Abstract
Breast cancer risk is a polygenic trait. To identify breast cancer modifier alleles that have a high population frequency and low penetrance we used a comparative genomics approach. Quantitative trait loci (QTL) were initially identified by linkage analysis in a rat mammary carcinogenesis model followed by verification in congenic rats carrying the specific QTL allele under study. The Mcs5a locus was identified by fine-mapping Mcs5 in a congenic model. Here we characterize the Mcs5a locus, which when homozygous for the Wky allele, reduces mammary cancer risk by 50%. The Mcs5a locus is a compound QTL with at least two noncoding interacting elements: Mcs5a1 and Mcs5a2. The resistance phenotype is only observed in rats carrying at least one copy of the Wky allele of each element on the same chromosome. Mcs5a1 is located within the ubiquitin ligase Fbxo10, whereas Mcs5a2 includes the 5′ portion of Frmpd1. Resistant congenic rats show a down-regulation of Fbxo10 in the thymus and an up-regulation of Frmpd1 in the spleen. The association of the Mcs5a1 and Mcs5a2 human orthologs with breast cancer was tested in two population-based breast cancer case-control studies (≈12,000 women). The minor alleles of rs6476643 (MCS5A1) and rs2182317 (MCS5A2) were independently associated with breast cancer risk. The minor allele of rs6476643 increases risk, whereas the rs2182317 minor allele decreases risk. Both alleles have a high population frequency and a low penetrance toward breast cancer risk.
Keywords: breast cancer genetics, cancer epidemiology, comparative genomics, noncoding elements, rat models
Despite immense efforts, the search for modifier genes underlying complex diseases has not been highly productive. Alleles of modifier genes that influence common disease risk have a moderate-to-high population frequency with a low penetrance. It has been suggested that alleles acting in this manner comprise the majority of genetic risk for many common diseases such as breast cancer (1, 2). It is estimated that if most risk alleles were identified, it would become possible to assign ≈90% of breast cancer risk to 50% of women (3). In most studies, candidate modifier genes are selected based on function, such as DNA repair or estrogen metabolism for breast cancer. Over 100 such candidate modifier genes have been tested in breast cancer case-control association studies (>400 SNPs); few show a consistent and significant association with risk in large sample populations (4). These results suggest the need for an alternative strategy to identify breast cancer modifier genes. Our laboratory has pursued the identification of candidate loci by using whole-genome linkage studies in inbred rat mammary cancer models, followed by fine-mapping in congenic rats. The rat was chosen because, similar to humans, it develops mammary carcinomas that are hormone-responsive and of ductal origin (5, 6). Using a backcross of [Wistar–Kyoto (WKy) × Wistar–Furth (WF)]F1 × WF rats, we identified four mammary carcinoma susceptibility QTL, Mcs5, Mcs6, Mcs7, and Mcs8, on rat chromosomes 5, 7, 10, and 14, respectively (7). The WKy allele of Mcs5 acts to suppress mammary tumor multiplicity in a susceptible WF genetic background and has been shown to consist of at least three clustered loci; among these, Mcs5a confers a phenotype of resistance to mammary cancer (8). Here we show that Mcs5a is a compound QTL located in a noncoding genomic region. We identified polymorphisms within the human genomic region orthologous to rat Mcs5a that significantly associate with breast cancer risk in women.
Results
To further analyze the Mcs5a locus, WF.WKy congenic rat lines with different segments of the WKy allele were established and phenotyped for resistance to 7,12-dimethylbenzanthracene (DMBA)-induced carcinogenesis (Fig. 1). Mammary carcinoma susceptibility in DMBA-treated rats was reduced ≈50% for each congenic line O, WW, and XX (Fig. 1). The boundaries of the Mcs5a locus are given by the overlapping WKy sequences of congenic lines WW and XX, which define a genomic interval of ≈116 kb containing Mcs5a. By incorporating phenotypic data from additional congenic lines within the interval (lines LL and B3) we demonstrated that at least two genetic elements exist within Mcs5a. These two lines are essentially overlapping. The sequence (313 bp) in the interval between the last informative SNPs at the respective adjacent ends of lines B3 and LL is identical between WKy and WF strains. Interestingly, there is no independent phenotype of resistance in WKy-homozygous rats from either line LL or B3, suggesting that Mcs5a is a compound locus requiring two genetic elements, Mcs5a1 and Mcs5a2, to be WKy alleles to confer resistance (Fig. 1).
Fig. 1.
Positional mapping of interacting loci within the rat Mcs5a compound QTL. Mcs5a (≈116 kb, pink) is a compound QTL located on rat chromosome 5. Adjacent Mcs5a1 (32 kb, black) and Mcs5a2 (84 kb, gray) are interacting loci within Mcs5a and were identified by using congenic lines. Rat transcripts are displayed as exons connected by the intronic sequence, with arrows indicating the direction of transcription. Mammary carcinoma susceptibility phenotypes of congenic lines with different segments of the Mcs5a WKy allele contained in line O were determined by using female rats administered DMBA (65 mg/kg) at 50–55 days of age. The congenic lines had the same pedigree at the N12 generation. Tumor multiplicity phenotypes are presented as the average number of mammary carcinomas ≥3 × 3 mm per rat ± SD after 15 weeks of initiation. Phenotypes for each genotype were compared within the congenic line by using the Mann–Whitney nonparametric test. Percent reduction in mammary carcinomas per rat compared with WF/WF littermates is shown at the center for lines determined to be significantly different (P < 0.05) than the WF/WF rats. The resistant (orange) and susceptible (yellow) congenic lines that define Mcs5a are shown. The relevant end of the WKy sequence in each line is indicated by a genetic marker and a dashed vertical line. Polymorphisms SNP-61634906, SNP-61666918, SNP-61667232, and gUwm23–29 mark the ends of congenic lines and are at base positions 61,634,906; 61,666,918; 61,667,232; and 61,751,595–61,751,793, respectively. The x axis represents rat chromosome 5 and the designated marks are SNPs (red), indels (purple), microsatellites (blue), CpG islands (green), and the UCSC Genome Browser June 2003 rat assembly base position (black). The 36.4-kb Mcs5a2 subregion marked by brackets contains the segment in which the human/rat CNS (60% identity over 90 bp) was resequenced.
Resistance is conferred only when the Mcs5a1 and Mcs5a2 WKy alleles are in cis (on the same chromosome). Heterozygous rats having both WKy elements in cis developed fewer carcinomas per rat, which significantly differed from both the WF and transchromosomal heterozygotes (P < 0.05) (Fig. 2). In contrast, heterozygous rats carrying one WKy copy of each Mcs5a1 and Mcs5a2 in trans developed a similar number of carcinomas as did WF rats (P = 0.33).
Fig. 2.
Mcs5a1 and Mcs5a2 elements interact only in a cis-chromosomal configuration. Heterozygous rats (n = 38 rats and 456 mammary glands) having both Mcs5a1 and Mcs5a2 WKy elements in cis developed fewer mammary carcinomas per rat ± SD 15 weeks after initiation than both the WF (n = 35 rats and 420 mammary glands, P = 0.005) and transchromosomal heterozygotes (n = 43 rats and 516 mammary glands, P = 0.048). Heterozygous rats with Mcs5a1 and Mcs5a2 WKy alleles in trans developed a similar number of mammary carcinomas as WF rats (P = 0.334). Bars in the graph that are marked with different lowercase letters are significantly different. Trans females were F1 offspring from a B3 × LL reciprocal cross. Cis animals were line O heterozygous F1 females from a line O × WF.WKy reciprocal cross. P values are from Conver–Inman multiple comparisons following a significant Kruskal–Wallis nonparametric test (P = 0.018).
Mcs5a1 is defined by the 32-kb genomic region of overlap between congenic lines WW and B3 (Fig. 1). Mcs5a1 consists mostly of the intronic sequence of the uncharacterized Fbxo10 gene, which contains an F-box domain (F-box proteins are components of SCF ubiquitin ligase complexes; ref. 9). The Mcs5a1 region contains a small section of intron 1, exon 2, and most of intron 2 of this gene. Resequencing the WF and WKy alleles of Fbxo10 exon 2 revealed no nucleotide differences. However, resequencing of the entire Mcs5a1 region identified 90 SNPs and 28 indels within the Fbxo10 intronic sequence of the Mcs5a1 locus.
The overlapping genomic regions of congenic lines LL and XX, which cover ≈84 kb, define Mcs5a2 (Fig. 1). This locus spans a region from within the first intron of Fbxo10, including its first exon and proximal promoter, to a location between the 5′ UTR and the first coding exon of the uncharacterized gene, Frmpd1 (FERM and PDZ domain-containing 1; refs. 10 and 11). There was no difference in the amino acid sequence encoded by the first exon of Fbxo10, and the nucleotide sequence of the transcribed Frmpd1 5′ UTR was identical between the WF and WKy rat strains. Using the University of California, Santa Cruz (UCSC), genome browser and miRBase (The Wellcome Trust Sanger Institute), we found no annotated microRNAs in the Mcs5a interval (12).
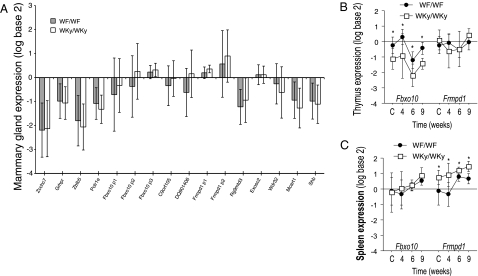
The transcript levels of Fbxo10 and Frmpd1 were found to be similar in mammary tissue from Mcs5a WF-homozygous and Mcs5a Wky-homozygous congenic rats, as was the expression of all other genes within an ≈1-megabase (Mb) region surrounding Mcs5a [Fig. 3 and supporting information (SI) Fig. 5). The expression of Fbxo10 and Frmpd1 also was quantified in several mammary abscopal tissues, including spleen, thymus, ovary, and brain. Among the Mcs5a WF-homozygous and WKy-homozygous congenic rat tissues tested, Fbxo10 was found to be differentially expressed only in thymus and Frmpd1 was differentially expressed only in spleen tissues of both control and DMBA-treated rats (Fig. 3 and SI Fig. 6). Neither gene was differentially expressed in ovary or brain (SI Fig. 6).
Fig. 3.
Organ-specific differential expression of Mcs5a genes. (A) Similar transcript levels of genes within and flanking (±500 kb) the Mcs5a1 and Mcs5a2 loci were measured in mammary glands of Mcs5a resistant (WKy/WKy) and susceptible (WF/WF) female congenic rats by using QPCR. (B and C) Differential expression between resistant and susceptible rats (P < 0.05) was found in rat thymus and spleen tissues for the Mcs5a candidate genes, Fbxo10 (B) and Frmpd1 (C), respectively. The differences in Fbxo10 (thymus) and Frmpd1 (spleen) expression levels were in 12-week-old virgin female rats (C = control) and tissues collected after initiation with carcinogen (time: 4, 6, and 9 weeks). Expression levels are presented as the log2 ± SD of the gene transcript level determined by real-time QPCR adjusted for Gapdh expression. Mann–Whitney comparisons with P < 0.05 are marked with asterisks.
To determine whether women carry alleles at the MCS5A1 and MCS5A2 loci that modulate breast cancer risk, we conducted population-based association studies on the human orthologous regions, which cover ≈93 kb on human chromosome 9 (Fig. 4). The human haplotype block structure of a 1-Mb region surrounding this area was determined (SI Fig. 7). Haplotypes from a single block (SI Fig. 7, block 4) in the MCS5A locus were tested for association with breast cancer risk in a Wisconsin case-control study (1,500 cases and 1,405 controls). Allele 4 of block 4.2, a subregion of this block (SI Fig. 8, block 4.2) that spanned SNPs rs10758440, rs999988, rs2182317, and rs2381718 was found to be marginally significant (SI Table 2) in the Wisconsin population. These four SNPs were further evaluated in the larger U.K. population (4,364 cases and 4,547 controls). The minor allele of rs2182317 (SNP-3) demonstrated a highly significant association, with a reduction in breast cancer risk (Table 1 and SI Table 3). The Wisconsin and U.K. populations combined (n = 11,779 women) yielded a heterozygous odds ratio of 0.86 [95% confidence interval (C.I.), 0.79–0.94], and a homozygous odds ratio of 0.77 (0.57–1.04). The uncorrected χ2 trends test, stratified by study, yielded a P value of 0.0003 (Table 1), and, when corrected for multiple comparisons by using the conservative Bonferroni method, the P value was 0.001. It is emphasized that this limited correction for multiple comparisons resulted from needing to evaluate only a small number of human polymorphisms within a precise genomic region defined by a global linkage study using the rat, which unlike human populations has few stratifying variables.
Fig. 4.
Human MCS5A1 (30.1 kb) and MCS5A2 (63.4 kb) loci contain breast-cancer-associated polymorphisms. The genomic regions shown on human chromosome 9 are orthologous to the rat Mcs5a1 (black) and Mcs5a2 (gray) regions. MCS5A transcripts are mapped as exons connected by the intronic sequence. Arrows indicate the direction of transcription. Haplotype blocks (blue horizontal bars) that overlap MCS5A contain the breast cancer risk-associated polymorphisms SNP-A1 (rs6476643) in MCS5A1 and SNP-3 (rs2182317) in MCS5A2. Association of these SNPs to breast cancer risk was determined by using two population-based samplings that evaluated ≈12,000 women. The minor allele frequency of SNP-A1 (MCS5A1) is 0.25. SNP-A1 associates with a 19% increased risk of breast cancer for women that are homozygous for the minor allele. The minor allele frequency of SNP-3 (MCS5A2) is 0.13. SNP-3 associates with a 14% decreased risk of breast cancer for the 24% of women that carry at least one minor allele. The three polymorphisms that correlated with SNP-A1 are in the 5.7-kb region marked by brackets within the MCS5A1 region. The SNPs correlated with SNP-3 lie within the human/rat MCS5A2 orthologous region and are within a 16.8-kb region (marked by brackets). The x axis is the human chromosome 9 base position based on the May 2004 UCSC human genome assembly. The x axis marks designate SNPs (red), indels (purple), CpG islands (green), and base positions (black).
Table 1.
Breast cancer risk-associated SNPs in MSC5A1 and MCS5A2, genotype frequencies, and genotype-specific risks in 5,848 women with breast cancer and 5,931 controls from Wisconsin and the U.K.
| Allele | P values trend, 1 df | Heterogeneity, 2 df | Major allele frequency | Minor allele frequency | Major allele homozygote, n/% | Heterozygote, n/% | Minor allele homozygote, n/% | No. genotyped |
|---|---|---|---|---|---|---|---|---|
| MCS5A1* | ||||||||
| Wisconsin | 0.058 | 0.143 | ||||||
| Cases | 0.72 | 0.28 | 778/53 | 590/40 | 113/8 | 1,481 | ||
| Controls | 0.75 | 0.25 | 765/55 | 532/39 | 84/6 | 1,381 | ||
| OR | 1 (ref.) | 1.12 (1.03–1.22) | 1 (ref.) | 1.09 (0.93–1.27) | 1.32 (0.98–1.78) | |||
| U.K. | 0.123 | 0.230 | ||||||
| Cases | 0.74 | 0.26 | 2,373/55 | 1,657/38 | 316/7 | 4,346 | ||
| Controls | 0.75 | 0.25 | 2,517/56 | 1,704/38 | 290/6 | 4,511 | ||
| OR | 1 (ref.) | 1.05 (1.01–1.11) | 1 (ref.) | 1.03 (0.94–1.13) | 1.16 (0.98–1.37) | |||
| Combined | 0.022 | 0.049 | ||||||
| Cases | 0.73 | 0.27 | 3,151/54 | 2,247/39 | 429/7 | 5,827 | ||
| Controls | 0.75 | 0.25 | 3,282/56 | 2,236/38 | 374/6 | 5,892 | ||
| OR | 1 (ref.) | 1.07 (1.01–1.13) | 1 (ref.) | 1.05 (0.97–1.13) | 1.19 (1.03–1.38) | |||
| MCS5A2† | ||||||||
| Wisconsin | 0.140 | 0.126 | ||||||
| Cases | 0.89 | 0.11 | 1,166/79 | 291/20 | 15/1 | 1,472 | ||
| Controls | 0.88 | 0.12 | 1,074/78 | 284/21 | 26/2 | 1,384 | ||
| OR | 1 (ref.) | 0.89 (0.75–1.04) | 1 (ref.) | 0.94 (0.78–1.13) | 0.53 (0.28–1.01) | |||
| U.K. | 0.001 | 0.003 | ||||||
| Cases | 0.89 | 0.11 | 3,436/79 | 878/20 | 62/1 | 4,376 | ||
| Controls | 0.87 | 0.13 | 3,430/75 | 1,045/23 | 72/2 | 4,547 | ||
| OR | 1 (ref.) | 0.86 (0.81–0.92) | 1 (ref.) | 0.84 (0.76–0.93) | 0.86 (0.61–1.21) | |||
| Combined | 0.0003 | 0.0016 | ||||||
| Cases | 0.89 | 0.11 | 4,602/79 | 1,169/20 | 77/1 | 5,848 | ||
| Controls | 0.87 | 0.13 | 4,504/76 | 1,329/22 | 98/2 | 5,931 | ||
| OR | 1 (ref.) | 0.86 (0.80–0.94) | 1 (ref.) | 0.86 (0.79–0.94) | 0.77 (0.57–1.04) | |||
Values in parentheses represent 95% C.I.; df, degree of freedom; OR, odds ratio; ref., reference genotype.
*SNP-A1, rs6476643.
†SNP-3, rs2182317.
SNP-3 and/or polymorphisms highly correlated to SNP-3 could be responsible for the observed association to breast cancer risk. To locate correlated polymorphisms, the human–rat conserved sequence (CNS) segments within MCS5A2 (an ≈27-kb total sequence, ≈30% CNS) and the entire MCS5A1 region (≈30 kb) were resequenced in 24 (12 cases and 12 controls) women from the Wisconsin case-control population (SI Data Set 1). These samples were chosen to be representative of the haplotype allele frequencies observed in the population. Resequencing data provided more extensive coverage of the region and resulted in the separation of haplotype block 4 into haplotype blocks A, B, and C (Fig. 4) based on r2 estimates. We found no polymorphisms in the MCS5A1 region that correlated to SNP-3 in the MCS5A2 region. We did find 12 SNPs, highly correlated to SNP-3, in the MCS5A2 region that localized to an ≈17-kb interval, which is ≈40 kb distal to the MCS5A1/MCS5A2 boundary (Fig. 4). This ≈17-kb sequence covers a region of FRMPD1 that includes the 5′ UTR exon (Fig. 4). It is likely that the MCS5A2 causative genetic polymorphism resides either within, or in close proximity to, this reduced interval (SI Text). Like the rat model, no polymorphisms were identified within the FRMPD1 5′ UTR in the human samples. Interestingly, resequencing of the distal ≈36-kb CNS of Mcs5a2 in the rat strains revealed that polymorphisms between the WKy and WF sequence occur only in the region orthologous to the SNP-3 correlated SNP region of humans (Figs. 1 and 4).
We tested the additional genetic variation, revealed after resequencing the human regions, to determine whether a polymorphism within the MCS5A1 region associated independently with breast cancer risk in women. Seventy-two (19 not listed in dbSNP) MCS5A1 SNPs with the minor allele observed in more than a single individual were documented. These SNPs were “binned” into highly correlated (r2) groups of polymorphisms. Bins were screened for an association with breast cancer risk in the Wisconsin case-control population. The results (Table 1 and SI Table 4) identified one bin (rs6476643, rs10758441, and rs7042509; and an indel, 138-9899, on chromosome 9:37563886–37563887) that qualified for further testing in the second population. One SNP from this bin (rs6476643, SNP-A1) was evaluated further in the U.K. breast cancer case-control population. The combined results for SNP-A1 yielded an odds ratio of 1.05 (95% C.I., 0.97–1.13) for heterozygotes and 1.19 (95% C.I., 1.03–1.38) for minor allele homozygotes (Table 1). A trends test, stratified by study, was significant for the minor allele (P = 0.02). The polymorphisms in this bin are not highly correlated to any other variation (SI Data Set 1); therefore, any of these four polymorphisms are likely causative (SI Text). The human sequence variation correlated to SNP-A1 (rs6476643) in MCS5A1 is located at the beginning of haplotype block B and is localized to a 5.7-kb interval (Fig. 4).
Discussion
This study illustrates the potential genetic complexity underlying the common diseases of rodents and humans and provides empirical support for the common disease/common variant hypothesis, which suggests that common diseases such as breast cancer have, for the most part, simple allelic spectra (1). Complexity is demonstrated by the rat compound Mcs5a locus that consists of at least two cis-only interacting elements Mcs5a1 and Mcs5a2. An analysis of the human orthologous regions identified alleles from both MCS5A1 and MCS5A2 that associated with breast cancer risk. The minor allele marked by SNP-A1 in the MCS5A1 region acts in a recessive manner, similar to the rat Mcs5a WF susceptibility allele, to increase risk by 19% in 6% of women homozygous for this allele. Within the MCS5A2 region, the minor allele marked by SNP-3 acts in a dominant manner, similar to the rat WKy allele, to decrease risk by 14% in 24% of women. Because of the apparent linkage across the MCS5A region and the inability to determine the chromosomal phase of the double heterozygous individuals, we were not able to compare MCS5A1 and MCS5A2 in cis and trans chromosomal configurations to test for an interaction. We observed only two of 11,659 samples that were homozygous for the MCS5A1 increased-susceptibility minor allele and homozygous for the MCS5A2 decreased-susceptibility allele. There were only 22 samples that were homozygous for the MCS5A1 minor allele and heterozygous for MCS5A2.
In both rats and humans the elements responsible for altering cancer risk at these loci do not reside in protein coding sequence. Noncoding elements have previously been associated with evolutionary biological differences (13) as well as risk for complex diseases (14, 15). Mcs5a modulates the expression of both surrounding, uncharacterized genes in an immune tissue-specific manner. Thus, the identification of these two breast cancer risk modifier alleles illustrates the utility of mining natural genetic variation in model organisms to identify novel genetic polymorphisms and mechanisms associated with the etiology of common polygenic diseases in humans.
Methods
Animal Experiments.
Congenic lines were established and maintained in an Association for Assessment and Accreditation of Laboratory Animal Care-approved facility as previously published (16). All animal protocols were approved by the University of Wisconsin Medical School Animal Care and Use Committee. Congenics are defined as genetic lines developed on a WF genome and carrying the selected WKy alleles shown in Fig. 1. The congenic generations (number of backcrosses) used to determine the mammary carcinoma multiplicity phenotypes of lines O, LL, WW, XX, and B3 were N12, N14, N14, N15, and N15, respectively. Congenic lines had the same pedigree at the N12 generation. Genetic marker primer sequences and the SNP base positions defining the ends of the WKy allele carried by each congenic line are listed in SI Text. The Mcs5a1–Mcs5a2 cis vs. trans experiment used line O heterozygous rats (cis) and B3 × LL F1 females (trans). Line O contains ≈4.45 Mb of WKy chromosome 5 sequence from 57,848,415–62,299,478. Line WW was expanded from an N13 line O recombinant that retained ≈0.66 Mb of WKy chromosome 5 sequence from 61634906–62299478. For gene expression analyses, female rats from congenic lines O and WW were compared with inbred WF and WF.WKy females. The WF.WKy line (N11F8–N11F10) is WF-homozygous at all known Mcs loci and was used to provide the Mcs5a WF comparison allele.
Female rats, 50–55 days old, were administered a single dose of DMBA (65 mg per kilogram of body weight) (ACROS Organics; Fisher Scientific, Pittsburgh, PA) in sesame oil by gastric intubation. Mammary carcinomas ≥3 × 3 mm were counted after 15 weeks of induction. Mammary carcinoma multiplicity data were analyzed by using the Mann–Whitney nonparametric test of StatView (SAS Institute, Cary, NC) for congenic line comparisons and the Kruskal–Wallis nonparametric test of the R statistical package (R Foundation for Statistical Computing, Wein, Austria) followed by Conover–Inman multiple comparison procedure (17) for the cis vs. trans experiment.
Comparative Genomics.
The Mcs5a rat chromosome 5 genomic region is based on the June 2003 genome assembly (UCSC, Genome Browser: www.genome.ucsc.edu) of SNP at chromosome 5:61634886C>T base position and the microsatellite marker (gUwm23-29) at the respective ends of the WKy Mcs5a allele delimited by congenics. The Mcs5a proximal and distal rat chromosome 5 base positions (chromosome 5:61634886–61751793) and the VISTA browser (http://pipeline.lbl.gov) were used to locate the human orthologous region on human chromosome 9 (37550841–37644624). Transcripts were found by using the UCSC and VISTA Rat and Human Genome browsers. Human/rat CNS in the MCS5A2/Mcs5a2 regions were identified by using the VISTA browser with settings of 90 bp and 60% identity. Resequencing methods are given in the SI Text. Polymorphisms observed in human resequencing data are listed in SI Data Set 1. Polymorphisms in SI Tables 5 and 6 (MCS5A1 and MCS5A2, respectively) have minor alleles that were observed in more than one individual and were not listed on the National Center for Biotechnology Information web site. Ninety-one SNPs that spanned a 1-Mb region on human chromosome 9 orthologous to the rat Mcs5a region were used to make the initial haplotype block map (SI Fig 7, SI Table 7, and SI Text). Subsequent human haplotype blocks A, B, and C (Fig. 4 and SI Figs. 9 and 10), based on the distribution of minor allele patterns (r2) observed after obtaining resequencing data, were defined in the initial haplotype blocks 4 and 5 (SI Fig. 7).
Quantitative RT-PCR (QPCR).
TaqMan QPCR primers and probes (Applied Biosystems, Foster City, CA) were designed by using Primer Express v 2.0 (Applied Biosystems) according to developer's specifications. Primer and probe sequences are given in SI Table 8. cDNA was synthesized from 2 μg of total RNA treated with TURBO-free DNase (Ambion, Austin, TX) and diluted 1:8 for QPCR. A microliter of the dilution (≈12.5 ng of RNA-equivalent cDNA) was used in a 16-μl TaqMan QPCR with Applied Biosystems 7900 default cycling conditions. The reaction components were 1× TaqMan Buffer A (Applied Biosystems); 5.5 mM MgCl2; dATP, dCTP, dGTP, and dTTP at 400 μM each; each experimental primer at 300 nM; 200 nM TaqMan experimental probe (Applied Biosystems); each Gapdh primer at 100 nM, 200 nM rodent Gapdh probe; and 0.4 units of Taq Gold DNA Pol (Applied Biosystems). The real-time QPCR cycling conditions were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. FAM (Mcs5a region gene locus probe) and VIC (Rodent Gapdh probe; Applied Biosystems) fluorescence values were measured by using Applied Biosystems SDS v 2.2 software; quantities of transcripts were measured by comparison of cycle threshold values with a standard curve calculated from serial dilutions. Sample measurements are an average of four replicates per sample and were standardized by dividing the quantity of rodent Gapdh. Data were analyzed by using the Mann–Whitney tests of StatView.
Breast Cancer Risk Association Study.
Population-based cases of incident invasive breast cancer and community controls from Wisconsin were recruited according to a protocol approved by the University of Wisconsin Health Sciences Institutional Review Board (18). Cases 20–69 years of age were identified through the Wisconsin statewide tumor registry. Controls were randomly sampled from driver's license files (ages, 20–64 years) and Medicare beneficiary lists (ages, 65–69 years). Controls (n = 1,790) were frequency-age-matched in 5-year intervals to have an age distribution similar to the cases (n = 1,737). Only women of European descent were included to limit stratification. Human DNA extraction, genotyping, and quality control methods are listed in SI Text. The U.K. samples have been described (19).
The tag SNPs that characterize the common haplotypes in block 4.2 (rs2381718, rs10758440, rs999988, and rs2182317) (SI Fig. 8) were subjected to Cochran Armitage tests for trend and normal approximations of log odds ratios for heterozygotes and minor allele homozygotes using a population of 4,376 breast cancer cases and 4,547 controls from the U.K. samples. The tests were applied retroactively to the Wisconsin population and the combined stratified sample. Seven polymorphisms from five additional SNP bins located in MCS5A1 were genotyped in the Wisconsin samples after the resequencing of 24 Wisconsin samples. Primer and probe sequences are listed in SI Table 9. Of these polymorphisms, only SNP-A1 (rs6476643) was tested in the Wisconsin and U.K. populations.
Supplementary Material
Acknowledgments
We thank Drs. William Dove and Norman Drinkwater for reviewing this manuscript; Dr. Patrick Remington and the staff and participants of the Wisconsin Women's Health Study; and Louis Gardner, Andrew Hitt, Craig Luccarini, Katie Nelson, Jordy Waller, Don Wigington, and Yu-Rong Wang for technical assistance. This work was supported by National Institutes of Health Grants CA077494 and CA123272. The Wisconsin Women's Health Study was supported by National Institutes of Health Grants CA82004 and CA047147 and a gift from the Fraternal Order of Eagles Arie no. 1502. D.J.S. and S.E.H. were supported by Department of Defense Grants DAMD17-03-1-0280 and W81XWH-04-1-0397.
Abbreviations
- C.I.
confidence interval
- CNS
conserved sequence
- DMBA
7,12-dimethylbenzanthracene
- Mb
megabase
- QPCR
quantitative PCR
- QTL
quantitative trait locus
- WF
Wistar–Furth
- WKy
Wistar–Kyoto.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ901406).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701687104/DC1.
References
- 1.Reich DE, Lander ES. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 2.Smith P, McGuffog L, Easton DF, Mann GJ, Pupo GM, Newman B, Chenevix-Trench G, Szabo C, Southey M, Renard H, et al. Genes Chrom Cancer. 2006;45:646–655. doi: 10.1002/gcc.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA. Nat Genet. 2002;31:33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- 4.Ponder BAJ, Cebrian A, Dunning AM, Easton DF, Lesueur F, Luccarini C, Pharoah PDP. Breast Cancer Res. 2005;7(Suppl 2):S14. doi: 10.1186/bcr1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russo J, Russo IH. J Mammary Gland Biol Neoplasia. 2000;5:187–200. doi: 10.1023/a:1026443305758. [DOI] [PubMed] [Google Scholar]
- 6.Russo J, Tait L, Russo IH. Am J Pathol. 1983;113:50–66. [PMC free article] [PubMed] [Google Scholar]
- 7.Lan H, Kendziorski CM, Haag JD, Shepel LA, Newton MA, Gould MN. Genetics. 2001;157:331–339. doi: 10.1093/genetics/157.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samuelson DJ, Aperavich BA, Haag JD, Gould MN. Cancer Res. 2005;65:9637–9642. doi: 10.1158/0008-5472.CAN-05-1498. [DOI] [PubMed] [Google Scholar]
- 9.Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Curr Biol. 1999;9:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- 10.Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, et al. Trends Biochem Sci. 1998;8:281–282. doi: 10.1016/s0968-0004(98)01237-7. [DOI] [PubMed] [Google Scholar]
- 11.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. Nucl Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King MC, Wilson AC. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 14.Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Cutler DJ, Green ED, Chakravarti A. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 15.Loots GG, Kneissel M, Keller H, Baptist M, Chang J, Collette NM, Ovcharenko D, Plajzer-Frick I, Rubin EM. Genome Res. 2005;15:928–935. doi: 10.1101/gr.3437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samuelson DJ, Haag JD, Lan H, Monson DM, Shultz MA, Kolman BD, Gould MN. Carcinogenesis. 2003;24:1455–1460. doi: 10.1093/carcin/bgg112. [DOI] [PubMed] [Google Scholar]
- 17.Conover WJ. Wiley B, O'Sullivan M. Practical Nonparametric Statistics. 3rd Ed. New York: Elm Street; 1999. p. 290. [Google Scholar]
- 18.Nelson SE, Gould MN, Hampton JM, Trentham-Dietz A. Breast Cancer Res. 2005;7:R357–R364. doi: 10.1186/bcr1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anglian Breast Cancer Study Group. Br J Cancer. 2000;83:1301–1308. doi: 10.1054/bjoc.2000.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.