Abstract
Human papillomaviruses (HPVs) have been implicated in the pathogenesis of a subset of squamous cell carcinoma of the head and neck (SCCHN). The goal of this study was to compare the cellular gene expression profiles of HPV-positive and HPV-negative oropharyngeal carcinomas with those of the normal oral epithelium. Using Affymetrix Human U133A GeneChip, our results showed that 397 genes were differentially expressed in HPV-positive SCCHN compared to the normal oral epithelium. The up-regulated genes included those involved in cell cycle regulation (CDKN2A), cell differentiation (SFRP4) and DNA repair (RAD51AP1), while the down-regulated genes included those involved in proteolysis (PRSS3). We also found 162 differentially expressed genes in HPV-negative SCCHN compared to the normal oral mucosa. The up-regulated genes included those involved in cell proliferation (AKR1C3) and transcription regulation (SNAPC1), while down-regulated genes included those involved in apoptosis (CLU) and RNA processing (RBM3). Our studies also identified a subgroup of 59 differentially expressed genes in HPV-positive SCCHN as compared to both HPV-negative SCCHN and normal oral tissues. Such up-regulated genes included those involved in nuclear structure and meiosis (SYCP2), DNA repair (RFC5), and transcription regulation (ZNF238). Genes involved in proteolysis (KLK8) and signal transduction (CRABP2) were found to be down-regulated in HPV-positive SCCHN. The results of GeneChip experiments were validated by quantitative real-time RT-PCR analysis of a few representative genes. Our results reveal specific gene expression patterns in HPV-positive and HPV-negative oropharyngeal squamous carcinomas that may serve as potential biomarkers for the development of SCCHN.
Keywords: Human papillomavirus, oral cancer, microarray, gene expression
1. Introduction
There are approximately 500,000 new cases of squamous cell carcinoma of the head and neck (SCCHN) worldwide and 45,000 cases in the United States per year.1–3 SCCHN is usually associated with such risk factors as heavy consumption of alcohol and/or tobacco. The survival in SCCHN patients is still poor and has not improved recently despite the advances in detection and therapies. It is well-established that human papillomaviruses (HPVs) are involved in the pathogenesis of cervical cancer.4–7 Recently, molecular epidemiologic studies have shown a strong correlation between oncogenic HPV infections and a subset of oropharyngeal cancers.8–12 It is not currently understood whether these HPV infections are an independent etiologic factor or a co-factor in the development of such tumours. Interestingly, a relative risk reduction from death of approximately 50% has been observed in HPV-associated tumours compared to those without detectable HPV DNA.11
HPVs are small double-stranded DNA viruses of approximately 7900 base pairs (bp). At present, more than 150 different types of HPV have been identified, and the high risk HPV types 16 and 18 are associated with a majority of cases of cervical cancer.6, 7 Replication of HPVs requires the viral E1 and E2 proteins as well as host replication factors.13–14 The E2 protein also downregulates the transcription of the viral E6 and E7 oncogenes, which are transcribed from the p97 promoter.15–17 The E6 and E7 oncoproteins of high-risk HPVs are involved in cellular transformation.15–19 Most benign and low-grade cervical lesions contain HPV DNA in an extrachromosomal state.20 However, in most cases of cervical carcinomas the HPV DNA is usually found integrated into the host chromosomes, frequently disrupting the E1 and E2 genes.6, 7, 20, 21 This results in increased expression of the viral E6 and E7 oncogenes.22 The E6 protein promotes ubiquitination and consequently proteasomal degradation of the cellular tumour suppressor proteins p53 and PDZ domain-containing disc large protein (DLG).4, 7, 23–25 E6 is also known to interact with a number of other cellular proteins and activates telomerase.6, 7 The E7 protein binds to and inactivates the function of pRB and related p107 and p130 proteins.4, 7 E7 also interacts with additional cellular proteins such as TBP, histone H1 kinase, cyclin E, etc.14–16, 18, 19 The E6 and E7 proteins are also known to alter cellular gene expression but the precise molecular mechanisms involved are not well-understood.16, 18, 19 In addition, E6/E7 expression promotes chromosomal instability, foreign DNA integration and other mutagenic events in the cell.4, 6, 15, 26 Although several studies have suggested a possible role for HPV infection in a subset of SCCHN, very little is known about the molecular events involved in carcinogenesis. Although the distribution of episomal and integrated HPV forms in precancerous and cancerous lesions of the head and neck has not been determined, limited evidence suggests a similar physical state as observed in cervical carcinoma. Molecular studies of HPV-associated SCCHN are necessary for a better understanding of the physical state and potential role of this virus in carcinogenesis, and for the development of new, more targeted therapeutic strategies.
Recently, DNA microarrays have been successfully used to identify global patterns of gene expression in different human neoplasias, including head and neck cancers.27–30 In the case of breast cancer studies, a set of 70 genes correctly predicted the nature and progression of the disease as well as the outcome.29, 30 These studies support the idea that changes in the molecular profiles of gene expression are early events during carcinogenesis and such global expression profiles can be used effectively to predict the course of the disease. Recently, investigators have used microarrays to analyze gene expression changes in SCCHN tissues and cell lines, but little is known about the gene expression changes in HPV-associated SCCHN.31–35 The identification of molecular portrait of gene expression profiles in HPV-positive and HPV-negative SCCHN, including their differences, could result in a better understanding of critical events during carcinogenesis.
This study was undertaken to identify changes in cellular gene expression profiles in HPV-positive and HPV-negative SCCHN as compared to normal tissues as well as to each other. Our microarray analysis showed considerable differences in the gene expression profiles that were specifically associated with these two types of cancers. Several of the differentially expressed genes were found to be involved in cell cycle regulation (CDKN2A), nuclear structure and meiosis (SYCP2), DNA replication and repair (RFC5), transcription regulation (ZNF238), cell differentiation (KLK8) and epidermis development (CRABP2). Unsupervised clustering analysis also showed that genes located in specific chromosomal regions such as 1p31–p36 and 12q21–24 were specifically overexpressed in HPV-positive oropharyngeal squamous carcinomas.
2. Material and methods
2.1. Tissue samples and cell lines
A total of 11 samples were analysed in this study: Three HPV-positive (SK20, SK30, SK31), four HPV-negative (SK32, SK33, SK34, SK35), and four normal oral mucosa (SK16, SK17, SK36, SK37). Tumours and normal mucosal specimens were snap-frozen and stored at −80°C until further use. Collection of tissues was performed under an IRB-approved Tissue Banking protocol, and written informed consent was obtained from each patient prior to sample collection. The SCCHN samples were from the oropharynx (tonsil and base of tongue), while the normal oral mucosa specimens were obtained from patients undergoing removal of the oropharyngeal tissues: tonsils, soft palate and uvula for sleep apnea. None of the patients received prior chemotherapy or radiotherapy. Cervical carcinoma cell lines were obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco’s modified Eagle medium (C-33A) or RPMI 1640 (CaSki) supplemented with 10% fetal bovine serum (FBS) at 37°C in the presence of 5% CO2.
2.2. Isolation of RNA and RT-PCR analysis
Total RNA from all frozen tumour samples and oropharyngeal mucosal tissues was isolated using the RNeasy Mini Kit (Qiagen Inc.) according to the manufacturer’s protocol. The mucosa was carefully dissected from any subjacent muscle or lymphoid tissue before RNA extraction, so that only normal squamous epithelium was studied. The RNA pellet was dried under a vacuum and resuspended in 30–50 μl of DEPC treated water. The integrity of RNA samples was confirmed by analysing 1 μg of total RNA on 1.2% (w/v) denaturing formaldehyde-agarose gels. Before DNA synthesis, RNA was treated with deoxyribonuclease I, Amplification Grade (Invitrogen) for 15 min at room temperature to avoid DNA contamination. DNase I was inactivated by incubation with 25 mM EDTA at 65ºC for 10 min. We examined the expression of the HPV-16 E6 and E7 genes in the above samples by RT-PCR36 using the following primers: HPV-16 E6 (forward) 5′- ATGCACCAAAAGAGAACTGC -3′ and (reverse) 5′- TTACAGCTGGGTTTCTCTAC -3′ (477-bp PCR product); HPV-16 E7 (forward) 5′-GTAACCTTTTGTTGCAAGTGTGACT-3′ and (reverse) 5′-GATTATGGTTTCTGAGAACAGATGG-3′ (137-bp RT-PCR product). The expression of the cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) house-keeping gene was used as a positive control using the following primers: GAPDH (forward) 5′- ACCACAGTCCATGCCATCAC-3′ and (reverse) 5′- TCCACCACCCTGTTGCTGTA-3′ (452-bp RT-PCR product). All RT-PCR reactions were done using the Thermoscript One-step System kit (Invitrogen Corp.). The cDNA synthesis was performed at 37°C for 1 hour in a final volume of 20 μl using 1 μg of total RNA template, 0.5 μg of oligo (dT)15, 10 mM dNTPs, 30 U of RNase inhibitor and 200 units of MMLV reverse transcriptase. PCR was performed in a 50 μl volume containing 50 mM KCl, 10 mM Tris (pH 8.3), 1.5 mM MgCl2, 0.01 % gelatin (w/v), 200 μM deoxynucleoside triphosphate (dNTP) mix, 0.4 μM of each primer and 2.5 units of the Taq DNA polymerase. The DNA was denatured at 94°C for 5 min, followed by 40 PCR amplification cycles that consisted of denaturation (94°C, 1 min), annealing (58–60°C, 1 min) and extension (72°C, 2 min). An additional extension step of 72°C for 5 min was included at the end of the reaction.
2.3. Quantitative real-time RT-PCR analysis
We validated the microarray data by quantitative real-time RT-PCR (QRT-PCR) analysis of a few representative genes using the QuantiTect SYBR Green one-step RT-PCR kit (QIAGEN, Valencia, CA). Approximately 400 ng of DNase I-treated total RNA from the samples was mixed with 25 μl of 2 X QuantiTect SYBR Green RT-PCR Master Mix, 0.5 μM of specific forward and reverse primers and RNase-free water in a final volume of 50 μl. The amount of fluorescence emitted by SYBR Green I was quantified by the ABI Prism 7700 system software (Applied Biosystems, Foster City, CA). The genes and primers used for this analysis are shown in Table 1. The following RT-PCR cycle parameters were used: reverse transcription at 50°C for 30 min, hot-start DNA polymerase activation 95°C for 15 min, 40 cycles of denaturation at 94°C for 15 sec each, annealing at 58°C for 30 sec and extension at 72°C for 30 sec. Each reaction was run in triplicate in a 96-well plate. Relative expression of the target gene was calculated using the 2 delta CT method described previously:37 (Relative expression = 2−ΔCT; where ΔCT = CT (Target gene) − CT (endogenous control gene)) where GAPDH was used as the endogenous control gene.
Table 1.
Primer sequences for quantitative RT-PCR
| Gene title | Forward primer 5′-3′ | Reverse primer 5′-3′ |
|---|---|---|
| cellular retinoic acid binding protein 2 (CRABP2) | TGCAGGGTCTTGCTTTCTTT | GGGCTAGGACTGCTGACTTG |
| cyclin-dependent kinase inhibitor 2A (CDKN2A) | AGTGAGCACTCACGCCCTAA | CCCCTGAGCTTCCCTAGTTC |
| kallikrein 8 (KLK8) | ACCAGTCCCCGAGAGAATTT | ACAGACCATGCCATCTGTGA |
| replication factor C (activator 1) 5, 36.5kDa (RFC5) | GTCACCAGCAGGTTCCAAAT | TCCTGCATGAAAACAAGTGC |
| synaptonemal complex protein 2 (SYCP2) | CATGTCACCGAAGCAAGTGT | GAAGTCTTCTGGGCTTGGACT |
| zinc finger protein 238 (ZNF238) | TTTCCTTTGAGGGGATAGGG | TCAAAAGACAATGCAGTGTTGA |
| glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | CAGCCTCAAGATCATCAGCA | TGTGGTCATGAGTCCTTCCA |
2.4. cRNA synthesis and microarray analysis
Microarray analysis was carried out using the high-density oligonucleotide U133A GeneChip® expression microarray (Affymetrix, Santa Clara, CA) that contains 22,215 transcript sets representing 14,820 human genes. RNA isolated from the samples described above was further purified by using the RNeasy Total RNA Isolation Kit (QIAGEN, Valencia, CA) to remove contaminating DNA. Twenty micrograms of total RNA from each sample were used to generate double-stranded (ds) cDNA using the Superscript II Choice system (GIBCO-BRL, Rockville, MD). First-strand synthesis was carried out using a T7-(dT)24 primer (Sigma-GenoSys, The Woodlands, TX) and Superscript II reverse transcriptase. This primer includes the promoter sequence for the T7 RNA polymerase. The second-strand synthesis was performed using RNase H and DNA polymerase I. The resulting ds cDNA was used to synthesize cRNA using biotin-labeled ribonucleotides and T7 RNA polymerase utilizing the BioArray HighYield RNA transcript labelling kit (ENZO Life Sciences, Inc. Farmingdale, NY). The cRNA was fragmented to 35–200 bases length and hybridized to the Human Genome U133A GeneChip® expression microarray in the Affymetrix Hybridization Oven (Affymetrix, Santa Clara, CA). The GeneChips were washed, and then stained with streptavidin-phycoerythrin (SAPE) to generate fluorescent signals from biotin-labeled cRNA. The GeneChips were scanned using an Affymetrix GeneArray® scanner (Affymetrix, Santa Clara, CA). Each probed array was scanned twice to calculate an average of two images, define the probe cells and compute intensity for each cell. Fragmentation, hybridization, staining and scan were performed by the Pitt Array Facility, University of Pittsburgh.
2.5. Microarray data analysis
The CEL image files obtained from the Affymetrix Microarray Suite 5.0 were converted to DCP files using the program dChip version 1.2 (www.dchip.org). Two different statistical tests, Significance Analysis of Microarray (SAM) program version 1.21 (www.stat.stanford.edu/~tibs/SAM/) and Gene Expression Data Analysis (GEDA) program, were used to analyze the microarray data in order to identify differentially expressed genes that were robust enough to appear in both the analyses. For SAM analysis, the dChip program was used to generate normalized intensity signal data. The invariant set normalization method used by dChip program is one of the most commonly used normalization method for analysing the data in the SAM program.38 In SAM analysis, a false discovery rate (FDR) of less than 5 percent and a difference of at least 2-fold was chosen to identify the total number of differentially expressed genes. We also utilized the GEDA tool developed at the University of Pittsburgh (James Lyons-Weiler, University of Pittsburgh Cancer Institute, http://bioinformatics.upmc.edu) for statistical analysis. For this, non-normalized intensity signal data from dChip (PM-only model) were normalized (log-base2 transformation, median within arrays, Global Mean Adjustment among arrays) and analysed with a t-Test at level alpha = 0.05. This normalization method is specifically used by the GEDA program. Genes with an expression ratio of at least 2-fold difference relative to the controls were considered to be differentially expressed in GEDA analysis. After analyzing the data with the two statistical tests, we overlapped the lists of differentially expressed genes in order to generate a list of differentially expressed genes using software developed by Dr. Jim Lund at the University of Kentucky (http://elegans.uky.edu/MA/progs/Compare.html). All the microarray experiments were designed in accordance with the MIAME guidelines.39
For unsupervised hierarchical clustering analysis, we first filtered the genes using a threshold of 0.05 in the ratio of the standard deviation and the mean of a gene’s expression values across all samples (Coefficient of Variation), and then applied an algorithm for similarity measurements using Pearson’s correlation coefficient.40
3. Results
3.1. Expression of the HPV-16 early genes in SCCHN samples by RT-PCR analysis
RT-PCR analysis was carried out to test for the presence and expression of HPV early genes in all the SCCHN samples and the control normal oral tissues. RNA from CasKi and C-33A cervical carcinoma cell lines was used as positive and negative control, respectively, for HPV-16 gene expression. The E6 (with different splicing variants) and E7 viral oncogene transcripts were present in the HPV-16 positive SCCHN samples SK20, SK30 and SK31 but not in the HPV-negative SCCHN samples SK32, SK33, SK34, and SK35 or the normal oral mucosa samples SK16, SK17, SK36 and SK37 (Fig. 1). The above results confirmed the appropriate expression of the HPV-16 oncogenes in the three HPV-positive SCCHN samples.
Fig. 1.
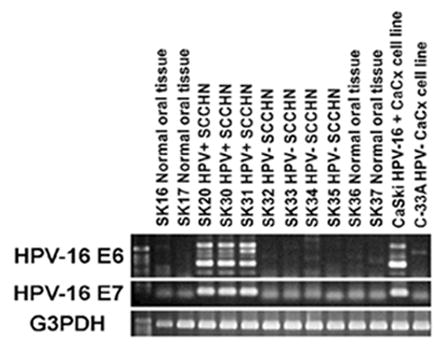
Analysis of HPV16 E6 and E7 gene expression in oropharyngeal tissue samples by RT-PCR. CaSki (HPV-16 positive) and C-33A (HPV-negative) cervical cell lines were used as controls. Expression of E6 splicing variants (E6, 477 bp, E6*I, 293 bp, E6*II, 176 bp) and E7 (137 bp) genes was found only in SK20, SK30 and SK31 SCCHN samples. The integrity of all the RNA samples was validated by amplification of the house-keeping gene GAPDH (452 bp).
3.2. Microarray analysis of genes differentially expressed in the HPV-positive SCCHN samples compared to the normal oral tissue
In order to identify differentially expressed genes in HPV-16 positive SCCHN samples (SK20, SK30 and SK31) as compared to the normal oral tissue samples (SK16, SK17, SK36 and SK37), we utilized the Affymetrix U133A GeneChip. Normalization and differential gene expression analyses were carried out as described in material and methods. A total of 1,329 and 1,431 genes showed up- and down-regulation, respectively, using a pooled variance t-Test analysis when all the HPV-16 positive SCCHN samples were compared to the normal oral tissues (supplementary data, Table A). On the other hand, 470 and 424 genes were found to be up- and down-regulated, respectively, using the SAM-Test analysis (supplementary data, Table B). When the results of the above two statistical tests were overlapped, 228 genes were found to be upregulated and 169 downregulated in HPV-16 positive SCCHN samples as compared to the normal oral tissues (Fig. 2). A list of differentially expressed genes representing various cellular pathways is shown in Table 2. A complete list of genes is shown in the supplementary data, Table C. Our analysis identified genes that have previously been shown to be overexpressed in HPV-infected cervical or oral epithelial cells such as those involved in DNA replication and cell cycle regulation: cyclin-dependent kinase inhibitor 2A (CDKN2A), minichromosome maintenance deficient genes (MCM2, MCM3), topoisomerase (DNA) II alpha 170-kDa (TOP2A) and proliferating cell nuclear antigen (PCNA).41–43 These findings suggest that some carcinogenic pathways are frequently affected in HPV-positive cervical as well as HPV-positive oral cancers. Our experiments also identified several genes that were previously implicated in tumourigenesis of different tissues but not in SCCHN. Examples of these include the upregulation of genes involved in cell differentiation or DNA repair such as secreted frizzled-related protein 4 (SFRP4), and RAD51 associated protein 1 (RAD51AP1).44, 45 Our results also showed the downregulation of genes involved in proteolysis such as protease serine 3 (PRSS3) and chemotaxis, such as chemokine (C-C motif) ligand 14 (CCL14), for the first time in HPV-positive SCCHN (Table 2).
Fig. 2.
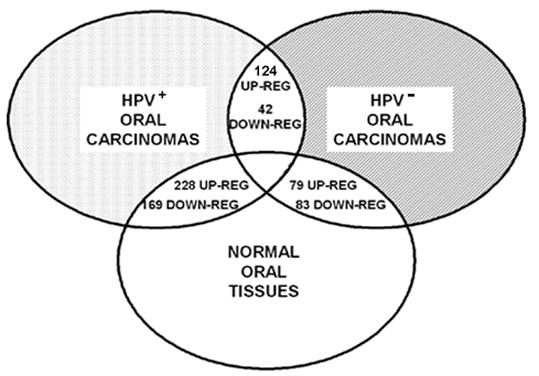
Summary of differentially expressed genes identified by microarray analysis using two different statistical analyses. The ovals represent the 3 groups of samples used in our study. The overlapping regions indicate the number of genes found to be up- or down-regulated between two different groups of samples.
Table 2.
Genes found differentially expressed in HPV-positive SCCHN versus normal oral epithelium
| Gene Title | Gene Symbol | Pooled variance t- Test fold change | SAM- Test fold change |
|---|---|---|---|
| Up-regulated genes | |||
| Apoptosis | |||
| lectin, galactoside-binding, soluble, 1 (galectin 1) | LGALS1 | 3.22 | 2.44 |
| Cell Adhesion | |||
| collagen, type XI, alpha 1 | COL11A1 | 6.29 | 8.84 |
| Cell Cycle Regulation | |||
| cyclin E2 | CCNE2 | 4.23 | 2.23 |
| cyclin-dependent kinase inhibitor 2A | CDKN2A | 16.59 | 8.04 |
| proliferating cell nuclear antigen | PCNA | 7.78 | 2.04 |
| retinoblastoma-like 1 (p107) | RBL1 | 6.09 | 2.37 |
| Chromatin assembly and modification | |||
| centromere protein F, 350/400ka (mitosin) | CENPF | 3.60 | 2.00 |
| SWI/SNF related, matrix associated, member 3 | SMARCA3 | 5.54 | 2.45 |
| Development | |||
| IGF-II mRNA-binding protein 3 | IMP-3 | 3.28 | 5.06 |
| DNA Repair | |||
| flap structure-specific endonuclease 1 | FEN1 | 3.3 | 2.28 |
| RAD51 associated protein 1 | RAD51AP1 | 3.65 | 2.14 |
| topoisomerase (DNA) II alpha 170kDa | TOP2A | 4.07 | 3.60 |
| DNA Replication | |||
| CDC7 cell division cycle 7 (S. cerevisiae) | CDC7 | 5.38 | 2.34 |
| MCM2 minichromosome maintenance deficient 2, mitotin | MCM2 | 4.39 | 3.36 |
| MCM3 minichromosome maintenance deficient 3 | MCM3 | 3.86 | 2.00 |
| Miscellaneous | |||
| lumican | LUM | 4.18 | 3.41 |
| mesoderm specific transcript homolog (mouse) | MEST | 8.88 | 3.01 |
| squamous cell carcinoma antigen recognised by T cells 3 | SART3 | 8.44 | 2.10 |
| Transcription Regulation | |||
| interferon regulatory factor 1 | IRF1 | 6.28 | 2.66 |
| Wnt Receptor Signaling Pathway | |||
| secreted frizzled-related protein 4 | SFRP4 | 4.45 | 5.61 |
|
| |||
| Down-regulated genes | |||
| Cell Adhesion | |||
| alpha-2-glycoprotein 1, zinc | AZGP1 | −3.07 | −13.77 |
| Cell Cycle Regulation | |||
| Ras association (RalGDS/AF-6) domain family 1 | RASSF1 | −3.96 | −2.51 |
| Cell Differentiation | |||
| deleted in malignant brain tumors 1 | DMBT1 | −4.04 | −9.31 |
| NDRG family member 2 | NDRG2 | −2.64 | −2.56 |
| transglutaminase 3 | TGM3 | −7.14 | −7.55 |
| Cellular Metabolism | |||
| aldehyde dehydrogenase 1 family, member A1 | ALDH1A1 | −4.60 | −4.85 |
| sulfotransferase family, cytosolic, 2B, member 1 | SULT2B1 | −2.33 | −3.00 |
| UDP glucuronosyltransferase 1 family, A10 / A8 / A7 / A6 | UGT1A | −3.71 | −2.56 |
| DNA Repair | |||
| mutL homolog 3 (E. coli) | MLH3 | −2.53 | −2.06 |
| Immune response | |||
| chemokine (C-C motif) ligand 14 / chemokine ligand 15 | CCL14/CCL15 | −5.23 | −2.10 |
| interleukin 18 (interferon-gamma-inducing factor) | IL18 | −4.47 | −4.18 |
| trefoil factor 3 (intestinal) | TFF3 | −2.61 | −10.42 |
| Miscellaneous | |||
| envoplakin | EVPL | −3.26 | −3.55 |
| huntingtin interacting protein-1-related | HIP1R | −3.72 | −2.65 |
| kallikrein 11 | KLK11 | −3.70 | −7.28 |
| kallikrein 13 | KLK13 | −4.41 | −12.15 |
| protease, serine, 3 (mesotrypsin) | PRSS3 | −3.88 | −7.21 |
| Signal Transduction | |||
| fibroblast growth factor binding protein 1 | FGFBP1 | −2.12 | −3.27 |
| insulin-like growth factor binding protein 6 | IGFBP6 | −3.69 | −7.74 |
| Transcription Regulation | |||
| BarH-like homeobox 2 | BARX2 | −2.96 | −3.71 |
3.3. Genes differentially expressed in HPV-negative SCCHN compared to the normal oral mucosa
Using the pooled variance t-Test analysis, we found that 1,052 genes were upregulated and 850 downregulated in HPV-negative SCCHN samples compared to the normal oral tissues (supplementary data, Table D). The SAM test identified 244 upregulated and 310 downregulated genes in the HPV-negative SCCHN samples (supplementary data, Table E). A comparison of the differentially expressed genes revealed that 79 upregulated and 83 downregulated genes were common in both the pooled variance t-Test and SAM analyses (supplementary data, Table F) (Fig. 2). A list of differentially expressed genes representing various cellular pathways is shown in Table 3. Upregulated genes involved in cell-cell signaling such as parathyroid hormone-like hormone (PTHLH ) or angiogenesis such as endothelial cell growth factor 1 (ECGF1), as well as downregulated genes involved in signal transduction or apoptosis such as insulin-like growth factor binding protein 5 (IGFBP5) and programmed cell death 4 (neoplastic transformation inhibitor) (PDCD4) were identified in our studies similar to those found in previous SCCHN studies, providing a good correlation with our results.46, 47 We also found differentially expressed genes that were not previously reported in SCCHN, although they were implicated in other types of tumours. Such upregulated genes included those involved in cell proliferation and transcription regulation such as aldo-keto reductase family 1, member C3 (AKR1C3), and small nuclear RNA activating complex, polypeptide 1, 43-kDa (SNAPC1), while downregulated genes included those involved in apoptosis or RNA processing such as clusterin (CLU) and RNA binding motif protein 3 (RBM3).48–52
Table 3.
Genes found differentially expressed in HPV-negative SCCHN versus normal oral epithelium
| Gene Title | Gene Symbol | Pooled variance t- Test fold change | SAM- Test fold change |
|---|---|---|---|
| Up-regulated genes | |||
| Angiogenesis | |||
| endothelial cell growth factor 1 (platelet-derived) | ECGF1 | 2.90 | 2.23 |
| Cell Adhesion | |||
| Cadherin, EGF LAG seven-pass G-type receptor 1 | CELSR1 | 2.67 | 2.21 |
| cadherin 3, type 1, P-cadherin (placental) | CDH3 | 2.51 | 3.72 |
| calcium/calmodulin-dependent serine protein kinase | CASK | 3.47 | 2.05 |
| laminin, beta 1 | LAMB1 | 3.36 | 2.18 |
| Cell-Cell Signaling | |||
| parathyroid hormone-like hormone | PTHLH | 2.61 | 9.26 |
| Cell Cycle Regulation | |||
| cyclin A1 | CCNA1 | 2.16 | 2.03 |
| exostoses (multiple) 1 | EXT1 | 3.25 | 2.99 |
| Development | |||
| S100 calcium binding protein A7 (psoriasin 1) | S100A7 | 2.58 | 4.15 |
| DNA Replication | |||
| DNA replication complex GINS protein PSF1 | PSF1 | 2.03 | 2.27 |
| Immune Response | |||
| CCAAT/enhancer binding protein (C/EBP), beta | CEBPB | 2.11 | 2.24 |
| Fc fragment of IgG, low affinity IIa, receptor (CD32) | FCGR2A | 2.89 | 2.39 |
| interferon-induced protein with tetratricopeptide repeats 1 | IFIT1 | 2.81 | 2.07 |
| Miscellaneous | |||
| aldo-keto reductase family 1, member B10 (aldose reductase) | AKR1B10 | 3.05 | 4.47 |
| aldo-keto reductase family 1, member C3 | AKR1C3 | 2.69 | 2.90 |
| chromosome 5 open reading frame 13 | C5orf13 | 2.16 | 2.09 |
| phosphoribosylglycinamide formyltransferase | GART | 2.55 | 2.13 |
| Transcription Regulation | |||
| basonuclin 1 | BNC1 | 2.83 | 2.27 |
| polymerase (RNA) II (DNA directed) polypeptide H | POLR2H | 2.59 | 2.03 |
| small nuclear RNA activating complex, polypeptide 1, 43kDa | SNAPC1 | 2.16 | 2.42 |
|
| |||
| Down-regulated genes | |||
| Apoptosis | |||
| programmed cell death 4 | PDCD4 | −2.617 | −2.00 |
| clusterin | CLU | −3.27 | −4.43 |
| Cell Adhesion | |||
| chemokine (C-X-C motif) ligand 12 | CXCL12 | −2.18 | −2.84 |
| claudin 10 | CLDN10 | −2.62 | −4.50 |
| Cell-Cell Signaling | |||
| sprouty homolog 2 (Drosophila) | SPRY2 | −2.65 | −2.37 |
| Cell Cycle Regulation | |||
| transforming, acidic coiled-coil containing protein 1 | TACC1 | −2.58 | −2.01 |
| Cell Differentiation | |||
| four and a half LIM domains 1 | FHL1 | −2.74 | −2.03 |
| Cellular Metabolism | |||
| glucosamine (UDP-N-acetyl)-2-epimerase/N acetylmannosamine kinase | GNE | −2.46 | −2.00 |
| glycine amidinotransferase | GATM | −3.71 | −3.72 |
| Development | |||
| chordin-like 1 | CHRDL1 | −2.44 | −2.44 |
| dystrophin (muscular dystrophy, Duchenne and Becker types) | DMD | −2.45 | −2.14 |
| Miscellaneous | |||
| cytochrome P450, family 3, subfamily A, polypeptide 5 | CYP3A5 | −2.13 | −4.45 |
| heat shock 70kDa protein 1A | HSPA1A | −4.49 | −2.04 |
| lymphoid-restricted membrane protein | LRMP | −3.06 | −4.24 |
| RNA Processing | |||
| RNA binding motif (RNP1, RRM) protein 3 | RBM3 | −2.32 | −2.56 |
| Signal Transduction | |||
| insulin-like growth factor binding protein 5 | IGFBP5 | −3.04 | −2.36 |
| protein kinase, cAMP-dependent, catalytic, beta | PRKACB | −3.36 | −2.29 |
| RAP1A, member of RAS oncogene family | RAP1A | −3.77 | −2.27 |
| Transcription Regulation | |||
| single-stranded DNA binding protein 2 | SSBP2 | −2.77 | −2.02 |
| thioredoxin interacting protein | TXNIP | −2.43 | −2.03 |
3.4. Identification of genes differentially expressed between HPV-positive and HPV-negative SCCHN
To identify pathways unique to the pathogenesis of HPV positive and negative SCCHN, we compared the gene expression profiles in these two types of cancers. The pooled t-Test analysis revealed that a total of 1,040 genes were upregulated and 1,363 downregulated in the HPV-positive SCCHN samples compared to the HPV-negative SCCHN samples (supplementary data, Table G). Using the SAM test, we identified 347 upregulated and 175 downregulated genes in the HPV-positive SCCHN compared to HPV-negative SCCHN (supplementary data, Table H). A comparison of the results obtained using the above two tests was done to identify genes present in both of these analyses. This showed that 124 genes were upregulated and 42 downregulated in HPV-16 positive SCCHN as compared to the HPV-negative SCCHN (Fig.2). A list of differentially expressed genes representing various cellular pathways is shown in Table 4 (a complete list of genes is shown in the supplementary data, Table I).
Table 4.
Genes found to be differentially expressed in HPV-positive SCCHN versus HPV-negative SCCHN
| Gene Title | Gene Symbol | Pooled variance t- Test fold change | SAM- Test fold change |
|---|---|---|---|
| Up-regulated genes | |||
| Apoptosis | |||
| baculoviral IAP repeat-containing 1 | BIRC1 | 3.08 | 2.11 |
| baculoviral IAP repeat-containing 3 | BIRC3 | 4.65 | 4.56 |
| CD2 antigen (p50), sheep red blood cell receptor | CD2 | 3.56 | 2.60 |
| Cell Adhesion | |||
| vascular cell adhesion molecule 1 | VCAM1 | 4.63 | 3.96 |
| Cell Cycle | |||
| B-cell CLL/lymphoma 2 | BCL2 | 2.43 | 2.53 |
| cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | CDKN2C | 2.83 | 3.04 |
| dual specificity phosphatase 4 | DUSP4 | 3.56 | 2.07 |
| synaptonemal complex protein 2 | SYCP2 | 13.58 | 12.54 |
| DNA Repair | |||
| RecQ protein-like (DNA helicase Q1-like) | RECQL | 3.90 | 2.16 |
| Immune response | |||
| lymphotoxin beta (TNF superfamily, member 3) | LTB | 2.56 | 2.22 |
| major histocompatibility complex, class II, DP beta 1 | HLA-DPB1 | 2.51 | 2.40 |
| major histocompatibility complex, class II, DQ alpha 1/ 2 | HLA-DQA1/A2 | 2.47 | 3.29 |
| Miscellaneous | |||
| ecotropic viral integration site 2A | EVI2A | 3.33 | 2.11 |
| tubulin, gamma complex associated protein 3 | TUBGCP3 | 4.92 | 2.14 |
| Signal Transduction | |||
| phosphoinositide-3-kinase, catalytic, gamma polypeptide | PIK3CG | 2.87 | 2.35 |
| RAS guanyl releasing protein 1 | RASGRP1 | 2.50 | 2.09 |
| Transcription Regulation | |||
| BCL2-associated transcription factor 1 | BCLAF1 | 7.29 | 2.70 |
| Splicing factor proline/glutamine rich | SFPQ | 3.64 | 2.57 |
| thymus high mobility group box protein TOX | TOX | 3.64 | 2.16 |
| zinc finger protein 238 | ZNF238 | 6.28 | 2.04 |
|
| |||
| Down-regulated genes | |||
| Cell Cycle | |||
| cyclin A1 | CCNA1 | −2.18 | −2.31 |
| Cell Differentiation | |||
| S100 calcium binding protein A7 (psoriasin 1) | S100A7 | −2.7 | −6.69 |
| small proline-rich protein 1B (cornifin) | SPRR1B | −3.40 | −2.28 |
| Cell Proliferation | |||
| block of proliferation 1 | BOP1 | −2.48 | −2.19 |
| Cellular Metabolism | |||
| aldo-keto reductase family 1, member B10 | AKR1B10 | −3.73 | −7.30 |
| aldo-keto reductase family 1, member C3 | AKR1C3 | −3.23 | −4.44 |
| solute carrier family 7, member 5 | SLC7A5 | −2.25 | −2.08 |
| UDP glucuronosyltransferase 2 family, polypeptide B4 | UGT2B4 | −4.97 | −2.71 |
| Chromatin assembly and modification | |||
| histone 1, H2bg | HIST1H2BG | −2.63 | −2.70 |
| Immune response | |||
| interleukin 1 receptor accessory protein | IL1RAP | −2.24 | −2.01 |
| killer cell lectin-like receptor subfamily G, member 1 | KLRG1 | −5.70 | −2.18 |
| serine (or cysteine) proteinase inhibitor, clade B, member 4 | SERPINB4 | −2.0 | −2.38 |
| Miscellaneous | |||
| corneodesmosin | CDSN | −2.97 | −2.62 |
| seven transmembrane domain protein | NIFIE14 | −5.28 | −2.05 |
| Protein modification | |||
| carboxypeptidase A4 | CPA4 | −2.58 | −2.14 |
| Signal Transduction | |||
| inositol(myo)-1(or 4)-monophosphatase 2 | IMPA2 | −2.05 | −2.05 |
| Transcription Regulation | |||
| achaete-scute complex-like 2 (Drosophila) | ASCL2 | −4.19 | −2.08 |
| bolA-like 2 (E. coli) | BOLA2 | −2.09 | −2.02 |
| Translation Regulation | |||
| eukaryotic translation initiation factor 3, subunit 8, 110kDa | EIF3S8 | −2.27 | −2.14 |
| eukaryotic translation initiation factor 5A | EIF5A | −6.05 | −2.79 |
Interestingly, a subgroup of 40 upregulated and 19 downregulated genes identified in HPV-positive versus HPV-negative SCCHN analysis, was also found in a comparison between HPV-positive SCCHN and normal oral tissues. Thus, it is likely that the expression of this group of genes (Table 5) is specifically affected by the presence of HPV. Such upregulated genes included those involved in nuclear structure and meiosis (synaptonemal complex protein 2 [SYCP2]), DNA repair (replication factor C 5 [RFC5]), and DNA methylation (DNA [cytosine-5-]-methyltransferase 1 [DNMT1]). Genes involved in proteolysis (kallikrein 7, 8, 10 [KLK7, KLK8, KLK10]) and signal transduction (cellular retinoic acid binding protein 2 [CRABP2]) were found to be downregulated in HPV-positive SCCHN compared to both HPV-negative SCCHN and normal oral tissues in these analyses.
Table 5.
Genes found differentially expressed exclusively in HPV-positive SCCHN versus HPV-negative SCCHN or normal oral epithelium
| Gene Title | Gene Symbol | Pooled variance t- Test fold change | SAM- Test fold change |
|---|---|---|---|
| Up-regulated genes | |||
| Apoptosis | |||
| tumor necrosis factor, alpha-induced protein 3 | TNFAIP3 | 2.40 | 2.58 |
| Cell Adhesion | |||
| collagen, type VII, alpha 1 | COL7A1 | 2.61 | 3.35 |
| collagen, type XXI, alpha 1 | COL21A1 | 15.43 | 3.80 |
| thrombospondin 4 | THBS4 | 3.09 | 3.30 |
| Cell Cycle | |||
| synaptonemal complex protein 2 | SYCP2 | 13.58 | 12.54 |
| Cell Differentiation | |||
| B cell RAG associated protein | GALNAC4S-6ST | 2.47 | 2.62 |
| Cell Proliferation | |||
| chemokine (C-X-C motif) ligand 10 | CXCL10 | 2.18 | 2.55 |
| cysteine and glycine-rich protein 2 | CSRP2 | 3.36 | 2.34 |
| pim-2 oncogene | PIM2 | 2.44 | 2.07 |
| Chromatin assembly and modification | |||
| SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | SMARCA2 | 3.36 | 2.06 |
| SMC5 structural maintenance of chromosomes 5-like 1 | SMC5L1 | 2.88 | 2.14 |
| DNA Repair | |||
| replication factor C (activator 1) 5, 36.5kDa | RFC5 | 5.01 | 2.18 |
| thymidylate synthetase | TYMS | 3.01 | 2.09 |
| Immune response | |||
| CD200 antigen | CD200 | 3.10 | 3.00 |
| chemokine (C-X-C motif) ligand 11 | CXCL11 | 2.05 | 3.52 |
| chemokine (C-X-C motif) ligand 9 | CXCL9 | 2.87 | 3.93 |
| lymphocyte antigen 75 | LY75 | 4.19 | 4.41 |
| phospholipase A2, group IVC | PLA2G4C | 4.12 | 2.27 |
| Miscellaneous | |||
| butyrophilin, subfamily 3, member A3 | BTN3A3 | 2.04 | 2.20 |
| chromosome 18 open reading frame 1 | C18orf1 | 4.05 | 3.89 |
| chromosome X open reading frame 45 | CXorf45 | 5.59 | 3.53 |
| Endothelin converting enzyme 1 | ECE1 | 2.68 | 2.30 |
| fatty acid desaturase 2 | FADS2 | 9.05 | 2.23 |
| inositol polyphosphate-5-phosphatase F | INPP5F | 3.67 | 2.92 |
| spastin | SPAST | 4.91 | 2.15 |
| stress 70 protein chaperone, microsome-associated, 60kDa | STCH | 2.57 | 2.33 |
| Protein modification | |||
| CDC-like kinase 4 | CLK4 | 7.15 | 3.21 |
| glutaminyl-peptide cyclotransferase (glutaminyl cyclase) | QPCT | 2.37 | 3.43 |
| proteasome subunit, beta type, 9 | PSMB9 | 2.15 | 2.07 |
| tyrosine kinase 2 | TYK2 | 3.68 | 2.66 |
| Signal Transduction | |||
| chemokine (C-X-C motif) receptor 4 | CXCR4 | 3.37 | 4.18 |
| chemokine orphan receptor 1 | CMKOR1 | 2.34 | 2.67 |
| inositol 1,4,5-trisphosphate 3-kinase B | ITPKB | 2.83 | 2.14 |
| lymphoid enhancer-binding factor 1 | LEF1 | 3.27 | 3.87 |
| phosphoinositide-3-kinase, regulatory subunit 3 | PIK3R3 | 3.15 | 2.37 |
| Ras association (RalGDS/AF-6) domain family 4 | RASSF4 | 3.06 | 3.45 |
| SMAD, mothers against DPP homolog 5 (Drosophila) | SMAD5 | 2.88 | 2.67 |
| Transcription Regulation | |||
| DNA (cytosine-5-)-methyltransferase 1 | DNMT1 | 6.22 | 2.22 |
| nuclear factor (erythroid-derived 2)-like 3 | NFE2L3 | 2.85 | 2.67 |
| sin3-associated polypeptide, 30kDa | SAP30 | 2.23 | 2.12 |
| suppressor of zeste 12 homolog (Drosophila) | SUZ12 | 2.86 | 2.17 |
|
| |||
| Down-regulated genes | |||
| Cell Adhesion | |||
| lymphocyte antigen 6 complex, locus D | LY6D | −2.24 | −4.29 |
| plakophilin 3 | PKP3 | −3.08 | −2.57 |
| Cell Differentiation | |||
| glucosaminyl (N-acetyl) transferase 2, I-branching enzyme | GCNT2 | −3.32 | −2.51 |
| kallikrein 7 (chymotryptic, stratum corneum) | KLK7 | −2.49 | −4.68 |
| kallikrein 8 (neuropsin/ovasin) | KLK8 | −10.36 | −4.6 |
| kallikrein 10 | KLK10 | −4.23 | −6.75 |
| keratin, hair, acidic, 1 | KRTHA1 | −2.49 | −2.29 |
| Cell Proliferation | |||
| amphiregulin (schwannoma-derived growth factor) | AREG | −3.12 | −2.55 |
| CD5 antigen (p56-62) | CD5 | −4.31 | −2.15 |
| Cellular Metabolism | |||
| cytidine deaminase | CDA | −2.07 | −5.50 |
| monoamine oxidase A | MAOA | −2.69 | −2.34 |
| UDP glucuronosyltransferase 1 family, polypeptide A6 | UGT1A6 | −2.04 | −3.85 |
| Chromatin assembly and modification | |||
| H2B histone family, member S | H2BFS | −2.91 | −2.67 |
| histone 1, H2bd | HIST1H2BD | −2.59 | −2.47 |
| Miscellaneous | |||
| kinesin family member 1C | KIF1C | −2.62 | −2.04 |
| solute carrier family 24, member 3 | SLC24A3 | −3.01 | −2.04 |
| Signal Transduction | |||
| cellular retinoic acid binding protein 2 | CRABP2 | −4.32 | −6.28 |
| wingless-type MMTV integration site family, member 4 | WNT4 | −3.91 | −2.13 |
| Transcription Regulation | |||
| paired-like homeodomain transcription factor 2 | PITX2 | −3.08 | −6.26 |
3.5. Identification of differentially expressed genes present in all SCCHN samples in comparison to normal oral tissues
We also identified genes whose expression is similarly affected in both HPV- positive and HPV-negative SCCHN as compared to the normal oral mucosa. Based on the pooled t-Test and SAM test analyses, we found that 55 genes were upregulated and 42 downregulated in HPV-positive as well as HPV-negative SCCHN as compared to the normal oral mucosa. A list of differentially expressed genes representing various cellular pathways is shown in Table 6 (a complete list of genes is shown in the supplementary data, Table J). These genes may represent common pathways altered during SCCHN irrespective of the presence or absence of HPVs. The upregulated genes included those involved in cell cycle regulation, epidermis development and cell adhesion such as signal transducer and activator of transcription 1 (STAT1), keratin 14, 16 (KRT14, KRT16) and transforming growth factor, beta-induced, 68-kDa (TGFBI). Also, downregulated genes found in both types of SCCHN samples included those involved in chromatin remodeling, cell-cell adhesion and apoptosis such as p300/CBP-associated factor (PCAF), annexin A9 (ANXA9) and mal, T-cell differentiation protein (MAL).
Table 6.
Genes found differentially expressed in all SCCHN samples versus normal oral epithelium
| Gene Title | Gene Symbol | Pooled variance t- Test fold change | SAM- Test fold change |
|---|---|---|---|
| Up-regulated genes | |||
| Cell Adhesion | |||
| matrix metalloproteinase 3 (stromelysin 1, progelatinase) | MMP3 | 5.02 | 2.55 |
| periostin, osteoblast specific factor | POSTN | 5.47 | 10.09 |
| transforming growth factor, beta-induced, 68kDa | TGFBI | 2.76 | 2.05 |
| Cell Proliferation | |||
| hepatoma-derived growth factor, related protein 3 | HDGFRP3 | 6.32 | 2.87 |
| keratin 16 (focal non-epidermolytic palmoplantar keratoderma) | KRT16 | 2.78 | 3.51 |
| Development | |||
| chondroitin sulfate proteoglycan 2 (versican) | CSPG2 | 2.85 | 3.59 |
| collagen, type I, alpha 1 | COL1A1 | 2.23 | 3.39 |
| keratin 14 (epidermolysis bullosa simplex) | KRT14 | 6.93 | 3.12 |
| DNA Replication | |||
| CDC6 cell division cycle 6 homolog (S. cerevisiae) | CDC6 | 3.75 | 2.07 |
| Immune Response | |||
| Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | FCGR3B | 2.63 | 3.29 |
| interferon, alpha-inducible protein 27 | IFI27 | 2.21 | 2.68 |
| interferon-induced protein 44 | IFI44 | 2.88 | 2.51 |
| Miscellaneous | |||
| cathepsin L2 | CTSL2 | 2.20 | 6.18 |
| plasminogen activator, urokinase | PLAU | 4.01 | 3.33 |
| secreted phosphoprotein 1 (osteopontin, bone sialoprotein I) | SPP1 | 7.00 | 18.90 |
| serine (or cysteine) proteinase inhibitor, clade H | SERPINH1 | 5.74 | 2.43 |
| Signal Transduction | |||
| CD14 antigen | CD14 | 2.15 | 2.23 |
| insulin-like growth factor 2 receptor | IGF2R | 4.72 | 2.52 |
| secreted protein, acidic, cysteine-rich (osteonectin) | SPARC | 6.22 | 4.18 |
| Transcription Regulation | |||
| signal transducer and activator of transcription 1, 91kDa | STAT1 | 5.01 | 2.94 |
|
| |||
| Down-regulated genes | |||
| Apoptosis | |||
| clusterin | CLU | −3.20 | −4.54 |
| growth arrest and DNA-damage-inducible, beta | GADD45B | −3.10 | −2.55 |
| mal, T-cell differentiation protein | MAL | −2.14 | −2.50 |
| Cell adhesion | |||
| annexin A9 | ANXA9 | −2.54 | −2.17 |
| multimerin 1 | MMRN1 | −3.70 | −4.51 |
| Cellular Metabolism | |||
| amylase, alpha 1A; salivary / 1B / 1C / 2A/ 2B | AMY1 | −5.15 | −7.67 |
| glycerol-3-phosphate dehydrogenase 1-like | GPD1L | −3.25 | −3.43 |
| Development | |||
| keratin 4 | KRT4 | −2.72 | −2.82 |
| sciellin | SCEL | −2.28 | −2.55 |
| Immune response | |||
| cysteine-rich protein 1 (intestinal) | CRIP1 | −7.21 | −5.16 |
| cysteine-rich secretory protein 3 | CRISP3 | −5.75 | −69.68 |
| interleukin 2 | IL2 | −2.97 | −3.16 |
| prostate stem cell antigen | PSCA | −4.63 | −8.65 |
| Miscellaneous | |||
| ATP-binding cassette, sub-family A (ABC1), member 8 | ABCA8 | −2.54 | −3.05 |
| dual specificity phosphatase 5 | DUSP5 | −2.54 | −2.97 |
| nucleobindin 2 | NUCB2 | −3.50 | −3.12 |
| Protein Modification | |||
| ubiquitin-like 3 | UBL3 | −2.28 | −2.67 |
| Signal Transduction | |||
| proline-rich protein HaeIII subfamily 1-2 | PRH1-2 | −3.17 | −20.84 |
| Transcription Regulation | |||
| p300/CBP-associated factor | PCAF | −2.51 | −2.43 |
| paternally expressed 3 | PEG3 | −6.73 | −6.01 |
3.6. Identification of several gene expression clusters in HPV-positive, HPV-negative SCCHN and normal oral tissues
Unsupervised hierarchical cluster analysis was developed using all SCCHN specimens as well as normal oral tissues. After normalization and filtering with different criteria (described in materials and methods), we obtained a set of 8,286 genes with significant variation in their expression in one or more of the total of three types of samples (HPV-positive SCCHN, HPV-negative SCCHN, and control oral mucosa). From these filtered genes, we identified five clusters that showed differences in gene expression not only between all the SCCHN samples and the normal oral epithelium but also between HPV-positive SCCHN and HPV-negative SCCHN (Fig. 3). Interestingly, some of the genes found in this analysis were also found in the supervised statistical analysis such as ZNF238, DNMT1, MCM3, aldo-keto reductase family 1, member C3 (AKR1C3), growth factor receptor-bound protein 10 (GRB10), tyrosine kinase 2 (TYK2) and squamous cell carcinoma antigen recognized by T cells 3 (SART3) (underlined in Fig. 3). After this analysis, we mapped the chromosomal location of the genes found in these clusters in order to visualize genomic regions that could contain a significant number of genes altered in their expression in HPV-positive SCCHN as compared to HPV-negative SCCHN as well as the normal oral epithelium. Fig. 4 shows that several such genes are present in chromosomal regions 1p34–p36 and 12q21–q24 (a complete list of the clustered genes is shown in the supplementary data, Table K). The genes found in the 1p34–p36 region are involved in repression of splicing (FUS interacting protein 1 [FUSIP1]), histone and nucleosomal binding (nuclear autoantigenic sperm protein [NASP], high-mobility group nucleosomal binding domain 2 [HMGN2]) and intracellular vesicle traffic (taxilin alpha [TXLNA]). On the other hand, the genes found in the chromosome region 12q21–q24 are involved in JAK/STAT-signaling pathway (cysteine and glycine-rich protein 2 [CSRP2]), regulation of nuclear architecture (thymopoietin [TMPO/LAP2]), myotonic dystrophy kinase cascade (citron [CIT]) and regulation of mRNA splicing and HIV gene expression and replication (squamous cell carcinoma antigen recognized by T cells 3 [SART3]).
Fig. 3.
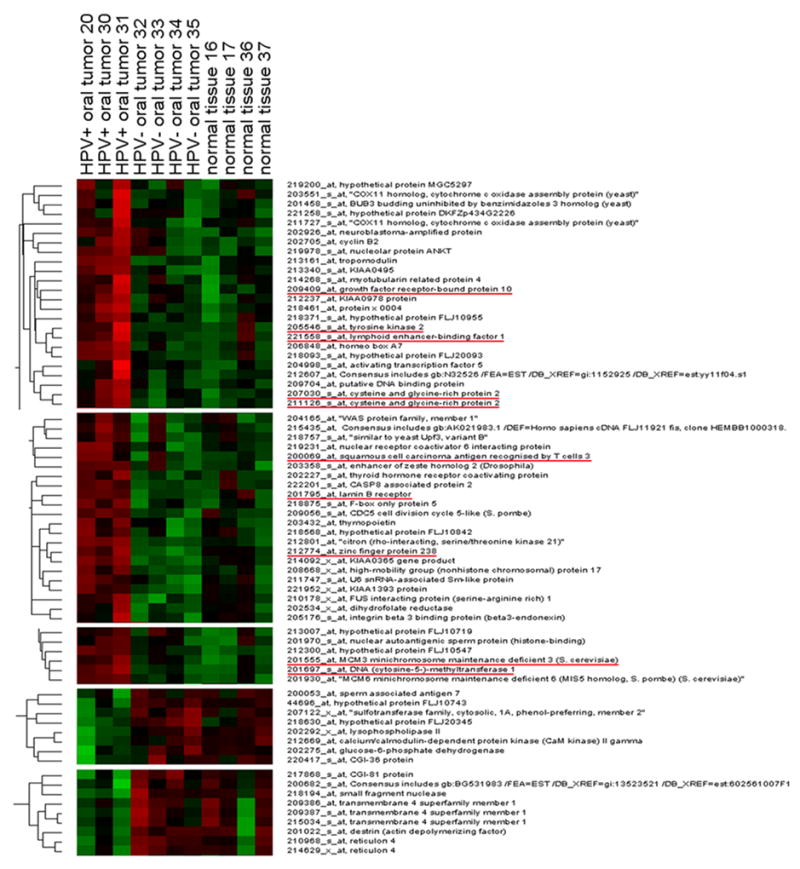
Unsupervised hierarchical cluster analysis in oropharyngeal tissue samples. From a total of 22,215 transcripts on the microarray, 8,286 genes showed variations in expression across all the samples. From this group of filter genes, we identified 7 clusters of genes that show differential expression between the 3 groups of samples (HPV-positive SCCHN, HPV-negative SCCHN and normal oral mucosa). Genes also found in the supervised statistical analysis are underlined.
Fig. 4.

Chromosomal location of genes found to be differentially expressed in the hierarchical cluster analysis. Squares to the right of the chromosomes indicate genes with increased expression in HPV-positive oropharyngeal squamous carcinomas as compared to HPV-negative carcinomas or the normal oral epithelium (clusters 1, 2 and 3). Circles to the left of the chromosomes indicate genes with reduced expression in HPV-positive oropharyngeal squamous carcinomas as compared to the HPV-negative and normal samples (clusters 4 and 5). The chromosome map was obtained and modified from the Department of Pathology, University of Washington web page http://www.pathology.washington.edu/research/cytopages/idiograms/human/ (Idiogram Album: Human copyright © 1994 David Adler).
3.7. Validation of microarray expression data by real-time quantitative RT-PCR analysis
We carried out quantitative real-time RT-PCR (QRT-PCR) analysis of a few representative genes in order to validate the differential gene expression profiles obtained by microarray analysis. Four genes found to be upregulated (CDKN2A, SYCP2, RFC5, ZNF238) and two genes found to be downregulated (KLK8, CRABP2) in HPV-positive SCCHN samples as compared to both the HPV-negative SCCHN as well as normal oral tissues were selected for validation (Fig. 5). The QRT-PCR analysis confirmed that CDKN2A and SYCP2 were up-regulated in all the HPV-positive SCCHN samples as compared to the HPV-negative SCCHN and normal epithelium, whereas RFC5 and ZNF238 were found to be up-regulated in two out of three HPV-positive SCCHN (Fig. 4). Similarly, the KLK8 and CRABP2 genes were downregulated in HPV-positive SCCHN (Fig. 5). However, there was variation in the extent of differential expression of the above genes in individual samples (Fig. 5). The above data showed that the results of QRT-PCR were consistent with those of the microarray analysis, although variations were observed in the fold-difference between these two types of analyses.
Fig. 5.
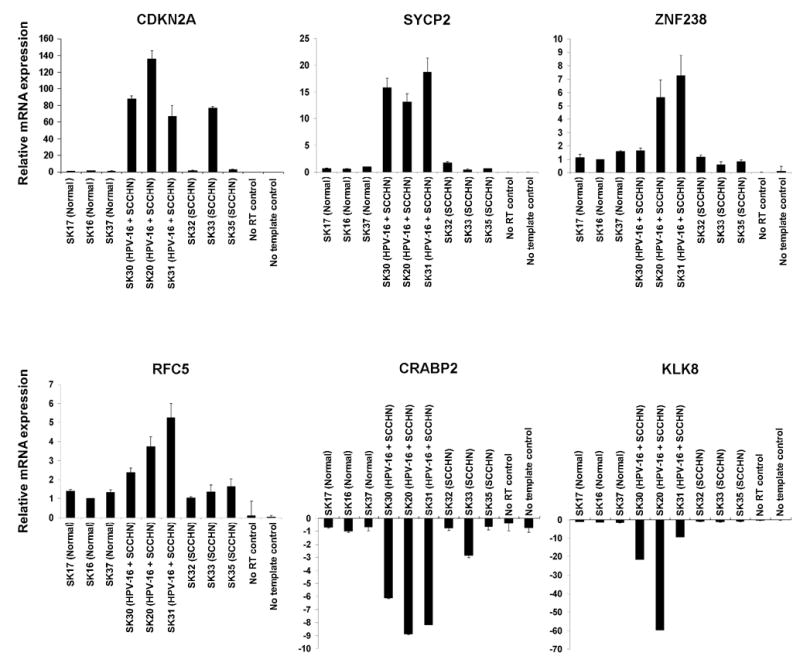
Validation of microarray data in oropharyngeal tissue samples by quantitative RT-PCR. Total RNA from tissue samples was used to verify four upregulated genes (cyclin-dependent kinase inhibitor 2A [CDKN2A], synaptonemal complex protein 2 [SYCP2], zinc finger protein 238 [ZNF238] and replication factor C 5 [RFC5]), and two downregulated genes (cellular retinoic acid binding protein 2 [CRABP2], kallikrein 8 [KLK8]) that were differentially expressed based on the microarray data. All reactions were performed in triplicates and the error bar represents the standard deviation. Relative expression of the target gene was calculated using the 2 delta CT method, where GAPDH was used as the endogenous control gene.
Discussion
Recently, an improved understanding of SCCHN has resulted from studies on carcinogenic (tobacco, alcohol, etc.) versus viral (HPV, EBV, etc.) exposures. Several investigators have compared these subgroups in order to identify the pathway (s) and potential therapeutic targets which are unique to these tumours, which are often localized to distinct head and neck sub sites. Indeed, our study is unique since we focused specifically on tumours from the oropharyngeal site, in order to compare gene expression profiles between HPV-16 positive SCCHN tumours and normal squamous oropharyngeal mucosa.
To date, no study has described such a tri-partite analysis, including the epithelial squamous cells from which both tumours were derived. This significantly strengthens our ability to conclude that oncogenic signaling identified in HPV-positive and HPV-negative SCCHN is appropriately attributed to the transformation process and not to the genes expressed in oropharyngeal squamous cells. Although the number of specimens used in our study is small, a unique feature of this study is that we have exclusively utilized tissues derived from the oropharyngeal site. While HPV infection can be identified throughout tumours of the head and neck, a disproportionate frequency is found in the oropharynx (approximately 50%). Thus, identifying tumours in this region eliminates other sub site causes of tumour heterogeneity which have been observed by others. Finally, detailed statistical analysis using multiple tests of significance enhances our ability to identify genes that are expressed uniquely in HPV-16 positive tumours. We have also carried out quantitative RT-PCR analysis to validate the expression of some of the genes found to be differentially expressed in the microarray analysis (Fig. 5). This provides additional confidence in the genes identified by the microarray analysis.
After confirming the expression of the HPV-16 E6 and E7 genes in HPV-positive, HPV-negative SCCHN and normal squamous mucosal samples, we proceeded with the global gene expression analysis. After analyzing the microarray data with two different normalizations and statistical tests (pooled variance t-Test and SAM-Test) in order to identify differentially expressed genes that were robust enough to appear in both analyses, we found that 228 genes were upregulated and 169 downregulated in HPV-positive SCCHN samples as compared to the normal oral tissues. The upregulated genes included those involved in DNA replication and cell cycle regulation such as CDKN2A (p16INK4a), MCM2, PCNA, TOP2A and RBL1 that have been previously identified to be affected in HPV-infected cervical and oral carcinomas.41–43 Several studies have shown either direct or indirect regulation of these genes by the tumour suppressor protein p53 (RBL1, PCNA, TOP2A) or by E2F transcription factors (CDKN2A and MCM2).43, 53 One possible explanation for the transcriptional deregulation of this group of genes may be the degradation of p53 and pRB (that bind and repress the activity of the family of E2F transcription factors) by the viral oncogenes E6 and E7, respectively. Interestingly, we also found several genes differentially expressed in our microarray analysis in HPV-positive SCCHN compared to the normal tissue that have been shown to be regulated by p53 or E2F such as the tumour necrosis factor receptor superfamily, member 10b (TNFRSF10B), mutS homolog 6 (MSH6), caldesmon 1 (CALD1), p300/CBP-associated factor (PCAF), collagen, type IV, alpha 1 (COL4A1), nidogen 2 (NID2), astrotactin 2 (ASTN2), cytochrome P450, family 4, subfamily F, polypeptide 3 (CYP4F3), DNA-damage-inducible transcript 4 (DDIT4), chondroitin sulfate proteoglycan 2 (CSPG2), insulin-like growth factor binding protein 6 (IGFBP6), keratin 17 (KRT17), CDC6 cell division cycle 6 homolog (CDC6), replication factor C4, 37kDa (RFC4), flap structure-specific endonuclease 1 (FEN1) and SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 3 (SMARCA3)43, 53–55 (see supplementary data, table C). These data are consistent with the results of previous studies and support the notion that HPV-positive SCCHN has a specific transcription profile depends partly on E6 and E7 expression. Specific E6 or E7 knockdown studies will clarify the validity of this conclusion. After comparing the HPV-positive SCCHN samples against the normal oral tissues, we also identified a group of differentially expressed genes previously implicated in tumourigenesis of different tissues but not in SCCHN. These included the upregulated genes SFRP4 and RAD51AP1 and the downregulated gene PRSS3 (Table 2). SFRP4 has been found to be upregulated in prostate cancers and in microsatellite stable endometrial cancers.44, 56 This protein has also been implicated in the inhibition of the Wnt-signaling cascade that is involved in the regulation of cellular proliferation.44 RAD51AP1 is an S phase cell cycle checkpoint gene that has recently been found to be directly induced by E2F1.45 The overexpression of RAD51AP1 further supports the idea that dysregulation of E2F inducible transcripts occurs in HPV-positive SCCHN due to the inactivation of pRB by E7 expression. PRSS3 is a member of the serine protease family that shares significant homology with trypsinogen 1 and 2 and has been classified as a tumour suppressor gene in several types of cancers such as esophageal squamous cell carcinomas, gastric adenocarcinomas and non-small cell lung cancers.57 There is also some evidence that PRSS3 is downregulated by promoter hypermethylation in lung cancers.57 Interestingly our results show that DNMT1, which is responsible for transcriptional silencing through DNA methylation, is upregulated in HPV-positive SCCHN suggesting possible hypermethylation of genes including PRSS3 by this enzyme.
After statistical analysis of the data obtained with HPV-negative SCCHN, we found 79 upregulated and 83 downregulated genes as compared to the normal oropharyngeal squamous mucosa (supplementary data, Table F). The upregulated genes such as PTHLH and ECGF1, and downregulated genes such as IGFBP5 and PDCD4 were also reported in previous SCCHN studies46, 47 providing a validation of our results. At the same time, we found novel upregulated genes such as AKR1C3 and SNAPC1 and downregulated genes such as CLU and RBM3 that were previously not known to be differentially expressed in SCCHN. AKR1C3 is a hydroxysteroid dehydrogenases involved in the regulation of local concentration of androgens and estrogens in hormone dependent tissues like prostate, breast and endometrium.58 AKR1C3 has been found overexpressed at the mRNA and protein level in prostate cancer specifically in the stromal cells.58 SNAPC1 is a subunit of the TBP-TAF complex involved in the transcription of both RNA polymerase II and III dependent small nuclear RNA genes. SNAPC1 has been linked to the tumourigenesis of breast cancers.49 CLU is a multifunctional gene which is involved in spermatogenesis and in the control of the immunological complement cascade.50 It has recently been shown that CLU is downregulated in low- and high-grade human prostate cancers and it may be involved in supporting cell survival and the induction of programmed cell death.50 RBM3, a glycine-rich RNA-binding protein induced by cold-stress, has recently been found to have apoptotic modulatory capabilities with a possible role as a tumour suppressor gene.51 Interestingly, there is evidence that RBM3 can change the expression of some microRNAs and enhance global protein synthesis suggesting an involvement of a homeostatic mechanism that regulates global levels of protein synthesis under normal and cold-stress conditions.52
We found 124 upregulated and 42 downregulated genes in HPV-positive SCCHN as compared to HPV-negative SCCHN (supplementary data, Table I). Among this group of genes, we found a subset of genes (41 upregulated and 19 downregulated) that were also differentially expressed in HPV-positive SCCHN as compared to the normal oropharyngeal mucosa. It is likely that the differential expression of these 60 genes (Table 5) is a consequence of the presence of HPV-16. This group of genes included the upregulated genes SYCP2, RFC5, ZNF238 and DNMT1, and the downregulated genes KLK8 and CRABP2. SYCP2 is part of the synaptonemal complex involved in forming lateral elements and cross bridges that contribute to pairing of sister chromatids during meiosis and probably are involved in the interaction between chromatin and nuclear envelope.59 There is evidence that HPV infection in cervical cells promotes morphological changes in the nuclear membrane as well as condensation of chromatin attached to the inner part of the nuclear membrane.60 Thus, it is possible that SYCP2 may be involved in chromatin-nuclear envelope interactions in HPV-positive SCCHN. A recent study by Slebos and colleagues also reported up-regulation of the SYCP2 and cyclin-dependent kinase inhibitor 2C (CDKN2C) in HPV-positive SCCHN as compared to HPV-negative SCCHN,61 similar to our data obtained using the two statistical methods. Since the data presented by Slebos and colleagues were based solely on the use of the SAM statistical method, we also compared the differentially expressed genes in our study identified only by using this method (supplementary Table H). This analysis showed that the CDKN2A, NEFH and FLJ12973 genes were similarly affected in both the studies. This further strengthens the likelihood that the above genes play important roles in the carcinogenesis of HPV-related SCCHN. Based on the use of SAM statistical method alone, we found that the SMARCA2 gene was up-regulated in HPV-positive SCCHN as compared to the HPV-negative SCCHN (Table H). This gene belongs to a family of proteins involved in transcriptional regulation by conformational changes of nucleosomes.62 The closely-related and functionally similar SMARCA3 gene was found to be up-regulated in the study of Slebos and colleagues.61 Similarly, while replication factor C 5 (RFC5) was found to be overexpressed in HPV-positive SCCHN in our study (Table 5), Slebos and colleagues reported overexpression of RFC4. Theses genes encode part of the RFC protein complex required, in conjunction with PCNA, in chromosomal DNA replication by DNA polymerase delta and epsilon. RFC5 protein not only interacts directly with PCNA but also with RAD24 which is involved in DNA damage checkpoint control in Saccharomyces cerevisiae.63, 64 These findings suggest a possible role of RFC5 upon infection of the oropharyngeal tissue with HPVs that may promote a DNA damage response.
The transcription factor ZNF238 (also known as RP58) is a component of a C2H2-type DNA-binding zinc finger protein that acts as a repressor of transcriptionally silent heterochromatin regions. Interestingly, the DNA methyltransferase protein Dnmt3a directly interacts with ZNF238 and promotes the repression of specific genes by histone deacetylation independently of its methyltransferase activity.65 The overexpression of ZNF238 could explain the downregulation of several genes in HPV-positive SCCHN as a consequence of chromatin condensation by histone deacetylation. KLK8, which is overexpressed in HPV-positive SCCHN, is a member of the human kallikrein gene family of serine proteases with a diverse physiologic function in many tissues. KLK8 is implicated in terminal differentiation of keratinocytes and in the modulation of interaction between cells and fibronectin in extracellular matrix required for tumour growth and invasion. Several studies show different patterns of KLK8 expression such as its downregulation in breast cancers, and in contrast, its overexpression in cervical cancers.66, 67 CRABP2 is a transcription factor that functions as a cytoplasmic retinoic acid binding protein that is highly expressed in human skin. Similar to our results showing its downregulation in HPV-positive SCCHN, CRABP2 is also downregulated in prostate cancer.68 Finally, in the unsupervised hierarchical cluster analysis, we were able to identify a group of genes in five different clusters with significant variation between HPV-positive SCCHN as compared to HPV-negative SCCHN as well as normal oropharyngeal epithelium (Fig.3). Some of the genes found to be differentially expressed in our statistical analysis were also found in these clusters such as ZNF238, DNMT1, MCM3, AKR1C3, GRB10, TYK2 and SART3 (underlined in Fig. 3). The absence of other clustered genes in our supervised statistical analysis could be explained by differences in expression levels below the 2-fold cut-off or by a value below the statistical significance that is considered by every test and overlapping processes. We also identified the localization of the various differentially expressed genes to particular chromosomes (Fig.4). Two regions in chromosome 1 (p34–p36) and 12 (q21–q240) showed a significant number of differentially expressed genes. This may suggest common regulatory pathways involved in the regulation of some of these genes.
Our studies also identified genes whose expression is similarly affected in both HPV-positive and HPV-negative SCCHN as compared to the normal oropharyngeal tissue. For example, STAT1 is overexpressed while PCAF is downregulated in both types of SCCHN. These genes may represent common pathways that are altered in oropharyngeal carcinogenesis. STAT1 is a transcription factor that participates in cellular events such as IFN signaling, development of the mammary gland and embryogenesis. It is considered a tumour suppressor gene since its activation is associated with growth arrest, but studies in head and neck cancers showed an overexpression of STAT1 in well or moderately differentiated tumours in vivo.69, 70 Our findings contrast with the recent publication of Xi and colleagues71 showing the downregulation of STAT1 in SCCHN through the hypermethylation of its promoter.
In summary, our data demonstrates specific changes in cellular gene expression profiles in HPV-positive SCCHN which also express the viral oncogenes. We have identified several genes such as those involved in nuclear structure and meiosis (SYCP2), cell differentiation (CRABP2), DNA repair (RFC5), transcription regulation (ZNF238) and epidermis development (KLK8) that were differentially expressed specifically in HPV-positive SCCHN as compared to both the HPV-negative SCCHN and normal oropharyngeal mucosa that could be used as potential biomarkers for the development of HPV-associated SCCHN.
Supplementary Material
Acknowledgments
We thank James Lyons-Weiler for help with the analysis of the microarray data. This work was supported by National Institutes of Health Grant DC016406 and pilot grants from the Hillman Foundation and University of Pittsburgh Cancer Institute. SAK and RLF are Hillman Fellows in Innovative Cancer Research.
Footnotes
Conflict of interest
None.
Novelty and impact of the paper: Using a 22,200 human transcript oligonucleotide microarray platform, we have examined the gene expression profiles in HPV-positive and HPV-negative oropharyngeal squamous cell carcinomas as compared to the normal oropharyngeal mucosa. Our study has identified several new genes whose expression is specifically associated with the presence of HPV-16 in the oropharyngeal squamous mucosa. Furthermore, we have identified several genes whose expression is altered in HPV-negative SCCHN as compared to the normal oral mucosa. Results of our study could contribute to the understanding of pathogenic mechanisms involved in the development of HPV-positive and HPV-negative oropharyngeal cancers and consequently in the identification of potential biomarkers associated with these two subtypes of squamous cell carcinomas.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics. CA Cancer J Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 2.Vikram B, Strong EW, Shah JP, Spiro R. Second malignant neoplasms in patients successfully treated with multimodality treatment for advanced head and neck cancer. Head Neck Surg. 1984;6:734–7. doi: 10.1002/hed.2890060306. [DOI] [PubMed] [Google Scholar]
- 3.Ferris RL, Thekdi AA. Diagnostic assessment of squamous cell carcinoma of the larynx. Otolaryngol Clin North Am. 2002;5:953–69. doi: 10.1016/s0030-6665(02)00041-5. [DOI] [PubMed] [Google Scholar]
- 4.Dell G, Gaston K. Contributions in the domain of cancer research: Review Human papillomaviruses and their role in cervical cancer. Cell Mol Life Sci. 2001;58:1923–42. doi: 10.1007/PL00000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–9. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 6.zur Hausen H. Papillomaviruses causing cancer: Evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690–8. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
- 7.zur Hausen H. Papillomaviruses and cancer: From basic studies to clinical application. Nature Rev Cancer. 2002;2:342–50. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz S, Daling J, Doody D, et al. Oral cancer risk in relation to sexual history and evidence of human papillomavirus infection. J Natl Cancer Inst. 1998;90:1626–36. doi: 10.1093/jnci/90.21.1626. [DOI] [PubMed] [Google Scholar]
- 9.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–20. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 10.Steinberg BM, DiLorenzo TP. A possible role for human papillomaviruses in head and neck cancer. Cancer Metastasis Rev. 1996;15:91–112. doi: 10.1007/BF00049489. [DOI] [PubMed] [Google Scholar]
- 11.Ritchie JM, Smith EM, Summersgill KF, et al. Human papillomavirus infection as a prognostic factor in carcinomas of the oral cavity and oropharynx. Int J Can. 2003;3:36–45. doi: 10.1002/ijc.10960. [DOI] [PubMed] [Google Scholar]
- 12.Wong DTW, Munger K. Association of human papillomaviruses with a subgroup of head and neck squamous cell carcinomas. J Natl Cancer Inst. 2000;92:675–7. doi: 10.1093/jnci/92.9.675. [DOI] [PubMed] [Google Scholar]
- 13.Chow LT, Broker TR. Papillomavirus DNA replication. Intervirology. 1994;37:150–8. doi: 10.1159/000150373. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs PG, Pfister H. Transcription of papillomavirus genomes. Intervirology. 1994;37:159–67. doi: 10.1159/000150374. [DOI] [PubMed] [Google Scholar]
- 15.McCance DJ. Transcriptional regulation by human papillomaviruses. Curr Opin Genet Dev. 2005;15:515–9. doi: 10.1016/j.gde.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Mansur CP, Androphy EJ. Cellular transformation by papillomavirus oncoproteins. Biochim et Biophys Acta. 1993;1155:323–45. doi: 10.1016/0304-419x(93)90013-3. [DOI] [PubMed] [Google Scholar]
- 17.McBride AA, Romanczuk H, Howley PM. The papillomavirus E2 regulatory proteins. J Biol Chem. 1991;266:18411–4. [PubMed] [Google Scholar]
- 18.Munger K, Phelps WC. The human papillomavirus E7 protein as a transforming and transactivating factor. Biochim et Biophys Acta. 1993;1155:111–23. doi: 10.1016/0304-419x(93)90025-8. [DOI] [PubMed] [Google Scholar]
- 19.Stoppler H, Stoppler MC, Schlegel R. Transforming proteins of papillomaviruses. Intervirology. 1994;37:168–79. doi: 10.1159/000150375. [DOI] [PubMed] [Google Scholar]
- 20.Durst M, Kleinheinz A, Hotz M, Gissmann L. The physical state of human papillomavirus type 16 DNA in benign and malignant genital tumors. J Gen Virol. 1985;66:1515–22. doi: 10.1099/0022-1317-66-7-1515. [DOI] [PubMed] [Google Scholar]
- 21.Meissner JD. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol. 1999;80:1725–33. doi: 10.1099/0022-1317-80-7-1725. [DOI] [PubMed] [Google Scholar]
- 22.Yee C, Krishnan HI, Baker CC, Schlegel R, Howley PM. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am J Pathol. 1985;119:361–6. [PMC free article] [PubMed] [Google Scholar]
- 23.Lechner MS, Mack DH, Finicle AB, Crook T, Vousden KH, Laimins LA. Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J. 1992;11:3045–52. doi: 10.1002/j.1460-2075.1992.tb05375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantovani F, Banks L. The interaction between p53 and papillomaviruses. Semin Cancer Biol. 1999;9:387–95. doi: 10.1006/scbi.1999.0142. [DOI] [PubMed] [Google Scholar]
- 25.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 26.Jeon S, Allen-Hoffman BL, Lambert PF. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol. 1995;69:2989–97. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 28.Jeon GA, Lee JS, Patel V, et al. Global gene expression profiles of human head and neck squamous carcinoma cell lines. Int J Cancer. 2004;112:249–58. doi: 10.1002/ijc.20399. [DOI] [PubMed] [Google Scholar]
- 29.Van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 30.Van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 31.Kuo WP, Jenssen TK, Park PJ, Lingen MW, Hasina R, Ohno-Machado L. Gene expression levels in different stages of progression in oral squamous cell acrcinoma. Proc AMIA. 2002;Annual Symposium:415–9. [PMC free article] [PubMed] [Google Scholar]
- 32.Leethanakul C, Knezevic V, Patel V, et al. Gene discovery in oral squamous cell carcinoma through the Head and Neck Cancer Genome Anatomy Project: confirmation by microarray analysis. Oral Oncology. 2003;39:248–58. doi: 10.1016/s1368-8375(02)00107-0. [DOI] [PubMed] [Google Scholar]
- 33.Ibrahim SO, Aarsaether N, Holsve MK, et al. Gene expression profile in oral squamous cell carcinomas and matching normal oral mucosal tissues from black Africans and white Caucasians: the case of the Sudan vs. Norway Oral Oncology. 2003;39:37–48. doi: 10.1016/s1368-8375(02)00018-0. [DOI] [PubMed] [Google Scholar]
- 34.Guo X, Lui WO, Qian CN, et al. Identifying cancer-related genes in nasopharyngeal carcinoma cell lines uding DNA and mRNA expression profiling analyses. Int J Oncology. 2002;21:1197–1204. [PubMed] [Google Scholar]
- 35.Gillison ML, Shah KV. Human papillomavirus-associated head and neck squamous cell carcinoma: mounting evidence for an etiologic role for human papillomavirus in a subset of head and neck cancers. Curr Opin Oncol. 2001;13:183–8. doi: 10.1097/00001622-200105000-00009. [DOI] [PubMed] [Google Scholar]
- 36.Ferris RL, Martinez I, Sirianni N, et al. Human papillomavirus-16 associated squamous cell carcinoma of the head and neck (SCCHN): a natural disease model provides insights into viral carcinogenesis. Eur J Cancer. 2005;41:807–15. doi: 10.1016/j.ejca.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Saviozzi S, Calogero RA. Microarray probe expression measures, data normalization and statistical validation. Comp Funct Genom. 2003;4:442–6. doi: 10.1002/cfg.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;4:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 40.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Thompson CH, Cossart YE, et al. The expression of key cell cycle markers and presence of human papillomavirus in squamous cell carcinoma of the tonsil. Head Neck. 2004;26:1–9. doi: 10.1002/hed.10335. [DOI] [PubMed] [Google Scholar]
- 42.Santin AD, Zhan F, Bignotti E, et al. Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology. 2005;331:269–91. doi: 10.1016/j.virol.2004.09.045. [DOI] [PubMed] [Google Scholar]
- 43.Ren B, Cam H, Takahashi Y, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–56. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horvath LG, Henshall SM, Kench JG, et al. Membranous expression of secreted frizzled-related protein 4 predicts for good prognosis in localized prostate cancer and inhibits PC3 cellular proliferation in vitro. Clin Cancer Res. 2004;10:615–25. doi: 10.1158/1078-0432.ccr-0707-03. [DOI] [PubMed] [Google Scholar]
- 45.Iwanaga R, Komori H, Ishida S, et al. Identification of novel E2F1 target genes regulated in cell cycle-dependent and independent manners. Oncogene. 2006;25:1786–98. doi: 10.1038/sj.onc.1209210. [DOI] [PubMed] [Google Scholar]
- 46.Kornberg LJ, Villaret D, Popp M, et al. Gene expression profiling in squamous cell carcinoma of the oral cavity shows abnormalities in several signaling pathways. Laryngoscope. 2005;115:690–8. doi: 10.1097/01.mlg.0000161333.67977.93. [DOI] [PubMed] [Google Scholar]
- 47.Lin SC, Wang CP, Chen YM, et al. Regulation of IGFBP-5 expression during tumourigenesis and differentiation of oral keratinocytes. J Pathol. 2002;198:317–25. doi: 10.1002/path.1220. [DOI] [PubMed] [Google Scholar]
- 48.Penning TM, Steckelbroeck S, Bauman DR, et al. Aldo-keto reductase (AKR) 1C3: role in prostate disease and the development of specific inhibitors. Mol Cell Endocrinol. 2006;248:182–91. doi: 10.1016/j.mce.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 49.Xie D, Jauch A, Miller CW, Bartram CR, Koeffler HP. Discovery of overexpressed genes and genetic alterations in breast cancer cells using a combination of suppression subtractive hybridization, multiplex FISH and comparative genomic hybridization. Int J Oncol. 2002;21:499–507. [PubMed] [Google Scholar]
- 50.Scaltriti M, Brausi M, Amorosi A, et al. Clusterin (SGP-2, ApoJ) expression is downregulated in low- and high-grade human prostate cancer. Int J Cancer. 2004;108:23–30. doi: 10.1002/ijc.11496. [DOI] [PubMed] [Google Scholar]
- 51.Sutherland LC, Rintala-Maki ND, White RD, Morin CD. RNA binding motif (RBM) proteins: a novel family of apoptosis modulators? J Cell Biochem. 2005;94:5–24. doi: 10.1002/jcb.20204. [DOI] [PubMed] [Google Scholar]
- 52.Dresios J, Aschrafi A, Owens GC, Vanderklish PW, Edelman GM, Mauro VP. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc Natl Acad Sci USA. 2005;102:1865–70. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei CL, Wu Q, Vega VB, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–19. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 54.Kannan K, Amariglio N, Rechavi G, et al. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene. 2001;20:2225–34. doi: 10.1038/sj.onc.1204319. [DOI] [PubMed] [Google Scholar]
- 55.Zhao R, Gish K, Murphy M, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–93. [PMC free article] [PubMed] [Google Scholar]
- 56.Risinger JI, Maxwell GL, Chandramouli GV, et al. Gene expression profiling of microsatellite unstable and microsatellite stable endometrial cancers indicates distinct pathways of aberrant signaling. Cancer Res. 2005;65:5031–7. doi: 10.1158/0008-5472.CAN-04-0850. [DOI] [PubMed] [Google Scholar]
- 57.Marsit CJ, Okpukpara C, Danaee H, Kelsey KT. Epigenetic silencing of the PRSS3 putative tumor suppressor gene in non-small cell lung cancer. Mol Carcinog. 2005;44:146–50. doi: 10.1002/mc.20125. [DOI] [PubMed] [Google Scholar]
- 58.Penning TM, Steckelbroeck S, Bauman DR, et al. Aldo-keto reductase (AKR) 1C3: role in prostate disease and the development of specific inhibitors. Mol Cell Endocrinol. 2006;248:182–91. doi: 10.1016/j.mce.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 59.Shakib K, Norman JT, Fine LG, Brown LR, Godovac-Zimmermann J. Proteomics profiling of nuclear proteins for kidney fibroblasts suggests hypoxia, meiosis, and cancer may meet in the nucleus. Proteomics. 2005;5:2819–38. doi: 10.1002/pmic.200401108. [DOI] [PubMed] [Google Scholar]
- 60.Bollmann M, Bankfalvi A, Trosic A, et al. Can we detect cervical human papillomavirus (HPV) infection by cytomorphology alone? Diagnostic value of non-classic cytological signs of HPV effect in minimally abnormal Pap tests. Cytopathology. 2005;1:13–21. doi: 10.1111/j.1365-2303.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- 61.Slebos RJ, Yi Y, Ely K, et al. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12:701–9. doi: 10.1158/1078-0432.CCR-05-2017. [DOI] [PubMed] [Google Scholar]
- 62.Yamamichi N, Yamamichi-Nishina M, Mizutani T, et al. The Brm gene suppressed at the post-transcriptional level in various human cell lines is inducible by transient HDAC inhibitor treatment, which exhibits antioncogenic potential. Oncogene. 2005;24:5471–81. doi: 10.1038/sj.onc.1208716. [DOI] [PubMed] [Google Scholar]
- 63.Kim J, MacNeill SA. Genome stability: a new member of the RFC family. Curr Biol. 2003;13:R873–875. doi: 10.1016/j.cub.2003.10.048. [DOI] [PubMed] [Google Scholar]
- 64.Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in cooperation with rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5888–96. doi: 10.1128/mcb.20.16.5888-5896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–44. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yousef GM, Yacoub GM, Polymeris ME, Popalis C, Soosaipillai A, Diamandis EP. Kallikrein gene downregulation in breast cancer. Br J Cancer. 2004;90:167–72. doi: 10.1038/sj.bjc.6601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cane S, Bignotti E, Bellone S, et al. The novel serine protease tumor-associated differentially expressed gene-14 (KLK8/Neuropsin/Ovasin) is highly overexpressed in cervical cancer. Am J Obstet Gynecol. 2004;190:60–6. doi: 10.1016/j.ajog.2003.07.020. [DOI] [PubMed] [Google Scholar]
- 68.Okuducu AF, Janzen V, Ko Y, et al. Cellular retinoic acid-binding protein 2 is down-regulated in prostate cancer. Int J Oncol. 2005;27:1273–82. [PubMed] [Google Scholar]
- 69.Calo V, Migliavacca M, Bazan V, et al. STAT proteins: from normal control of cellular events to tumourigenesis. J Cell Physiol. 2003;197:157–68. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 70.Arany I, Chen SH, Megyesi JK, et al. Differentiation-dependent expression of signal transducers and activators of transcription (STATs) might modify responses to growth factors in the cancers of the head and neck. Cancer Lett. 2003;199:83–9. doi: 10.1016/s0304-3835(03)00345-8. [DOI] [PubMed] [Google Scholar]
- 71.Xi S, Dyer KF, Kimak M, et al. Decreased STAT1 expression by promoter methylation in squamous cell carcinogenesis. J Natl Cancer Inst. 2006;98:181–9. doi: 10.1093/jnci/djj020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


