Abstract
The systemic inflammatory response syndrome (SIRS) is a life-threatening medical condition characterized by a severe and generalized inflammatory state that can lead to multiple organ failure and shock. The CNS regulates many features of SIRS such as fever, cardiovascular, and neuroendocrine responses. Central and systemic manifestations of SIRS can be induced by LPS or IL-1β administration. The crucial role of IL-1β in inflammation has been further highlighted by studies of mice lacking caspase 1 (casp1, also known as IL-1β convertase), a protease that cleaves pro-IL-1β into mature IL-1β. Indeed, casp1 knockout (casp1−/−) mice survive lethal doses of LPS. The key role of IL-1β in sickness behavior and its de novo expression in the CNS during inflammation led us to test the hypothesis that IL-1β plays a major role modulating the brain transcriptome during SIRS. We show a gene–environment effect caused by LPS administration in casp1−/− mice. During SIRS, the expression of several genes, such as chemokines, GTPases, the metalloprotease ADAMTS1, IL-1RA, the inducible nitric oxide synthase, and cyclooxygenase-2, was differentially increased in casp1−/− mice. Our findings may contribute to the understanding of the molecular changes that take place within the CNS during sepsis and SIRS and the development of new therapies for these serious conditions. Our results indicate that those genes may also play a role in several neuropsychiatric conditions in which inflammation has been implicated and indicate that casp1 might be a potential therapeutic target for such disorders.
Keywords: IL-1β, LPS, mouse
Systemic inflammatory response syndrome (SIRS) is an aggressive and multifactorial pathophysiological state that has a high mortality rate of up to 51% and has been ranked as the 10th leading cause of death in intensive care units (1). SIRS diagnosis includes a requirement for two or more of the following symptoms: fever or hypothermia, tachycardia, and leukocytosis or leucopenia (2). The major difference between SIRS and sepsis is that sepsis is produced by infectious microorganisms, whereas SIRS may occur in the absence of a documented source of infection (3). Although causation is difficult to ascertain in complex and life-threatening medical illnesses, there is ample clinical and experimental evidence to support the concept that the severity of the clinical inflammatory response is the critical determinant of outcome and survival (4). In fact, the inflammatory response produced during sepsis is within the spectrum of SIRS and involves rapid amplification of numerous signals and responses that spread beyond the invaded tissue (3). Septic shock may happen when failure of major organs occurs because counterregulatory control mechanisms are overwhelmed and homeostasis is jeopardized (5). One of the major components that underlines the pathophysiology of sepsis is the exacerbated activation of the innate immune response with the consequent increase of proinflammatory cytokines such as IL-1β and TNF-α (3). These cytokines cause clear biological effects mediated through the CNS such as fever (6, 7), anorexia (7), and activation of the hypothalamic–pituitary–adrenal axis (8) resulting in increased production of adrenal corticoids, which in turn increase the cardiovascular and respiratory rates.
The pivotal role of the innate immune system during SIRS is documented by the increase of proinflammatory factors after administration of LPS from Gram-negative bacteria (9–16), which produces a generalized inflammation similar to that observed during SIRS and sepsis (17).
In rodent models of SIRS, we have previously shown that peripheral administration of LPS increased synthesis of proinflammatory factors occurring first in peripheral systems and organs and then in brain areas that are within the blood–brain barrier (BBB) (13, 14). Specifically, we showed that specific and similar spatial–temporal changes in the expression of IL-1β and inducible nitric oxide synthase [nitric oxide synthase 2 (NOS2)] occurred in the rat brain during LPS-induced SIRS (13, 14). Their expression initiated in areas that were predominantly outside of the BBB such as the pituitary, the pineal gland, and the choroid plexus and were subsequently evident in brain regions that display leaky BBB such as the subfornical organ and arcuate nucleus and other brain regions such as the paraventricular nucleus (13, 14).
The important role exerted by IL-1β during LPS-induced SIRS was previously demonstrated in knockout (KO) and transgenic mice with altered components of the IL-1 network (18, 19). Mice with a KO of the casp1 gene, a cysteine-protease that cleaves biologically inactive pro-IL-1β to render the biologically active 18 kDa IL-1β, are resistant to a lethal dose of LPS (18). In contrast, mice with a KO of the endogenous IL-1 receptor antagonist (IL-1RA) were shown to be more sensitive to the lethal effects of LPS (19). Altogether those results support the notion that IL-1β has a major proinflammatory role during SIRS. Because many of the changes that occur during inflammation require de novo synthesis and because IL-1β is an important proinflammatory cytokine that we showed to be synthesized within the brain (13), we hypothesized that casp1, a key modulator of IL-1β bioactivity, could regulate the brain transcriptome during LPS-induced SIRS. Thus, to understand the participation of casp1 and IL-1β within the CNS during SIRS, we used microarrays to compare CNS transcriptional changes between casp1−/− and wild-type mice after peripheral LPS administration.
Results
Microarray.
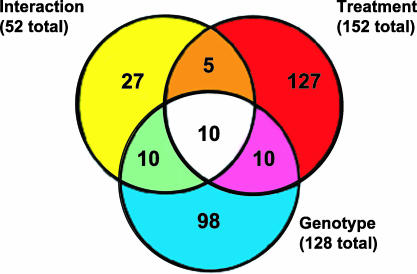
Peripheral administration of LPS induced transcriptional changes within the mouse brain after 6 h of administration. ANOVA analysis showed that the expression of a total of 287 transcripts was significantly altered (P < 0.01) (Fig. 1). These transcripts were sorted according to their main effect, namely genotype, treatment, or interaction as illustrated in the Venn diagram (Fig. 1). Transcripts that were differentially expressed had predominantly an LPS (treatment) effect (152 transcripts) followed by genotype (128 transcripts) and finally by interaction (52 transcripts). Thirty-five transcripts showed significant changes in at least two of these categories, and 10 of them had significant changes in all three categories (Fig. 1). Another important criterion in microarray analysis is the fold change (FC) of transcription expression. It is now accepted that FC has to be at least 1.8 to be considered as a potential relevant alteration in gene expression (20–22). In our studies, the FC represents changes in the mRNA expression of the whole brain, which dilutes changes that are region-specific. Therefore, an FC ±1.8 or greater is in this study a highly stringent criterion. Our FC analysis revealed that from the total 287 transcripts identified in our analysis, 32 transcripts displayed an FC ±1.8 or greater and had an LPS treatment effect regardless of the mice genotype.
Fig. 1.
Summary of microarray results. Within the Venn diagram, circles represent the main effects of our ANOVA analysis: LPS treatment (red circle), genotype (yellow circle), and interaction (blue circle). Numbers inside each compartment represent the number of transcripts that are significant for that effect. The intersections of the sets represent genes with P < 0.01 for each of the effects involved in the intersection.
Cluster Analysis.
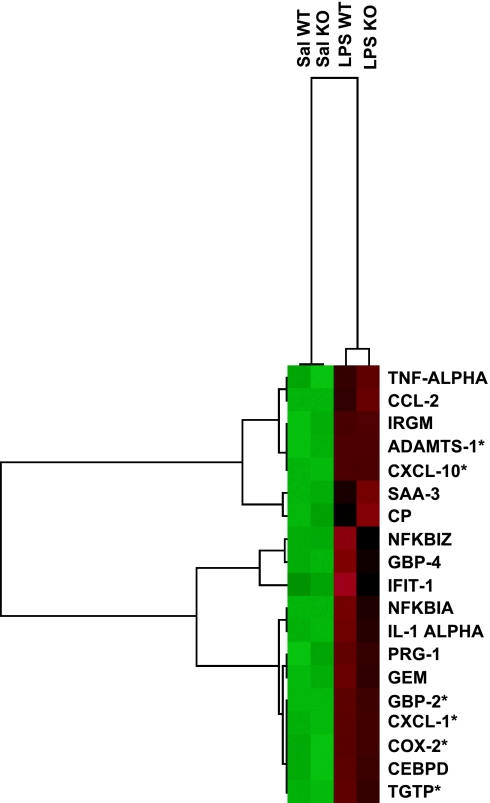
Fig. 2 shows the cluster analyses of 19 genes of the 32 transcripts mentioned previously that we selected for confirmation studies by using RT-PCR. Regardless of the genotype, the expression level of all of these transcripts was relatively low in saline-treated mice (green). LPS treatment produced variable increases in gene expression, which are reflected in the intensity of the color red. At least 12 of these genes are related to inflammation and/or immune response. From the remaining seven, four of them are related to intracellular pathways such as signal transduction and/or transcription regulation, one with transport, and one with metalloprotease activity.
Fig. 2.
Transcriptional patterns within the CNS during LPS-induced SIRS. Cluster analysis of genes that displayed a treatment FC 1.8 or greater and that were confirmed by RT-PCR. In each treatment group, green squares represent a low mRNA concentration during resting conditions, whereas red squares represent up-regulation after 6 h of administration of LPS. These genes can be classified according to their biological process as follows: (i) immune, inflammatory, and/or acute phase response: TNF-α, CCL-2, IRGM, CXCL-10, SAA-3, NFkbiz, GBP-4, IFIT-1, NFKBIa, IL-1α, CXCL-1, COX-2, and TGTP; (ii) cell proliferation: GBP-2; (iii) proteolysis: ADAMTS-1; (iv) transcription: CEBPD; (v) ion transport: CP; (vi) signal transduction: GEM; (vii) undetermined: PRG-1. Sal WT, saline-injected wild-type mice; Sal KO, saline-injected KO; LPS WT, LPS-injected wild-type, LPS KO, LPS-injected KO; CCL-2, chemokine (C-C motif) ligand 2; IRGM, immunity-related GTPase family, M; ADAMTS-1, a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1, motif 1; CXCL-10, chemokine (C-X-C motif) ligand 10; SAA-3, serum amyloid A 3; CP, ceruloplasmin; NFKBIZ, nuclear factor of κ light polypeptide gene enhancer in B-cells inhibitor, ζ; GBP-4, guanylate nucleotide binding protein 4; IFIT-1, IFN-induced protein with tetratricopeptide repeats 1; NFKBIA, nuclear factor of κ light chain gene enhancer in B-cells inhibitor, α; PRG-1, proteoglycan 1, secretory granule; GEM, GTP binding protein (gene overexpressed in skeletal muscle); GBP-2, guanylate nucleotide binding protein 2; CXCL-1, chemokine (C-X-C motif) ligand 1; COX-2, cyclooxygenase 2; CEBPD, CCAAT/enhancer binding protein (C/EBP), δ; TGTP, T cell-specific GTPase. ∗, genes that showed genotype effect after performing semiquantitative PCR.
Confirmation of Change of Gene Expression Within the LPS Treatment Group.
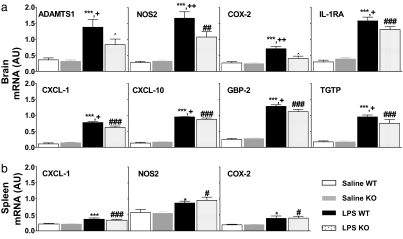
Using semiquantitative RT-PCR, we assessed gene expression in RNA samples from individual mouse tissue that were pooled for the microarray experiment. We confirmed the treatment effect of 19 genes that showed an FC ±1.8 or greater (Fig. 2). We also analyzed the expression level of other genes that are known to be increased during LPS-induced inflammation such as NOS2, IL-6, IL-1β, IL-1RA, and IL-10. Interestingly, our RT-PCR results showed that there was a significant genotype effect in eight of these genes, six that were selected from the microarray (Figs. 2 and 3) and two from the literature, NOS2 and IL-1RA (Fig. 3). Mice of the different genotype showed significant differences after LPS treatment: casp1−/− mice had significantly smaller induction in the expression (10–50% lower when compared with the wild type) in six genes, and they showed no induction in the expression levels of two genes [ADMTS1 and cyclooxygenase-2 (COX-2)] (Fig. 3).
Fig. 3.
Semiquantitative RT-PCR of genes induced 6 h after LPS. We have confirmed by RT-PCR the expression of the 19 genes shown in Fig. 2. All of these genes displayed a significant LPS effect when compared with the saline-treated group (P < 0.001). (a) Genes that were differentially increased by LPS within the brain are as follows: ADAMTS-1, NOS2, COX-2, IL-1RA, CXCL-1, CXCL-10, GBP-2, and TGTP. (b) Lack of genotype effect in the spleen. ∗∗∗, P < 0.001 vs. Sal WT group; ∗, P < 0.05 vs. Sal WT group; +, P < 0.05 vs. LPS KO group; ++, P < 0.01 vs. LPS KO group; #, P < 0.05 vs. Sal KO group; ##, P < 0.01 vs. Sal KO group; ###, P < 0.001 vs. Sal KO group; ⋀, P value is not significant vs. Sal KO group.
Peripheral Tissues.
Spleen tissue, an immune organ, was used to test whether the CNS transcriptional changes also occurred in the periphery. RT-PCR experiments of three genes that had significant genotype effect in the CNS, namely CXCL-1, NOS2, and COX-2, showed no significant differences between expression levels in wild-type and casp1−/− mice either at baseline or after LPS treatment in the spleen (Fig. 3).
Discussion
The pathophysiology of SIRS is characterized by a number of changes that alter the cross-talk among the immune, endocrine, and neural systems (23). Several of these events are regulated at the genomic level, resulting in changes in gene expression of proinflammatory and antiinflammatory factors both within the CNS (13, 14) and the periphery (24). We focused on casp1 because this is a key regulator of IL-1β, a cytokine that is activated in a specific spatial–temporal manner within the rat brain (7), and it also plays a crucial role during LPS-induced inflammation (18, 19).
Our microarray studies have shown that 6 h after i.p. injection of LPS, the expression of 152 transcripts was significantly altered within the brain. However, only a fraction of these transcripts showed a FC 1.8 or greater, probably because we used whole brain tissue and changes in the level of mRNA expression reflect the net change that occurs in various cell types; therefore, it is not surprising that most of our changes are relatively small, because large FCs would reflect very large transcription increases in a significant proportion of cells. Thus, the level of FC shown should be interpreted as a minimum level of expression that would be greatly enhanced if the discrete brain areas were to be studied.
Although our microarray results showed that the LPS (treatment) effect was the most significant effect for the genes whose expression was significantly altered by FC 1.8 or greater, our semiquantitative RT-PCR results obtained in individual mice samples revealed that some of these genes also displayed a genotype effect. Therefore, our results suggest that LPS-induced IL-1β plays an important stimulatory role in the regulation of gene expression within the brain. Some of these genes such as IL-1RA (25), NOS2 (8), COX-2 (26), CXCL-1 (27), and CXCL-10 (27) were already described to be increased in the brain after peripheral administration of endotoxin. We previously described that, within the brain, LPS-induced SIRS elicited a pattern of IL-1β mRNA expression that was similar to that of NOS2, suggesting that IL-1β modulated NOS2 expression. The fact that casp1−/− mice had lower levels of LPS-induced NOS2 expression when compared with wild-type mice further strengthens the data supporting the critical role of IL-1β in inducing NOS2 increase after LPS administration. It is well known that NO increases the synthesis of prostaglandins by stimulating the expression of the inducible COX-2, thus, it appears that in the lower stimulation of the DNA-directed synthesis of NOS2 in casp1−/− mice led to decreased expression of COX-2.
The chemokines CXCL-1 and CXCL-10 had smaller gene expression activation in casp1−/− mice after LPS. These chemokines belong to a group of small molecules that control cell trafficking of various types of leukocytes through interactions with G protein-coupled receptors (28). Both CXCL-1 and CXCL-10 have already been described to be induced by LPS in areas of the brain that lack BBB. CXCL-1 was found to be up-regulated in the choroid plexus, whereas CXCL-10 was up-regulated in circumventricular organs, including the subfornical organ and area postrema, with an expression pattern that was consistent with the biological action of chemokines to recruit leukocytes from the circulation into the CNS (29). Therefore, our data support the notion that deficiency of IL-1β decreases leukocyte trafficking into the brain parenchyma.
Two other genes that were shown to be differentially induced by LPS between wild-type and casp1−/− mice were TGTP and GBP-2, which may have antiviral effects. In the periphery, TGTP expression was found in T-lymphocytes (30) and macrophages (31), whereas in the brain, TGTP was found to be induced by murine cytomegalovirus, suggesting that it is involved in the antiviral response triggered by IFN-γ (32). With regard to GBP-2, it was reported that it is involved in the IFN-induced antiviral activity (33) and that in vitro it increased fibroblast proliferation (34). To the best of our knowledge, its expression and role in the CNS has not been previously reported.
Gene expression of ADAMTS1 was also smaller in casp1−/− mice. ADAMTS1 is an enzyme that belongs to the family of the metalloproteases, which promote the proteolytic degradation of the extracellular matrix of maternal decidua, a biological process that is stimulated by IL-1β (35). Because during the inflammatory stress produced by LPS, several metalloproteases have been shown to degrade components of the BBB (36), it might be possible that IL-1β-induced ADAMTS1 participates in this process, too.
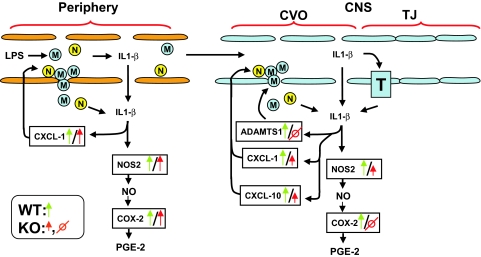
Fig. 4 summarizes our results and presents a hypothetical pathophysiological mechanism as a consequence of IL-1β deficiency during inflammation. LPS binds to its CD14 receptors present on peripheral macrophages and monocytes (37). Thereafter, the LPS-CD14 complex interacts with Toll-like receptors to trigger the synthesis (37), posttranslational processing through activation of casp1 (38), and release of IL-1β together with other proinflammatory cytokines such as TNF-α and IL-6 (39). Then, proinflammatory cytokines may enter into the brain parenchyma either through a saturable transport system or through areas of the brain that display leaky BBB such as the choroid plexus and circumventricular organs (40). This process is facilitated by chemokines (CXCL-1 and CXCL-10), which attract macrophages and neutrophiles to the circumventricular organs, and metalloproteases such as ADAMTS1, which degrade extracellular matrix and allow extravasation of these leukocytes from blood to the brain parenchyma. Increased IL-1β within the brain parenchyma would trigger its own synthesis and release from glial cells, which in turn trigger synthesis of NOS2 with the consequent increase of NO. Thus, increased NO will propagate through the brain altering the activity of preformed enzymes that display heme-oxygenase groups and triggering de novo synthesis of target genes that play important roles in the inflammatory cascade such as COX-2. Rise in COX-2 levels will lead to the increase of prostaglandin E2, a major proinflammatory molecule. A similar mechanism would also occur in casp1−/− mice, but they would display a smaller degree of inflammation than the wild-type mice. Indeed, deficiency of IL-1β would lead to reduced LPS-induced stimulatory effect of several genes such as CXCL-1, CXCL-10, ADAMTS1, NOS2, and, ultimately COX-2. This could be relevant in protecting casp1−/− mice against lethal doses of LPS (18, 19).
Fig. 4.
Summary diagram. It is hypothesized that peripherally injected LPS acts on receptors present on macrophages, monocytes, and neutrophils to increase synthesis and release of proinflammatory cytokines such as IL-1β. IL-1β might enter into the brain parenchyma through different routes; IL-1β or leukocytes producing IL-1β might enter through brain areas that display leaky BBB such as arcuate nucleus, subfornical organ, and choroid plexus or through specific IL-1β transporters. Metalloproteases such as ADAMTS1 increase endothelial permeability that allow infiltration and entrance of proinflammatory cytokines. IL-1β induces its own synthesis and also NOS2 acting mainly in glial cells. Increased NOS2 expression leads to the consequent increase of NO that in turn triggers the synthesis of COX-2, which in turn leads to increased synthesis and release of prostaglandin E2, a potent proinflammatory prostanoid that modulates fever. A similar mechanism is depicted in the periphery with the genes that were assayed in the spleen. Red arrows and symbols refer to casp1−/− mice, whereas green arrows and symbols refer to wild-type mice. M, macrophage or monocyte; N, neutrophil; T, IL-1β specific transporter; CVO, circumventricular organs; TJ, tight junction.
Because casp1 also cleaves IL-18 and the recently characterized IL-33 (41), the participation of these two cytokines in the biological processes described here have to be considered. We interpreted that IL-1β is the major cytokine that accounts for the differences between wild-type and casp1−/− mice because the synthesis and release of this proinflammatory cytokine is highly increased by LPS, whereas IL-18 was not altered in our studies. However, IL-18 was shown to be essential for Shigella Flexneri-induced inflammation (42) and trauma-induced brain damage (43). To the best of our knowledge, there are no data reporting that LPS alters synthesis and/or release of IL-33. The fact that IL-33 was reported to drive a TH2 response (41) decreases the likelihood that LPS would alter its expression. A major biological action of this cytokine was recently described as a crucial intracellular nuclear factor with transcriptional regulatory properties associated with Crohn's disease (44).
In conclusion, our studies of the interactions of genetic background (casp1 deficiency) and environment (LPS-induced systemic inflammation) provided new insights into the role of casp1 and IL-1β in modulating the CNS gene expression profile that occurs during SIRS. Based on our findings, we hypothesized a mechanism through which IL-1β modulates the inflammatory cascade within the brain that ultimately leads to the increase in the synthesis and release of prostaglandin E2. Studies carried out in humans emphasized the major role that IL-1β plays in brain inflammation in patients undergoing neonatal-onset multisystem inflammatory disease, a pathological condition caused by a dysfunctional regulation of caspase-1 (45). Indeed, their brain inflammatory condition was highly improved by the daily administration of s.c. injections of the IL-1RA anakinra. The pathological role of proinflammatory factors within the brain might also be linked to neuropsychiatric disorders. In fact, sickness-like behavior can be elicited by both proinflammatory cytokines and major depression, suggesting that common molecular pathways are shared by inflammation and neuropsychiatric disorders. Interestingly, it was recently shown that the COX-2 inhibitor celecoxib has therapeutic effects both in major depression (46) and schizophrenia (47). Thus, reduction of casp1 bioactivity (48, 49) might be a possible therapeutic strategy to treat both SIRS and neuropsychiatric disorders with prominent inflammatory components. It is noteworthy that casp1 inhibitors are the first orally active agents that target cytokines. The casp1 inhibitor pranalcasan is in clinical trials. Such drugs reduce not only IL-1β, but also the proinflammatory actions of IL-18 and IL-33 (44). Therefore, a better understanding of the molecular events that occur during inflammation within the CNS could lead to the development of novel therapeutic strategies to treat disorders associated with activation of inflammatory pathways in the brain.
Materials and Methods
Mice and Sample Preparation.
Studies were carried out in accordance with animal protocols approved by the National Institutes of Health and UCLA. Experiments were designed to avoid confounding variables such as infection, stress, and circadian variation in mRNA levels. We used virus and antibody-free, 8-week-old, male C57BL/6 mice housed in a light- (12-h on/12-h off) and temperature-controlled environment with food and water ad libitum. Injections were timed so that all tissue collection occurred at 1000 to 1200. Different groups of animals were given i.p. injections of either saline or 25 mg/kg Escherichia coli (serotype 055:B5) LPS (Sigma–Aldrich, St. Louis, MO) and killed 6 h later. Brains were extracted, snap-frozen, and stored at −80°C until processing. Total RNA was extracted from each of the brains by using TRIzol (Invitrogen, Carlsbad, CA). The RNA was cleaned by using RNeasy columns (Qiagen, Valencia, CA), its concentration measured by spectrophotometry, and the quality assessed by running a Nano-Chip assay in an Agilent 2100 electrophoresis bioanalyzer (Life Sciences Chemical Analysis, Foster City, CA). The same amount of high-quality RNA from each of the five mouse samples was used to make LPS and saline RNA sample pools.
Probe Preparation and Hybridization.
Following the protocol recommended by Affymetrix (Santa Clara, CA), 10 μg of each RNA pool was processed in duplicate as follows: T7-(dT) 24-primed RT and second-strand synthesis was done by using the SuperScript Choice System (Invitrogen), and in vitro transcription yielding biotin-labeled cRNA was performed with the Enzo BioArray HighYield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY). Labeled cRNA was purified by using RNeasy columns (Qiagen) and then fragmented at 94°C for 35′ with a Tris-acetate buffer containing magnesium and potassium acetate. Fragmented cRNA (15 μg) was used for hybridization in a 2-(N-morpholino) ethanesulfonic acid-based (Sigma–Aldrich) buffer containing BSA, herring sperm DNA, control oligonucleotide B2, and eukaryotic hybridization controls BioB, BioC, BioD, and cre (Affymetrix). Samples were processed in duplicate on separate occasions and hybridized to Affymetrix Murine Genome Mu11KA and B microarrays at 45°C for 16 h under constant rotation. After hybridization, the microarrays were washed, stained with streptavidin phycoerythrin (Molecular Probes, Eugene, OR), washed again, and scanned in a Hewlett-Packard GeneChip scanner (Palo Alto, CA). Affymetrix Mu 11KA and B chips provided the capacity of the use of built-in positive controls; thus, the expression levels of proinflammatory cytokines such as TNF, IL-1β, IL-6, IL-12, and INFγ were used as positive controls.
Statistical Analysis of Microarray Data.
Microarrays were scanned and the array image data files (CEL) were loaded into Dchip Ver1.1 software package (50). This package computed two summary measures for each gene. The first measure was the model-based expression value that summarized the approximate expression level over the 20 individual probes. The second measure was the present/absent call. This call was a decision rule using a number of details of the probe pairs to determine whether there was any mRNA corresponding to that particular gene in the sample. Dchip was used to normalize the data and compute the model-based expression index and standard error for the model-based expression index for each gene. Next, Dchip identified outliers, which are treated as missing in the subsequent analyses. Our procedure for identifying differentially expressed genes relied on a set of four metrics as follows: (i) absent/present calls: We required that the model-based expression index was significantly different from zero in at least one of the treatment groups; (ii) absolute difference: We required that the absolute difference in expression exceeded a certain threshold. This threshold was one-fifth of the median expression, which gave a value of 100; (iii) FC: We required that the FC between the treatment conditions exceeded a certain threshold: (±1.8 or 1.6 statistically based on the ±3 SD of the null FC distribution); and (iv) lower bound of the 95% confidence interval: We required that the lower bound (in absolute value) of the 95% confidence interval did not include 1. Genes that passed all four metrics were identified as differentially expressed.
Hierarchical clustering was performed by using CLUSTER and TREEVIEW software (51). Genes and arrays were clustered by using Average Linkage Clustering. Expression values in the cluster diagrams were standardized by subtracting the mean expression and dividing by the standard deviation.
Semiquantitative RT-PCR.
First-strand cDNAs were prepared with SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) with 1 μg of RNA and random hexamers primers. The cDNAs were diluted five times after synthesis and 0.5 μl was used in each PCR. We used Quantum mRNA 18S Internal Standards (Ambion, Austin, TX) to perform multiplex PCR with the following modifications to the manufacturer's protocol. For all of the genes tested, a ratio of 0.5:9.5 for 18S primer:competimer was used. The optimal cycle number was determined to be the one in which reactions for the specific gene and 18S in saline- and LPS-treated samples were in the linear range. For a given gene tested, all samples were assayed concomitantly using aliquots of the same PCR mixture and first-strand cDNA derived from individual samples; PCR products were loaded in the same agarose gel; gel was SYBR-green stained and analyzed by using AlphaImager System (Alpha Innotech, San Leandro, CA). Ratio of signal intensities of the interested gene versus the internal control was calculated for every sample. We selected all of the genes that in the microarray showed a FC >1.8 and genes that were previously shown to be expressed within the brain such as NOS2.
Statistical Analysis.
Differences were analyzed by one-way ANOVA followed by the Student-Neuman-Keuls multiple comparison test for unequal replications. Data are expressed as mean ± SEM. We set the threshold of P < 0.05 for each of these effects.
Acknowledgments
We thank the University of California at Los Angeles Microarray Center and Dr. Stanley Nelson for their help and expertise in the conduction of array experiments. This work was supported by National Institutes of Health Grants MH062777, RR017365, and DK063240 (to M-L.W.) and RR017611, RR016996, HL004526, DK058851, HG002500, and GM061394–04 (to J.L.). J.H. was supported by a National Institutes of Health postgraduate training grant in psychoneuroimmunology (University of California, Los Angeles) and institutional funds from Department of Psychiatry and Behavioral Sciences, University of Miami Miller School of Medicine.
Abbreviations
- BBB
blood–brain barrier
- COX-2
cyclooxygenase-2
- FC
fold change
- IL-1RA
IL-1 receptor antagonist
- KO
knockout
- NOS2
nitric oxide synthase 2
- SIRS
systemic inflammatory response syndrome.
Footnotes
The authors declare no conflict of interest.
References
- 1.Martin GS, Mannino DM, Eaton S, Moss M. N Engl J Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 2.Bone RC. J Am Med Assoc. 1992;268:3452–3455. [Google Scholar]
- 3.Takala A, Nupponen I, Kylanpaa-Back ML, Repo H. Ann Med. 2002;34:614–623. doi: 10.1080/078538902321117841. [DOI] [PubMed] [Google Scholar]
- 4.Sasse KC, Nauenberg E, Long A, Anton B, Tucker HJ, Hu TW. Crit Care Med. 1995;23:1040–1047. doi: 10.1097/00003246-199506000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Cohen J. Nature. 2002;420:885–891. doi: 10.1038/nature01326. [DOI] [PubMed] [Google Scholar]
- 6.Dinarello CA. J Endotoxin Res. 2004;10:201–222. doi: 10.1179/096805104225006129. [DOI] [PubMed] [Google Scholar]
- 7.Maier SF, Goehler LE, Fleshner M, Watkins LR. Ann NY Acad Sci. 1998;840:289–300. doi: 10.1111/j.1749-6632.1998.tb09569.x. [DOI] [PubMed] [Google Scholar]
- 8.Licinio J, Wong ML. Mol Psychiatry. 1997;2:99–103. doi: 10.1038/sj.mp.4000251. [DOI] [PubMed] [Google Scholar]
- 9.Wong ML, Xie B, Beatini N, Phu P, Marathe S, Johns A, Gold PW, Hirsch E, Williams KJ, Licinio J, Tabas I. Proc Natl Acad Sci USA. 2000;97:8681–8686. doi: 10.1073/pnas.150098097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mastronardi CA, Srivastava V, Yu WH, Les Dees W, McCann SM. Neuroimmunomodulation. 2005;12:182–188. doi: 10.1159/000084851. [DOI] [PubMed] [Google Scholar]
- 11.Mastronardi CA, Yu WH, McCann S. Neuroimmunomodulation. 2001;9:148–156. doi: 10.1159/000049019. [DOI] [PubMed] [Google Scholar]
- 12.Mastronardi CA, Yu WH, Rettori V, McCann S. Neuroimmunomodulation. 2000;8:91–97. doi: 10.1159/000026458. [DOI] [PubMed] [Google Scholar]
- 13.Wong ML, Bongiorno PB, Rettori V, McCann SM, Licinio J. Proc Natl Acad Sci USA. 1997;94:227–232. doi: 10.1073/pnas.94.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong ML, Rettori V, al-Shekhlee A, Bongiorno PB, Canteros G, McCann SM, Gold PW, Licinio J. Nat Med. 1996;2:581–584. doi: 10.1038/nm0596-581. [DOI] [PubMed] [Google Scholar]
- 15.Mastronardi CA, Yu WH, Srivastava VK, Dees WL, McCann SM. Proc Natl Acad Sci USA. 2001;98:14720–14725. doi: 10.1073/pnas.251543598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong ML, O'Kirwan F, Khan N, Hannestad J, Wu KH, Elashoff D, Lawson G, Gold PW, McCann SM, Licinio J. Proc Natl Acad Sci USA. 2003;100:14241–14246. doi: 10.1073/pnas.2336220100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dantzer R, Bluthe RM, Gheusi G, Cremona S, Laye S, Parnet P, Kelley KW. Ann NY Acad Sci. 1998;856:132–138. doi: 10.1111/j.1749-6632.1998.tb08321.x. [DOI] [PubMed] [Google Scholar]
- 18.Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnston C, McDowell J, Paskind M, Rodman L, Salfeld J, et al. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch E, Irikura VM, Paul SM, Hirsh D. Proc Natl Acad Sci USA. 1996;93:11008–11013. doi: 10.1073/pnas.93.20.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jagoe RT, Lecker SH, Gomes M, Goldberg AL. Faseb J. 2002;16:1697–1712. doi: 10.1096/fj.02-0312com. [DOI] [PubMed] [Google Scholar]
- 21.Sokolov BP, Polesskaya OO, Uhl GR. J Neurochem. 2003;84:244–252. doi: 10.1046/j.1471-4159.2003.01523.x. [DOI] [PubMed] [Google Scholar]
- 22.Franco S, Canela A, Klatt P, Blasco MA. Carcinogenesis. 2005;26:1613–1626. doi: 10.1093/carcin/bgi107. [DOI] [PubMed] [Google Scholar]
- 23.McCann SM, Mastronardi C, de Laurentiis A, Rettori V. Ann NY Acad Sci. 2005;1057:64–84. doi: 10.1196/annals.1356.064. [DOI] [PubMed] [Google Scholar]
- 24.Pitossi F, del Rey A, Kabiersch A, Besedovsky H. J Neurosci Res. 1997;48:287–298. doi: 10.1002/(sici)1097-4547(19970515)48:4<287::aid-jnr1>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 25.Eriksson C, Nobel S, Winblad B, Schultzberg M. Cytokine. 2000;12:423–431. doi: 10.1006/cyto.1999.0582. [DOI] [PubMed] [Google Scholar]
- 26.Quan N, Whiteside M, Herkenham M. Brain Res. 1998;802:189–197. doi: 10.1016/s0006-8993(98)00402-8. [DOI] [PubMed] [Google Scholar]
- 27.Reyes TM, Walker JR, DeCino C, Hogenesch JB, Sawchenko PE. J Neurosci. 2003;23:5607–5616. doi: 10.1523/JNEUROSCI.23-13-05607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cartier L, Hartley O, Dubois-Dauphin M, Krause KH. Brain Res Brain Res Rev. 2005;48:16–42. doi: 10.1016/j.brainresrev.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 29.Liu MT, Keirstead HS, Lane TE. J Immunol. 2001;167:4091–4097. doi: 10.4049/jimmunol.167.7.4091. [DOI] [PubMed] [Google Scholar]
- 30.Carlow DA, Marth J, Clark-Lewis I, Teh HS. J Immunol. 1995;154:1724–1734. [PubMed] [Google Scholar]
- 31.Lafuse WP, Brown D, Castle L, Zwilling BS. J Leukoc Biol. 1995;57:477–483. doi: 10.1002/jlb.57.3.477. [DOI] [PubMed] [Google Scholar]
- 32.van den Pol AN, Robek MD, Ghosh PK, Ozduman K, Bandi P, Whim MD, Wollmann G. J Virol. 2007;81:332–348. doi: 10.1128/JVI.01592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlow DA, Teh SJ, Teh HS. J Immunol. 1998;161:2348–2355. [PubMed] [Google Scholar]
- 34.Gorbacheva VY, Lindner D, Sen GC, Vestal DJ. J Biol Chem. 2002;277:6080–6087. doi: 10.1074/jbc.M110542200. [DOI] [PubMed] [Google Scholar]
- 35.Ng YH, Zhu H, Pallen CJ, Leung PC, MacCalman CD. Human Reprod. 2006;21:1990–1999. doi: 10.1093/humrep/del108. [DOI] [PubMed] [Google Scholar]
- 36.Rosenberg GA. Glia. 2002;39:279–291. doi: 10.1002/glia.10108. [DOI] [PubMed] [Google Scholar]
- 37.Guha M, Mackman N. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 38.Schumann RR, Belka C, Reuter D, Lamping N, Kirschning CJ, Weber JR, Pfeil D. Blood. 1998;91:577–584. [PubMed] [Google Scholar]
- 39.Reichlin S. J Endocrinol Invest. 2004;27:48–61. [PubMed] [Google Scholar]
- 40.McCann SM, Kimura M, Karanth S, Yu WH, Mastronardi CA, Rettori V. Ann NY Acad Sci. 2000;917:4–18. doi: 10.1111/j.1749-6632.2000.tb05368.x. [DOI] [PubMed] [Google Scholar]
- 41.Dinarello CA. Immunity. 2005;23:461–462. doi: 10.1016/j.immuni.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Sansonetti PJ, Phalipon A, Arondel J, Thirumalai K, Banerjee S, Akira S, Takeda K, Zychlinsky A. Immunity. 2000;12:581–590. doi: 10.1016/s1074-7613(00)80209-5. [DOI] [PubMed] [Google Scholar]
- 43.Sifringer M, Stefovska V, Endesfelder S, Stahel PF, Genz K, Dzietko M, Ikonomidou C, Felderhoff-Mueser U. Neurobiol Dis. 2007;25:614–622. doi: 10.1016/j.nbd.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 44.Carriere V, Roussel L, Ortega N, Lacorre DA, Americh L, Aguilar L, Bouche G, Girard JP. Proc Natl Acad Sci USA. 2007;104:282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldbach-Mansky R, Dailey NJ, Canna SW, Gelabert A, Jones J, Rubin BI, Kim HJ, Brewer C, Zalewski C, Wiggs E, et al. N Engl J Med. 2006;355:581–592. doi: 10.1056/NEJMoa055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller N, Schwarz MJ, Dehning S, Douhe A, Cerovecki A, Goldstein-Muller B, Spellmann I, Hetzel G, Maino K, Kleindienst N, et al. Mol Psychiatry. 2006;11:680–684. doi: 10.1038/sj.mp.4001805. [DOI] [PubMed] [Google Scholar]
- 47.Riedel M, Strassnig M, Schwarz MJ, Muller N. CNS Drugs. 2005;19:805–819. doi: 10.2165/00023210-200519100-00001. [DOI] [PubMed] [Google Scholar]
- 48.Dinarello CA. Curr Opin Pharmacol. 2004;4:378–385. doi: 10.1016/j.coph.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Loher F, Bauer C, Landauer N, Schmall K, Siegmund B, Lehr HA, Dauer M, Schoenharting M, Endres S, Eigler A. J Pharmacol Exp Ther. 2004;308:583–590. doi: 10.1124/jpet.103.057059. [DOI] [PubMed] [Google Scholar]
- 50.Li C, Wong WH. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisen MB, Spellman PT, Brown PO, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]