Summary
Rab GTPases, the largest subgroup in the superfamily of Ras-like GTPases, play regulatory roles in multiple steps of intracellular vesicle trafficking. They are activated by guanine nucleotide exchange factors (GEFs), which catalyze the interconversion of the GDP-bound, or inactive, form of Rab to the GTP-bound, or active, form. Relatively little is known of the mechanisms by which GEFs activate Rabs. Here we present the crystal structure of the GEF domain of Sec2p in complex with its Rab partner Sec4p. The Sec2p GEF domain is a 220 Å long coiled-coil, striking in its simplicity and in the use of the coiled-coil motif for catalysis. The structure suggests a mechanism whereby Sec2p induces extensive structural rearrangements in the Sec4p switch regions and phosphate binding loop that are incompatible with nucleotide binding. We show that Sec2p is specific for Sec4p and that specificity determinants reside in the two switch regions of Sec4p.
Introduction
Rab GTPases represent the largest subgroup in the superfamily of Ras-like GTPases (Zerial & McBride, 2001). Different Rabs regulate membrane traffic through various stages along the exocytic or endocytic pathways. One Rab can modulate and control an entire stage of transport by controlling several of its steps—such as vesicle budding, delivery, docking, and fusion (Grosshans et al, 2006b).
Like all small GTPases, Rab proteins are active in their GTP bound form and inactive in their GDP bound form, with the forms differing primarily in two structural regions, switches I and II, surrounding the nucleotide-binding site. GTPase specific guanine nucleotide-exchange factors (GEFs) activate GTPases by catalyzing the exchange of GDP for GTP. These GEFs are classified into families on the basis of sequence similarity and type of interacting GTPase, and the families characterized so far are structurally unrelated (Cherfils & Chardin, 1999). While GEFs for the Rho, Ras, ARF and Ran GTPases have been extensively studied (Boriack-Sjodin et al; 1997; Goldberg, 1998, Worthylake et al., 2000; Renault et al., 2001; Snyder et al; 2002; Kristelly et al; 2004), little is known regarding the mechanisms by which Rab GEFs recognize their targets and effect nucleotide exchange. The structure of the GEF domain of Rabex-5, which acts on Rab5, has been determined (Delprato et al., 2004), but details of recognition and nucleotide exchange cannot be understood based on the GEF structure alone, in the absence of a Rabex-5/Rab5 structure. Additionally, the mechanism by which the Mss4 protein catalyzes guanine nucleotide exchange for a number of Rab family GTPases has been explored both structurally and kinetically (Itzen et al, 2006). However, Mss4 is an atypical GEF in its unusually low catalytic efficiency (Esters et al, 2001) and in its broad range of Rab partners (see Itzen et al, 2006). Its proposed cellular role is as a chaperone for guanine nucleotide-free Rabs rather than as a GEF (Nuoffer et al, 1997; Collins et al, 1997; Itzen et al, 2006).
The N-terminal domain of Sec2p is a prototype for a structurally distinct family of Rab GEFs, which contain coiled-coil motifs. Sec2p is a highly efficient exchange factor 100–1000 more effective than Mss4 or Rabex-5 (Itzen et al., 2006). It is critical in activating the Rab GTPase Sec4p (Walch-Solimena et al, 1999) during the last stage of polarized exocytosis in budding yeast as secretory vesicles are delivered to exocytic sites and then dock and fuse with the plasma membrane (Grosshans et al, 2006, 2006b). Two sequence-related Rab GEFs, the mammalian proteins Rabin3/8 and GRAB, are also predicted to contain coiled-coil motifs and like Sec2p function in the exocytic pathway (Luo et al, 2001; Hattula et al, 2002).
To address how Sec2p and other coiled-coil GEFs catalyze nucleotide exchange for the Rab GTPases, we have determined the structure of the Sec2p GEF domain in complex with the nucleotide-free form of Sec4p, an intermediate in the reaction pathway after GDP release and before GTP binding. We find that two Sec2p GEF domains form a single, long, parallel coiled-coil, to which Sec4p binds. While interactions between Rab GTPases and coiled-coil proteins have been reported (Zhu et al, 2004; Beard et al, 2005), the direct involvement of a coiled-coil in catalysis, in this case of guanine nucleotide exchange, is to our knowledge unprecedented. A comparison of our Sec2p/Sec4p structure with nucleotide-bound structures of Sec4p (Stroupe & Brunger, 2000) suggests that Sec2p functions by inducing extensive structural reorganizations in the Sec4p switch regions and the phosphate-binding loop (P-loop), perturbing much of the nucleotide binding pocket. We further demonstrate that Sec2p acts specifically on Sec4p and show that the Sec4p switch regions are determinants for Sec2p GTPase specificity.
Results and Discussion
Overview of the structure
The guanine exchange activity of the 85 kDa Sec2p resides in the N-terminal 160 residues (Ortiz et al, 2002), and when overexpressed behind a GAL10 promoter this fragment is sufficient to complement a SEC2 disruption (M. Medkova and P. Novick, unpublished data). We have crystallized a catalytically active fragment of Sec2p (residues 17–167) in complex with the nucleotide free form of Sec4p (residues 18–187). The structure of the complex was determined to 3.3 Å by the single wavelength anomalous diffraction method using crystals grown from selenomethionine substituted protein. The complex consists of one Sec4p and two Sec2p GEF domain monomers, dimerized into a 220 Å long parallel coiled-coil (Figure 1). Because the dimer deviates from two-fold symmetry, it provides only a single Sec4p recognition surface. Dynamic light scattering data confirm that the same heterotrimeric complex observed in the crystal structure also forms in solution (Supplemental Methods).
Figure 1.
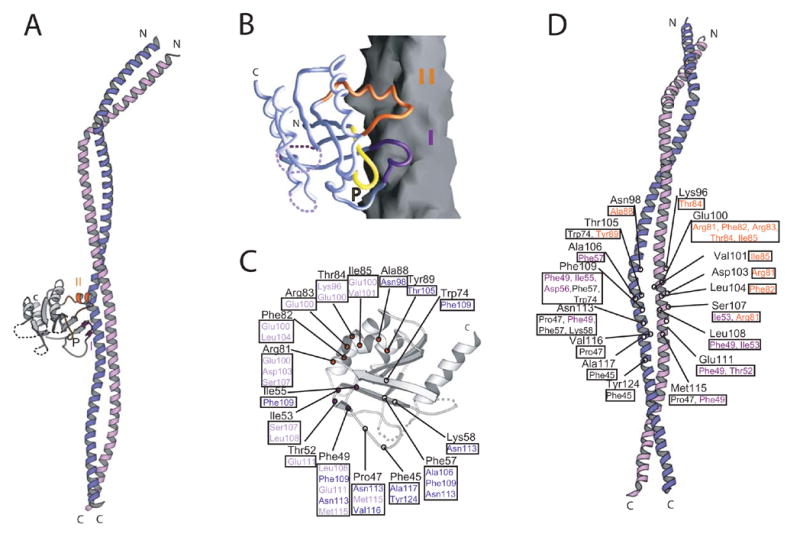
Structure of the Rab GTPase Sec4p in complex with the GEF domain of Sec2p. (A) A ribbons diagram. Sec4p is grey and Sec2p purple/lilac. Switches I (indigo) and II (orange) and the P-loop of Sec4p are labeled. (B) Sec4p as a Cα worm on the Sec2p surface. (C) Residues of Sec4p within 4 Å of Sec2p are labeled. Residues contacted in Sec2p are boxed and purple or lilac, depending to which Sec2p monomer they belong. (D) Residues of Sec2p within 4 Å of Sec4p are labeled. Contacted residues in Sec4p are boxed; residues in Switch I and II are indigo and orange, respectively.
The Sec2p coiled-coil is kinked near the N-terminus, where three residues (53–55sc2), a so-called stammer (Lupas & Gruber, 2005), are inserted into the heptad repeat of mostly hydrophobic residues that form the coiled-coil core. The hydrophobic side chain packing is also disrupted between residues 94sc2 and 119sc2 of both monomers. In this region, which is where Sec4p binds, the “knob-into-holes” packing characteristic of coiled-coils (Lupas & Gruber, 2005) is discontinued, and helices from the two Sec2p monomers interact so that the ridges of one pack against the groove of the second. Likely the interruption of the heptad repeat pattern in this region rather than Sec4p binding causes this distortion from a coiled-coil geometry. The Sec2p/Sec4p interface is mostly hydrophobic and buries ~3060 Å2 of solvent accessible surface. The residues from both monomers of Sec2p that contribute to this interface are highly conserved in the mammalian Rabin3/Rabin8 and GRAB, which act on Rab8 and Rab3a, respectively (Luo et al, 2001; Hattula et al, 2002) (Figure 2). We therefore expect Rabin3/Rabin8 and GRAB to function similarly to Sec2p.
Figure 2.
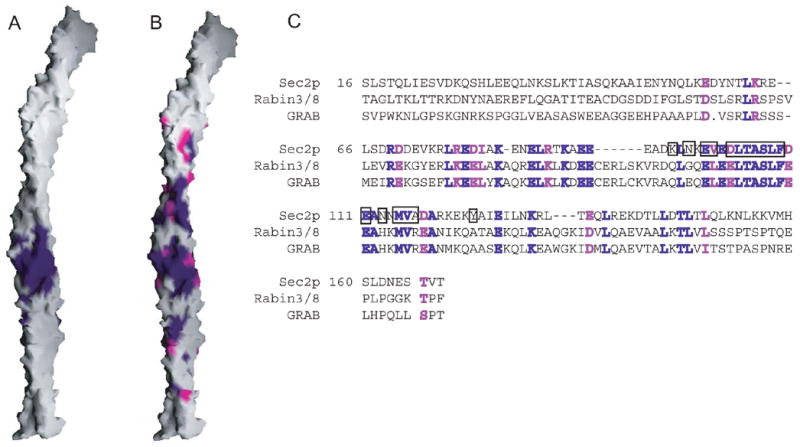
The Sec4p binding surface and conservation. (A) Surface representation of Sec2p. Residues within 4 Å of Sec4p are purple. (B) Residues identical in Sec2p and the mammalian Rabin3/8 and GRAB proteins are purple; similar residues (R=K, D=E, I=L=V) are magenta. (C) Sequence alignment of the Sec2p GEF domain with Rabin3/8 (human isoform β1) and GRAB (Rattus norvegicus). Identical and similar residues are purple and magenta, respectively. The Rabin3 and Rabin8 sequences are almost identical. Residues in Sec2p within 4 Å of Sec4p are boxed.
Sec2p induces a reorganization of the Sec4p nucleotide binding pocket
Sec4p residues (45–58sc4 and 81–89sc4) in and around switch regions I and II are involved in interactions with the Sec2p dimer (Figure 1B). Comparison with crystal structures of Sec4p complexed either with GDP or the non-hydrolyzable GTP analog guanosine-5’-(β,γ) imidotriphosphate (GppNHp) (Stroupe & Brunger, 2000) suggests that the interaction with Sec2p leads directly to structural reorganization of the switch regions. These regions along with the P-loop (residues 27–33sc4), which normally functions in binding the nucleotide β-phosphate, are the most extensively reordered in the Sec2p/Sec4p structure when compared with crystal structures of nucleotide bound Sec4p (Stroupe & Brunger, 2000) (Figure 3). In GDP-bound structures of Sec4p, switch I (residues 48–56sc4) is completely or largely disordered, whereas it adopts a stable conformation to interact with the nucleotide γ-phosphate group in the GppNHp bound structures. On binding Sec2p, switch I adopts a conformation that differs from the GppNHp bound form (Figure 3C) and places Ile50sc4 as close as 0.9 Å to the site occupied by Mg++ in the nucleotide bound forms. Ile50sc4 may therefore have a role in preventing the magnesium cation from binding, thus lowering the affinity of Sec4p for guanine nucleotide. As would be expected if Ile50sc4 is indeed important for nucleotide exchange, a Sec4p mutant in which Ile50sc4 has been replaced by alanine is no longer efficiently activated by Sec2p (Figure 4). In the course of switch I reorganization, the nearby residues 42–47sc4 also move dramatically from their position in the nucleotide structures. In particular, Phe45sc4 is displaced by ~8 Å from its position in the Sec4p/GDP and Sec4p/GppNHp structures (Figure 3), where it stabilizes the guanosine base through an edge-to-face aromatic-aromatic interaction. A Sec4p Phe45Ala mutant in which this interaction has been eliminated no longer stably binds GDP, as determined in the course of our exchange assays (data not shown). Similarly, eliminating this interaction by displacing Phe45sc4 from the nucleotide binding pocket would lower the affinity of Sec4p for nucleotide and likely plays a key role in nucleotide exchange. The displacement of equivalent phenylalanines is also important in the activation of several other GTPases, including Ras and the Rho GTPase Tiam1 (Boriack-Sjodin et al, 1997; Worthylake et al, 2000).
Figure 3.
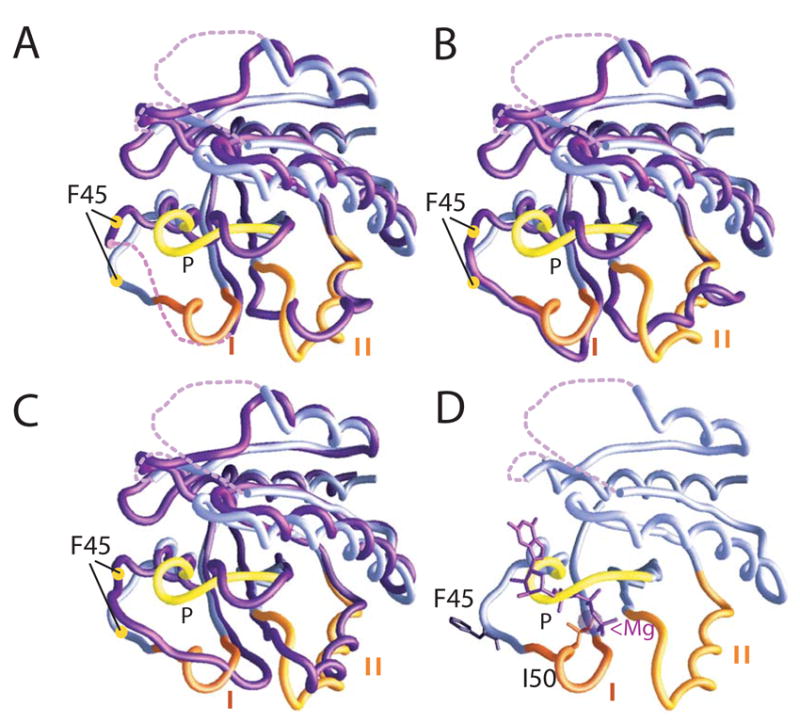
Superpositions of the Cα backbones of nucleotide/Mg++-free Sec4p from the Sec2p/Sec4p complex (lilac) with nucleotide bound forms of Sec4p (purple) (Stroupe & Brunger, 2000). In the nucleotide-free Sec4p from the complex, switches I and II and the P-loop are labeled, and it is superimposed with: (A) the GDP-bound form. Yellow circles indicate the Cα positions of Phe45. (B) an additional GDP-bound form of Sec4p. (C) the GppNHp-bound form. The conformation of switch II in nucleotide-free Sec4p from the complex more closely resembles that of the GppNHp-bound form than that of the GDP-bound form. (D) Nucleotide-free Sec4p with the positions of GppNHP and Mg++ derived from the GppNHp-bound Sec4p structure indicated. Phe45 and Ile50 are indicated.
Figure 4.
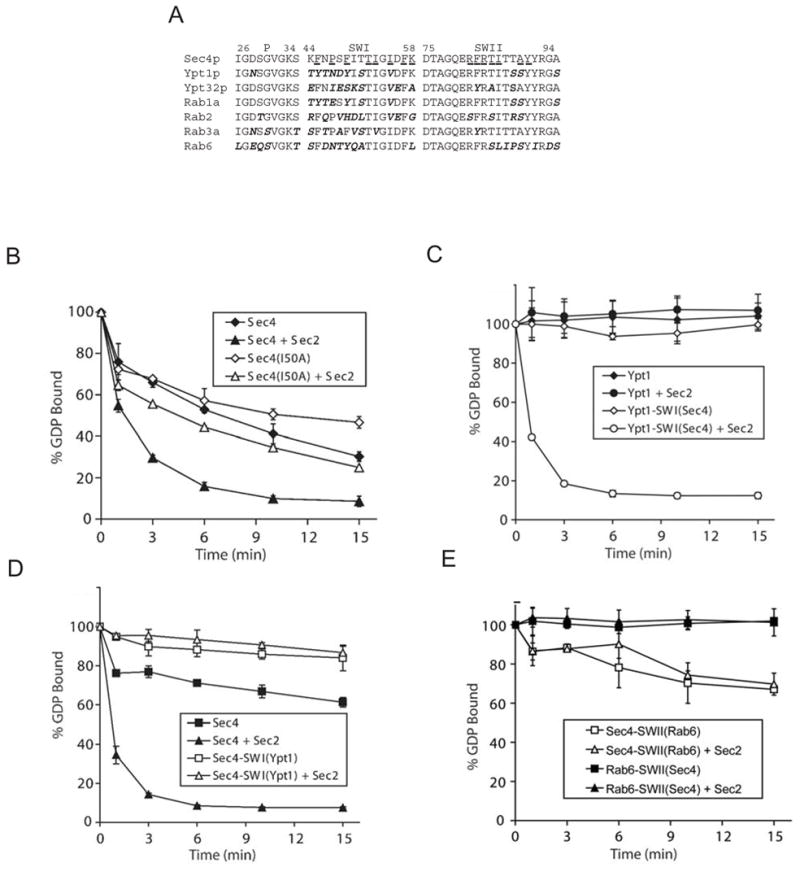
Importance of the Sec4p switch regions for GEF activity. (A) Sequences of the switch regions and P-loops of different Rabs. Sec4p residues within 4 Å of Sec2p in the complex are underlined. (B) Comparison of the release of [3H]GDP from 200 nM wild type Sec4p and from the Ile50Ala Sec4p mutant in the absence and presence of 100 nM Sec2p exchange domain. Sec2p catalyzed exchange is impaired for the mutant as compared to wild type Sec4p. Here Sec4p constructs are expressed as GST-fusion proteins. (C) Comparison of the release of [3H]GDP from 200 nM wild type Ypt1p and Ypt1p with switch I replaced by the Sec4p sequence, Ypt1-SWI(Sec4), in the absence or presence of 100 nM Sec2p exchange domain. While Sec2p does not act on Ypt1p, Ypt1-SWI(Sec4) is a substrate. (D) Comparison of the release of [3H]GDP from 200 nM wild type Sec4p and Sec4p with switch I replaced by the Ypt1p sequence, Sec4-SWI(Ypt1), in the absence or presence of 100 nM Sec2p exchange domain. Sec2p does not stimulate GDP exchange for Sec4-SWI(Ypt1). (E) Comparison of the release of [3H]GDP from 200 nM Sec4p with switch II replaced by the Rab6 sequence, Sec4p-SWII(Rab6), and Rab6 with a Sec4p switch Rab6-SWII(Sec4), in the absence or presence of 100 nM Sec2p exchange domain. Sec2p does not act on Sec4p-SWII(Rab6) or Rab6-SWII(Sec4). Sec4p and Rab6 constructs are expressed as GST-fusions. Error bars represent standard deviation.
Switch II (residues 76–93sc4) has been found in two different conformations in Sec4p/GDP complexes and a third in the Sec4p/GppNHp complex (Stroupe & Brunger, 2000). In the Sec2p/Sec4p complex, residues 82–90sc4 form a helix, stabilized by hydrogen bonds between the Sec2p Glu100 carboxylate and the amide nitrogens of Ala84sc4 and Ile85sc4. This switch II conformation more closely resembles that observed in the GppNHp-bound form of Sec4p. Residues 76–91sc4 are shifted in the nucleotide-free structure as compared with the GppNHp-bound form, however (Figure 3C).
Although residues 27–34sc4 in the P-loop do not interact with Sec2p directly, they must also reorganize in the complex to accommodate switch I and II changes (Figure 3). Otherwise Asp28sc4, Ser29sc4, Lys33sc4, and Ser34sc4 would clash with residues in switches I and II. The reorganized P-loop in turn would clash with the guanosine base, the ribose moiety, and all phosphates of the nucleotide in the GDP/GTP bound forms of Sec4p. Thus the P-loop largely fills the nucleotide binding site, preventing any binding interaction.
Most of Sec4p, excluding the regions that interact with Sec2p, is poorly ordered in the crystals, as indicated by the high thermal factors assigned during refinement (Supplemental Table 1 and Supplemental Figure 1). Likely Sec4p is free to move within a Sec2p crystal lattice since it is constrained by few crystal contacts, and no rigid structural elements connect the Sec2p-bound regions to the rest of Sec4p. Several regions of Sec4p, including residues 64–69sc4, 133–144sc4 and 164–167sc4, have no electron density at all. Residues 136sc4 and 162–163sc4 normally form hydrogen bonds with the guanosine base (Stroupe & Brunger, 2000) of the bound nucleotide, so that the disorder in and around these regions may result from loss of nucleotide interaction rather than any direct effects of Sec2p binding. The lack of order in regions surrounding the binding site, and possibly the overall lack of order in Sec4p (if it represents thermal motion on an appropriate time scale), may aid the initial entry of the guanine nucleotide and facilitate the extensive refolding of the switch and P-loop regions, as well as loops 133–144sc4 and 164–167sc4, that accompany GTP binding.
The mechanism for nucleotide exchange
Our structure and accompanying biochemical studies suggest that Sec2p binding results in an ordering of the switch I region, which is disordered in the GDP-bound form of Sec4p, and a change in the conformation in switch II. These changes result in removal of Phe45sc4, critical in stabilizing the nucleotide, from the binding pocket and a subsequent decrease in affinity for the nucleotide. Ile50sc4 in switch I is reoriented, likely leading to a loss of the magnesium and again contributing to a decrease in affinity of Sec4p for nucleotide. Finally, the reordering of the switch regions leads to a rearrangement of the P-loop, which adopts a position normally occupied by nucleotide, also leading to reduction in nucleotide affinity.
Thus, Sec2p catalyzes guanine nucleotide exchange by the same general strategy of all known GEFs, that of stabilizing the GTPase binding pocket in a conformation incompatible with nucleotide binding. The details of the exchange mechanisms, however, differ. The mechanism for Sec4p activation used by Sec2p varies most dramatically from that deduced from the Mss4/Rab8 structure. Mss4/Rab8 complex formation disorders the P-loop and adjacent residues, switch II, and additional loops of the Rab8 nucleotide binding pocket. This Mss4-induced GTPase unfolding was proposed as a key element in the nucleotide exchange mechanism (Itzen et al, 2006). Unfolding of the nucleotide binding pocket in the absence of guanine nucleotide may be a characteristic of the Rab GTPases as the nucleotide pocket is also partially disordered in the Sec2p/Sec4p complex. However, the unfolding is more extensive in Rab8 than in Sec4p, and loops in the Sec4p nucleotide binding pocket likely are disordered as a consequence of nucleotide removal rather than as the cause. Sec2p is further distinct from Mss4 and also most other GEFs (Boriack-Sjodin et al, 1997; Goldberg, 1998; Renault et al, 2001; Kawashima et al, 1996) in that the reconfiguration of the Sec4p guanine binding site does not involve the insertion of a portion of Sec2p into the binding site. In this respect Sec2p resembles the Dbl homology (DH) domains that catalyze nucleotide exchange for the Rho family GTPases (Worthylake et al, 2000; Snyder et al, 2002; Kristelly et al, 2004). The detailed mechanisms differ, however, since in the DH-Rho interaction alterations in the GTPase switch regions alone destabilize nucleotide binding, but the P-loop remains essentially undisturbed.
Specificity of Sec2p/Sec4p interactions
Our structure of the Sec2p/Sec4p complex indicates that determinants of Sec2p specificity for Sec4p likely reside in and around the two switch regions of Sec4p. Additionally, since the P-loop also appears to play a role in nucleotide displacement, its sequence may also be important for nucleotide exchange by Sec2p. Sec2p is specific for Sec4p as it does not stimulate GDP exchange for other members of the Rab family such as Ypt1p, Ypt32 (Ortiz et al, 2002), Rab1a, Rab2, Rab3a, or Rab6 (Figure 4C, D and data not shown). Since the Ypt1p and Sec4p sequences differ significantly in and around switch I and are almost identical in switch II and the P-loop (Figure 4A), our structure predicts that Sec2p discriminates between these two proteins via the switch I region. Consistent with these predictions, replacing Ypt1 residues around switch I by residues 44–55sc4 of Sec4p confers activation by Sec2p (Figure 4C). Further, a reciprocal construct, where residues 44–55sc4 of Sec4p have been replaced by their Ypt1p counterparts, is no longer a substrate for Sec2p (Figure 4D). Sec2p also failed to activate Sec4p constructs in which switch II residues had been replaced by corresponding residues from Rab6 (which differs in sequence over this region (Figure 4A)), illustrating the importance of switch II for recognition (Figure 4E). A Rab6 construct with the switch II sequence from Sec4p also is not a substrate for Sec2p (Figure 4E). This result is expected as the switch I and P-loop sequences of Rab6 and Sec4p differ significantly (Figure 4A). A construct in which the Sec4p P-loop was replaced by the Rab6 sequence failed to bind GDP efficiently, as determined in exchange assays, and could therefore not be tested for activation by Sec2p (data not shown).
Sec4p recognition via both switch regions contributes to a picture for Rab recognition gradually emerging from biochemical analyses and structural studies (Grosshans et al, 2006b). Perhaps not surprisingly, since effectors must discriminate between the GDP and GTP bound forms of Rabs, the Rab-effector interface commonly includes the two switches as well as additional regions (Ostermeier & Brunger, 1999; Zhu et al, 2004, Eathiraj et al, 2005). The Rab GEF Rabex-5 also requires sequences in both switch regions for recognition (Delprato et al, 2004). In contrast, Mss4, which is less specific and activates an array of Rabs, interacts with switch I, interswitch regions, and the N-terminus but not with switch II (Itzen et al, 2006).
Concluding Remarks
Our structure lends insight into mechanism by which a class of coiled-coil GEFs can distinguish between different Rab proteins to activate specific partners. Most remarkable is that a structure as simple as a coiled-coil can facilitate the same guanine nucleotide exchange process as GEFs with significantly more complex topologies. The simplicity of the Sec2p GEF domain scaffold raises the intriguing possibility of protein engineering studies directed at retargeting Sec2p to other Rabs, with possible applications in the manipulation of signal transduction pathways and/or disease therapies.
In vivo, the exchange activity of Sec2p is normally concentrated near its substrate Sec4p on the surface of the secretory vesicles, thereby facilitating the exchange reaction (Walch-Solimena et al, 1999). This localization is achieved through a Rab cascade mechanism in which Sec2p initially binds to the Golgi-associated rab Ypt32p in its GTP-bound state (Ortiz et al, 2002). Activation of Sec4p then leads to the recruitment of one of its downstream effectors Sec15p, a subunit of the exocyst tethering complex. Sec15p also binds to Sec2p, establishing a rab GEF-effector complex that may serve to maintain a metastable micro-domain of Sec4p-GTP bound to the exocyst. Both Ypt32-GTP and Sec15p bind to the region of Sec2p C-terminal to the exchange domain and compete against each other for binding (Medkova et al, 2006). To better understand the molecular mechanisms that regulate vesicle transport, it will also be important to analyze the structures of Sec2p complexed with either Ypt32-GTP or Sec15p.
Experimental Procedures
Cloning, Expression, and purification
The cloning, expression, and purification of Sec2p and Sec4p and the formation of the Sec2p/Sec4p complex are described in the Supplemental Methods. The cloning, expression, and purification of the Sec4p mutants (Phe45Ala, Ile50Ala), other Rab GTPases (Rab1a, Rab2, Rab3a, Rab6, Ypt1), and chimeras of Sec4p and either Ypt1 or Rab6 that were used in exchange assays are also described there. Some Rabs were expressed with N-terminal GST fusions. GST does not inhibit Sec2p mediated nucleotide exchange (Figure 4, compare B, D). GST tags also do not interfere with exchange mediated by the GEF rabex-5 (Delprato et al. 2004).
Crystallization, data collection, and structure determination
Crystals of the selenomethionine substituted Sec2p/Sec4p complex were grown at 22 °C by the hanging drop method against a well liquor containing 100 mM sodium acetate (pH 5.0), 200 mM calcium acetate, 20 % (v/v) PEG 400, and 10 mM DTT. For data collection, crystals were transferred over a period of ~20 minutes to solutions supplemented with increasing concentrations of glycerol and finally 20% (v/v) glycerol. The crystals were loop mounted and flash frozen by plunging into liquid nitrogen. They diffracted to 3.3 Å at beamline 24ID (NE-CAT) at the Advanced Photon Source, Argonne National Laboratory. The crystals belong to space group P6122, with unit cell dimensions of a = b = 93.00 Å and c = 295.91 Å.
An anomalous data set collected at the selenium edge was used in phasing. Data were integrated and scaled using the program HKL2000 (Otwinowski & Minor, 1997). Selenium sites were located using SHELXD (Schneider & Sheldrick, 2002) and refined with autoSHARP/Sushi (La Fortelle & Bricogne, 1997). The SAD phases together with solvent flattening as implemented in SHARP produced interpretable experimental maps at 3.3 Å. The electron density for Sec2p is very clear, but portions of Sec4p, in particular the helical regions that surround the central beta-sheet of Sec4p, are not as well defined (Supplemental Figure 1). The initial model was built in Coot (Emsley & Cowtan, 2004) and O (Kleywegt & Jones, 1997). Sec4p from the GppNHp bound structure (PDBid 1G17) was placed into the experimental map using the 6D phased rotation/translation program BRUTEPTF (Strokopytov & Almo, 2001). Portions of Sec4p without density in the experimental map were omitted. Residues in the switch regions and the P-loop, which were well defined and had shifted from their positions in the GppNHp-bound structure, were also omitted at this stage. We were guided by the position of selenomethionine side chains in aligning the sequence of Sec2p to the model of its polyalanine backbone. Improved maps were calculated by combining phases from a partial model (Sec2p and Sec4p, lacking the switch regions and P-loop) with experimental phases prior to density modification. The switch regions and the P-loop of Sec4p were rebuilt into the improved maps. The model was further improved by cycles of torsion angle dynamics and B-factor refinement, as implemented in the CNS software suite (Brunger et al, 1998), and manual rebuilding. No non-crystallographic symmetry restraints were applied to the two copies of Sec2p in the asymmetric unit since initial maps revealed significant differences between the two copies and test applications of symmetry restraints led to increased values of both R and Rfree. For the final model, R=25.0% and Rfree=32.5% for all data between 20–3.3 Å. It contains two copies of Sec2p (chain A: residues 17–162; chain B: residues 17–163) and one Sec4p molecule (chain C: residues 18–186, with three disordered loops spanning residues 64–68, 133–144, and 162–167). Side chains without density in 2Fo-Fc maps have been assigned an occupancy of 0. B-factors for Sec4p are high, likely reflecting that Sec4p has few crystal contacts beyond the complex interface. The final model has good geometry: 75.1% and 24.9% of residues are in the most favored and additionally allowed regions, respectively. Data and refinement statistics are reported in Supplemental Table 1.
GDP exchange assays
GDP displacement activity was monitored as described (Ortiz et al, 2002) with minor modifications. Rab proteins were preloaded with [3H]GDP (10 Ci/mmol) by incubation in buffer A (50 mM Tris, pH 8.0, 100 mM KCl, 1 mM EDTA, 1mg/ml BSA, 1 mM DTT) containing 2 μM [3H]GDP for 20 minutes at 30 °C. The MgCl2 concentration was then adjusted to 6 mM, and the proteins were incubated for an additional 45 minutes. Reactions were initiated by adding 30 μl of the [3H]GDP loaded Rab to an equal volume of Sec2p (residues 17–167) at 200 nM in buffer A with 1 mM GDP and 6 mM MgCl2. The rate of intrinsic GDP release was measured by omitting Sec2p from the reaction mixture. To lower the high intrinsic GDP release rate of Sec4p, experiments with Sec4p were conducted at 10 °C, as were experiments with Ypt1p, Sec4p/Ypt1p, and Sec4p/Rab6 chimeras. Experiments with other Rabs (Rab1a, Rab2, Rab3a, Rab6), which have lower intrinsic GDP release rates, were conducted at 30 °C. 10 μl aliquots were withdrawn at the times indicated in Figure 4 and placed into 1 ml of ice-cold stop buffer solution (25 mM Tris, pH 8.0, 20 mM MgCl2). Rab associated radioactivity was determined by filter binding followed by scintillation counting. All experiments were repeated at least three times.
Figures (except Figure 4) were prepared with the programs Molscript (Kraulis, 1991) and GRASP (Nicholls, 1993).
Supplementary Material
Acknowledgments
We thank C. Fu for help in purifying Rab6 and Sec4p/Rab6 chimeras. We are grateful to D.W. Rodgers and G. Warren for discussions regarding this manuscript. We thank the staff, especially R. Kanagalaghatta, at beamline ID-24 at the Advanced Photon Source for help with data collection. Numerous data sets not used in the final structure determination were also collected at beamlines X25 and X29 at Brookhaven National Lab, and we thank the staff for their help. G.D. has been supported by a fellowship from the American Heart Association. This work was also funded by grants from the G. Harold and Leila Y. Mathers Foundation, the Pew Charitable Trust (KMR), and the NIH (CA46128 to PN).
Footnotes
Accession numbers: Coordinates and data have been deposited in the Protein Data Bank with ID code 2OCY.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beard M, Satoh A, Shorter J, Warren G. A cryptic Rab1-binding site in the p115 tethering protein. J Biol Chem. 2005;280:25840–25848. doi: 10.1074/jbc.M503925200. [DOI] [PubMed] [Google Scholar]
- Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1997;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- Brunger AT, et al. Crystallography and NMR system (CNS): a new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:904–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Chardin P. GEFS: structural basis for their activation of small GTP-binding proteins. Trends Biochem Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- Collins RN, Brennwald P, Garrett M, Lauring A, Novick P. Interactions of nucleotide release factor Dss4p with Sec4p in the post-secretory pathway of yeast. J Biol Chem. 1997;272:18281–18289. doi: 10.1074/jbc.272.29.18281. [DOI] [PubMed] [Google Scholar]
- Delprato A, Merithew E, Lambright DG. Structure, exchange determinants, and family-wide Rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell. 2004;118:607–617. doi: 10.1016/j.cell.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Eathiraj S, Pan X, Ritacco C, Lambright DG. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature. 2005;436:415–419. doi: 10.1038/nature03798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Goldberg J. Structural basis for bctivation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- Grosshans BL, Andreeva A, Gangar A, Niessen S, Yates JR, 3rd, Brennwald P, Novick P. The yeast lgl family member Sro7p is an effector of the secretory Rab GTPase Sec4p. J Cell Biol. 2006;172:55–66. doi: 10.1083/jcb.200510016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. PNAS. 2006b;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattula K, Furuhjelm J, Arffman A, Peranen J. A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol Biol Cell. 2002;13:3268–3280. doi: 10.1091/mbc.E02-03-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzen A, Pylypenko O, Goody RS, Alexandrov K, Rak A. Nucleotide exchange via local protein unfolding—structure of Rab8 in complex with Mss4. EMBO J. 2006;25:1445–1455. doi: 10.1038/sj.emboj.7601044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzen A, Rak A, Goody RS. Sec2 is a highly efficient exchange factor for the Rab Protein Sec4. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.10.096. Doi 10/1016/j.jmb.2006.10.096. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Berthet-Colominas C, Wulff M, Cusack S, Leberman R. The Structure of the Escherichia coli EF-Tu. EF-Ts complex at 25 Å resolution. Nature. 1996;379:511–518. doi: 10.1038/379511a0. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Model building and refinement practice. Methods Enzymol. 1997;277:208–230. doi: 10.1016/s0076-6879(97)77013-7. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Kristelly R, Gao G, Tesmer JJG. Structural Determinants of RhoA Binding and Nucleotide Exchange in Leikemia-associated Rho guanine nucleotide exchange factor. J Biol Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- Luo HR, Saiardi A, Nagata E, Ye K, Yu H, Jung TS, Luo X, Jain S, Sawa A, Snyder SH. GRAB: a physiologic guanine nucleotide exchange factor for Rab3A, which interacts with inositol hexakisphosphate kinase. Neuron. 2001;31:439–51. doi: 10.1016/s0896-6273(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Gruber M. The structure of α-helical coiled coils. Advances in Protein Chemistry. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- Medkova M, France YE, Coleman J, Novick P. The rab exchange factor Sec2p reversibly associates with the exocyst. Mol Biol Cell. 2006;17:2757–2769. doi: 10.1091/mbc.E05-10-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A. GRASP: graphical representation and analysis of surface properties (computer program) Columbia University; New York: 1993. [Google Scholar]
- Nuoffer C, Wu SK, Dascher C, Balch WE. Mss4 does not function as an exchange factor for Rab in endoplasmic reticulum to Golgi transport. Mol Biol Cell. 1997;8:1305–1316. doi: 10.1091/mbc.8.7.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz D, Medkova M, Walch-Solimena C, Novick P. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J Cell Biol. 2002;157:1005–1015. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier C, Brunger AT. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3a complexed with the effector domain of Rabphilin-3A. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1) Cell. 2001;105:245–255. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Cryst. 2002;D58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Snyder JT, Worthylake DK, Rossman KL, Betts L, Pruitt WM, Siderovski DP, Der CJ, Sondek J. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat Struct Biol. 2002;9:468–475. doi: 10.1038/nsb796. [DOI] [PubMed] [Google Scholar]
- Strokopytov B, Almo S. American Crystallographic Association Annual Meeting; July 21–26, 2001; Los Angeles, CA. 2001. P218 (abstract) [Google Scholar]
- Stroupe C, Brunger AT. Crystal Structure of a Rab protein in its inactive and active conformation. J Mol Biol. 2002;304:585–598. doi: 10.1006/jmbi.2000.4236. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Collins RN, Novick PJ. Sec2 mediates nucleotide exchange on Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthylake DK, Rossman KL, Sondek J. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature. 2002;408:682–688. doi: 10.1038/35047014. [DOI] [PubMed] [Google Scholar]
- Zerial M, Mcbride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhu G, Zhai P, Liu J, Terzyan S, Li G, Zhang XC. Structural basis of Rab5-Rabaptin5 interaction in endocytosis. Nat Struct Mol Biol. 2004;11:975–983. doi: 10.1038/nsmb832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


