Abstract
Conjugal transfer from Escherichia coli to Alcaligenes eutrophus of the A. eutrophus genes coding for plasmid-borne resistance to cadmium, cobalt, and zinc (czc genes) was investigated on agar plates and in soil samples. This czc fragment is not expressed in the donor strain, E. coli, but it is expressed in the recipient strain, A. eutrophus. Hence, expression of heavy metal resistance by cells plated on a medium containing heavy metals represents escape of the czc genes. The two plasmids into which this DNA fragment has been cloned previously and which were used in these experiments are the nonconjugative, mobilizable plasmid pDN705 and the nonconjugative, nonmobilizable plasmid pMOL149. In plate matings at 28 to 30 degrees C, the direct mobilization of pDN705 occurred at a frequency of 2.4 x 10(-2) per recipient, and the mobilization of the same plasmid by means of the IncP1 conjugative plasmids RP4 or pULB113 (present either in a third cell [triparental cross] or in the recipient strain itself [retromobilization]) occurred at average frequencies of 8 x 10(-4) and 2 x 10(-5) per recipient, respectively. The czc genes cloned into the Tra- Mob- plasmid pMOL149 were transferred at a frequency of 10(-7) to 10(-8) and only by means of plasmid pULB113. The direct mobilization of pDN705 was further investigated in sandy, sandy-loam, and clay soils. In sterile soils, transfer frequencies at 20 degrees C were highest in the sandy-loam soil (10(-5) per recipient) and were enhanced in all soils by the addition of easily metabolizable nutrients.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF


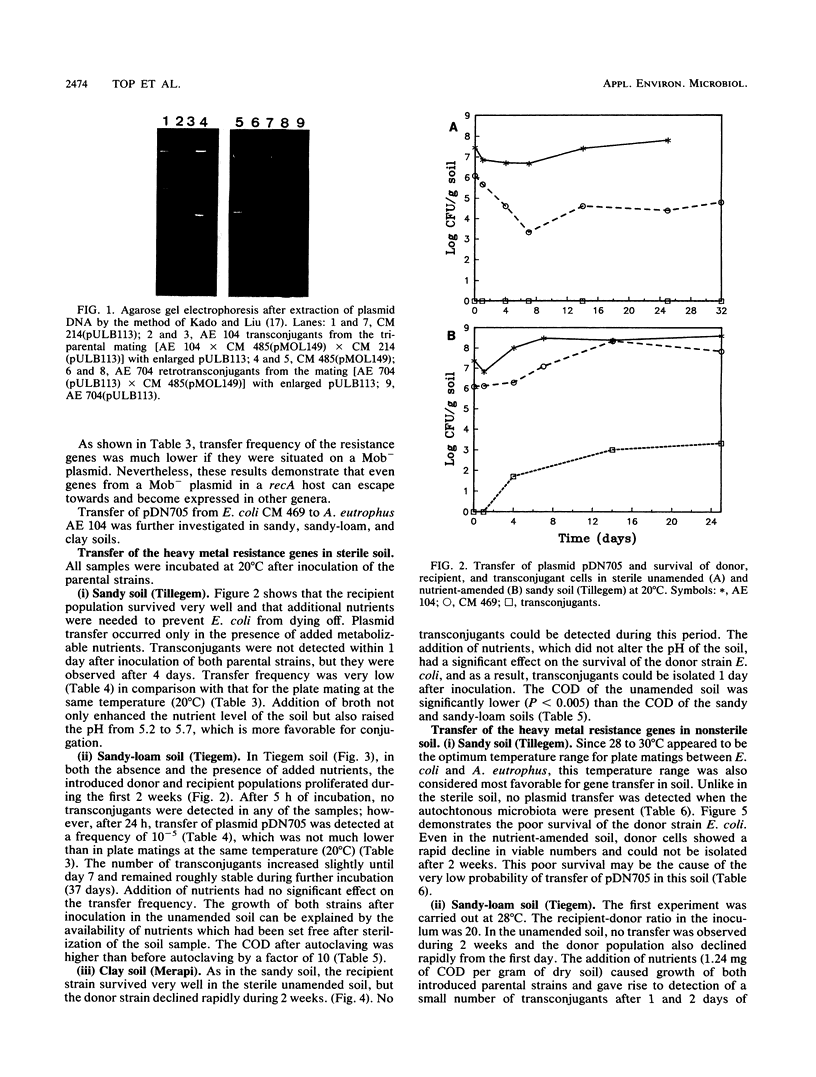
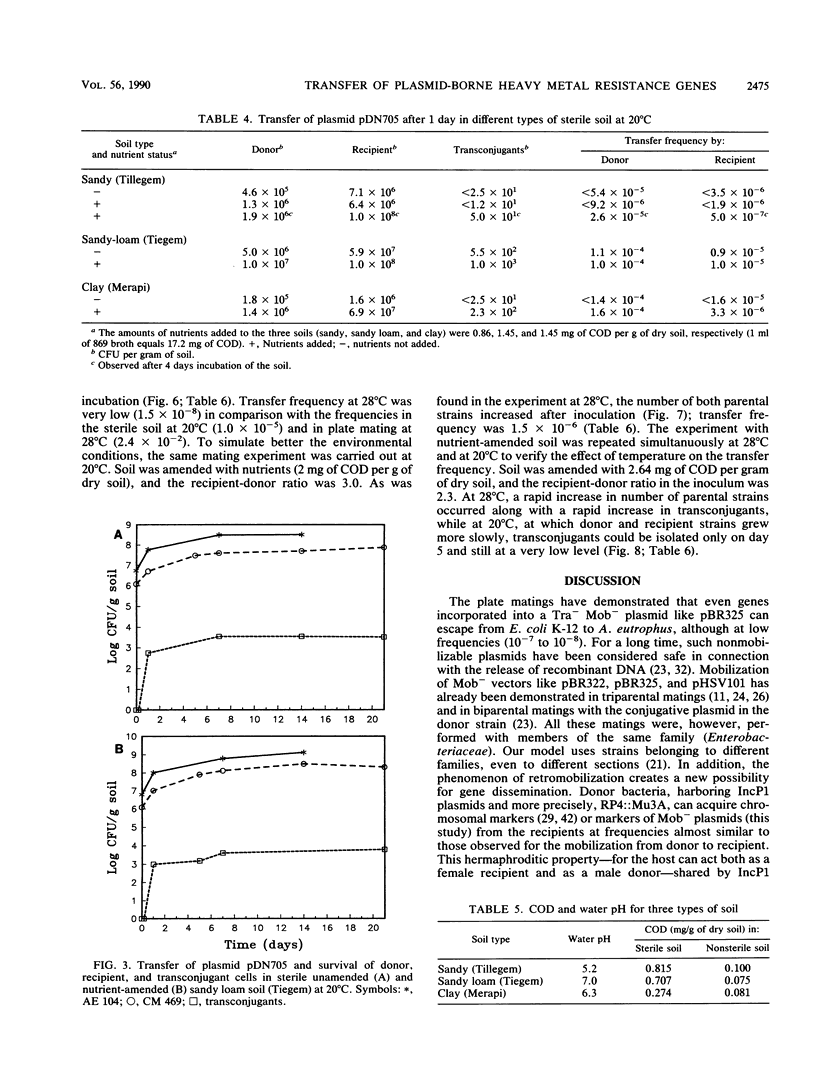
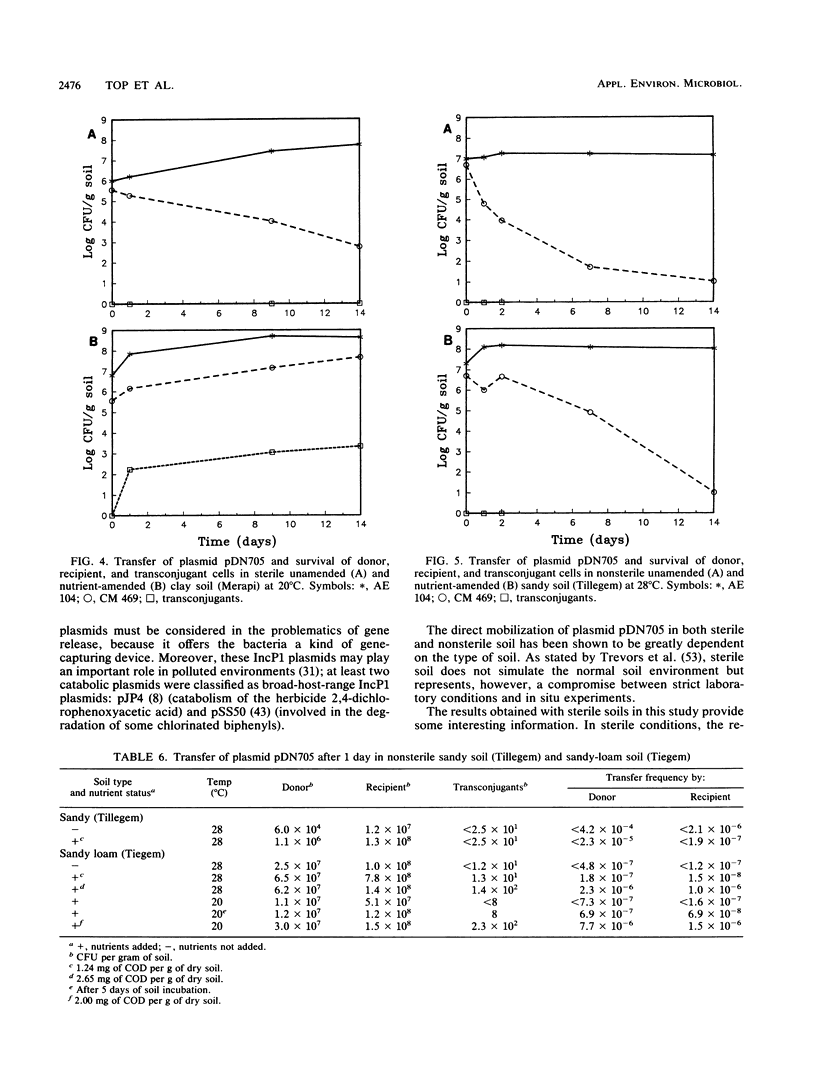



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Chaudhry G. R., Toranzos G. A., Bhatti A. R. Novel method for monitoring genetically engineered microorganisms in the environment. Appl Environ Microbiol. 1989 May;55(5):1301–1304. doi: 10.1128/aem.55.5.1301-1304.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W., Shaw E. J., Sykes R. B., Richmond M. H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971 Dec;108(3):1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diels L., Mergeay M. DNA probe-mediated detection of resistant bacteria from soils highly polluted by heavy metals. Appl Environ Microbiol. 1990 May;56(5):1485–1491. doi: 10.1128/aem.56.5.1485-1491.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Pemberton J. M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981 Feb;145(2):681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelen M., Resibois A., Toussaint A. Mini-mu: an insertion element derived from temperate phage mu-1. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1169–1177. doi: 10.1101/sqb.1979.043.01.132. [DOI] [PubMed] [Google Scholar]
- Fredrickson J. K., Bezdicek D. F., Brockman F. J., Li S. W. Enumeration of Tn5 mutant bacteria in soil by using a most- probable-number-DNA hybridization procedure and antibiotic resistance. Appl Environ Microbiol. 1988 Feb;54(2):446–453. doi: 10.1128/aem.54.2.446-453.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gealt M. A., Chai M. D., Alpert K. B., Boyer J. C. Transfer of plasmids pBR322 and pBR325 in wastewater from laboratory strains of Escherichia coli to bacteria indigenous to the waste disposal system. Appl Environ Microbiol. 1985 Apr;49(4):836–841. doi: 10.1128/aem.49.4.836-841.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman D. J., Gowland P. C., Slater J. H. Large plasmids from soil bacteria enriched on halogenated alkanoic acids. Appl Environ Microbiol. 1986 Jan;51(1):44–51. doi: 10.1128/aem.51.1.44-51.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D. A., Casida L. E., Jr Escherichia coli die-out from normal soil as related to nutrient availability and the indigenous microflora. Can J Microbiol. 1967 Nov;13(11):1461–1470. doi: 10.1139/m67-194. [DOI] [PubMed] [Google Scholar]
- Kozyrovskaya N. A., Gvozdyak R. I., Muras V. A., Kordyum V. A. Changes in properties of phytopathogenic bacteria effected by plasmid pRD1. Arch Microbiol. 1984 Apr;137(4):338–343. doi: 10.1007/BF00410731. [DOI] [PubMed] [Google Scholar]
- Lejeune P., Mergeay M., Van Gijsegem F., Faelen M., Gerits J., Toussaint A. Chromosome transfer and R-prime plasmid formation mediated by plasmid pULB113 (RP4::mini-Mu) in Alcaligenes eutrophus CH34 and Pseudomonas fluorescens 6.2. J Bacteriol. 1983 Sep;155(3):1015–1026. doi: 10.1128/jb.155.3.1015-1026.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. M., Kaper J. B., Lockman H., Black R. E., Clements M. L., Falkow S. Recombinant DNA risk assessment studies in humans: efficacy of poorly mobilizable plasmids in biologic containment. J Infect Dis. 1983 Oct;148(4):699–709. doi: 10.1093/infdis/148.4.699. [DOI] [PubMed] [Google Scholar]
- Mancini P., Fertels S., Nave D., Gealt M. A. Mobilization of plasmid pHSV106 from Escherichia coli HB101 in a laboratory-scale waste treatment facility. Appl Environ Microbiol. 1987 Apr;53(4):665–671. doi: 10.1128/aem.53.4.665-671.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson P., Gealt M. A. Isolation of indigenous wastewater bacterial strains capable of mobilizing plasmid pBR325. Appl Environ Microbiol. 1986 May;51(5):904–909. doi: 10.1128/aem.51.5.904-909.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M., Gerits J. F'-plasmid transfer from Escherichia coli to Pseudomonas fluorescens. J Bacteriol. 1978 Jul;135(1):18–28. doi: 10.1128/jb.135.1.18-28.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M., Houba C., Gerits J. Extrachromosomal inheritance controlling resistance to cadmium, cobalt, copper and zinc ions: evidence from curing in a Pseudomonas [proceedings]. Arch Int Physiol Biochim. 1978 May;86(2):440–442. [PubMed] [Google Scholar]
- Mergeay M., Lejeune P., Sadouk A., Gerits J., Fabry L. Shuttle transfer (or retrotransfer) of chromosomal markers mediated by plasmid pULB113. Mol Gen Genet. 1987 Aug;209(1):61–70. doi: 10.1007/BF00329837. [DOI] [PubMed] [Google Scholar]
- Mergeay M., Nies D., Schlegel H. G., Gerits J., Charles P., Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985 Apr;162(1):328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D. H., Nies A., Chu L., Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D., Mergeay M., Friedrich B., Schlegel H. G. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol. 1987 Oct;169(10):4865–4868. doi: 10.1128/jb.169.10.4865-4868.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford A. J., Oliver J., Kelly W. J., Reanney D. C. Translocatable resistance to mercuric and phenylmercuric ions in soil bacteria. J Bacteriol. 1981 Sep;147(3):1110–1112. doi: 10.1128/jb.147.3.1110-1112.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii F., Crawford D. L. Transfer of conjugative plasmids and mobilization of a nonconjugative plasmid between Streptomyces strains on agar and in soil. Appl Environ Microbiol. 1988 Jun;54(6):1334–1340. doi: 10.1128/aem.54.6.1334-1340.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richaume A., Angle J. S., Sadowsky M. J. Influence of soil variables on in situ plasmid transfer from Escherichia coli to Rhizobium fredii. Appl Environ Microbiol. 1989 Jul;55(7):1730–1734. doi: 10.1128/aem.55.7.1730-1734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak D. B., Colwell R. R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987 Sep;51(3):365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHATZ A., BOVELL C., Jr Growth and hydrogenase activity of a new bacterium, Hydrogenomonas facilis. J Bacteriol. 1952 Jan;63(1):87–98. doi: 10.1128/jb.63.1.87-98.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilf W., Klingmüller W. Experiments with Escherichia coli on the dispersal of plasmids in environmental samples. Recomb DNA Tech Bull. 1983 Sep;6(3):101–102. [PubMed] [Google Scholar]
- Schoonejans E., Toussaint A. Utilization of plasmid pULB113 (RP4::mini-Mu) to construct a linkage map of Erwinia carotovora subsp. chrysanthemi. J Bacteriol. 1983 Jun;154(3):1489–1492. doi: 10.1128/jb.154.3.1489-1492.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields M. S., Hooper S. W., Sayler G. S. Plasmid-mediated mineralization of 4-chlorobiphenyl. J Bacteriol. 1985 Sep;163(3):882–889. doi: 10.1128/jb.163.3.882-889.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotzky G., Babich H. Survival of, and genetic transfer by, genetically engineered bacteria in natural environments. Adv Appl Microbiol. 1986;31:93–138. doi: 10.1016/s0065-2164(08)70440-4. [DOI] [PubMed] [Google Scholar]
- Stotzky G. Influence of clay minerals on microorganisms. II. Effect of various clay species, homoionic clays, and other particles on bacteria. Can J Microbiol. 1966 Aug;12(4):831–848. doi: 10.1139/m66-111. [DOI] [PubMed] [Google Scholar]
- Stotzky G., Rem L. T. Influence of clay minerals on microorganisms. I. Montmorillonite and kaolinite on bacteria. Can J Microbiol. 1966 Jun;12(3):547–563. doi: 10.1139/m66-078. [DOI] [PubMed] [Google Scholar]
- Trevors J. T. Use of microcosms to study genetic interactions between microorganisms. Microbiol Sci. 1988 May;5(5):132–136. [PubMed] [Google Scholar]
- Van Gijsegem F., Toussaint A. Chromosome transfer and R-prime formation by an RP4::mini-Mu derivative in Escherichia coli, Salmonella typhimurium, Klebsiella pneumoniae, and Proteus mirabilis. Plasmid. 1982 Jan;7(1):30–44. doi: 10.1016/0147-619x(82)90024-5. [DOI] [PubMed] [Google Scholar]




