduced circulating insulin levels, and overt diabetes mellitus (38). The basis for these metabolic anomalies is not known. Mice with targeted mutations in Pitx2 have been previously described and manifest multiple developmental malformations (7, 39–42), but pancreatic islet defects have not yet been described, to our knowledge. Thus, further studies are needed to show whether Pitx2 is sufficient or required for pancreatic β cell proliferation.
Another study (14) provided evidence that the timing or extent of Wnt pathway modulation can significantly alter the outcome of mouse embryonic and perinatal pancreas development. Thus, the range of nonendocrine and endocrine pancreatic phenotypes reported by us and others (11–14) likely reflects the distinct experimental strategies and transgenic strains used to disrupt in vivo pancreatic Wnt signaling in these studies.
Proliferation of postnatal islet cells is relatively low compared with tissues such as intestines or bone marrow, except in states favoring robust β cell growth, such as pregnancy, insulin resistance, and obesity (3). Could Wnt signaling regulate adult β cell growth? Fujino et al. (43) reported that older mice lacking LRP5, a Wnt coreceptor, had impaired β cell function and impaired glucose tolerance when challenged with a high-fat diet (43). This finding suggests that Wnt signaling may be required to maintain islet functions in physiologic conditions demanding adaptive β cell responses. Multiple Wnt ligands, receptors, and signal transduction factors are expressed in the embryonic and adult pancreas (12, 13, 33), and it is unclear which endogenous factors might mediate Wnt signaling in β cell proliferation. Further studies to clarify these issues may prove useful for generating methods to control pancreatic islet cell proliferation and to test whether Wnt signaling dysregulation might underlie some forms of neuroendocrine tumor pathogenesis.
Materials and Methods
Animal Breeding.
RIP-Cre (24), β-catactive (26), TRE-Axin (27), and Pdx1-tTA mice (28) have been previously described. Animals were maintained on a standard light/dark cycle and handled in accordance with Stanford University Animal Care and Use Guidelines. Dox (2 mg/ml; Sigma, St. Louis, MO) was administered through drinking water as described in ref. 29.
Immunohistochemistry.
Tissue was fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Fort Washington, PA) or in Z-Fix (Anatech, Battle Creek, MI). Immunohistochemistry was performed on paraffin-embedded tissue, and immunofluorescence studies were performed on either paraffin- or cryo-preserved tissue, depending on antibody requirements. Antigen retrieval, antisera, and detection methods used here are described in SI Materials and Methods. Images were captured with a Leica (Deerfield, IL) SP2 AOBS confocal microscope (Cell Sciences Imaging Facility, Stanford Medical Center, Stanford, CA). Light images were captured with a Zeiss (Oberkochen, Germany) Axioplan 2 microscope and software. Cell counting, point-counting morphometry, and β cell quantification were performed by using standard morphometric techniques (14, 44, 45) or as described in SI Materials and Methods. For quantification of immunostained cells, pancreas tissue was obtained from at least four mice per genotype. All data are presented as the average ± SEM. Two-tailed t tests were conducted to determine statistical significance.
Physiologic Studies and Islet Isolation.
Glucose challenge studies were performed as described in ref. 45. Pancreatic insulin content was measured with a Mouse Insulin ELISA kit (Alpco Diagnostics, Windham, NH).
Islets were isolated from C57BL/6 mice on P4–8. Pancreata were digested with collagenase (Sigma–Aldrich, St. Louis, MO), and islets were purified by using a multilayer islet-specific ficoll gradient (Mediatech, Washington, DC).
Isolation of RNA, Protein, and Quantitative PCR Analysis.
The murine insulinoma cell line MIN6 (18) was used for in vitro studies. MIN6 cells or postnatal islets were treated with purified Wnt3a (final concentration 100 ng/ml, equal to 2.4 nM) (16), vehicle only, or Wnt3a plus Fz8-CRD (final concentration 200 ng/ml, equal to 3.3 nM). After treatment, RNA was isolated with TRIzol (Invitrogen, Carlsbad, CA). By using the RetroScript Kit (Ambion, Austin, TX), 1.5 μg of total RNA was then reverse transcribed into cDNA. Quantitative PCR analysis was performed on an Applied Biosystems (Foster City, CA) 7300 Real Time Machine PCR machine with TaqMan probe sets for Pitx2, cyclin D2, cyclin D1, CDK4. All transcript levels were normalized against β-actin. ChIP assays were performed on MIN6 cells and islets as described in ref. 19. Before ChIP analysis and proliferation analysis, islets were maintained for 24 h in RPMI medium 1640, penicillin/streptomycin, Hepes, fungizone, and l-glutamine (Invitrogen-GIBCO, Carlsbad, CA) containing 10% FBS (HyClone, Logan, UT) at 37°C. We used monoclonal antibody specific for Pitx2 (1:100, Santa Cruz Biotechnology, Santa Cruz, CA). Primers specific for cyclin D2 proximal promoter sequences and regions of the cyclin D1 and cdk4 promoters are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. F. Costantini (Columbia University, New York, NY), P. Herrera (University of Geneva, Geneva, Switzerland), and R. J. MacDonald (University of Texas Southwestern Medical Center, Dallas, TX) for mice; A. Mikels and J. Van Der Velde (Stanford University) for purified Wnt3a and Fz8-CRD; Dr. Magali Fontaine (Stanford University) for assistance in procuring human islet samples; Drs. C. Logan, D. L. Jones, K. Willert, E. Rulifson, M. Yen, and I. Weissman for discussions; and members of the S. K. Kim Laboratory for comments on earlier versions of this manuscript. I.R. was supported by a fellowship from the Walter V. and Idun Berry Postdoctoral Program at Stanford University School of Medicine. S. K. Karnik was supported by a Stanford Cancer Council Award, a gift from Dr. Raymond Sackler and Beverly Sackler through the Verto Institute, and a Ruth Kirschstein Postdoctoral Fellowship from the U.S. National Institutes of Health. P.H. was a student in the University of California, San Francisco, Biomedical Science graduate program. This research was supported by National Institutes of Health Grants DK60533, CA112537 (both to M.H.), and DK56709 (to S. K. Kim), the American Diabetes Association (M.H.), the Larry L. Hillblom Foundation (M.H.), funds from the Peterson Family Foundation (to S. K. Kim), and Program Project Grant 4-2004-345 from the Juvenile Diabetes Research Foundation (to R.N. and S. K. Kim).
Abbreviations
- CDK4
cyclin-dependent kinase 4
- Dox
doxycycline
- Fz
Frizzled
- Fz8-CRD
Fz 8-cysteine-rich domain
- Pn
postnatal day n
- RIP-Cre
Cre-recombinase in β cells from the rat insulin promoter
- TRE
tetracycline response element
- tTA
tetracycline-regulated transactivator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701509104/DC1.
References
- 1.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 2.Ryan EA, Paty BW, Senior PA, Bigam D, Alfadhli E, Kneteman NM, Lakey JR, Shapiro AM. Diabetes. 2005;54:2060–2069. doi: 10.2337/diabetes.54.7.2060. [DOI] [PubMed] [Google Scholar]
- 3.Heit JJ, Karnik SK, Kim SK. Annu Rev Cell Dev Biol. 2006;22:311–338. doi: 10.1146/annurev.cellbio.22.010305.104425. [DOI] [PubMed] [Google Scholar]
- 4.Cozar-Castellano I, Fiaschi-Taesch N, Bigatel TA, Takane KK, Garcia-Ocana A, Vasavada R, Stewart AF. Endocr Rev. 2006;27:356–370. doi: 10.1210/er.2006-0004. [DOI] [PubMed] [Google Scholar]
- 5.Logan CY, Nusse R. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 6.Baek SH, Kioussi C, Briata P, Wang D, Nguyen HD, Ohgi KA, Glass CK, Wynshaw-Boris A, Rose DW, Rosenfeld MG. Proc Natl Acad Sci USA. 2003;100:3245–3250. doi: 10.1073/pnas.0330217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohqi KA, Lin C, Gleiberman A, Wang J, et al. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh K, Krupnik VE, Sokol SY. Curr Biol. 1998;8:591–594. doi: 10.1016/s0960-9822(98)70229-5. [DOI] [PubMed] [Google Scholar]
- 10.Sakanaka C, Weiss JB, Williams LT. Proc Natl Acad Sci USA. 1998;95:3020–3023. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dessimoz J, Bonnard C, Huelsken J, Grapin-Botton A. Curr Biol. 2005;15:1677–1683. doi: 10.1016/j.cub.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 12.Murtaugh LC, Law AC, Dor Y, Melton DA. Development (Cambridge, UK) 2005;132:4663–4674. doi: 10.1242/dev.02063. [DOI] [PubMed] [Google Scholar]
- 13.Papadopoulou S, Edlund H. Diabetes. 2005;54:2844–2851. doi: 10.2337/diabetes.54.10.2844. [DOI] [PubMed] [Google Scholar]
- 14.Heiser PW, Lau J, Taketo MM, Herrera PL, Hebrok M. Development (Cambridge, UK) 2006;133:2023–2032. doi: 10.1242/dev.02366. [DOI] [PubMed] [Google Scholar]
- 15.Heller RS, Dichmann DS, Jensen J, Miller C, Wong G, Madsen OD, Serup P. Dev Dyn. 2002;225:260–270. doi: 10.1002/dvdy.10157. [DOI] [PubMed] [Google Scholar]
- 16.Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, Nusse R. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 17.Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Development (Cambridge, UK) 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 18.Miyazaki J, Araki K, Yamato E, Ikegami H, Asano T, Shibasaki Y, Oka Y, Yamamura K. Endocrinology. 1990;127:126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 19.Karnik SK, Hughes CM, Gu X, Rozenblatt-Rosen O, McLean GW, Xiong Y, Meyerson M, Kim SK. Proc Natl Acad Sci USA. 2005;102:14659–14664. doi: 10.1073/pnas.0503484102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rane SG, Dubus P, Mettus RV, Galbreath EJ, Boden G, Reddy EP, Barbacid M. Nat Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- 21.Georgia S, Bhushan A. J Clin Invest. 2004;114:963–968. doi: 10.1172/JCI22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kushner JA, Ciemerych MA, Sicinska E, Wartschow LM, Teta M, Long SY, Sicinski P, White MF. Mol Cell Biol. 2005;25:3752–3762. doi: 10.1128/MCB.25.9.3752-3762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelengaris S, Khan M, Evan GI. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 24.Herrera PL. Development (Cambridge, UK) 2000;127:2317–2322. 2000. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 25.Heit JJ, Apelqvist ÅA, Gu X, Winslow MM, Neilson JR, Crabtree GR, Kim SK. Nature. 2006;443:345–349. doi: 10.1038/nature05097. [DOI] [PubMed] [Google Scholar]
- 26.Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu W, Shakya R, Costantini F. J Cell Biol. 2001;155:1055–1064. doi: 10.1083/jcb.200107066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holland AM, Hale MA, Kagami H, Hammer RE, MacDonald RJ. Proc Natl Acad Sci USA. 2002;99:12236–12241. doi: 10.1073/pnas.192255099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smart NG, Apelqvist ÅA, Gu X, Harmon EB, Topper JN, MacDonald RJ, Kim SK. PLoS Biol. 2006;4:e39. doi: 10.1371/journal.pbio.0040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu G, Dubauskaite J, Melton DA. Development (Cambridge, UK) 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 31.Jonsson J, Carlsson L, Edlund T, Edlund H. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 32.Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. Development (Cambridge, UK) 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 33.Heller RS, Klein T, Ling Z, Heimberg H, Katoh M, Madsen OD, Serup P. Gene Expr. 2003;11:141–147. doi: 10.3727/000000003108749035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pedersen AH, Heller RS. Biochem Biophys Res Commun. 2005;333:961–968. doi: 10.1016/j.bbrc.2005.05.189. [DOI] [PubMed] [Google Scholar]
- 35.Laybutt DR, Weir GC, Kaneto H, Lebet J, Palmiter RD, Sharma A, Bonner-Weir S. Diabetes. 2002;51:1793–1804. doi: 10.2337/diabetes.51.6.1793. [DOI] [PubMed] [Google Scholar]
- 36.Briata P, Ilengo C, Corte G, Moroni C, Rosenfeld MG, Chen CY, Gherzi R. Mol Cell. 2003;12:1201–1211. doi: 10.1016/s1097-2765(03)00407-6. [DOI] [PubMed] [Google Scholar]
- 37.Hardikar AA, Marcus-Samuels B, Geras-Raaka E, Raaka BM, Gershengorn MC. Proc Natl Acad Sci USA. 2003;100:7117–7122. doi: 10.1073/pnas.1232230100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aarskog D, Ose L, Pande H, Eide N. Am J Med Genet. 1983;15:29–38. doi: 10.1002/ajmg.1320150104. [DOI] [PubMed] [Google Scholar]
- 39.Gage PJ, Suh H, Camper SA. Development (Cambridge, UK) 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- 40.Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y, et al. Development (Cambridge, UK) 1999;126:5749–5758. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- 41.Lin CR, Kioussi C, O'Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 42.Liu C, Liu W, Lu MF, Brown NA, Martin JF. Development (Cambridge, UK) 2001;128:2039–2048. doi: 10.1242/dev.128.11.2039. [DOI] [PubMed] [Google Scholar]
- 43.Fujino T, Asaba H, Kang MJ, Ikeda Y, Sone H, Takada S, Kim DH, Ioko RX, Ono M, Tomoyori H, et al. Proc Natl Acad Sci USA. 2003;100:229–234. doi: 10.1073/pnas.0133792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harmon EB, Apelqvist AA, Smart NG, Gu X, Osborne DH, Kim SK. Development (Cambridge, UK) 2004;131:6163–6174. doi: 10.1242/dev.01535. [DOI] [PubMed] [Google Scholar]
- 45.Kim SK, Hebrok M, Li E, Oh SP, Schrewe H, Harmon EB, Lee JS, Melton DA. Genes Dev. 2000;14:1866–1871. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.