Abstract
The cottontail rabbit papillomavirus (CRPV)/rabbit model has been used to study oncogenicity and immunogenicity of different antigens from the papillomavirus genome and has therefore served as a preclinical model for the development of preventive and therapeutic vaccines against papillomavirus infections. One unique property of the CRPV model is that infection can be initiated using viral DNA. This property allows for the functional testing of viral mutants in vivo. We have introduced point mutations, insertions and deletions into all of the different coding and non-coding regions of the CRPV genome and have tested their infectivity in this model. We found that the majority of the mutant genomes retained viability and could induce papillomas in domestic rabbits. These data indicated that the CRPV genome is tolerant of many modifications without compromising its ability to initiate skin papillomas. In combination with our recently established HLA-A2.1 transgenic rabbit model, this plasticity allows us to extend the utility of the CRPV/rabbit model to the screening of HLA-A2.1 restricted epitopes from other human viral and tumor antigens.
Keywords: CRPV, modification, mutants, DNA challenge, functional study
Introduction
Human papillomaviruses (HPVs) are small DNA tumor viruses which cause skin and genital warts in the human population (zur Hausen 2000). More than 100 different genotypes have been identified to date. These viruses cause infections in both cutaneous and mucosal tissues and their infections are highly species and tissue specific (Bosch, Lorincz et al., 2002). Therefore, no in vivo animal model is available to study HPV infection in immunocompetent hosts.
An effective animal model is essential to study viral-host interaction in vivo. Several animal models are utilized extensively in papillomavirus research (Campo, 2002). Cottontail rabbit papillomavirus (CRPV) was the first identified animal papillomavirus and the CRPV/rabbit model system has been one of the most widely used (Christensen, 2005). The CRPV/rabbit model has added value because CRPV- induced cancer mimics the progression and metastases of HPV induced tumors in the human population (Christensen, Han et al., 2000).
We have worked with the CRPV/New Zealand White (NZW) rabbit model system for many years and have developed refinements including an improved DNA infectivity technique which has allowed us to study viral mutants in the context of the domestic rabbit without the requirement for infectious virions. This approach allows us to test different mutants for infectivity studies in vivo based on the monitoring of tumor development and the histological assessment of lesions. Our infectivity assay is based predominantly on whether or not the constructs are capable of inducing papillomas on animals. Any constructs which are able to induce papilloma growth are identified as infectious constructs while those that are unable to induce papillomas on animals are identified as dysfunctional or non-viable constructs.
The rationale for the choice and construction of the different mutations described in this study, mutations that are scattered throughout the CRPV genome, can be defined by several broad research goals. First of all, we wished to identify those genes essential for CRPV function in vivo using ATG knockouts or early stop codon mutations. Second, we wanted to explore two major fundamental questions of papillomavirus biology: 1) What are the factors controlling the tissue specificity of papillomavirus infection? and 2) What gene or gene region contributes to the benign versus malignant phenotypes of the papillomas ? We have two distinct types of rabbit papillomaviruses for our studies; one is CRPV which induces skin tumors and the other is rabbit oral papillomavirus (ROPV) which induces mucosal tumors. These two viruses share the highest homology in their genome coding proteins when compared with other papillomaviruses (Hu, Cladel et al., 2004). We are interested in determining which region controls the tissue specificity of these viruses, and these studies provide the rationale for construction of numerous fragment replacements using CRPV and ROPV sequences (Table 1). In addition to having two distinct types of rabbit papillomaviruses, we also have two distinct phenotypes or strains of CRPV that are naturally occurring variants (Salmon, Nonnenmacher et al., 2000). One strain is designated a progressive strain (CRPVp) in which papillomas persist and progress. The second strain is termed a regressive strain (CRPVr) in which papillomas regress at high rates in both inbred and outbred rabbits. Our goal with these two viral strains was to define the region of the genome that plays a dominant role in discriminating between the two outcomes of infection. In our previous publications, we reported that the carboxy-terminal region of CRPV E6 controlled regression (Hu, Cladel et al., 2002). However, since ROPV infection generates benign papillomas which quickly regress, we therefore constructed CRPV/ROPV hybrid E6 genes in the CRPV backbone to test whether such hybrid genes could function in vivo and lead to regression. (Hu, Cladel et al., 2004).
Table 1.
Representative CRPV mutants list
| Construct name | Mutation site | Mutation information |
|---|---|---|
| E1m#1 | E1 | CRPVE1 sequence 3128–3170bp replaced by corresponding ROPVE1 |
| E1m#2 | CRPVE1 sequence 1999–2017 replaced by corresponding ROPVE1 | |
| E2m#1 | E2 | CRPVE2 sequence 4021–4283bp replaced by corresponding ROPVE2 |
| E2m#2 | CRPVE2 sequence 3128–3382bp replaced by corresponding ROPVE2 | |
| E5m#1 | E5 | CRPVE5/E8 double ATG mutant with wild type E8 at NsiI |
| E5m#2 | CRPVE5 ATG mutant with 314bp fragment of HSV-tk gene in the sense orientation at NsiI | |
| E5m#3 | CRPV with a deletion of 4300–4344bp in E5 | |
| E6m#1 | E6 | CRPVE6 with new enzyme sites engineered at 326bp/MscI and 476bp/HpaI |
| E6m#2 | CRPV E6 and E7 replaced by ROPVE6 and E7 | |
| E6m#3 | CRPVE6 fragment 326–476bp replaced by corresponding ROPV fragment | |
| E6m#4 | CRPVE6 fragment 326–973bp replaced by corresponding ROPV fragment | |
| E6m#5 | CRPVE6 fragment 476–973bp replaced by corresponding ROPV fragment | |
| E7m#1 | E7 | CRPVE7 point mutation resulting in S21 to G |
| E7m#2 | CRPVE7 point mutation resulting in S21 to D | |
| E7m#3 | CRPVE7 point mutation resulting in R57 to S | |
| E7m#4 | CRPVE7 replaced by CRPV regressive strain E7 | |
| E7m#5 | CRPVE7 containing HPV16E7/82–90 epitope LLMGTLGIV | |
| URRm#1 | URR | CRPV with 7732–7786 deletion and a mutation in the 8th E2 binding site |
| URRm#2 | Replacement of CRPVURR with ROPVURR at engineered enzyme sites BsiW1/7380 and SacII/152 deletion of URR7671–7798 fragment) | |
| URRm#3 | ||
| URRm#4 | CRPVURR with four additional E2 binding sites at Bg lII | |
| URRm#5 | CRPVURR with two copies of ROPVURR 7458–7556bp fragment in sense orientation at BglII | |
| URRm#6 | CRPVURR with full length of ROPVURR inserted at Bg lII | |
| URRm#7 | CRPVURR with a deletion of most 5′ E2 binding site 7420–7466bp | |
| URRm#8 | CRPVURR with insertion of a 314bp fragment of the HSV-tk gene at BglII | |
| URRm#9 | CRPVURR with two additional E2 binding sites at BglII | |
| URRm#10 | CRPVURR with six additional E2 binding sites at BglII |
We have created point mutants, insertions and deletions and have tested their infectivity and, in some cases, their malignant potential in rabbits. Using ATG knockout technology and introducing early stop codons, we have demonstrated that E1, E2, E6 and E7 are essential for papilloma outgrowth in rabbits. These findings are consistent with earlier reports (Wu, Xiao et al., 1994;Jeckel, Huber et al., 2002). In addition, we confirmed that the L2 protein is dispensable for papilloma growth (Nasseri, Meyers et al., 1989). On the other hand and contrary to earlier reports, we found that the L1 protein is also dispensable for papilloma formation (Nasseri, Meyers, & Wettstein, 1989). We found that both E4 and E5 are dispensable for papilloma outgrowth confirming earlier reports. (Peh, Brandsma et al., 2004;Brandsma, Yang et al., 1992). However, in contrast to previous reports (Meyers & Wettstein, 1991;Nonnenmacher, Salmon et al., 2006), E8 and SE6 were not essential for tumor growth but such mutations led to smaller and slower-growing tumors (Christensen, 2005). We also observed that many deletions, insertions and point mutations in individual genes could be introduced without destroying the ability of the genome to generate papillomas. Our findings have also demonstrated that portions of foreign genes can be inserted into the CRPV genome and that these mutants can still induce tumors in rabbits. Our system allows us, therefore, to study both oncogenicity and immunogenicity of modified genomes in the context of the live animal and to investigate the mechanism of papillomavirus induced malignancy in vivo. The purpose of this study is to present the collective observations of our studies on CRPV mutations in the unifying context of (a) the same CRPV genetic strain; (b) a series of genetic mutations in all ORF and non-coding regions; and (c) identical infection procedures for comparative studies within the same research laboratory. These findings demonstrate that significant mutations are tolerated in the CRPV genome, and provide opportunities for studies on host/virus interactions, immunogenicity, oncogenicity and tissue tropism in vivo in immunocompetent animals.
Methods and materials
Constructs
Hershey progressive strain CRPV (CRPVp, Figure 5) cloned into PUC19 at SalI was identified as wild type CRPV and used as the backbone for most mutants. Hershey regressive CRPV strain (CRPVr) and rabbit oral papillomavirus (ROPV) genes were cloned into PUC19 as previously described (Hu, Cladel, Pickel, & Christensen, 2002)(Hu, Peng et al., 2005). Herpes Simplex virus Thymidine kinase (HSV-tk) gene was provided by Dr. Kristin Eckert of Pennsylvania State Hershey Medical Center. Mutant constructs were categorized as site mutations, hybrids, insertions or deletions. For the convenience of cloning and handling, each individual gene was cloned into PUC19 for subsequent modification. Point mutations and deletions were introduced into each gene by using a QuickChange™ site-directed mutagenesis kit (Stratagene, La Jolla, CA.). Other insertions and replacements were made with routine restriction digest and ligation technology strategy. All the mutations were confirmed by DNA sequence analysis at the Core Facility of Pennsylvania State University Hershey Medical Center. Representative mutants reported in this paper are shown in table 1.
Figure 5.
Genetic map of the circular CRPV genome. E6, SE6, E7, E2 and L2 are coded from the open reading frame 1, E8, E4, E5 and L1 are coded from the open reading frame 2 and E1 is from the open reading frame 3. The CRPV genome used for DNA challenge and mutant generation is cloned in PUC19 at single enzyme site in E5 (SalI 4572). To standardized the results, all the mutants were based on Hershey progressive CRPV strain reported in our previous studies. Every mutation was confirmed by DNA sequence analysis.
DNA challenge and monitoring of tumors
The constructs were purified by cesium chloride ultracentrifugation and adjusted to 200μg/ml in 1× TE buffer (Hu, Cladel, Pickel, & Christensen, 2002) for challenge on animals. New Zealand White (NZW) rabbits and EIII/JC inbred rabbits were maintained in the animal facility of the Pennsylvania State University College of Medicine. The studies were approved by the Institutional Animal Care and Use Committee of the Pennsylvania State University. For application of virus and viral DNA, rabbits were sedated using Ketamine/xylazine anesthesia. Back skin of the animals was scarified with a scalpel blade and superficially scratched. Three days later, the wounded sites were scratched 20 times with a 21G needle to introduce small abrasions into the scab. Each site was then challenged with 10μgDNA in 50μl of 1×TE buffer (Hu, Cladel, Pickel, & Christensen, 2002). Monitoring of papilloma outgrowth began three weeks later and continued until week 12. In some cases, the animals were kept to monitor cancer development. Papilloma biopsies were collected after animal sacrifice and H&E staining was conducted for histological examination.
Statistics
Papilloma size was determined by calculating the cubic root of the product of length × width × height of individual papillomas in millimeters to obtain a geometric mean diameter (GMD). Data were represented as the means (+ SEMs) of the GMDs for each test group. Statistical significance was determined by unpaired t-test comparison (P<0.05 was considered significant).
Results
CRPV DNA challenge induced papillomas comparable to those induced by virus infection in domestic rabbits
Hershey CRPV cloned into PUC19 at SalI/4572 (wild type CRPV) has been demonstrated to be infectious in domestic rabbits. We conducted a comparative study with rabbits challenged with viral DNA and infectious virus stock (1:100, a dose used for most of our previous infection studies). Three NZW rabbits were challenged at four left back sites with virus stock (1:100) and at four right back sites with viral DNA (10μg). Papilloma outgrowth was monitored and recorded.
All challenge sites grew papillomas from both viral DNA and virus infection. Tumors induced by viral DNA were significantly smaller when compared with those induced by virus infection (P<0.05, unpaired student t test) in the first several weeks. But no significant difference in tumor size was found after week 9 of infection (P>0.05, unpaired student t test, Figure 1). Therefore, our method of viral DNA infection was efficient in domestic rabbits.
Figure 1.
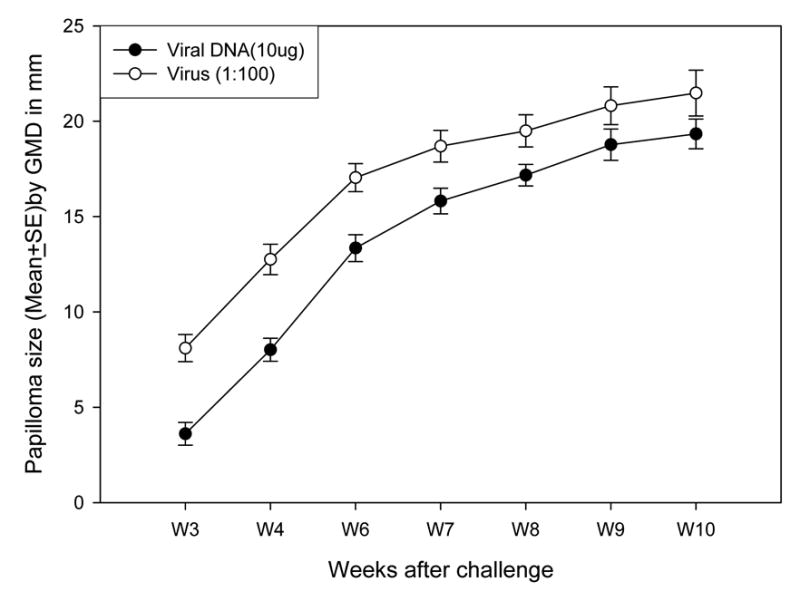
Papilloma outgrowth after CRPV infectious virions (1:100) and DNA (10μg) challenge on three ourbred rabbits. Four left and right back skin sites were challenged with infectious virions and viral DNA respectively (N=12). Papillomas induced by virions were significantly larger when compared with those induced by CRPV DNA infection before week 9 after challenge (P<0.05, unpaired student t test). No significant difference was found in the papilloma size between these two challenge methods after week 10 (P>0.05, unpaired student t test).
E1, E2, E6 and E7 but not other early and late genes are essential for papilloma outgrowth in rabbits
To determine which genes were critical to the induction of papilloma growth in rabbits, we generated ATG knockout and early stop codon mutants for each individual gene. Our results are summarized in table 2. E1, E2, E6 and E7 were essential for tumor growth. In contrast, short E6 (SE6), E8, E4, E5, L1 and L2 were not required for papilloma outgrowth (Table 2). However, papilloma growth rates were diminished by SE6 (Figure 2) and E8 ATGko mutations (Hu, Han et al., 2002). E8ATGko mutant infectious virus was generated in wild cottontail rabbits and the mutant virus showed slower papilloma growth when compared with that of wild type virus but all challenge sites grew papillomas (Christensen, 2005). No significant difference in growth rates was found between E4 mutant and E5 ATGko and wild type CRPV induced papillomas (data not shown).
Table 2.
Function and tumor growth of CRPVATG knockout and early stop mutants
| Gene | ATG knock out | Early stop codon | Animal Numbers | Papillomas/challenge sites |
|---|---|---|---|---|
| E1 | + | 4 | 0/8 | |
| E2 | + | 4 | 0/8 | |
| E6 | + | 4 | 0/8 | |
| E7 | + | 8 | 0/16 | |
| SE6 | + | 3 | 2/6 | |
| E8 | + | (Hu, Han, Cladel, Pickel, & Christensen, 2002) | ||
| E4 | + | 4 | 8/8 | |
| E5 | + | 4 | 10/12 | |
| E5/E8 | + | 4 | 12/12 | |
| L1 | + | 2 | 10/10 | |
| L2 | + | 4 | 12/12 | |
Figure 2.
Papilloma outgrowth after wild type CRPV, CRPV hybrid with regressive strain E7 (E7r) and CRPVSE6ATGko DNA challenge in four EIII/JC inbred rabbits. Two sites/per construct/per animals were challenged with wild type CRPV, CRPVE7r and CRPVSE6ATGko respectively (N=8). CRPVE7r hybrid infection showed a similar papilloma growth pattern to that of the wild type CRPV (P>0.05, unpaired student t test). Significantly smaller papillomas were induced by CRPVSE6ATGko infection when compared to those induced by the wild type CRPV (P<0.01, unpaired student t test).
E1, E2, E6 and E7 could be modified without loss of papilloma formation
The ATG knockout and early stop codon mutants showed that E1, E2, E6 and E7 were essential for papilloma outgrowth. We next wanted to test if we could introduce mutations into these individual genes without loss of papilloma formation. Point mutations (changing one or two amino acid residues) and hybrids (replacement of a defined region of amino acids with those from a different papillomavirus) were generated in these genes (Table 3). Although we have noted some replacements that were lethal to CRPV genome function, many changes in these genes did not affect the ability of the genome to induce papillomas.
Table 3.
Function and tumor outgrowth of CRPV early gene hybrids
| Construct | Animal Numbers | Papillomas/challenge sites | Cancer development (>14 month) |
|---|---|---|---|
| E1m#1 | 2 | 6/6 | ND |
| E1m#2 | 2 | 7/8 | ND |
| E2m#1 | 4 | 12/12 | ND |
| E2m#2 | 2 | 6/6 | ND |
| E5m#1 | 2 | 7/8 | ND |
| E5m#2 | 2 | 7/8 | ND |
| E5m#3 | 2 | 8/8 | ND |
| E6m#1 | 3 | 6/6 | ND |
| E6m#2 | 2 | 0/6 | |
| E6m#3 | 2 | 0/6 | |
| E6m#4 | 4 | 0/8 | (Hu, Cladel, Budgeon, & Christensen, 2004) |
| E6m#5 | 4 | 0/8 | |
| E7m#1 | 2 | 6/6 | 0/6 |
| E7m#2 | 2 | 6/6 | 4/6 |
| E7m#3 | 2 | 6/6 | 2/6 |
| E7m#4 | 4 | 6/6 | ND |
| E7m#5 | 5 | 15/16 | ND |
ND-------not determined
A panel of mutants in the CRPV progressive strain (wild type CRPV) E6 gene has been reported previously (Hu, Cladel, Pickel, & Christensen, 2002). The compatibility between wild type CRPVE6 and a regressive CRPV strain (CRPVr) E6 was very high and CRPV regressive strain E7 replacement in CRPV also produced papillomas at wild type rates (Figure 2, Table 3). However, the E6 and E7 genes of CRPV and rabbit oral papillomavirus (ROPV) were not interchangeable. Several genomes containing hybrid CRPV-ROPV E6 and E7 genes were also dysfunctional (Hu, Cladel et al., 2004) (Table 3).
We had made several point mutants in CRPV E7 based on the findings that E7 amino acid position 21 played an important role in transforming activity in vitro (Edmonds, C. and Vousden, K. H, 1989). We hypothesized that changing S21 of CRPV E7 to G or D would result in a more benign (G) or malignant (D) phenotype. Our results correlated with this prediction although no significant difference was found between these mutants when compared with the wild type genome because only limited numbers of animals was used. A second point mutation (R57S) which resulted in a large side- chain change at this position showed a phenotype similar to that of the wild type CRPV. In addition, we generated HPV16 E7/82–90 (LLMGTLGIV, an HLA-A2.1 restricted epitope) (Kast, Brandt et al., 1993) hybrid in the CRPV genome and this mutant genome generated papillomas. The papillomas were significantly smaller when compared to those induced by wild type CRPV (Figure 3, P<0.05, unpaired student t test). These data indicate that it is possible to embed an HLA-A2.1 restricted epitope from another virus or antigen into a CRPV gene in the context of the whole CRPV genome and retain the function of papilloma induction.
Figure 3.
Papilloma outgrowth after wild type CRPV and a CRPV mutant containing an HLA-A2.1 epitope of HPV16E7 (LLMGTLGIV, amino residues82–90) DNA infection in four outbred rabbits. Four left and right back skin sites were challenged with wild type CRPV and CRPV-HPV16E7/82–90 (N=16). Significantly smaller papillomas were induced by CRPV-HPV16E7/82–90 when compared with those induced by the wild type CRPV (P<0.05, unpaired student t test).
The CRPV E5 gene can be modified extensively
The CRPV E5 gene was previously identified as an oncogene (Han, Cladel et al., 1998). However, our studies of the ATG mutant as well as the studies of other investigators have indicated that E5 is dispensable for CRPV infection (Brandsma, Yang, DiMaio, Barthold, Johnson, & Xiao, 1992). In addition to these findings, we have found that E5 may contain regulatory elements. For example, the insertion of a SnaB1 site at 4292bp with no change in amino acid sequence to E5 resulted in a non-viable genome. We were also able to insert the CRPV E8 gene and a fragment of the HSV-tk gene into E5 without disrupting papilloma formation by these constructs (Table 3).
L1 and L2 proteins are dispensable but the DNA sequences are required for tumor outgrowth
Neither the L1 nor the L2 ATGko mutations prevented induction of tumors in rabbits (Table 2). Interestingly, the L1ATGko induced significantly larger papillomas when compared to those induced by wild type CRPV (Figure 4, P<0.05, unpaired student t test) in EIII/JC inbred rabbits. No significant difference in growth was found between the L2ATGko and wild type CRPV papillomas based on the limited numbers of animals in which this construct was tested. To further delineate the role played by these two genes in papilloma formation, we made a mutant in which the entire region containing the L1 and L2 genes was deleted. This double deletion mutant was non-viable. Therefore, although L1 and L2 proteins are not critical to papilloma induction in rabbits, these genes appear to contain essential elements. Next, we exchanged the CRPV L1 gene for that of ROPV, which shares significant homology with its CRPV counterpart. 11 out of 12 sites infected with the CRPV/ROPVL1 hybrid genome grew papillomas in 4 NZW outbred rabbits. In contrast, 50% of the sites challenged by this CRPV/ROPVL1 hybrid grew tumors in EIII/JC inbred rabbits and these tumors were significantly smaller than those generated by wild type CRPV (Figure 4, P<0.01, unpaired student t test). Regression was also noticed in these tumors.
Figure 4.
Papilloma outgrowth after wild type CRPV, CRPVL1ATGknockout mutant and CRPV/ROPVL1 hybrid DNA challenge in four EIII/JC inbred rabbits. Four sites/per animal were challenged with each constructs respectively (N=16). The CRPVL1ATGko mutant induced significantly larger papillomas when compared with those induced by wild type CRPV (P<0.05, unpaired student t test). Significantly smaller and fewer papillomas were induced by CRPV/ROPVL1 hybrid infection when compared to those induced by wild type CRPV (P<0.01, unpaired student t test).
Upstream Regulatory Region (URR) was tolerant of different modifications
In view of our findings that there was considerable plasticity in the CRPV coding regions, we were interested to determine whether the upstream regulatory region (URR) exhibited similar tolerance to mutation. The Hershey CRPV URR contains eight E2 binding sites (E2BS) as well as numerous other putative regulatory elements. A panel of deletions or insertions of different E2 binding sites did not influence tumor development in animals (Table 4). One hybrid in which we replaced the CRPV URR with that of ROPV was dysfunctional (Table 5). However, a hybrid with an insertion of the entire ROPV URR into the CRPV URR produced papillomas. We also found that the region of the CRPV URR between 7671 and 7798 was essential because deletion of this sequence prevented papilloma formation. A final observation was that the CRPV URR remained functional following an insertion of foreign non-viral DNA (Table 4). These insertions were retained in the DNA isolated from papillomas as determined by PCR amplification and subsequent sequencing of the PCR products. Interestingly, we also found sequences in the amplimers that indicated that the foreign element had been eliminated. These bands indicating the latter scenario were always a minor component of the amplification.
Table 4.
Function of papilloma outgrowth of CRPV URR mutants
| Construct | Animal Numbers | Papillomas/challenge sites |
|---|---|---|
| URRm#1 | 2 | 1/4 |
| URRm#2 | 1 | 0/4 |
| URRm#3 | 2 | 0/6 |
| URRm#4 | 1 | 6/6 |
| URRm#5 | 1 | 6/6 |
| URRm#6 | 2 | 6/6 |
| URRm#7 | 2 | 6/6 |
| URRm#8 | 2 | 12/12 |
| URRm#9 | 2 | 6/6 |
| URRm#10 | 2 | 6/6 |
Deletion and insertions in the “non-coding” region between the E6 and E7 genes
We observed that a region between CRPVE6 and E7 contained a small coding sequence for a putative 40aa protein in a different reading frame. We named this candidate gene E10. We made an ATG knockout of this putative gene in an attempt to determine if it had any significance in the papillomavirus life cycle. Our results showed that no change in papilloma formation and outgrowth occurred following infection with this E10ATGko mutant. A second mutation was made which represented a deletion of about 90bp of the putative E10 coding region (CRPVE10del). Five out of 10 papillomas in 4 NZW outbred rabbits and 6 out of 12 papillomas in 3 EIII/JC inbred rabbits induced by CRPVE10del regressed. In addition to higher rates of regression, this mutant showed a reduced growth rate when compared to wild type CRPV induced papillomas. A mutant genome with an insertion of EGFP in this region was dysfunctional.
Discussion
The CRPV/Rabbit model has been widely used in different laboratories. Data accumulated in our laboratory over a number of years have indicated that the CRPV genome can tolerate significant modifications. These observations have provided valuable insights into the genetics of the papillomavirus and have provided new opportunities for the application of this model system. Our data have shown that the early genes E1, E2, E6 and E7 are essential for tumor development in rabbits which is consistent with previous reports (Wu, Xiao, & Brandsma, 1994). These genes, however, were shown to be tolerant of some modifications without compromising function. Other early genes including E4, SE6 and E8 are not essential but phenotypic changes were induced (Peh, Brandsma, Christensen, Cladel, Wu, & Doorbar, 2004; Hu, Han, Cladel, Pickel, & Christensen, 2002). Our data showed that the E5 ATGko exhibited the same phenotype as wild type. Consistent with others, however, we also found that a regulatory element within this putative gene was critical for CRPV genome function (Brandsma, Yang, DiMaio, Barthold, Johnson, & Xiao, 1992). Late gene products (L1 and L2) were not required for papilloma development but the DNA sequence had to be present for successful infections. The URR could not be replaced but could be modified extensively without destroying CRPV genome function. Taken together, these results show that the CRPV genome is very tolerant to modification and therefore has significant potential as a surrogate vector to screen epitopes from other viral genomes when combined with our recently developed HLA-A2.1 transgenic rabbit system.
We and others have demonstrated that CRPV DNA cloned into a plasmid is infectious in domestic rabbits. Different delivery methods have been utilized and varying efficiencies noted (Lin, Borenstein et al., 1993; Brandsma, Yang et al., 1991; Brandsma & Xiao, 1993; Kreider, Cladel et al., 1995; Xiao & Brandsma, 1996; Salmon, Nonnenmacher et al., 2000). In many cases, low infection rates and high variations in papilloma sizes were found in these early studies. In our previous DNA infection studies we used a pre-treatment strategy to achieve very high efficiency of papilloma induction (Hu, Cladel, Pickel, & Christensen, 2002). However, variations in papilloma size and growth rates were still observed. We noted also that the same construct could show different growth rates and regression profiles depending upon the strain of animals used (Hu, Cladel, Pickel, & Christensen, 2002). The modified infection method recently established in our laboratory improves the consistency of the results in both inbred and outbred strains of animals used in our experiments and has the added advantage of being safer to use. The method consists of wounding sites three days prior to the delivery of DNA. The establishment of a wound-healing environment at the time of viral DNA challenge has been shown to greatly enhance efficiency of papilloma induction and has resulted in highly consistent results (manuscript in preparation).
Many early studies using genetically modified CRPV were hampered by technological limitations and primarily involved deletions, which could be generated with technologies existing at the time. In the present studies, we tested the function of each gene by generating ATG knockout or early stop codon mutants, which did not otherwise perturb the DNA sequence of each gene. We noticed that these two mutational approaches resulted in different outcomes that could lead to different interpretations. Dysfunction resulting from deletion of a portion of a gene could result from the inadvertent deletion of a cryptic regulatory element as well as a disruption of the integrity of the gene product. An example of this phenomenon can be seen in studies with the L1 gene. We demonstrated in this paper that CRPVL1 was dispensable for CRPV infection; that replacement of CRPV L1 with ROPV L1 reduced papilloma growth rates, and that deletion of the entire L1 gene resulted in dysfunction. Similar results were found for the putative gene E10. Although an ATG knockout did not prevent papilloma formation, deletion of this region resulted in a significant phenotypic change (slower growth rate and regression of papillomas). We conclude that our modified infection method provides a unique opportunity to investigate the function of different papillomavirus genes in vivo in a more reproducible manner.
Demonstration of the considerable plasticity of the CRPV genome will aid our continuing efforts to understand the life cycle of this virus. The flexibility of the genome will also provide opportunities for the development of therapeutic targets for treatment against viral infection. Furthermore, we can also use this model to test the expression potential of specific epitopes of human pathogens and can potentially use the CRPV genome as a vector to test immunogenicity of the epitopes in our in vivo system. We have recently developed an HLA-A2.1 transgenic rabbit model that will help us to screen host immunity targets from the HPV genome (manuscript in revision). With the successful engineering of candidate epitopes into the CRPV genome, testing in the context of our HLA-A2.1 rabbits will become a very promising preclinical model system for immunotherapeutic studies.
Acknowledgments
We thank Martin Pickel for excellent help with the animals. This work was supported by the National Cancer Institute grant R01 CA47622 from the National Institutes of Health and the Jake Gittlen Memorial Golf Tournament.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Bosch FX, Lorincz A, Munoz N, Meijer CJLM, Shah KV. The causal relation between human papillomavirus and cervical cancer. Journal of Clinical Pathology. 2002;55:244–265. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsma JL, Xiao W. Infectious virus replication in papillomas induced by molecularly cloned cottontail rabbit papillomavirus DNA. Journal of Virology. 1993;67:567–571. doi: 10.1128/jvi.67.1.567-571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsma JL, Yang ZH, Barthold SW, Johnson EA. Use of a rapid, efficient inoculation method to induce papillomas by cottontail rabbit papillomavirus DNA shows that the E7 gene is required. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:4816–4820. doi: 10.1073/pnas.88.11.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsma JL, Yang ZH, DiMaio D, Barthold SW, Johnson E, Xiao W. The putative E5 open reading frame of cottontail rabbit papillomavirus is dispensable for papilloma formation in domestic rabbits. Journal of Virology. 1992;66:6204–6207. doi: 10.1128/jvi.66.10.6204-6207.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo MS. Animal models of papillomavirus pathogenesis. Virus Research. 2002;89:249–261. doi: 10.1016/s0168-1702(02)00193-4. [DOI] [PubMed] [Google Scholar]
- Christensen ND. Cottontail rabbit papillomavirus (CRPV) model system to test antiviral and immunotherapeutic strategies. Antivir Chem Chemother. 2005;16:355–362. doi: 10.1177/095632020501600602. [DOI] [PubMed] [Google Scholar]
- Christensen ND, Han R, Kreider JW. Cottontail Rabbit Papillomavirus (CRPV) In: Ahmed R, Chen I, editors. Persistent Viral Infections. John Wiley & Sons Ltd.; Sussex, England: 2000. pp. 485–502. [Google Scholar]
- Edmonds C, Vousden KH. A Point Mutational Analysis of Human Papillomavirus Type 16 E7 Protein. Journal of Virology. 1989;63:2650–2656. doi: 10.1128/jvi.63.6.2650-2656.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R, Cladel NM, Reed CA, Christensen ND. Characterization of transformation function of cottontail rabbit papillomavirus E5 and E8 genes. Virology. 1998;251:253–263. doi: 10.1006/viro.1998.9416. [DOI] [PubMed] [Google Scholar]
- Hu J, Cladel NM, Budgeon LR, Christensen ND. Characterization of three rabbit oral papillomavirus oncogenes. Virology. 2004;325:48–55. doi: 10.1016/j.virol.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Hu J, Cladel NM, Pickel MD, Christensen ND. Amino Acid residues in the carboxy-terminal region of cottontail rabbit papillomavirus e6 influence spontaneous regression of cutaneous papillomas. Journal of Virology. 2002;76:11801–11808. doi: 10.1128/JVI.76.23.11801-11808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Han R, Cladel NM, Pickel MD, Christensen ND. Intracutaneous DNA vaccination with the E8 gene of cottontail rabbit papillomavirus induces protective immunity against virus challenge in rabbits. Journal of Virology. 2002;76:6453–6459. doi: 10.1128/JVI.76.13.6453-6459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Peng X, Cladel NM, Pickel MD, Christensen ND. Large cutaneous rabbit papillomas that persist during cyclosporin A treatment can regress spontaneously after cessation of immunosuppression. Journal of General Virology. 2005;86:55–63. doi: 10.1099/vir.0.80448-0. [DOI] [PubMed] [Google Scholar]
- Jeckel S, Huber E, Stubenrauch F, Iftner T. A transactivator function of cottontail rabbit papillomavirus e2 is essential for tumor induction in rabbits. Journal of Virology. 2002;76:11209–11215. doi: 10.1128/JVI.76.22.11209-11215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast WM, Brandt RMP, Drijfhout JW, Melief CJM. Human-Leukocyte Antigen-A2.1 Restricted Candidate Cytotoxic T-Lymphocyte Epitopes of Human Papillomavirus Type-16 E6-Protein and E7-Protein Identified by Using the Processing-Defective Human Cell Line-T2. Journal of Immunotherapy. 1993;14:115–120. doi: 10.1097/00002371-199308000-00006. [DOI] [PubMed] [Google Scholar]
- Kreider JW, Cladel NM, Patrick SD, Welsh PA, DiAngelo SL, Bower JM, Christensen ND. High efficiency induction of papillomas in vivo using recombinant cottontail rabbit papillomavirus DNA. J Virol Methods. 1995;55:233–244. doi: 10.1016/0166-0934(95)00062-y. [DOI] [PubMed] [Google Scholar]
- Lin YL, Borenstein LA, Ahmed R, Wettstein FO. Cottontail rabbit papillomavirus L1 protein-based vaccines: Protection is achieved only with a full-length, nondenatured product. Journal of Virology. 1993;67:4154–4162. doi: 10.1128/jvi.67.7.4154-4162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers C, Wettstein FO. The late region differentially regulates the in vitro transformation by cottontail rabbit papillomavirus DNA in different cell types. Virology. 1991;181:637–646. doi: 10.1016/0042-6822(91)90897-k. [DOI] [PubMed] [Google Scholar]
- Nasseri M, Meyers C, Wettstein FO. Genetic analysis of CRPV pathogenesis: The L1 open reading frame is dispensable for cellular transformation but is required for papilloma formation. Virology. 1989;170:321–325. doi: 10.1016/0042-6822(89)90388-7. [DOI] [PubMed] [Google Scholar]
- Nonnenmacher M, Salmon J, Jacob Y, Orth G, Breitburd F. Cottontail rabbit papillomavirus E8 protein is essential for wart formation and provides new insights into viral pathogenesis. Journal of Virology. 2006;80:4890–4900. doi: 10.1128/JVI.80.10.4890-4900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peh WL, Brandsma JL, Christensen ND, Cladel NM, Wu X, Doorbar J. The viral E4 protein is required for the completion of the cottontail rabbit papillomavirus productive cycle in vivo. Journal of Virology. 2004;78:2142–2151. doi: 10.1128/JVI.78.4.2142-2151.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon J, Nonnenmacher M, Caze S, Flamant P, Croissant O, Orth G, Breitburd F. Variation in the nucleotide sequence of cottontail rabbit papillomavirus a and b subtypes affects wart regression and malignant transformation and level of viral replication in domestic rabbits. Journal of Virology. 2000;74:10766–10777. doi: 10.1128/jvi.74.22.10766-10777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Xiao W, Brandsma J. Papilloma formation by cottontail rabbit papillomavirus requires E1 and E2 regulatory genes in addition to E6 and E7 transforming genes. Journal of Virology. 1994;68:6097–6102. doi: 10.1128/jvi.68.9.6097-6102.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Brandsma JL. High efficiency, long-term clinical expression of cottontail rabbit papillomavirus (CRPV) DNA in rabbit skin following particle-mediated DNA transfer. Nucleic Acids Research. 1996;24:2620–2622. doi: 10.1093/nar/24.13.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]


