Abstract
Infection of the stomach by Helicobacter pylori is ubiquitous among humans. However, while H. pylori strains from different geographic areas are associated with clear phylogeographic differentiation1-4, the age of an association between these bacteria with humans remains highly controversial5, 6. Here we show, using sequences from a large dataset of bacterial strains that, as in humans, genetic diversity in H. pylori decreases with geographic distance from East Africa, the cradle of modern humans. We also observe similar clines of genetic isolation by distance (IBD) for both H. pylori and its human host at a worldwide scale. Like humans, simulations indicate that H. pylori seems to have spread from East Africa around 58,000 years ago. Even at more restricted geographic scales, where IBD tends to become blurred, principal component clines in H. pylori from Europe strongly resemble the classical clines for Europeans described by Cavalli-Sforza and colleagues7. Taken together, our results establish that anatomically modern humans were already infected by H. pylori prior to their migrations from Africa and demonstrate that H. pylori has remained intimately associated with their human host populations ever since.
Over half of all humans are infected by Helicobacter pylori, a Gram-negative bacterium that can cause peptic ulcers and constitutes a risk factor for stomach cancer8. Not only is H. pylori ubiquitous, but it also possesses strong phylogeographic structure1, suggesting that bacterial polymorphisms reflect human phylogeography and historical migrations2, 3, 5. In 2003, Falush et al. assigned 370 H. pylori strains to four main population clusters, two of which were subdivided into subpopulations2. The geographic sources of these strains reflected major events in human settlement history, such as the colonisation of Polynesia and the Americas and the African Bantu migrations. However, these discrete groupings seem to contradict an apparent continuity of the geographic component of genetic diversity in humans: Genetic differentiation between human populations increases linearly with geographic distance computed along landmasses9-12; and their genetic diversity declines with increasing geographic distance from East Africa13, 14.
There are several possible explanations why detailed population genetic patterns differ between H. pylori and their human hosts. Infection of humans by H. pylori might be too recent to have been affected by ancient events in human settlement history, e.g. it might date from a recently acquired zoonosis5. Or differences in population structure between bacteria and humans may reflect frequent horizontal transmission of H. pylori. Alternatively, apparent differences in the population genetic patterns may simply be a matter of perception due to differing analytical methodology: H. pylori population genetics has so far focused on the description of clusters, whereas human population genetics is influenced by a traditional emphasis on clines15.
We used an expanded dataset (769 H. pylori isolates from 51 ethnic sources, Table S1) to test whether patterns in their geographic distribution mimic those of humans. Bayesian MCMC cluster analyses identified the same five ancestral sources of nucleotides as found previously with a smaller dataset2 (Fig. 1a,b). These analyses also assigned the isolates to six populations containing various degrees of ancestry from the five ancestral sources (Fig. 1a, S1A). Four of the populations had been previously identified, and designated hpEurope, hpEastAsia, hpAfrica1 and hpAfrica2 due to their obvious geographical associations2. In agreement, almost all strains isolated from Europeans belong to hpEurope, including Basques in Spain, Russians and Kazakhs, and most isolates from East Asia belong to hpEastAsia (Table S1). The results also confirmed that hpAfrica2, previously represented by only few isolates, is common in South Africa. Two new populations were identified and designated hpAsia2 and hpNEAfrica. hpAsia2 was isolated in northern India, Thailand, Bangladesh, the Philippines and elsewhere in S. E. Asia (Fig. S1B). hpNEAfrica was predominant among isolates from Ethiopia, Somalia, Sudan and Nilo-Saharan speakers in northern Nigeria.
FIGURE 1.
Five ancestral populations in H. pylori. a) DISTRUCT24 plot of the proportions of ancestral nucleotides in 769 H. pylori isolates as determined by Structure V2.0 (linkage model)17. A thin line for each isolate indicates the estimated amount of ancestry from each ancestral population as five colored segments. The lines are grouped by (sub)population and ordered by ancestry. b) Neighbor joining tree (black lines) of the relationships between and diversity within five ancestral populations calculated as described2. Circle diameters are proportional to within-population genetic diversity and angles of filled arcs are proportional to the amount of ancestry attributable to each population among modern strains. scale bar: 0.01. c-g) Spatial distribution of five geographic sources of ancestral nucleotides. Dark-light gradients show clinal declines in proportions of ancestral nucleotides by distance from a geographic centre. Color-coding as in part b. AE1, AE2: ancestral Europe 1 and 2.
Matrices of pair-wise FST (a measure of genetic differentiation between populations) between paired groups of samples from analogous geographic locations were strongly correlated (Mantel regression coefficient = 0.86, p<0.001) between H. pylori sequences and human microsatellite data16; 73% of human variation can be explained by a linear relationship with microbial variation (Fig. S2). Thus, the geographic component of genetic diversity seems to be quantitatively comparable between H. pylori and humans, except that FST is considerably higher in H. pylori.
We next address evidence for a continuum of genetic ancestry in H. pylori. Whereas the assignments of individual isolates to populations are quite unambiguous with the no-admixture model in STRUCTURE17, its linkage model shows that the proportions of ancestry from the five ancestral sources almost form a continuum (Fig. 1a) between the five populations other than hpAfrica2, which is highly distinct. Similarly, although most concatenated sequences cluster according to their population assignment in a phylogenetic tree (Fig. S3), their relatedness is also almost continuous, again with the exception of hpAfrica2. A nearly continuous distribution of the proportions of ancestry suggests localized admixture due to recombination. Such admixture would blur differences between initially distinct populations in close geographical proximity, and could potentially lead to strong signals of IBD in H. pylori, as observed in humans9-12.
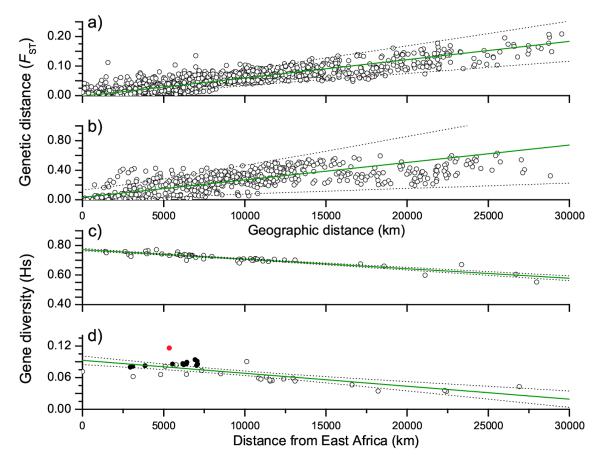
Diversity was only poorly correlated with geographical sources in initial analyses, which might reflect noise due to recent human migrations plus horizontal transmission of H. pylori between ethnically distinct groups. Therefore, we excluded 147 isolates from obvious recent human migrants and their admixed descendents as well as 31 isolates whose population assignments were highly incongruent with their sources of isolation (horizontal transmission) (see Methods). The resulting “non-migrant” dataset (Table S2) contained 532 H. pylori isolates and 1,405 polymorphisms. The results were compared with human diversity based on 783 autosomal microsatellites16. Similar to previous analyses11, 12, 14, 77% of the variance in FST between autosomal human markers from distinct geographic sources was accounted for by the shortest geographic distance along landmasses (Fig. 2a). For H. pylori, only 47% of the variance was accounted for by geographic distance (Fig. 2b, p ≤ 0.001), but this estimate rose to 72% when a standard conversion of genetic diversity was plotted against the logarithm of the geographic distance for the 442 haplotypes from geographic locations with at least ten isolates (Fig. S4). Thus, comparable proportions of the genetic diversity are due to IBD in H. pylori as in humans.
FIGURE 2.
Parallel geographic patterns of genetic diversity in humans (a,c) and H. pylori (b, d). a, b) Genetic distance between pairs of geographic populations (FST) versus geographic distance between the two populations. c, d) Average gene diversity within geographic populations (HS) versus geographic distance from East Africa. R2: a,0.77; b,0.47; c,0.85; d,0.59. In part d, samples that are predominantly composed of hpEurope isolates are indicated by filled circles while the red circle identifies the sample from South Africa. Confidence intervals are indicated by dotted lines.
Genetic diversity within modern humans decreases with distance from East Africa, reflecting their recent African origin12-14; 85% of this decrease in diversity could be accounted for by distance from East Africa (Fig. 2c). The non-migrant H. pylori dataset also showed a similar trend (Fig. 2d, p ≤ 0.001) and 59% of the decrease in diversity could be accounted for by distance from East Africa. Unlike IBD, where trends with H. pylori might mimic those of humans without a joint demography, parallel decreases in diversity with distance from East Africa indicate close associations between the two. Simulations with human data indicated that they migrated from East Africa 56,000 ± 5500 years ago14. Similar demographically explicit genetic simulations now indicate that an East African source for H. pylori is more probable than South Africa or China, and that H. pylori migrated from East Africa 58,000 ± 3500 years ago (Table 1, S3, Fig. S5; see Supplementary Information for details). Thus, we conclude that H. pylori accompanied anatomically modern humans during their migrations from Africa that have been estimated at 50,000-70,000 years ago14, 18-21. This implies that H. pylori was present in Africa prior to these migrations, suggesting that Africa is the source of both H. pylori and humans.
Table 1.
Simulations for three hypothetical scenarios for the origin of H. pylori.
| Estimated parameters | Model | ||||||
|---|---|---|---|---|---|---|---|
| Scenario | Ancestral population (K0) | Carrying capacity (K) | Migration (K*m) | Growth rate (r) | Time (years) | R 2 | AIC |
| East Africa | 561 ± 182 | 203 ± 87 | 65 ± 6 | 0.73 ± 0.14 | 57,955 ± 3748 | 0.68 | 83.3 |
| South Africa | 541 ± 214 | 185 ±76 | 86 ± 8 | 0.68 ± 0.13 | 61,746 ± 4436 | 0.57 | 88.1 |
| East Asia | 747 ± 207 | 467 ± 191 | 370 ± 169 | 0.70 ± 0.16 | 36,500 ± 6728 | 0.02 | 101.5 |
Additional details are in Supplementary Information. The five estimated parameters are the size of the founder population (K0) and the carrying capacity of any subsequently colonised demes (K), both of which are expressed as the number of effective strains that succeed in infecting the subsequent host generation. K*m represents the number of effective migrants sent by any deme, r is the growth rate within demes in a logistic growth model and time refers to the duration of the entire colonisation process. The time estimates presented here include an additional 2,500 year migration phase after the last deme had been colonised. Mean and standard deviations were computed for all simulations that fell within the 95% confidence interval. The fit of the models to the data is given by R2, the amount of variance explained, and AIC, the Akaike information criterion.
Are clines in genetic diversity truly contradictory to discrete clusters? Discrete clusters can be defined even within a perfectly continuous pattern due to sampling artefacts10. But for H. pylori, geographic isolation is only a marginally better predictor of genetic differentiation than discrete clusters based on genetic similarity. Within a generalised linear model framework, assignment of populations to the six clusters defined by STRUCTURE explains 70% of the variance of pair-wise FST for populations with ten or more isolates, versus up to 72% by geographical distance. This situation resembles previous results for the geographic apportionment of human genetic diversity11, 16, except that even when geography is first accounted for, clusters as defined by STRUCTURE still explain 11% of additional variance in genetic differentiation between H. pylori populations as compared to only 2% in humans. We therefore examined the modern geographic sources of the nucleotides associated with the five ancestral populations according to STRUCTURE (Fig. 1c-g). The spatial distribution of ancestral nucleotides indicated that AE2 originated in East Africa, AE1 in central Asia, ancestral EastAsia in East Asia and ancestral Africa1 and Africa2 in Africa. These data probably reflect extensive population expansions, subsequent to the global spread that accompanied migrations out of Africa, and may well reflect important episodes in human history during the Neolithic period and later.
If H. pylori were also a marker for human migrations at a more local scale, one would expect to find similar patterns between human and bacterial diversity within Europe, which was not one of the sources of ancestral nucleotides in H. pylori. Indeed, the first two principal components of spatial autocorrelation with hpEurope isolates (Fig. 3a-b) were very similar to those that had been obtained with human allozymes in classical work by Cavalli-Sforza and colleagues7 (Fig. 3d-e) and the third PC showed similar East-West clines. Such clines were originally interpreted as genetic signatures or indications of episodic migratory events7, although this interpretation has been questioned22. We note that the first principal component of the H. pylori data, PC1, is a cline from the South East that correlates (R2=0.35, p<0.01) with the proportion of ancestry from AE2 (Fig. S6A), which originated in North East Africa. For PC2, a cline from the North East, the correlation (R2=0.6, p<0.01) is with AE1 (Fig. S6B), which originated in Central Asia. These correlations show that in H. pylori, as previously suggested2, much of the spatial pattern observed in Europe can be attributed to admixture from different sources. It also supports the controversial hypothesis7 that similar clines in humans are also due to waves of migration of genetically distinct populations into Europe (demic diffusion), except that the spatial sources of ancestral nucleotides are assigned to northeast Africa and central Asia. We further conclude that there are highly striking, quantitative parallels in clines and IBD both at the global and the local scale between humans and H. pylori. These presumably reflect the dissemination of H. pylori by a variety of prehistoric human migrations, followed by admixture after horizontal transmission between human populations.
FIGURE 3.
Similar clinal gradients between principal components 1-3 from European H. pylori (a-c, concatenated sequences) and humans (d-f, allozymes). Figs d-f, reprinted with permission from citation 7(copyright 1995 National Academy of Sciences, U.S.A.).
In this paper we have shown that the key patterns in the distribution of H. pylori genetic diversity mirror those of its human host. At a worldwide scale, we recovered similar patterns of isolation by distance, though absolute genetic differentiation is higher in H. pylori. As in humans, we observed a continuous loss of genetic diversity with increasing geographic distance from East Africa, the likely cradle of anatomically modern humans. Even at the more restricted scale of Europe, we largely recreated the classical clines described by Cavalli-Sforza and colleagues. Finally, simulations predict that H. pylori has spread from East Africa over the same time scale as anatomically modern humans. These extraordinary parallel population genetic patterns between H. pylori and its human host all demonstrate an old association predating the “out of Africa” event4. The results further point to a scenario where H. pylori and human populations have been evolving intimately ever since, with limited long-range transmission by horizontal infections.
Methods
Bacterial isolates and sequencing
The expanded H. pylori dataset consists of 3,406 bp of unique, concatenated sequences of fragments of atpA, efp, mutY, ppa, trpC, ureI, yphC from 769 H. pylori isolates (Table S1). The dataset includes 347 novel isolates in addition to data from 422 other strains that have been described previously2, 3,23. The new bacteria were isolated from 25 additional ethnic sources in Asia (8 countries), Europe (4, including Basques), Africa and the Middle East (9) and South America (2), for a total of 51 ethnic sources (Table S1). The forward and reverse strands were sequenced as described1. Almost half (1,522 sites, 45%) of the nucleotides are polymorphic, resulting in a nucleotide diversity (π) of 4.2% for the entire data set.
The non-migrant dataset excluded bacteria that were isolated from the following migrant human populations: Europeans and Cape Coloureds from Cape Town; Mestizos from Colombia and Venezuela; Whites and African Americans from the USA; isolates in Thailand from Chinese or without ethnic association. hpAfrica2 isolates from Xhosas near Pretoria were excluded because they were a selective subset rather than a population-wide sample. The Philippines were also removed because almost all bacterial populations were found there, probably due to their colonial history. For isolates from Native Americans, only hspAmerind strains were considered non-migrant. The dataset was further restricted to geographic samples with at least four isolates, to avoid statistical noise, which resulted in the elimination of all Jewish and Russian isolates and singletons from locations in China and Japan.
Supplementary Material
Acknowledgments
We gratefully acknowledge the receipt of bacterial strains from Arie van der Ende, Martin J. Blaser, Nigel J. Saunders, Robert J. Owen, Francis Mégraud and sequences from Kuvat T. Momynaliev and Christian Kraft. We thank Jérôme Goudet for providing a modified version of FSTAT able to deal with the large dataset and help with R by Klaus-Peter Pleissner. Grant support was from the German Federal Ministry for Education and Research (BMBF) in the framework of the PathoGenoMik Network (MA, SS; Grant 03U213), the Biotechnology and Biological Sciences Research Council (FB), the Swedish Research council (TW; 16x-04723) and Lund University Hospital (TW; ALF).
Footnotes
Competing Interests statement: The authors have no competing interests.
EMBL Accession numbers: AM413111-418360.
Reference List
- 1.Achtman M, et al. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 2.Falush D, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 3.Wirth T, et al. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: Lessons from Ladakh. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4746–4751. doi: 10.1073/pnas.0306629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eppinger M, et al. Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLoS Genet. 2006;2:e120. doi: 10.1371/journal.pgen.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kersulyte D, et al. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 2000;182:3210–3218. doi: 10.1128/jb.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dailidiene D, et al. Helicobacter acinonychis: Genetic and rodent infection studies of a Helicobacter pylori-like gastric pathogen of cheetahs and other big cats. J. Bacteriol. 2004;186:356–365. doi: 10.1128/JB.186.2.356-365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piazza A, et al. Genetics and the origin of European languages. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5836–5840. doi: 10.1073/pnas.92.13.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suerbaum S, Michetti P. Helicobacter pylori infection. New England J. Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 9.Relethford JH. Global patterns of isolation by distance based on genetic and morphological data. Hum. Biol. 2004;76:499–513. doi: 10.1353/hub.2004.0060. [DOI] [PubMed] [Google Scholar]
- 10.Serre D, Paabo S. Evidence for gradients of human genetic diversity within and among continents. Genome Res. 2004;14:1679–1685. doi: 10.1101/gr.2529604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manica A, Prugnolle F, Balloux F. Geography is a better determinant of human genetic differentiation than ethnicity. Hum. Genet. 2005;118:366–371. doi: 10.1007/s00439-005-0039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran S, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prugnolle F, Manica A, Balloux F. Geography predicts neutral genetic diversity of human populations. Curr. Biol. 2005;15:R159–R160. doi: 10.1016/j.cub.2005.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am. J. Hum. Genet. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton, NJ: Princeton University Press; 1994. [Google Scholar]
- 16.Rosenberg NA, et al. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Underhill PA, et al. Y chromosome sequence variation and the history of human populations. Nat. Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 19.Ingman M, Gyllensten U. Analysis of the complete human mtDNA genome: methodology and inferences for human evolution. J. Hered. 2001;92:454–461. doi: 10.1093/jhered/92.6.454. [DOI] [PubMed] [Google Scholar]
- 20.Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am. J. Hum. Genet. 2003;72:1171–1186. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellars P. Why did modern human populations disperse from Africa ca. 60,000 years ago? A new model. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9381–9386. doi: 10.1073/pnas.0510792103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Currat M, Excoffier L. The effect of the Neolithic expansion on European molecular diversity. Proc. Biol. Sci. 2005;272:679–688. doi: 10.1098/rspb.2004.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Momynaliev KT, et al. Population identification of Helicobacter pylori isolates from Russia. Genetika. 2005;41:1182–1185. [PubMed] [Google Scholar]
- 24.Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.