Summary
Yeast cells arrest in the G1 phase of the cell cycle upon exposure to mating pheromones. As cells commit to a new cycle, G1 CDK activity (Cln/CDK) inhibits signaling through the mating MAPK cascade. Here, we show that the target of this inhibition is Ste5, the MAPK cascade scaffold protein. Cln/CDK phosphorylates a cluster of sites flanking a small, basic membrane-binding motif in Ste5, thereby disrupting Ste5 membrane localization. Effective inhibition of Ste5 signaling requires multiple phosphorylation sites and a substantial accumulation of negative charge, suggesting that Ste5 acts as a sensor for high G1 CDK activity. Thus, Ste5 is an integration point for both external and internal signals. When Ste5 cannot be phosphorylated, pheromone triggers an aberrant arrest of cells outside G1, either in the presence or absence of the CDK inhibitor protein Far1. These findings define a mechanism and physiological benefit of restricting antiproliferative signaling to G1.
Introduction
Cellular decisions are commonly regulated by external signals via mitogen activated protein (MAP) kinase cascades (Qi and Elion, 2005). Though widely appreciated to stimulate cell proliferation, MAP kinase pathways can also regulate cell differentiation. Relatively little is known about how differentiation and antiproliferative signals may be integrated with, or counteracted by, the cell division status of individual cells. In yeast, mating pheromones activate a MAP kinase cascade to trigger fusion between two haploid gamete cells (Dohlman and Thorner, 2001). This mating reaction exhibits fundamental hallmarks of differentiation, in that cells exit the cell cycle, induce a unique program of gene expression, and undergo morphogenetic changes that allow them to adopt a new fate. Mating pheromones cause cells to arrest specifically in the G1 stage of the cell cycle, prior to the G1 → S transition step known as “Start”. However, cells that have already passed Start become refractory to pheromone arrest, a property that was used to define Start as a unique point of commitment to a new round of division (Hartwell et al., 1974).
One contributor to the G1 specificity of pheromone arrest involves mutual antagonism between cyclin-dependent kinases (CDKs) and Far1, a CDK inhibitor (Chang and Herskowitz, 1990). Pheromone signaling in G1 cells allows the MAPK Fus3 to phosphorylate and activate Far1 (Chang and Herskowitz, 1992; Breitkreutz et al., 2001), which inhibits CDK activity associated with G1 cyclin (Cln) proteins by an unresolved mechanism (Peter and Herskowitz, 1994; Gartner et al., 1998; Jeoung et al., 1998). Conversely, as cells pass Start, Cln/CDKs phosphorylate Far1, targeting it for degradation (McKinney et al., 1993; Henchoz et al., 1997). However, other mechanisms may play an equally critical role in restricting pheromone arrest to G1, but they are poorly understood. In particular, signal transduction through the MAP kinase cascade is actively inhibited by G1 CDKs, such that pheromone-induced transcription of mating genes (e.g., FUS1) is minimized during periods of maximum G1 cyclin expression (Oehlen and Cross, 1994; Wassmann and Ammerer, 1997). This results in a period from Start through S phase in which cells are unresponsive to pheromone.
While G1 CDK inhibition of pheromone signaling has been recognized for many years, the target and mechanism have remained elusive. Previous studies suggest that the inhibited step of the signaling pathway lay somewhere between the heterotrimeric G protein βγ dimer (Gβγ) and the first kinase of the MAP kinase cascade, the MAPKKK Ste11 (Wassmann and Ammerer, 1997; Oehlen and Cross, 1998). Indeed, Cln2/CDK can phosphorylate the PAK-family kinase Ste20 (Oehlen and Cross, 1998; Wu et al., 1998), but removing CDK sites in Ste20 had no effect on the ability of Cln/CDK to inhibit pheromone signaling (Oda et al., 1999), thus failing to confirm Ste20 as a relevant target. In the interim, our understanding of this signaling pathway has advanced considerably.
An important step in activation of the mating pathway (see Figure 1A) is the plasma membrane recruitment of the MAP kinase cascade scaffold protein, Ste5, by the pheromone-activated Gβγ dimer (Pryciak and Huntress, 1998; Mahanty et al., 1999; van Drogen et al., 2001; Winters et al., 2005). Indeed, artificial targeting of Ste5 to the plasma membrane causes constitutive signaling (Pryciak and Huntress, 1998). Membrane recruitment of Ste5 serves two roles: (1) it promotes activation of Ste11 by its membrane-localized activator, Ste20 (Pryciak and Huntress, 1998; van Drogen et al., 2000); and (2) it amplifies signal transmission from active Ste11 through the remainder of the kinase cascade (Lamson et al., 2006). Recently, we found that although Gβγ is the usual trigger for Ste5 recruitment, it is not sufficient. Instead, Ste5 also binds directly to membranes, and the cooperative effect of these two weak interactions (Ste5-Gβγ and Ste5-membrane) controls membrane recruitment (Winters et al., 2005). The Ste5-membrane interaction requires an N-terminal “PM” (plasma membrane) domain, a short basic-rich amphipathic α-helix that binds acidic phospholipid membranes, and which can also target Ste5 to the nucleus when not engaged at the plasma membrane. Gain-of-function mutations in the PM domain cause increased membrane affinity, allowing Ste5 to localize to the plasma membrane and activate signaling even without Gβγ (Winters et al., 2005).
Figure 1.

- Pheromone response pathway, showing membrane recruitment of Ste5.
- Methods for activating membrane-localized signaling. From left to right: α factor (αf) treatment or Gβ overexpression (Whiteway et al., 1990); hyperactive membrane localization of Ste5 via an enhanced PM domain (Winters et al., 2005); membrane targeting of Ste5 via a foreign transmembrane domain (Pryciak and Huntress, 1998); membrane targeting of Ste11 via a prenylation/palmitoylation motif (Winters et al., 2005).
- Cln2/CDK inhibition correlates with dependence on the Ste5 PM domain. Pathway-activating components were expressed from the GAL1 promoter and compared for their ability to induce FUS1-lacZ transcription in ste4Δ strains ± PGAL1-CLN2 (n = 4).
- Ste20-independent signaling is sensitive to Cln2 inhibition. Wild-type Ste11 (WT) or a Ste20-independent mutant (Ste11-Asp3) was expressed in ste11Δ or ste11Δ ste20Δ strains ± PGAL1-CLN2. FUS1-lacZ induction was measured after α factor treatment (n = 6).
- PM domain mutations that disrupt nuclear targeting (NLSm) do not affect Cln2 inhibition. FUS1-lacZ was induced by α factor treatment in ste5Δ ± PGAL1-CLN2 strains expressing Ste5-WT or Ste5-NLSm (n = 9).
- Increased Ste5 membrane affinity causes increased resistance to Cln2. Left, Ste5 variants contained PM domain mutations that increase membrane affinity. Right, the native PM domain was replaced with 1 or 2 copies of the PLCδ PH domain. All forms were expressed from the native STE5 promoter in ste5Δ strains ± PGAL1-CLN2, and response to α factor was measured (n = 4-9).
Data in all bar graphs show the mean ± SD.
In this study, we report that G1 CDK activity inhibits pheromone signaling by inhibiting Ste5 membrane recruitment. The Ste5 PM domain is flanked by multiple CDK sites that are phosphorylated by G1 CDKs in vivo and in vitro, and the addition of multiple negatively-charged phosphates impedes binding to acidic phospholipid membranes. Furthermore, we show that when CDK regulation of Ste5 is disrupted, pheromone signaling blocks cell cycle progression even after cells pass Start, and even in the absence of Far1, providing a physiological rationale for antagonizing pheromone signaling as cells begin a new division cycle.
Results
G1 CDKs inhibit the function of the Ste5 PM domain
To identify the target of Cln/CDK inhibition we used several new tools to dissect early steps in the mating pathway. First, we expressed activated forms of various pathway components (Figure 1B) from a strong, galactose-inducible promoter (PGAL1), to bypass upstream signaling steps. We then compared signaling with and without overexpression of Cln2, the G1 cyclin that is most potent at inhibiting pheromone response (Oehlen and Cross, 1994). While each activation method caused strong signaling, only those that require the Ste5 PM domain could be inhibited by Cln2 (Figure 1C). Especially revealing is the comparison between Ste5-Q59L and Ste5-CTM. Both activate signaling by targeting Ste5 to the plasma membrane, and both bypass Gβγ but still require Ste20 (Pryciak and Huntress, 1998; Winters et al., 2005). Yet they behaved oppositely with regard to Cln2 sensitivity: Ste5-Q59L (which is targeted to the membrane by an enhanced PM domain; Winters et al., 2005) was sensitive to Cln2 inhibition, whereas Ste5-CTM (which is targeted to the membrane by a foreign transmembrane domain; Pryciak and Huntress, 1998) was resistant. Furthermore, when comparing two Ste11 derivatives, we found that Ste11-Cpr (which requires the Ste5 PM domain; Winters et al., 2005) remained sensitive to Cln2 inhibition, whereas Ste11ΔN (which bypasses Ste5 altogether; Pryciak and Huntress, 1998) was resistant. These results argue that G1 CDK inhibition does not act on Gβγ, Ste20, Ste11, or even the ability of Ste5 to facilitate signaling through the MAP kinase cascade. Instead, signaling is inhibited only when membrane localization by the Ste5 PM domain is required.
In addition to promoting the initial Ste20 → Ste11 step, membrane localization of Ste5 serves a second, Ste20-independent role in boosting signal transmission from active Ste11 through the MAP kinase cascade (Lamson et al., 2006). Because of this “amplification” effect, pheromone can stimulate signaling in ste20Δ cells that harbor a pre-activated Ste11 mutant, Ste11-Asp3 (Lamson et al., 2006). Cln2 still inhibited this Ste20-independent response (Figure 1D), indicating that there must be a target other than Ste20. Notably, however, partial Cln2 resistance was observed. Yet because this was true for both the Ste20-independent signaling by Ste11-Asp3 (Figure 1D) and the Ste20-dependent signaling by Ste11-Cpr (Figure 1C), it did not reflect the participation of Ste20 per se. Rather, we suggest that Cln2 inhibition is strongest when it can antagonize two successive signaling steps that each rely on Ste5 membrane localization, and becomes incomplete when the first step is bypassed by pre-localization or pre-activation of Ste11.
Further experiments showed that Cln2/CDK inhibition of pheromone response could be reversed by strengthening the Ste5-membrane interaction. First, mutations in the PM domain (Winters et al., 2005) that enhance membrane binding (T52L, Q59L, or a T52L Q59L double mutant) reduced Cln2 inhibition (Figure 1F, left), from 89% (WT) to 34% (T52L Q59L). Second, by replacing the native Ste5 PM domain with foreign membrane-binding motifs (Winters et al., 2005), we found that signaling remained Cln2-sensitive when using a relatively weak motif, the PH domain from PLCδ, but became Cln2-resistant when using two tandem copies of this same motif (Figure 1F, right). The PM domain can also target Ste5 into the nucleus (Winters et al., 2005), but Cln2 sensitivity was not changed by a PM domain mutation (NLSm; Figure 1E) that specifically disrupts nuclear localization (Winters et al., 2005). Collectively, our results suggest that Cln/CDK antagonizes the ability of the Ste5 PM domain to mediate membrane-localized signaling.
Multiple CDK sites flanking the Ste5 PM domain regulate signaling
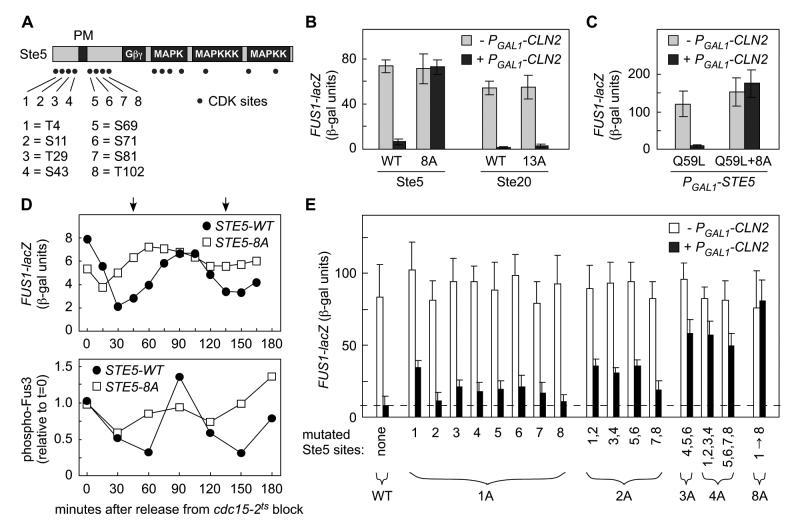
Of fifteen possible CDK sites (i.e., SP or TP) within Ste5, eight are concentrated around the PM domain (Figure 2A). This conspicuous clustering, coupled with results described above, suggested that phosphorylation at one or more of these sites might regulate Ste5 signaling. Indeed, small deletions on either side of the Ste5 PM domain conferred partial Cln2 resistance (Figure S1B). Therefore, we replaced the Ser or Thr residues at all eight SP/TP sites with nonphosphorylatable Ala residues. This “Ste5-8A” mutant remained fully capable of pheromone response but was now completely resistant to Cln2 inhibition (Figures 2B and S1C). This phenotype was specific to the Ste5-8A mutant, as a previously-described Ste20 mutant lacking 13 CDK sites, Ste20-13A (Oda et al., 1999), conferred no Cln2 resistance in parallel tests (Figure 2B). Furthermore, the Cln2 resistance displayed by Ste5-8A was separable from any possible effects on Ste5-Gβγ binding, because Gβγ-independent signaling by PGAL1-STE5-Q59L became resistant to Cln2 when Ste5 harbored the 8A mutations (Figure 2C).
Figure 2.
- Locations of potential CDK phosphorylation sites (SP or TP) in Ste5.
- Elimination of 8 N-terminal CDK sites in Ste5 (Ste5-8A) causes resistance to Cln2.
- Response to α factor was measured in ste5Δ ± PGAL1-CLN2 cells expressing Ste5 variants (WT or 8A) from the native STE5 promoter, or in ste20Δ ± PGAL1-CLN2 cells expressing Ste20 variants (WT or 13A) from the native STE20 promoter. Bars, mean ± SD (n = 8).
- CDK resistance caused by 8A mutations restores membrane signaling independent of Ste5-Gβγ interaction. Gβγ-independent signaling was activated by PGAL1-STE5-Q59L ± 8A in ste4Δ ste5Δ cells ± PGAL1-CLN2. Bars, mean ± SD (n = 7).
- The Ste5-8A mutant disrupts cell cycle periodicity of pheromone response. Cells (cdc15-2 or cdc15-2 STE5-8A) were synchronized in late M phase by arrest at 36°C, and then transferred to 25°C. At various times, response to brief treatment with α factor was monitored (see Experimental Procedures). Top, FUS1-lacZ induction (mean of 4 trials). Bottom, Fus3 activation (phospho-Fus3) was measured using phospho-specific antibodies (mean of 6 trials). Arrows mark the times of bud emergence (see Figures S1D and 5G).
- Ste5 phosphorylation sites were replaced with Ala residues either singly (1A) or in various combinations (2A, 3A, 4A, 8A). Response to α factor was tested in ste5Δ strains ± PGAL1-CLN2 (mean + SD, n = 8-16).
We also examined Ste5-8A signaling in synchronous cultures (Figure 2D). As cells progressed through the cell cycle, pheromone response was monitored by transcriptional induction of a FUS1-lacZ reporter and by phosphorylation of the MAPK Fus3. By either assay, Ste5-8A largely disrupted the normal cell cycle periodicity of pheromone response. Some fluctuation remained, especially at times immediately after release from cdc15 arrest, which could represent a minor effect of cell cycle position on other targets or a nonspecific effect of the temperature shift protocol. Overall, however, it is clear that the Ste5-8A mutant confers a very strong, if not complete, resistance to G1 CDK regulation of pheromone response.
To ask which of these eight Ste5 sites governs CDK regulation, we replaced individual Ser or Thr sites with Ala residues. Remarkably, none of the eight single (“1A“) mutants displayed the complete Cln2 resistance shown by Ste5-8A, and instead most conferred weak partial resistance (Figure 2E), with some variation in strength. By making combined mutants, we observed gradually increasing resistance to Cln2 as more sites were removed (Figure 2E), yet complete resistance was seen only when all eight sites were eliminated. Conversely, however, while Cln2 could inhibit signaling to a measurable degree when Ste5 retained 4 or 5 CDK sites, inhibition was much stronger when Ste5 retained 6, 7, or 8 CDK sites. Clearly then, no single site controls sensitivity to Cln2. Rather, multiple CDK sites are required to fully inhibit Ste5 signaling.
Inhibition is proportional to added negative charge
We hypothesized that the addition of anionic phosphates next to the basic-rich PM domain interferes with its electrostatic attraction to acidic phospholipid membranes. Although our findings hinted that full interference might require phosphorylation at multiple Ste5 sites, it was equally possible that multiple sites merely serve to increase the likelihood of phosphorylation at one or a few sites. To address these issues, we replaced the same Ser/Thr residues with negatively-charged Glu residues (Figure 3A). Placement of Glu residues at all 8 sites (Ste5-8E) did reduce pheromone response, but not as strongly as when Ste5-WT was inhibited by Cln2 (Figure 3B). To explain this partial effect, we reasoned that if electrostatic interference was the operative mechanism, then the net charge might dictate the level of inhibition. Because Ser/Thr phosphorylation introduces a charge of −2, phosphorylation at 8 sites would add a net charge of −16, whereas 8 Glu residues would add a net charge of only −8 and thus would be less inhibitory. To test this notion, we sought to better mimic the −2 charge of each phosphate by using two Glu residues, and so we replaced the SP or TP dipeptides at each of the 8 Ste5 sites with EE dipeptides (Figure 3A). Indeed, pheromone response by this “Ste5-16E” mutant was reduced to a level similar to when Ste5-WT was inhibited by Cln2 (Figure 3B). This strong effect required EE dipeptides at all 8 sites, because EE dipeptides at only 4 sites either before or after the PM domain (Ste5-up8E or Ste5-dn8E) caused only a partial reduction similar to when the eight Glu residues were distributed among all 8 sites (Ste5-8E), except that signaling could be further inhibited by Cln2 via the remaining 4 CDK sites.
Figure 3.
- Glu replacement mutations at CDK sites. S/T residues were replaced with E or A, and SP/TP dipeptides were replaced with EE or AA, as indicated.
- Inhibition of Ste5 signaling is proportional to added negative charge. Ste5 mutants were tested for α factor response in ste5Δ cells ± PGAL1-CLN2. Ste5 “up4A” and “dn4A” refer to Ala mutations at sites #1-4 and #5-8, respectively. Bars, FUS1-lacZ levels, relative to Ste5-WT (mean ± SD, n = 6).
- Anti-myc blot showing levels of Ste5-myc13 mutants expressed in ste5Δ cells.
- The Ste5-16E mutant can still bind Ste4. Extracts of ste4Δ ste5Δ cells coexpressing Ste5-myc and GFP-Ste4 (after 3 hr induction of PGAL1-GFP-STE4) were analyzed by immunoprecipitation (IP) and immunoblotting (blot) as indicated. Ste5-C180A served as a control that is defective at binding Ste4 (Feng et al., 1998).
- Ste5 Glu mutants are competent to mediate basal signaling (i.e., no α factor) activated by Ste11-4 in ste4Δ ste5Δ ste20Δ cells. Bars, mean ± SD (n = 4).
- The 16E mutations only inhibit signaling that requires the Ste5 PM domain. FUS1-lacZ (mean ± SD, n = 3-6) was induced in ste4Δ ste5Δ cells (without α factor) by PGAL1-driven expression of Ste5, Ste5-Q59L, or Ste5-CTM, each of which either contained the 16E mutations (+16E) or did not (−16E). Anti-GFP blots confirmed that protein levels were unaffected by the 16E mutations (data not shown).
- Glu mutants disrupt Gβγ-independent, membrane-localized signaling. Signaling (mean ± SD, n = 6) was activated in ste4Δ ste5Δ cells (without α factor) by coexpression of PGAL1-STE11-Cpr with the indicated Ste5 derivatives.
- Ste5 derivatives containing various Ala or Glu mutations were coexpressed in ste4Δ ste5Δ cells with either Ste4-WT or Ste4 mutants (K55E or N157H S175P) that weaken Ste5 binding (Leeuw et al., 1998; Winters et al., 2005). Response to α factor was measured (mean ± SD, n=6).
Several observations suggest that the strong signaling deficit of the Ste5-16E mutant reflects a specific effect on the Ste5 PM domain and not a complete inactivation of Ste5. First, the Ste5-16E mutant showed normal protein levels (Figure 3C), and it still bound Ste4 (Gβ) (Figure 3D). Second, the Ste5-16E mutant could still mediate basal signaling from an activated Ste11 derivative, Ste11-4 (Figure 3E), which does not require the Ste5 PM domain (Mahanty et al., 1999; Winters et al., 2005). Third, as with inhibition by Cln2 (see Figure 1C), the 16E mutation inhibited Ste5-Q59L but not Ste5-CTM (Figure 3F) and therefore only blocks membrane signaling that requires the Ste5 PM domain. A control mutant with AA dipeptides at all 8 sites was not informative because it was poorly expressed and completely defective in all assays, which we traced to the fact that an AA dipeptide was not tolerated at site #3 or #4 (data not shown). By allowing sites #3 and #4 to harbor only single replacements (either A or E) and replacing the six other sites with AA or EE dipeptides, generating “Ste5-14A” and “Ste5-14E”, we found that Ste5-14A remained functional (and fully resistant to Cln2), whereas Ste5-14E was strongly inhibited (Figure 3B). Therefore, the EE phenotypes reflect the addition of charge rather than the multiplicity of mutations.
As with inhibition by Cln2, the Glu replacements affected membrane-based signaling in general, rather than Gβγ-triggered signaling in particular, because they also disrupted Gβγ-independent signaling by Ste11-Cpr (Figure 3G). Again, the degree of inhibition was proportional to the added negative charge. Notably, Ste5-14E retained more function than Ste5-16E, suggesting that 16 negative charges are more inhibitory than 14, which agrees with our finding that removing single CDK sites confers partial Cln2 resistance. Because the Ste5 PM domain normally acts cooperatively with Ste5-Gβγ binding (Winters et al., 2005), we predicted that the inhibitory effect of a small number of negative charges would be enhanced when Ste5-Gβγ affinity is reduced. Indeed, when using Ste4 (Gβ) mutants with reduced binding to Ste5, we could now detect inhibition by 1 to 4 added Glu residues (Figure 3H). Thus, a small number of charges has measurable inhibitory potential, but multiple charges are necessary when the interactions governing Ste5 membrane recruitment occur with normal affinity. Collectively, the Glu replacement phenotypes show that added negative charges disrupt membrane signaling mediated by the Ste5 PM domain, and that the requirement for multiple CDK sites truly reflects a need for adding multiple phosphates.
G1 CDK activity disrupts Ste5 membrane localization
Using a Ste5-GFP×3 fusion expressed at native levels (Winters et al., 2005), we found that PGAL1-CLN2 inhibited pheromone-induced membrane recruitment of Ste5-WT, but not Ste5-8A (Figure S2A). We also examined membrane localization mediated specifically by the Ste5 PM domain, in isolation from upstream factors (e.g., pheromone, Gβγ) or downstream signaling consequences (e.g., transcriptional induction, cell cycle arrest), by using the hyperactive Ste5 variant Ste5-Q59L (Winters et al., 2005). Expression of Cln2 displaced Ste5-Q59L from the plasma membrane (Figure 4A), and this effect was blocked by the 8A mutations (Ste5-Q59L+8A). Results were similar in strains lacking Ste20 or the MAPKs Fus3 and Kss1 (Figure S2), ruling out any contribution from Cln2/CDK phosphorylation of Ste20 (Oehlen and Cross, 1998; Wu et al., 1998) or MAPK phosphorylation of Ste5 (Kranz et al., 1994; Flotho et al., 2004; Bhattacharyya et al., 2006). Because these experiments were performed in non-signaling strains, the effect of Cln2 was not due to a shift from G1-arrested cells to cycling cells. Thus, Cln2/CDK actively inhibits Ste5 membrane localization mediated by the PM domain.
Figure 4.

- Localization of GFP-Ste5-Q59L ± 8A, expressed from the GAL1 promoter in ste4Δ ste7Δ cells ± PGAL1-CLN2. Note that hyperpolarized bud growth is due to Cln2 overexpression, not mating signaling. Also see Figure S2B.
- The 16E mutations disrupt Ste5 membrane localization mediated by the PM domain (Q59L), but not that mediated by a foreign transmembrane domain (CTM). Top, membrane localization induces mating pathway signaling, causing pear-shaped “shmoo” morphology. Bottom, localization results were similar in a non-signaling strain (ste4Δ ste7Δ).
- Negative charge disrupts membrane localization of Ste5 N-terminal fragments. Localization was compared (in ste4Δ ste7Δ cells) for WT and mutant derivatives of GFP-Ste5(1-214) and GST-GFP-Ste5(1-125), which can localize to the membrane in the absence of pheromone, Gβγ, and other Ste5 sequences (Winters et al., 2005).
To address whether these localization effects were due to added negative charge, we examined the Glu replacement mutations. In agreement with their signaling phenotypes (see Figure 3F), the 16E mutation disrupted membrane localization of Ste5-Q59L, which depends on the PM domain, but not Ste5-CTM, which is independent of the PM domain (Figure 4B). Furthermore, displacement of Ste5 can be ascribed to a local effect on the PM domain (rather than more global changes in Ste5), because Glu mutations blocked membrane localization of N-terminal Ste5 fragments (Figure 4C). When membrane binding by an N-terminal fragment was enhanced using the Q59L mutation (Figure 4C, bottom), displacing it required greater negative charge (i.e., 16E, rather than 8E), suggesting that competition between attractive and repulsive interactions determines the net membrane affinity. Collectively, these results indicate that phosphorylation near the Ste5 PM domain disrupts plasma membrane binding.
Cln2-dependent phosphorylation of Ste5
To test if Cln2/CDK directly phosphorylates Ste5, we performed in vitro kinase assays. As a substrate we used a purified Ste5 fragment (residues 1-125) encompassing the PM domain and all eight N-terminal CDK sites. Purified Cln2/Cdc28 phosphorylated the wild type Ste5 fragment, and this was severely reduced by the 8A or 8E mutations (Figure 5A). Thus, the N-terminus of Ste5 can serve as a direct Cln2/CDK substrate, and the CDK sites are required for phosphorylation.
Figure 5.
- Phosphorylation of the Ste5 N-terminus by Cln2/Cdc28 in vitro. Bacterially-expressed GST-Ste51-125 fusions (WT, 8A, and 8E) were phosphorylated by recombinant Cln2-Cdc28. Histone H1 served as a control substrate.
- Cln2 expression in vivo alters Ste5 electrophoretic mobility. HA-tagged Ste5 (WT, 8A, and 16E) was immunoprecipitated from the indicated strains after 3 hr galactose induction (to express Cln2), resolved by SDS-PAGE, and analyzed by anti-HA immunoblot.
- The Ste5 mobility shift is due to phosphorylation. Ste5-HA3 was immunoprecipitated from ste5Δ ± PGAL1-CLN2 strains, and treated with calf intestinal phosphatase (CIP).
- Effects on Ste5 mobility are specific to Cln2. Ste5-HA3 derivatives (WT, 8A, ΔNLS, ΔNLS+8A) were analyzed as in panel B, using various PGAL1-cyclin strains.
- Ste5ΔNLS is more fully modified by Cln2. Ste5-HA3 derivatives were analyzed as in panel B.
- Ste5ΔNLS modification is elevated after Start and requires Cln1/2. WT and mutant strains (“cln1,2” = cln1 cln2) expressing Ste5-HA3 (ΔNLS or ΔNLS+8A) were incubated for 3 hr at 37°C.
- Modification of the Ste5 N-terminus is cell cycle dependent. Cells (cdc15-2 or cdc28-13) harboring Ste5-HA3 (ΔNLS or ΔNLS+8A) were arrested at 37°C for 3 hr, then transferred to 25°C to resume cycling. Samples were collected at 20 min intervals (0-180 min). As cdc15 cells arrest with large buds, emergence of small buds was used to follow cell cycle progression (c.f., Figure S1D).
In panels B-G, Ste5-HA3 derivatives were expressed from the native STE5 promoter.
In vivo, Cln2 overexpression promoted phosphorylation of Ste5, as evidenced by reduced electrophoretic mobility (Figure 5B) that was reversed by subsequent phosphatase treatment (Figure 5C). This agrees with a previous report that the phosphorylation status of Ste5 in vivo depends on Cdc28 (Flotho et al., 2004). The Ste5 mobility shift was consistent with multiply phosphorylated forms, as it was comparable to the Ste5-16E mutant, which mimics phosphorylation at all 8 N-terminal sites (Figure 5B). Furthermore, the effect of Cln2 on Ste5 mobility required the N-terminal CDK sites, because Ste5-8A and Ste5-16E were unaltered by Cln2 (Figure 5B). Results were similar in strains lacking Fus3 and Kss1 (Figure 5B, bottom), ruling out any contribution from Ste5-affiliated MAPKs (which can also phosphorylate SP/TP motifs). Unlike Cln2, overexpression of other cyclins (i.e., Cln3, Clb5, or Clb2) did not alter Ste5 mobility (Figure 5D), which agrees with their inability to inhibit pheromone signaling (Oehlen and Cross, 1994; Oehlen et al., 1998). Thus, Ste5 is a specific substrate of Cln2/CDK activity in vivo, and the relevant phosphorylation sites correspond to those that allow Cln2 to regulate Ste5 signaling.
Next, we wished to follow cell cycle-dependent changes in Ste5 phosphorylation, but our efforts were hindered by the low fraction of Ste5 showing a clear mobility shift. While a technical issue could be partly to blame (e.g., phosphatase activity in cell lysates), we wondered if Ste5 molecules in different subcellular locales might be modified to different extents. Indeed, when Ste5 was restricted to the cytoplasm by using the “ΔNLS” allele (Winters et al., 2005), Cln2 expression could modify nearly all Ste5 molecules (Figures 5D, 5E). In other respects phosphorylation of Ste5ΔNLS resembled that of wild-type Ste5, including a specific requirement for Cln2 and for the 8 N-terminal CDK sites (Figures 5D, 5E). Exploiting these favorable detection properties, we saw that Ste5ΔNLS phosphorylation was elevated in cells arrested immediately after Start (cdc4, cdc53, or cdc34), but not when Cln1/2 were absent (cdc34 cln1 cln2) or in cells arrested in G1 (cdc28-4, cdc28-13) (Figure 5F). Furthermore, in synchronous cultures, phosphorylation of Ste5ΔNLS fluctuated during the cell cycle, peaking at the onset of budding (Figure 5G). Thus, modification of the N-terminal CDK sites in Ste5 occurs as cells pass Start.
CDK-resistant signaling causes aberrant cell cycle arrest
To ask why it is beneficial to inhibit pheromone signaling as cells pass Start, we explored the physiological consequences of CDK-resistant Ste5 mutants. First, we examined the G1 specificity of pheromone arrest. Unlike wild-type cells, which arrested uniformly in G1, a significant fraction of STE5-8A cells (∼15%) arrested at a post-Start stage with 2N DNA content (Figure 6A). The Ste5-8A phenotype was dominant to wild type, and an increasing fraction of 2N arrest was observed as more CDK sites were removed from Ste5 (Figure 6B), suggesting that it reflects ectopic (non-G1) signaling by CDK-resistant Ste5 (rather than a leaky G1 arrest). Indeed, the post-Start arrest phenotype required the STE5-8A cells to be cycling at the time of pheromone addition, and was not observed when pheromone was added to a uniform population of G1 cells (Figures 6C and S3). Also, pheromone treatment of cycling STE5-8A cells induced a unique morphology in which mating projections appeared to emanate from cell buds (Figures 6C and S4), suggesting that signaling responses occurred during the budding phase of the cell cycle. Remarkably, elimination of only one or two CDK sites in Ste5 caused a measurable increase in 2N arrest (Figure 6B), showing that full inhibition of Ste5 via multiple CDK sites serves a physiologically important function. Accordingly, expression of Ste5-8E, which mimics the addition of 4 phosphates but cannot be inhibited further by Cln2/CDK, also allowed a substantial level of 2N arrest. We conclude that a failure of G1 CDKs to downregulate the mating MAP kinase pathway is detrimental, as it allows pheromone to arrest cells at an inappropriate cell cycle stage.
Figure 6.
- Pheromone arrests some STE5-8A cells at a post-Start (2N) stage. Cells were treated with α factor for the indicated times. FACS profiles show DNA content.
- The post-Start arrest phenotype is dominant and reflects the level of CDK-resistance of Ste5. Wild type cells harboring STE5 plasmids (1A#5 = site #5; 1A#1 = site #1; 2A = sites #5-6) were treated with α factor for 3.25 hrs. The percent of cells with 2N DNA content was quantified by FACS (mean ± SD; n = 4). The dashed line marks the %2N value observed when the STE5-WT plasmid is present in wild-type cells.
- STE5-8A allows near-uniform 2N arrest when G1 arrest is bypassed using PGPD1-CLB5. Left, DNA content of cells after 3 hr ± α factor. Right, halo assays showing growth arrest by α factor.
- Halo assays showing that STE5-8A restores pheromone arrest to far1Δ cells.
- Suppression of the far1Δ arrest defect increases as more CDK sites are eliminated from Ste5. Fivefold serial dilutions of strains harboring STE5 plasmids (1A = site #5; 2A = sites #5-6; 4A = sites #1-4) were incubated on –Ura plates ± 1 μM α factor.
- Pathway activation by CDK-resistant constructs causes Far1-independent arrest. Deletion strains harboring PGAL1-regulated activators of the mating pathway were grown on –Ura glucose or galactose plates.
- The post-Start arrest triggered by Ste5-8A signaling is independent of Far1. The indicated strains were analyzed in parallel with those in panel D.
Because STE5-8A cells can arrest at either G1 or post-Start stages, the percentage that arrest at the latter stage (usually ∼15% 2N) likely reflects the fraction of cells in the asynchronous culture that were between the two arrest points. Indeed, when pheromone was added to STE5-8A cultures at different times after leaving G1, the level of 2N arrest roughly correlated with the fraction of the initial cell population that were in S phase (Figure S3). Hence, cells outside this susceptible window likely arrest in G1. Consistent with this view, the majority of STE5-8A cells could be trapped at the 2N stage when G1 arrest mechanisms were bypassed. Specifically, overexpression of the B-type cyclin Clb5 can push cells through Start even in the presence of pheromone, thereby making wild-type cells pheromone-resistant (Oehlen et al., 1998, and references therein). In STE5-8A cells, however, Clb5 overexpression could still push cells through Start but the cells arrested in response to pheromone, and did so almost entirely at the 2N stage (Figure 6D). Therefore, signaling by CDK-resistant Ste5 has a dangerous potential to disrupt events during a post-Start window of the cell cycle.
CDK-resistant Ste5 permits Far1-independent arrest
To further explore the consequences of CDK-resistant signaling, we tested the role of Far1, which is ordinarily required for pheromone-induced arrest (Chang and Herskowitz, 1990). Strikingly, the CDK-resistant Ste5-8A mutant restored pheromone arrest to far1Δ cells (Figure 6E). Removal of as few as one or two CDK sites in Ste5 allowed significant suppression of the far1Δ arrest defect, with stronger suppression as more sites were removed (Figures 6F and S5A). Thus, Far1 becomes dispensable when Ste5 signaling cannot be inhibited. Indeed, far1Δ cells could be arrested (Figure 6G) by activating the mating pathway with CDK-resistant constructs (PGAL1-STE5-Q59L+8A, PGAL1-STE5-CTM), but not with CDK-sensitive constructs (PGAL1-STE4, PGAL1-STE5-Q59L). Far1-independent arrest has been observed in previous studies but remains poorly understood (Chang and Herskowitz, 1990; Valdivieso et al., 1993; Tyers, 1996; Oehlen et al., 1998; Cherkasova et al., 1999). Our results indicate that pheromone signaling is capable of robust growth arrest without Far1, but this is masked in far1Δ cells because the absence of Far1 allows Cln/CDK to downregulate Ste5.
Pheromone signaling in far1Δ STE5-8A cells caused arrest at more than one stage, because treated cultures showed a heterogeneous mix of 1N and 2N cells (Figure 6H and S5B) as well as unbudded and budded cells (data not shown), despite an immediate cessation of proliferation (Figure S5C). The G1 arrest appeared somewhat leaky and could be counteracted by G1 CDK activity (Figure S5B). Most notably, Far1 proved entirely dispensable for the post-Start arrest, because the near-uniform 2N arrest seen in STE5-8A cells overexpressing Clb5 (Figure 6D) was independent of Far1 (Figure 6H). Altogether, our results indicate that CDK-resistant signaling by Ste5-8A can impede cell cycle progression of both G1 and post-Start cells in a Far1-independent manner. Therefore, cell cycle control of Far1 (McKinney et al., 1993; Henchoz et al., 1997) is not sufficient for cells to escape the arrest effects of pheromone, and instead downregulation of Ste5 is also critical.
Discussion
Mechanism for cell cycle regulation of MAP kinase cascade signaling
In this study we define a mechanism by which G1 CDKs inhibit signaling through the yeast mating MAP kinase cascade. We show that the MAP kinase cascade scaffold protein Ste5 is the target of this inhibition, and that G1 CDK activity inhibits signaling by phosphorylating sites flanking a membrane-binding domain in Ste5. Our findings support a model in which these negatively-charged phosphates disrupt Ste5 membrane association by electrostatic interference (Figure 7A). Hence, through the use of two weak interactions that cooperatively control its membrane recruitment, Ste5 serves as an integration point for both external and internal regulatory cues (Figure 7B), such that signaling is activated only when two conditions are satisfied—i.e., when pheromone is present and the cell cycle stage is appropriate. The physiological benefit of this arrangement is that it restricts pheromone arrest to G1, thus preventing inappropriate disruption of cell cycle progression in cells that have passed Start.
Figure 7.
- As cells pass Start, G1 CDK activity inhibits pheromone signaling by phosphorylating CDK sites flanking the PM domain in Ste5. The negatively-charged phosphates interfere with binding between the basic PM domain and the anionic phospholipid membrane.
- Ste5 serves as an integration point for both external and internal regulatory cues, which act through the Gβγ-binding domain and the membrane-binding domain.
- Far1 promotes pheromone arrest by inhibiting Cln/CDK activity, but Far1-independent arrest pathways also exist and are revealed when Ste5 signaling cannot be downregulated by Cln/CDK activity.
The regulatory CDK sites in Ste5 lie in sequences flanking the PM domain that are dispensable for its normal signaling role (Winters et al., 2005; see also Figure S1B), and are predicted to be mostly random coil (Figure S6). Hence, rather than affecting a specific tertiary structure, the phosphorylated N-terminus of Ste5 may behave as an unstructured electronegative mass, making juxtaposition to its target energetically unfavorable. This mode of regulation may be generally applicable where phosphorylation serves to disrupt interactions. Phosphorylation of Ste5 provides a variation of “electrostatic switch” mechanisms seen in other signaling proteins such as Src or MARCKS, where membrane interactions are disrupted by phosphorylation within a membrane-binding domain (McLaughlin and Aderem, 1995). In Ste5, the use of sites distal to the membrane-binding domain may impose a requirement for multiple phosphorylations, which is likely to be advantageous to the regulatory circuit (see below).
Multisite phosphorylation as a sensor for high CDK activity
Inhibition of Ste5 signaling requires phosphorylation at multiple CDK sites, and maximal inhibition of Ste5 is required to avoid aberrant arrest. This behavior suggests that multisite phosphorylation of Ste5 serves as a sensor for high G1 CDK activity, so that signaling is not fully inhibited until CDK activity is high enough to promote cell cycle entry. Conceptually, this tactic is similar to that used by the B-type cyclin/CDK inhibitor protein Sic1, which must be phosphorylated on at least six CDK sites to trigger its degradation and the resultant progression into S and M phases (Nash et al., 2001). In each example, multisite phosphorylation likely enables a sharp phenotypic switch between distinct cell cycle stages. Unlike Sic1, we saw no indication that CDK phosphorylation (or added Glu residues) affects Ste5 protein stability. Moreover, CDK effects on Ste5 may be rapidly reversible, because chemical inactivation of Cdc28 can immediately restore pheromone response to post-Start cells (Colman-Lerner et al., 2005). Thus, robust dephosphorylation by cellular phosphatases may also help enforce a demand for high G1 CDK levels to inactivate Ste5 signaling.
Cell cycle arrest by pheromone
Cell cycle control of Ste5 signaling is necessary to avoid an aberrant pheromone arrest. The best-known mediator of pheromone arrest is the CDK inhibitor Far1, yet for years it has been evident that Far1-independent arrest pathways must exist, because Far1 is dispensible in cells lacking Cln2 (Chang and Herskowitz, 1990) or all 3 G1 cyclins (Tyers, 1996; Oehlen et al., 1998). Our results show that MAPK signaling can arrest growth without Far1, but this is ordinarily masked in far1Δ cells because Ste5 gets inactivated by G1 CDKs (Figure 7C). Hence, when Ste5 cannot be inhibited, the Far1-independent arrest is revealed. This can explain the preferential ability of cln2Δ to suppress far1Δ (Chang and Herskowitz, 1990), because Cln2 is the strongest of the 3 G1 cyclins at inhibiting pheromone response (Oehlen and Cross, 1994). Therefore, although cells eliminate Far1 as they pass Start (McKinney et al., 1993; Henchoz et al., 1997), this regulation is essentially futile without also downregulating Ste5 signaling.
The mechanisms by which CDK-resistant Ste5 signaling can impede cell cycle progression are largely unknown. The ability of Ste5-8A to trigger a G1 arrest (or delay) without Far1 is consistent with reports that pheromone can repress G1/S transcription (Valdivieso et al., 1993; Cherkasova et al., 1999). This repression may normally act in conjunction with Far1 to promote a robust, stable G1 arrest, yet may still allow a weak G1 arrest in far1Δ cells. The post-Start arrest is more enigmatic. A similar phenotype was seen in cells lacking G1 cyclins (Oehlen et al., 1998) or expressing a stable form of Far1 (McKinney and Cross, 1995). In light of our findings, these prior cases may result from disrupted Cln/CDK regulation of Ste5. The window of susceptibility to post-Start arrest roughly overlaps S phase, consistent with the period in which pheromone signaling is normally downregulated (Oehlen and Cross, 1994), and preliminary work suggests that the 2N cells cannot enter mitosis (S.C.S. and P.M.P., unpublished observations), but the molecular cause remains unknown. Possibly, signaling events that ordinarily help promote G1 arrest (e.g., CDK inhibition, transcriptional repression) can also block later cell cycle steps if signaling is unabated. Or MAPK signaling during S phase might induce DNA damage or replication errors, triggering a checkpoint arrest. Alternatively, physiological changes induced by the mating pathway (e.g., cytoskeletal rearrangements) may clash with cell division due to biochemical incompatibility or competition. In any scenario, downregulation of Ste5 would allow cells to properly commit to a new division cycle, by eliminating impediments to cell cycle progression.
Coordinating signaling with cell cycle stage
The discovery that cells sharply alter their sensitivity to extrinsic stimuli upon commitment to division played an important role in the formulation of early models for the cell cycle (Hartwell et al., 1974). Sharp transitions between distinct cell cycle stages can be ensured by feedback loops (Brandman et al., 2005). In the pheromone response pathway, mutual reinforcement between Ste5 and Far1 (in which Ste5-dependent signaling activates Far1, and Far1 blocks inactivation of Ste5) establishes a positive feedback loop. Conversely, Cln/CDK inhibition of both proteins can facilitate a decisive switch between conflicting states (i.e., arrest vs. proliferation).
During animal development, control of cell fate by external signals often occurs in the context of carefully orchestrated patterns of cell division (Vidwans and Su, 2001). Thus, it may be generally important to coordinate the response to differentiation signals with cell division status. While it is common for MAP kinase cascades to regulate the cell cycle, the reciprocal regulation is less well appreciated, yet its utility is clearly demonstrated by the behavior of the pheromone response pathway. Such mutual antagonism can also occur in pathways not involving MAP kinases, as in the TGFβ pathway where antiproliferative signaling by Smad3 is inhibited by G1 CDKs (Matsuura et al., 2004). Thus, while the mechanisms may vary, we expect that the beneficial role of coordinating signaling with cell cycle stage will be shared by other antiproliferative pathways.
Experimental Procedures
Strains and Plasmids
Yeast strains are listed in Table S1. Information on strain construction can be found in the Supplemental Experimental Procedures. Plasmids are listed in Table S2.
Signaling Assays
FUS1-lacZ induction and β-galactosidase assays were performed as described (Pryciak and Huntress, 1998; Lamson et al., 2002). Activation of FUS1-lacZ by galactose-inducible constructs in the presence and absence of PGAL1-CLN2 was measured 3 hrs after addition of 2% galactose to cultures grown in 2% raffinose media. To measure effects of PGAL1-CLN2 on FUS1-lacZ induction by pheromone, cells were grown in 2% raffinose media, then induced with 2% galactose for 1 hr, followed by 5 μM α factor for an additional 2 hr. For FUS1-lacZ experiments not involving PGAL1-CLN2, cells grown in glucose media were treated with 5 μM α factor for 2 hr.
For signaling in synchronous cultures, cdc15 strains AA2596 and PPY1761 harbored either pPP1044 (FUS1-lacZ) or pPP1513 (Fus3-myc13). Cultures in selective media were diluted into YPD and grown overnight at 25° C, shifted to 36° C for 3 hrs, then pelleted and resuspended in YPD at 25° C to release the mitotic block. To measure FUS1-lacZ induction, every 15 min aliquots were treated with 10 μM α factor for 22 min. Induction was stopped by transfer to an ice water bath. To measure Fus3 activation, every 30 min aliquots were treated with 5 μM α factor for 8 min. Cells were then pelleted, frozen in dry ice, and stored at −80° C. Fus3-myc13 was immunoprecipitated from cell extracts, and phospho-Fus3 was detected by rabbit anti-phospho-p44/42 blots (#9101; Cell Signaling Technology), quantified by densitometry, normalized to Fus3-myc13 levels measured in separate anti-myc blots, and expressed relative to time 0 for each experiment.
Halo assays (Lamson et al., 2002) used 20 μl of 1 mM (Figures 6E, S5A), 200 μM (Figure S5A), 100 μM (Figures 6D, 6H, S1C, S5B) or 20 μM (Figure S1C) α factor. Unless indicated otherwise, growth arrest was monitored after incubation for 2 days at 30°C.
FACS analysis
For FACS experiments, bar1Δ strains were treated with 1 μM α factor in order to minimize the possibility of leaky arrest. Results were similar using 0.1 μM α factor (data not shown). Cells were grown to OD660 = 0.2-0.3 in YPD, and incubated ± α factor at 30°C. For experiments involving plasmids, cells grown overnight in selective media were transferred to YPD for 1.5 hrs prior to α factor addition. Cells were analyzed by FACS as described (Haase and Reed, 2002); instead of pepsin, cells were treated with 0.2 ml Proteinase K solution (1 mg/ml in 50 mM Tris, pH 8.0) for 1 hr at 36°C.
Immunoblotting
Immunoprecipitation and yeast extract preparation used published methods (Lamson et al., 2002). Immunoprecipitation of Ste5-myc13, Fus3-myc13, Ste5-HA3, and GFP-Ste4 used mouse anti-myc (9E10; Santa Cruz Biotech), anti-HA (HA.11; Covance), and anti-GFP (clones 7.1 and 13.1; Roche) antibodies. Blots were probed with rabbit anti-myc (A-14; Santa Cruz Biotech), rabbit anti-HA (Y-11; Santa Cruz Biotech), or mouse anti-GFP (B34; Covance). Analysis of phosphorylation-dependent mobility of Ste5-HA3 used a lysis buffer with high salt and phosphatase inhibitors (Harvey et al., 2005) and 10% polyacrylamide (30:1 acryl:bis) gels. Phosphatase treatment of Ste5-HA3 was performed as described (Chang and Herskowitz, 1992).
Kinase assays
GST-Ste51-125 fusions (WT, 8A and 8E) were purified from E. coli strain BL21-Codon Plus (Stratagene) using glutathione-Sepharose beads. The Cln2/Cdc28 complex was purified from Sf9 insect cells infected with recombinant baculoviruses (Nash et al., 2001). Cln2/Cdc28 (100 ng) was mixed with GST-Ste51-125 (30 μg) or histone H1 (1 μg) in 10 μl of reaction buffer (50 mM Hepes pH 7.5, 10 mM MgCl2, 1 mM DTT, 10 μCi [32P]-γ-ATP, 100 μM cold ATP), and incubated for 30 min at 30°C. Products were resolved by SDS-PAGE and stained with coomassie blue. Gels were then dried and analyzed by autoradiography.
Microscopy
GFP fusions to Ste5 (full-length or fragments) were visualized without fixation, after induction from the GAL1 promoter with 2% galactose for 2-4 hr (Pryciak and Huntress, 1998; Winters et al., 2005). Results representative of multiple repeated experiments are shown.
Supplementary Material
Acknowledgements
We are indebted to N. Rhind for advice and use of his FACS analyzer, and to D. McCollum for help with elutriation. We also thank A. Amon, D. Lew, B. Oehlen, and C. Wittenberg for strains and plasmids, and S. Harvey and D. Kellogg for advice on detecting phosphorylation by SDS-PAGE. This work was supported by grants from the National Institutes of Health (GM57769) to P.M.P. and from the Canadian Institutes of Health Research to M.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental Data include two tables, six figures and Supplemental Experimental Procedures.
References
- Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- Brandman O, Ferrell JE, Jr., Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A, Boucher L, Tyers M. MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr Biol. 2001;11:1266–1271. doi: 10.1016/s0960-9822(01)00370-0. [DOI] [PubMed] [Google Scholar]
- Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- Chang F, Herskowitz I. Phosphorylation of FAR1 in response to alpha-factor: a possible requirement for cell-cycle arrest. Mol Biol Cell. 1992;3:445–450. doi: 10.1091/mbc.3.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V, Lyons DM, Elion EA. Fus3p and Kss1p control G1 arrest in Saccharomyces cerevisiae through a balance of distinct arrest and proliferative functions that operate in parallel with Far1p. Genetics. 1999;151:989–1004. doi: 10.1093/genetics/151.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Pesce CG, Brent R. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- Feng Y, Song LY, Kincaid E, Mahanty SK, Elion EA. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- Flotho A, Simpson DM, Qi M, Elion EA. Localized feedback phosphorylation of Ste5p scaffold by associated MAPK cascade. J Biol Chem. 2004;279:47391–47401. doi: 10.1074/jbc.M405681200. [DOI] [PubMed] [Google Scholar]
- Gartner A, Jovanovic A, Jeoung DI, Bourlat S, Cross FR, Ammerer G. Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28-Cln2 kinase, in vivo. Mol Cell Biol. 1998;18:3681–3691. doi: 10.1128/mcb.18.7.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Reed SI. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1:132–136. [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Harvey SL, Charlet A, Haas W, Gygi SP, Kellogg DR. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell. 2005;122:407–420. doi: 10.1016/j.cell.2005.05.029. [DOI] [PubMed] [Google Scholar]
- Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies RJ, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeoung DI, Oehlen LJ, Cross FR. Cln3-associated kinase activity in Saccharomyces cerevisiae is regulated by the mating factor pathway. Mol Cell Biol. 1998;18:433–441. doi: 10.1128/mcb.18.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz JE, Satterberg B, Elion EA. The MAP kinase Fus3 associates with and phosphorylates the upstream signaling component Ste5. Genes Dev. 1994;8:313–327. doi: 10.1101/gad.8.3.313. [DOI] [PubMed] [Google Scholar]
- Lamson RE, Takahashi S, Winters MJ, Pryciak PM. Dual role for membrane localization in yeast MAP kinase cascade activation and its contribution to signaling fidelity. Curr Biol. 2006;16:618–623. doi: 10.1016/j.cub.2006.02.060. [DOI] [PubMed] [Google Scholar]
- Lamson RE, Winters MJ, Pryciak PM. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol Cell Biol. 2002;22:2939–2951. doi: 10.1128/MCB.22.9.2939-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw T, Wu C, Schrag JD, Whiteway M, Thomas DY, Leberer E. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- Mahanty SK, Wang Y, Farley FW, Elion EA. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- McKinney JD, Chang F, Heintz N, Cross FR. Negative regulation of FAR1 at the Start of the yeast cell cycle. Genes Dev. 1993;7:833–843. doi: 10.1101/gad.7.5.833. [DOI] [PubMed] [Google Scholar]
- McKinney JD, Cross FR. FAR1 and the G1 phase specificity of cell cycle arrest by mating factor in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2509–2516. doi: 10.1128/mcb.15.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci USA. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlen LJ, Cross FR. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- Oehlen LJ, Cross FR. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J Biol Chem. 1998;273:25089–25097. doi: 10.1074/jbc.273.39.25089. [DOI] [PubMed] [Google Scholar]
- Oehlen LJ, Jeoung DI, Cross FR. Cyclin-specific START events and the G1-phase specificity of arrest by mating factor in budding yeast. Mol Gen Genet. 1998;258:183–198. doi: 10.1007/s004380050722. [DOI] [PubMed] [Google Scholar]
- Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- Pryciak PM, Huntress FA. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M, Elion EA. MAP kinase pathways. J Cell Sci. 2005;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivieso MH, Sugimoto K, Jahng KY, Fernandes PM, Wittenberg C. FAR1 is required for posttranscriptional regulation of CLN2 gene expression in response to mating pheromone. Mol Cell Biol. 1993;13:1013–1022. doi: 10.1128/mcb.13.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F, O'Rourke SM, Stucke VM, Jaquenoud M, Neiman AM, Peter M. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr Biol. 2000;10:630–639. doi: 10.1016/s0960-9822(00)00511-x. [DOI] [PubMed] [Google Scholar]
- van Drogen F, Stucke VM, Jorritsma G, Peter M. MAP kinase dynamics in response to pheromones in budding yeast. Nat Cell Biol. 2001;3:1051–1059. doi: 10.1038/ncb1201-1051. [DOI] [PubMed] [Google Scholar]
- Vidwans SJ, Su TT. Cycling through development in Drosophila and other metazoa. Nat Cell Biol. 2001;3:E35–39. doi: 10.1038/35050681. [DOI] [PubMed] [Google Scholar]
- Wassmann K, Ammerer G. Overexpression of the G1-cyclin gene CLN2 represses the mating pathway in Saccharomyces cerevisiae at the level of the MEKK Ste11. J Biol Chem. 1997;272:13180–13188. doi: 10.1074/jbc.272.20.13180. [DOI] [PubMed] [Google Scholar]
- Whiteway M, Hougan L, Thomas DY. Overexpression of the STE4 gene leads to mating response in haploid Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:217–222. doi: 10.1128/mcb.10.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters MJ, Lamson RE, Nakanishi H, Neiman AM, Pryciak PM. A membrane binding domain in the Ste5 scaffold synergizes with Gβγ binding to control localization and signaling in pheromone response. Mol Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Wu C, Leeuw T, Leberer E, Thomas DY, Whiteway M. Cell cycle- and Cln2p-Cdc28p-dependent phosphorylation of the yeast Ste20p protein kinase. J Biol Chem. 1998;273:28107–28115. doi: 10.1074/jbc.273.43.28107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.