Summary
The coordinated action of histone acetyltransferases (HATs) and ATP-dependent chromatin remodeling enzymes in promoter-dependent transcription initiation represents a paradigm for how epigenetic information regulates gene expression. However, little is known about how such enzymes function during transcription elongation. Here we investigated the role of RSC, a bromodomain-containing ATPase, in nucleosome transcription in vitro. Purified S. cerevisiae RNA polymerase II (pol II) arrests at two primary locations on a positioned mononucleosome. RSC stimulates passage of pol II through these sites. The function of RSC in elongation requires the energy of ATP hydrolysis. Moreover, the SAGA and NuA4 HATs strongly stimulated RSC’s effect on elongation. The stimulation correlates closely with Acetyl-CoA-dependent recruitment of RSC to nucleosomes. Thus, RSC can recognize acetylated nucleosomes and facilitate passage of pol II through them. These data support the view that histone modifications regulate accessibility of the coding region to pol II.
Introduction
Transcription initiation by RNA polymerase II involves the coordinated action of the general transcription machinery with chromatin modifying and remodeling enzymes (Roeder, 2005). ATP-dependent remodeling enzymes such as the multisubunit SWI/SNF play a critical role in altering the chromatin landscape to permit preinitiation complex assembly. The recruitment of SWI/SNF is achieved by its direct interaction with activators or by binding of its bromodomain to lysines modified by histone acetyltransferases (HATs) such as SAGA (Peterson and Workman, 2000). However, after pol II initiates and commences elongation little is known of the mechanism by which it transcribes through a nucleosome. A key issue is whether the ATP-dependent remodeling machines and HATs influence transcription elongation?
Biochemical studies have revealed that the nucleosome presents a potent barrier to pol II elongation (Studitsky et al., 2004). Pol II arrests at pause sites near the nucleosome and becomes locked in a catalytically inactive state. Studitsky and colleagues have shown that pol II elongation in vitro on a simple mononucleosome substrate displaces an H2A/H2B dimer generating a hexasome but otherwise does not change the position of the nucleosome on DNA (Kireeva et al., 2002). Elongation through the block is facilitated by salt concentrations that favor loss of the dimer. Enzymes that remove a dimer may also promote elongation. ACF/ISWI facilitates H2A-H2B removal in a manner dependent upon the p300 histone acetyltransferase. It has been speculated that this effect may underlie the ability of p300 to stimulate transcription in vitro (Ito et al., 2000).
Several known elongation factors including FACT and TFIIS stimulate pol II passage through a nucleosome. FACT stimulates elongation on mononucleosomes and nucleosomal arrays by promoting removal of H2A/H2B dimers (Belotserkovskaya et al., 2003; Reinberg and Sims, 2006). FACT-dependent transcription on arrays is further stimulated by the PAF elongation complex and monoubiquitination of H2B by RNF40/60 and UbcH6 (Pavri et al., 2006). TFIIS stimulates transcript cleavage and permits backtracking by pol II on mononucleosomes (Kireeva et al., 2005). The backtracking reactivates arrested pol II allowing it to resample the nucleosomal block until it eventually passes. TFIIS is a component of chromatin transcription enabling activity (CTEA), which stimulates elongation on chromatin arrays. TFIIS works in concert with p300-mediated acetylation to promote elongation (Guermah et al., 2006).
Despite considerable progress in recapitulating pol II elongation on chromatin substrates in cell free systems the role of HATs and ATP-dependent chromatin remodeling enzymes has remained elusive. Kingston and colleagues reported that SWI/SNF enhances HSF1-mediated stimulation of pol II paused near the start-site in vitro (Brown et al., 1996). Additionally, transcription of nucleosomal substrates is stimulated by p300 and SAGA in vitro (Kraus and Kadonaga, 1998; Steger et al., 1998). However, there is no direct evidence that ATPases and HATs coordinate their function during elongation. Moreover, it is clear that co-transcriptional histone acetylation occurs in vivo and a signaling mechanism involving histone methylation is used to subsequently deacetylate nucleosomes in transcribed regions (Workman, 2006). Yet the reasons for and functions of co-transcriptional acetylation are unknown.
To explore these issues we utilized an elongation assay employing a mononucleosome substrate preceded by a single-stranded stretch of dC residues. The dC-stretch facilitates initiation by pol II in the absence of the general transcription machinery. Using purified yeast pol II, RSC, NuA4 and SAGA we demonstrate that RSC facilitates pol II elongation on a nucleosome and that its effect is stimulated by HATs.
Results and Discussion
Nucleosome Elongation Assay
To study pol II elongation we employed a template bearing a 20-nucleotide, single-stranded stretch of dC residues ligated to the 601 mononucleosome positioning sequence (Thastrom et al., 2004) (Figure 1A). The so-called “C-tail” has been widely used to study transcription elongation on free DNA templates (Kadesch and Chamberlin, 1982) and has been shown to support transcription of mononucleosomes by pol II in vitro (Liu et al., 2003). On our template, pol II initiates at the border between the 20-nucleotide, single-stranded C-tail and the double-stranded DNA bearing the 601 sequence. The C-tailed template was assembled into chromatin using unmodified recombinant X. laevis octamers (Luger et al., 1997). The template was then utilized for in vitro transcription using purified S. cerevisiae pol II. We typically employ chromatin preparations where ∼90% of the DNA is assembled into chromatin as measured by EMSA. Figure 1B compares pol II elongation on equivalent amounts of free DNA and nucleosomal template. Pol II elongates efficiently on free DNA and generates a full-length (FL) transcript of the expected size. However, on two preparations of nucleosomal substrates, pol II arrests at several discreet locations and only a small amount of FL transcript is generated. As stated by others, we believe the FL transcript synthesized in this reaction is due to the small amount of contaminating free DNA in the nucleosome preparations (Kireeva et al., 2002).
Figure 1.
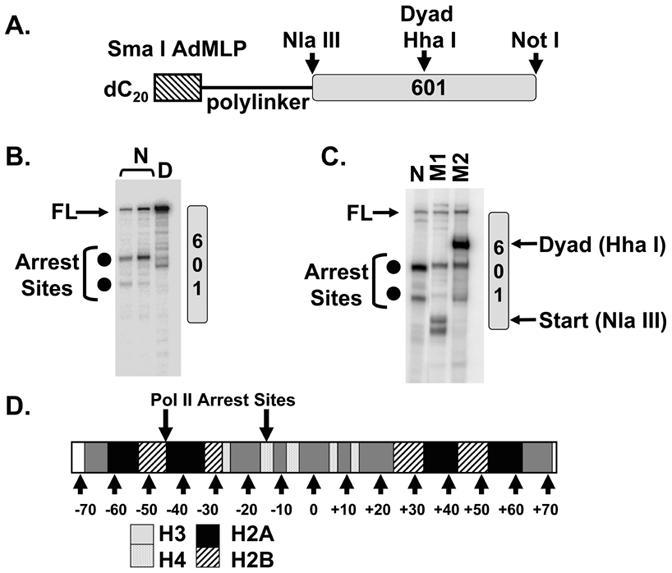
The Pol II Elongation Assay.
(A) Schematic of the template. The template contained single-stranded dC20 attached to double-stranded DNA bearing the 601 nucleosome positioning sequence. The position of 601 and restriction endonuclease cleavage sites used for RNA mapping are indicated.
(B) Transcription of Free DNA (D) and Nucleosome (N). Pol II was incubated with the template and nucleotides for 1 h. The products were fractionated on an 8% polyacrylamide/urea gel. FL is the 252 nucleotide full-length transcript. Closed circles adjacent to the gel indicate the major arrest products.
(C) The C-tailed 601 sequence was cleaved with either Nla III (M1) or Hha I (M2). The DNAs were mixed with the nucleosomal substrate (N) and subjected to transcription by pol II. The arrows indicate the run-off transcripts.
(D) An illustration of the DNA-histone contacts along a 147-bp nucleosome [adapted from (Luger et al., 1997)] with gray arrows above the bar indicating the approximate locations of the major pol II arrest products. ImageQuant TL software was used to map the mobilities of the arrest products relative to the markers.
The two major arrest sites were mapped roughly using RNAs generated from run-off transcription on DNA templates cleaved with restriction endonucleases located within the 601 sequence (Figure 1C). The arrest sites are located at positions where the H2A/H2B dimer and H3/H4 tetramer contact DNA within the first half of the nucleosome (Figure 1D). A similar result has been noted previously in a mononucleosome transcription assay in which pol II initiation was achieved on a template bearing an RNA primer (Kireeva et al., 2005; Kireeva et al., 2002). In conclusion, the data in Figure 1 show that nucleosomes form a strong barrier to elongation by pol II in our transcription system.
The Effects of ATP-dependent Remodeling Enzymes on Pol II Elongation
ATP-dependent chromatin remodeling complexes render the promoter accessible to the general transcription machinery. We wished to establish if remodeling complexes also assist pol II in overcoming the nucleosomal barrier during elongation. We initiated our analysis using RSC (Cairns et al., 1996) and SWI/SNF (Cote et al., 1994), two of the best-characterized yeast ATP-dependent remodeling enzymes. RSC and SWI/SNF were purified from yeast strains expressing TAP-tagged subunits. Preliminary titrations were performed to determine the optimal amounts of enzyme that would stimulate transcription. Figure 2 shows a titration of SWI/SNF and RSC within their active ranges. Both enzymes decrease the accumulation of arrested RNA products and increase the production of FL transcript. RSC exhibited a potent effect on elongation and stimulated production of FL transcript by 6-fold versus the background level. More importantly, when considering the sum of the two arrests products and FL transcript, in the absence of RSC the arrest products accounted for 86% of the transcription, while FL accounted for 14%. In the presence of RSC this ratio was essentially reversed as FL accounted for 79% of the product. The effect of SWI/SNF was weaker and FL transcript increased only 2-fold. We also tested CHD1 (Tran et al., 2000) and ISWI (Tsukiyama et al., 1995), two other ATP-dependent remodeling enzymes that influence transcription in vivo. At optimal concentrations these yielded stimulatory effects similar to SWI/SNF (data not shown).
Figure 2.
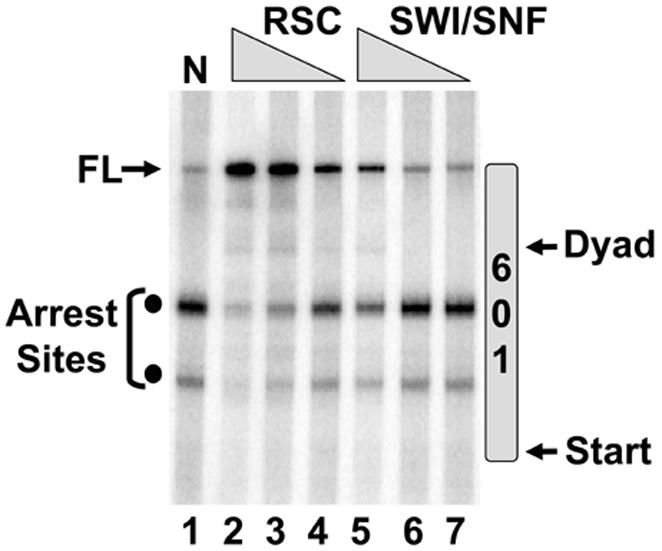
Effects of RSC and SWI/SNF on Nucleosomal Transcription. The nucleosomal templates were preincubated with the SWI/SNF (30, 10, 3 fmol) or RSC (100, 33, 11 fmol) for 30 min at 30°C. Pol II was added for 1 hour and the 32P-RNA products were fractionated on a gel. N represents reaction containing the nucleosome and pol II without the ATPase.
It is unclear why SWI/SNF elicits a weaker effect on elongation. SWI/SNF has been linked genetically to elongation factors such as TFIIS (Davie and Kane, 2000). However, the connection between RSC and pol II is more direct. RSC was shown to interact directly with Rpb5, a subunit shared among RNA polymerases I, II and III (Soutourina et al., 2006). It is plausible that this direct interaction facilitates elongation through the nucleosomal barrier.
ATP-Dependence of RSC in Elongation
The mechanism of RSC action is well studied and involves movement of the histone octamer along the DNA in an ATP-dependent manner (Saha et al., 2002; Saha et al., 2005). The catalytic subunit of RSC, Sth1, directs DNA translocation beginning two turns from the dyad. The enzyme pulls DNA in from one side of the dyad and moves it toward the opposite side (Saha et al., 2005). To determine whether RSC was stimulating transcription in an ATP-dependent manner we altered the reaction conditions because pol II utilizes ATP as a substrate. However, pol II can also employ AMPPNP (adenyl-5'-yl imidodiphosphate) for transcript elongation in vitro. AMPPNP bears an intact α-β phosphoanhydride bond necessary for transcript elongation but contains a non-hydrolyzable β-γ bond necessary for the function of ATP-dependent enzymes (Sawadogo and Roeder, 1984). We used dATP in place of ATP to provide the energy for remodeling by RSC.
Figure 3 illustrates the ATP-dependence of RSC on transcription. In the absence of dATP, RSC alters the arrest site preference such that the pol II favors the pause site located where the H2A-H2B dimer contacts DNA. Additionally, the amount of FL transcript decreases slightly. These effects may be due to ATP-independent binding of RSC to the nucleosome. Importantly, however, addition of dATP stimulates the appearance of FL transcript and the disappearance of arrest products. The disappearance correlates with the appearance of RNA bands that chase upwards from the arrest products to FL. Whereas the arrested RNAs represent 76% of the total transcript in reactions containing nucleosome alone (lane 1) they represent only 35% in the reaction containing the highest concentration of RSC and dATP (lane 5). We conclude that the stimulation of transcription by RSC requires ATP hydrolysis consistent with the idea that active chromatin remodeling is necessary for efficient pol II elongation. Indeed, we have also found that RSC increases the accessibility of the 601 nucleosome to restriction enzyme cleavage but pol II does not augment this effect (data not shown). Therefore, although pol II may directly interact with RSC (Soutourina et al., 2006), we have no compelling reason to believe that pol II fundamentally alters remodeling by RSC.
Figure 3.
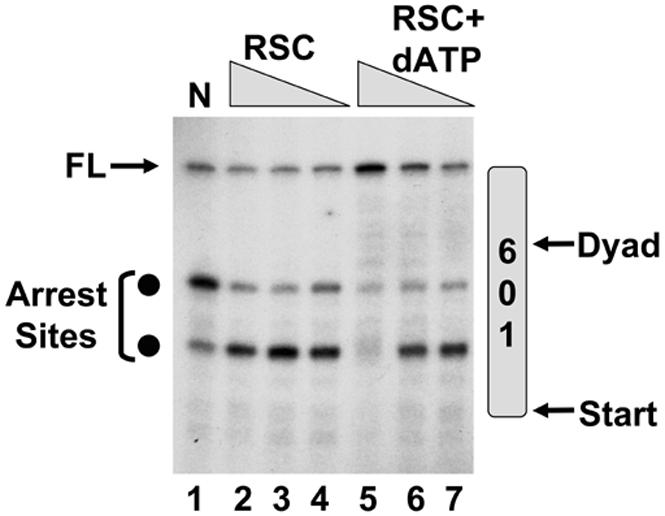
ATP-Dependence of RSC. The transcription mixtures contained 200 μM AMPPNP in place of ATP. Increasing concentrations of RSC were added in the presence and absence of 2 mM dATP for 30 min followed by 1-h incubation with pol II.
RSC Elongation Activity is Stimulated by SAGA and NuA4
The bromodomains of RSC4 recognize H3K14, the main target of the GCN5 HAT subunit of SAGA (Kasten et al., 2004). This feature, combined with the fact that RSC contains six additional bromodomains, suggested that the activity of RSC might be stimulated in vitro by acetylated nucleosomes. We therefore characterized the action of two well-studied HATs, SAGA and NuA4, on the ability of RSC to stimulate pol II elongation. Both HATS have been previously shown to stimulate recruitment of SWI/SNF to nucleosomal arrays (Hassan et al., 2001). However, unlike SAGA, NuA4 acetylates predominantly the four conserved lysines in Histone H4 (Allard et al., 1999). Figure 4A demonstrates that SAGA and Acetyl-CoA alone do not stimulate transcription. In contrast, when combined with RSC, SAGA and Acetyl-CoA increase the overall amount of FL transcript and decrease the appearance of the arrest products. The stimulatory effect is most evident at low RSC concentrations.
Figure 4.
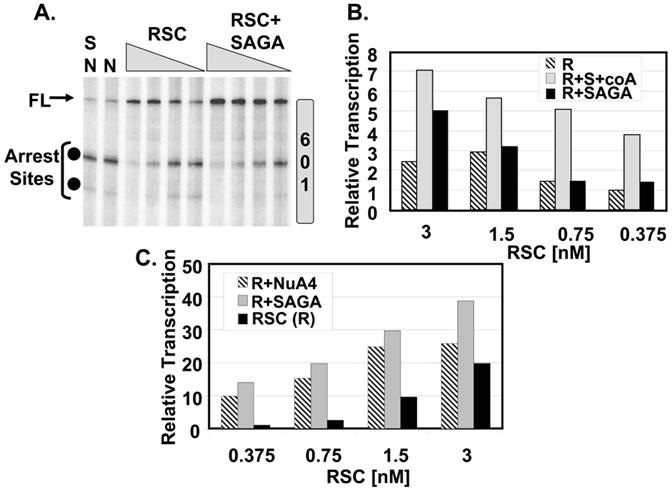
The Effects of SAGA and NuA4 on RSC-stimulated Transcription. (A) SAGA Stimulates RSC. The transcription mixtures, where indicated, contained 170 fmol SAGA (S) preincubated for 30 min with the nucleosome (N) in the presence of Acetyl-CoA, followed by a 30-min incubation with 100, 50, 25 and 12.5 fmol RSC in the presence of 2 mM ATP. Pol II was added for 1 h and the products were isolated and fractionated on an 8% polyacrylamide/urea gel.
(B) The effect of Acetyl-CoA (coA) on stimulation of RSC (R) by SAGA (S). The transcription mixtures contained SAGA and the indicated concentrations of RSC in the presence (gray bars) and absence (black bars) of 500 μM Acetyl-CoA (coA). The data were quantitated using ImageQuant TL. The FL signal from the nucleosome alone was subtracted and the relative transcription signals were plotted as a bar graph normalized to the transcription stimulated by 0.375 nM RSC alone (assigned a value of 1).
(C) The effects of SAGA and NuA4 were compared using the experimental scheme in (A). The transcription mixtures contained approximately SAGA or NuA4 and the indicated concentrations of RSC. The data were quantitated as above. The signal from the nucleosome alone was subtracted and the relative transcription signals were plotted as a bar graph normalized to the transcription stimulated by 0.375 nM RSC alone. The amount of pol II transcription in the presence of RSC alone and with SAGA and NuA4 is shown.
To determine if the stimulatory effect of SAGA was dependent upon its HAT activity we performed transcription reactions in the presence and absence of Acetyl-CoA. Figure 4B is a bar graph quantitating the results. The graph illustrates that transcription in the presence of Acetyl-CoA and SAGA is greater than SAGA alone. At low concentrations of RSC the stimulatory effect of SAGA is completely dependent upon Acetyl-CoA. However, at high concentrations there appears to be a 2-fold stimulation of RSC even in the absence of Acetyl-CoA. We conclude that the effect of SAGA on elongation is largely dependent upon histone acetylation but there appears to be a minor Acetyl-CoA independent contribution.
NuA4 also stimulated RSC-responsive transcription. Figure 4C is a bar graph quantitating an experiment in which NuA4 and SAGA were compared. The relative increase in transcription by RSC alone and in the presence of SAGA and NuA4 is plotted as described in the legend. The data reveal that NuA4 stimulates RSC-dependent transcription but not as well as SAGA. NuA4 had no effect on transcription in the absence of RSC (data not shown). The effect of both HATs was typically more evident when RSC was present at low concentrations. These effects ranged from 10-to 14-fold for NuA4 and SAGA, respectively. This observation raised the possibility that SAGA- and NuA4-mediated histone modifications could be recruiting RSC via its bromodomains to the 601 nucleosome.
Acetylation Enhances RSC Binding to the Nucleosome
To determine whether NuA4 and SAGA were enhancing recruitment of RSC we performed electrophoretic mobility shift assays (EMSA) on 32P-labeled C-tailed 601 nucleosome substrates. Figures 5A and Bshow that RSC binding to the nucleosome is significantly enhanced in the presence of SAGA or NuA4. Nucleosome binding by RSC can occur in the absence of ATP and it is stimulated only slightly by its presence. This result taken together with the ATP-dependence of its transcriptional effect implies that remodeling by RSC, and not simply binding per se, is required for stimulation of pol II elongation.
Figure 5.
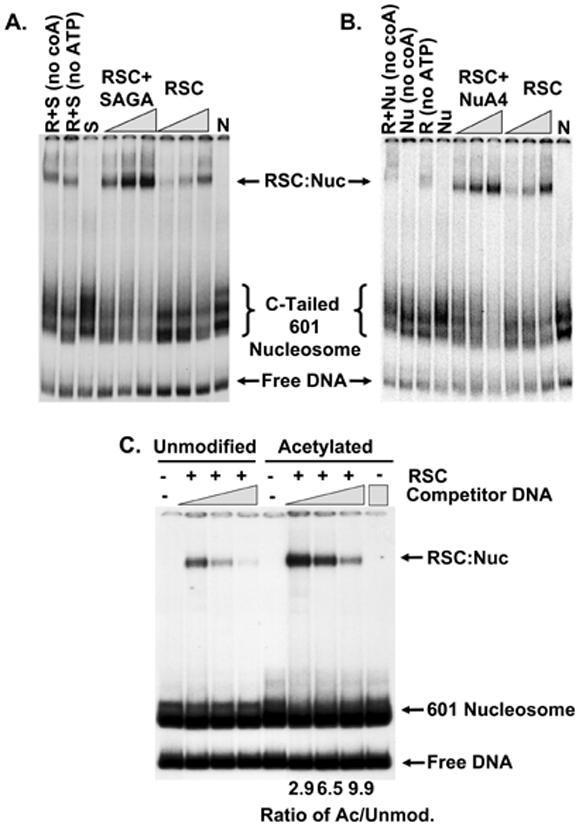
EMSA of RSC-Nucleosome Complexes.
(A) SAGA stimulates RSC recruitment. The EMSA reactions were essentially as described for the transcription reactions above. The mixtures contained nucleosome (N) and 60, 30 and 15 fmol RSC (R) in the presence and absence of SAGA, ATP and Acetyl-CoA. Reactions lacking Acetyl-CoA or ATP contained SAGA and 1.5 nM RSC. The locations of the various complexes are indicated to the right.
(B) NuA4 stimulates RSC recruitment. The reactions were as indicated except employed NuA4 (Nu) in place of SAGA.
(C) Acetylated nucleosomes recruit RSC. Unacetylated and acetylated 32P-labeled 601 nucleosomes were prepared as described in Experimental Procedures. The reaction mixtures contained 4, 8 and 16 ng competitor DNA and 0.25 nM RSC and were fractionated on 3.5% polyacrylamide gels.
The SAGA-stimulated recruitment of RSC appears to be largely dependent upon Acetyl-CoA although there is a minor stimulation in reactions lacking Acetyl-CoA. The data are consistent with the effect of SAGA on RSC-stimulated pol II elongation in Figure 4C. It is plausible that the Acetyl-CoA-independent component of the binding is due to interaction of RSC and SAGA. However, SAGA alone does not bind to the nucleosome under the conditions employed in the assay nor does it supershift the RSC-nucleosome complex. Since SAGA acetylates the nucleosome under the conditions employed here the interaction must be transient or too weak to observe by EMSA.
To determine if the histone modifications alone were sufficient to augment RSC binding we designed a strategy to remove SAGA from the nucleosome preparations after acetylation. We generated a 32P-labeled, biotinylated 601 nucleosome bearing an Eco RV restriction site on a flanking stretch of DNA. The nucleosomes were immobilized on streptavidin-coated magnetic beads and treated with SAGA and Acetyl-CoA. SAGA was removed by washing the beads in high salt buffer after which the nucleosomes were released from the beads by Eco RV digestion. The purified nucleosomes were then subjected to EMSA in the presence of a fixed concentration of RSC. Different amounts of carrier DNA were added to optimize RSC binding. The data in Figure 5C establish that RSC binds better to acetylated versus unmodified nucleosomes. The effect of the modifications is accentuated by the presence of carrier DNA. The ratio of RSC binding to the acetylated versus unmodified nucleosome increases with the amount of carrier. We conclude that histone modifications are sufficient to recruit RSC to a nucleosome even in the absence of SAGA.
We crafted a hypothesis based on emerging data that suggested RSC directly interacts with pol II (Soutourina et al., 2006). We reasoned that RSC would be required for elongation and that, based on the preponderance of bromodomains contained within its subunits, its activity would be stimulated by HATs. We found that both RSC and SWI/SNF stimulated pol II elongation on a simple nucleosomal substrate in an ATP-dependent manner. The effects of RSC were stronger than SWI/SNF in both the elimination of pol II arrest sites and the production of FL transcript. RSC-mediated pol II elongation through the nucleosomal barrier was enhanced by SAGA and NuA4. The majority of the SAGA stimulation was dependent upon Acetyl-CoA. The transcriptional stimulatory effects of NuA4 and SAGA correlated well with an Acetyl-CoA-dependent increase in RSC binding measured by EMSA. Importantly, the acetylated nucleosome was still able to recruit RSC after removal of SAGA. These observations imply that the acetylated nucleosome alone stimulates binding of RSC.
An emerging issue in the literature is the role of acetylation during transcription elongation. Coding regions are actively deacetylated by Rpd3S in a manner dependent upon methylation of H3K36 by Set2 [reviewed in (Lieb and Clarke, 2005)]. Deacetylation restricts initiation to the 5' ends of genes and mutations in Rpd3S subunits cause initiation within the coding region (Carrozza et al., 2005). Interestingly, mutations in other elongation factors such as FACT and Spt6 elicit similar effects on internal initiation [reviewed in (Lieb and Clarke, 2005)]. Therefore, one role of the machinery involved in elongation [reviewed in (Shilatifard et al., 2003; Sims et al., 2004)] is likely to be maintenance of chromatin stability during and after passage of pol II. The recruitment of RSC, SAGA and NuA4 may be restricted to the nucleosomes preceding the elongating pol II. This would permit acetylation and remodeling of chromatin to be transient so as to prevent nucleosome removal or remodeling after pol II passage.
The ability of individual factors like RSC, FACT and SII to promote pol II elongation through nucleosomes in vitro raises the issue of how they can function independently. The simplest explanation is that in vivo the process of elongation involves proteins that have context-dependent functions. In some cases these functions might be dependent upon the position of pol II on the gene. The use of a C-tail template could mask the effects of factors required for proper function at a promoter-initiated system but emphasize proteins involved in downstream events. Another explanation is that the RSC and FACT complexes have overlapping roles and stimulate the others activity much like the effect of SAGA and RSC or SII and FACT. It is likely that the most informative biochemical reconstitution of pol II elongation on a oligo-nucleosomal template will require a complex mixture of factors including Elongator, COMPASS, SptT4/5 and Spt6, along with a promoter-initiated pol II (Shilatifard et al., 2003).
In conclusion, our data provide a mechanism by which histone modification and recruitment of a remodeling enzyme can stimulate pol II elongation. These results provide one example of why co-transcriptional histone acetylation occurs and a reason for deacetylating nucleosomes following the passage of the polymerase.
Experimental Procedures
Assembly of the Nucleosomal Template
The template contained the Bam HI-Not I fragment from pGEM3Z601R ligated to an oligonucleotide bearing a 20-nucleotide single-stranded dC-tail, a 34-nucleotide double stranded sequence containing a Sma I site and AdMLP start sequence, and a Bgl II overhang. Typically, ∼1 μg of DNA was assembled into chromatin as previously described (Li et al., 2005) using recombinant X. laevis octamers (Luger et al., 1997). The chromatin preparations were analyzed on 5% polyacrylamide gels (29:1). Samples in which 80 to 90% of the DNA was assembled into chromatin were employed for in vitrotranscription or EMSA. At higher octamer concentrations the DNA aggregated and migrated in the well. For EMSA assays, the DNA was first labeled with γ32P-ATP and polynucleotide kinase prior to chromatin assembly.
In Vitro Transcription and EMSA
The 20-μl transcription reaction mixtures contained buffer A (20 mM HEPES, pH 7.9, 10 mM MgCl2, 50 mM NaCl, 10% glycerol, 1 mM DTT, 50 μg/ml BSA, 0.1% NP40), 3.3 ng of chromatin (0.6 nM), 10 ng of pGem3Z601R carrier DNA, 500 μM ATP, CTP, and GTP, 5 μM UTP and 5-10 μCi of α32P-UTP. Pol II (Rpb9-TAP), RSC (Rsc2-TAP), SWI/SNF (Snf6-TAP), SAGA (Spt7-TAP) and NuA4 (Epl1-TAP) were purified using a standard TAP tag protocol (Li et al., 2003). NuA4 and SAGA were added at the concentrations indicated in the legends. In a typical reaction the chromatin was modified for 30 min by the HAT in the presence of 500 μM Acetyl-CoA, followed by a 30-min incubation with RSC prior a 1-h incubation with RNA polymerase II (40-80 fmol). The reactions were terminated by addition of 100 μl stop buffer containing 0.3 M NaOAc, 0.1 % SDS and 5 mM EDTA supplemented with 1 μg of proteinase K. After 15 min at 55°C, the products were extracted once with phenol-chloroform and ethanol precipitated. The 32P-RNAs were fractionated on 18-cm 8% polyacrylamide/urea gels. The gels were dried, exposed to a Phosphorimager or XAR-5 film, and quantitated using ImageQuant TL software.
EMSAs in Fig. 5A and B were performed essentially as the transcription assays described above except that 32P-labeled C-tailed 601 mononucleosome was employed. Nucleotides were omitted with the exception of ATP when indicated. The products were separated on 4% native polyacrylamide gels (37.5:1 acrylamide-bis acrylamide) electrophoresed in 0.5 X TBE at 4°C. In Fig. 5C, the reactions contained 0.25 nM 601 nucleosome lacking a C-tail. The 601 nucleosome was assembled on a 32P-labeled biotinylated PCR fragment bearing an Eco RV site at one end. Nucleosomes were immobilized on streptavidin-conjugated Dynal beads and acetylated by SAGA. SAGA and Acetyl-CoA were removed by washing the beads three times in binding buffer containing 0.6 M KCl. The nucleosomes were released from the beads by Eco RV cleavage. Reactions contained the amounts of carrier DNA indicated in the figure legends.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. Embo J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Davie JK, Kane CM. Genetic interactions between TFIIS and the Swi-Snf chromatin-remodeling complex. Mol Cell Biol. 2000;20:5960–5973. doi: 10.1128/mcb.20.16.5960-5973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell. 2006;125:275–286. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Ito T, Ikehara T, Nakagawa T, Kraus WL, Muramatsu M. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 2000;14:1899–1907. [PMC free article] [PubMed] [Google Scholar]
- Kadesch TR, Chamberlin MJ. Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J Biol Chem. 1982;257:5286–5295. [PubMed] [Google Scholar]
- Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. Embo J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Kadonaga JT. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Clarke ND. Control of transcription through intragenic patterns of nucleosome composition. Cell. 2005;123:1187–1190. doi: 10.1016/j.cell.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Liu YV, Clark DJ, Tchernajenko V, Dahmus ME, Studitsky VM. Role of C-terminal domain phosphorylation in RNA polymerase II transcription through the nucleosome. Biopolymers. 2003;68:528–538. doi: 10.1002/bip.10302. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. J Biol Chem. 2006 doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- Sawadogo M, Roeder RG. Energy requirement for specific transcription initiation by the human RNA polymerase II system. J Biol Chem. 1984;259:5321–5326. [PubMed] [Google Scholar]
- Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu Rev Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, Werner M. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Eberharter A, John S, Grant PA, Workman JL. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc Natl Acad Sci U S A. 1998;95:12924–12929. doi: 10.1073/pnas.95.22.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studitsky VM, Walter W, Kireeva M, Kashlev M, Felsenfeld G. Chromatin remodeling by RNA polymerases. Trends Biochem Sci. 2004;29:127–135. doi: 10.1016/j.tibs.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Thastrom A, Bingham LM, Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J Mol Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Tran HG, Steger DJ, Iyer VR, Johnson AD. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. Embo J. 2000;19:2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]


