Abstract
Bacterial autotransporters are comprised of an N-terminal ‘passenger domain' and a C-terminal β barrel (‘β domain') that facilitates transport of the passenger domain across the outer membrane. Following translocation, the passenger domains of some autotransporters are cleaved by an unknown mechanism. Here we show that the passenger domain of the Escherichia coli O157:H7 autotransporter EspP is released in a novel autoproteolytic reaction. After purification, the uncleaved EspP precursor underwent proteolytic processing in vitro. An analysis of protein topology together with mutational studies strongly suggested that the reaction occurs inside the β barrel and revealed that two conserved residues, an aspartate within the β domain (Asp1120) and an asparagine (Asn1023) at the P1 position of the cleavage junction, are essential for passenger domain cleavage. Interestingly, these residues were also essential for the proteolytic processing of two distantly related autotransporters. The data strongly suggest that Asp1120 and Asn1023 form an unusual catalytic dyad that mediates self-cleavage through the cyclization of the asparagine. Remarkably, a very similar mechanism has been proposed for the maturation of eukaryotic viral capsids.
Keywords: autotransporters, enzyme mechanism, E. coli, protease, secretion
Introduction
Autotransporters are a superfamily of virulence factors secreted by Gram-negative bacteria that consist of two domains, a large (⩾100 kDa) N-terminal ‘passenger domain' and an ∼30 kDa C-terminal ‘β domain' (reviewed in Henderson et al, 2004). Passenger domains are highly diverse in sequence and mediate a wide range of different effector functions. β domains can be identified by sequence alignment algorithms as members of a single domain family (PFAM03797), but otherwise are also weakly related. After autotransporters are translocated across the inner membrane, the β domain forms a typical β barrel structure in the outer membrane (OM) and promotes the translocation of the passenger domain into the extracellular space. Although the mechanism by which the β domain promotes passenger domain translocation is unclear, structural and biochemical data imply that ∼30 amino acids of the passenger domain-β domain border region reside in a narrow ∼10 Å pore formed by the β domain after the translocation reaction is complete (Oomen et al, 2004; Skillman et al, 2005). Some autotransporters remain intact, but others undergo proteolytic processing that releases the passenger domain from the cell surface. The long-standing assumption that proteolytic processing occurs after the passenger domain is translocated across the OM has recently been confirmed (Skillman et al, 2005).
To date, the mechanism of passenger domain cleavage has been elucidated for only a few autotransporters. The Neisseria App, NalP and IgA1 protease proteins and the Haemophilus influenzae Hap protein are all processed by endogenous serine proteases located in the passenger domain. Proteolytic processing is abolished when the serine protease motif GDSG is deleted, when the catalytic serine is mutated or when serine protease inhibitors are used (Pohlner et al, 1987; Hendrixson et al, 1997; Turner et al, 2002; Serruto et al, 2003). In contrast, the Shigella flexneri IcsA passenger domain is excised by IcsP/SopA, an OM protease homologous to OmpT (Egile et al, 1997; Shere et al, 1997). Finally, NalP has been shown to act in trans on other autotransporters that undergo self-cleavage to mediate a second step of proteolytic processing (Van Ulsen et al, 2003). In all of these cases, cleavage occurs in the extracellular space and results in the retention of a small fragment of the passenger domain on the cell surface.
Neither the mechanism by which most autotransporter passenger domains are cleaved nor the intracellular compartment in which the cleavage occurs, however, is known. The Escherichia coli AIDA-1 protein, the Helicobacter pylori VacA protein and the Bordetella pertussis BrkA, pertactin and TcfA proteins all contain cleaved passenger domains, but their sequences do not suggest that they harbor endogenous protease activities. In contrast, the passenger domains of the SPATE (serine protease autotransporters of Enterobacteriaceae) family of autotransporters all contain a classical GDSG serine protease motif. The proteases encoded by these proteins have been shown to cleave several mammalian proteins (Dutta et al, 2002). Nevertheless, mutation of the catalytic serine residue of EatA or EspC or the use of serine protease inhibitors does not prevent passenger domain cleavage (Stein et al, 1996; Patel et al, 2004). Furthermore, deletion of most of the passenger domain (including the serine protease region) of EspP, an SPATE produced by E. coli O157:H7, does not affect proteolytic processing (Velarde and Nataro, 2004; Szabady et al, 2005). Finally, the passenger domains of SPATEs produced in E. coli K-12 strains are cleaved efficiently, but the processing of Pet, Pic and EspP is independent of OmpT, OmpP or DegP (Eslava et al, 1998; Henderson et al, 1999; Szabady et al, 2005). The data imply that SPATEs are cleaved either by an uncharacterized periplasmic or OM protease or by an autocatalytic mechanism that is not easily predicted from the protein sequence. Given that the cleavage always occurs between two asparagine residues and that the sequence of the region surrounding the cleavage site as well as the entire β domain is extremely highly conserved (Figure 1), it is likely in either case that the same processing mechanism is used by all of the SPATEs.
Figure 1.
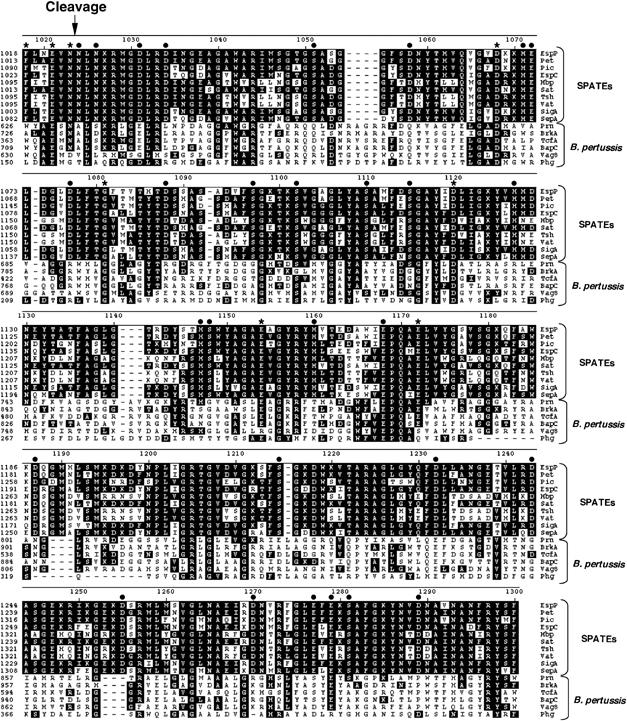
Sequence alignment of the β domains of SPATEs and B. pertussis autotransporters. The sequences of selected SPATEs and B. pertussis autotransporters were aligned using the DNAStar MegAlign program (ClustalW method). Amino acids that are identical to the corresponding EspP residues are highlighted in black. The arrow shows the passenger domain cleavage site. Mutations introduced into EspP at positions denoted with an asterisk impaired passenger domain cleavage, whereas those denoted with a filled circle did not.
In this study, we used EspP as a model protein to elucidate the mechanism by which SPATE passenger domains are released from the cell surface. Initial studies that examined the accessibility of tobacco etch virus (TEV) protease sites inserted near the cleavage junction provided evidence that cleavage occurs inside the β barrel. Subsequent site-directed mutagenesis studies showed that mutation of a single aspartate in the β domain (Asp1120), as well as the ‘P1' residue just upstream of the cleavage site (Asn1023), abolished cleavage without affecting translocation of the passenger domain across the OM. Interestingly, we also found that the same residues are essential for the cleavage of the passenger domains of two autotransporters that are unrelated to the SPATEs. These results, together with the observation that purified EspP undergoes self-cleavage, strongly suggested that Asp1120 and Asn1023 form a conserved catalytic dyad that mediates passenger domain release in an unprecedented intra-barrel protease reaction. Examination of the cleaved passenger domain by liquid chromatography (LC) and mass spectrometry (MS) provided strong evidence that polypeptide scission involves the cyclization of Asn1023 to form a succinimide. Remarkably, a similar autocatalytic mechanism is thought to be involved in the maturation of two classes of eukaryotic virus capsids. Our data suggest that the unique chemistry of asparagine accounts for the independent evolution of closely related autoproteolytic reactions that play important regulatory roles in self-assembly pathways.
Results
The EspP cleavage site likely resides in the pore formed by the β domain
Initially we found that the cleavage of the EspP passenger domain was unaffected by the elimination of most of the known periplasmic and OM proteases (K Williams, unpublished data). To narrow down the possible mechanisms by which processing might occur, we therefore decided to determine the location of the cleavage junction following passenger domain translocation by identifying the ∼30-amino-acid segment that is embedded within the pore formed by the β domain. To this end, we examined the accessibility of TEV protease sites engineered into a ‘minimal' version of EspP that has a non-cleavable passenger domain, but that undergoes all other steps of protein biogenesis normally (here designated ‘EspP*Δ1', but previously called ‘EspP*Δ1–851'; see Figure 2A). EspP*Δ1 contains the C-terminal 116 amino acids of the 968-residue passenger domain and a double mutation (N1023S/N1024S) at the cleavage site. After determining that the insertions do not impair translocation of the passenger domain across the OM (Supplementary Figure 1), we used an N-terminal His tag to affinity purify all of the EspP*Δ1 derivatives under native conditions, incubated them with TEV protease and analyzed the products by SDS–PAGE. We expected that TEV sites situated inside or very close to the β barrel would be protected from digestion.
Figure 2.
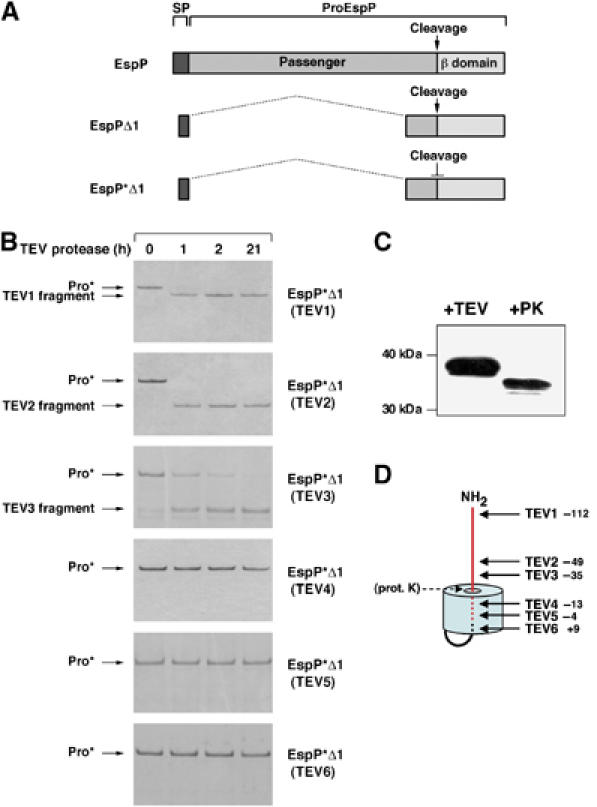
The EspP passenger domain cleavage site is embedded within the β barrel. (A) Illustration of EspP, EspPΔ1 and EspP*Δ1, a previously described non-cleavable version of EspPΔ1 that harbors the double mutation N1023S/N1024S. ProEspP is the form of the protein that contains covalently linked passenger and β domains but no signal peptide (SP). (B) Derivatives of EspP*Δ1 that contain a TEV protease site were purified and incubated with TEV protease for various lengths of time. Reaction products were then analyzed by SDS–PAGE on 4–12% NuPAGE gels using MES buffer (Invitrogen). (C) Purified EspP*Δ1(TEV3) was incubated with TEV protease or proteinase K (PK). The major cleavage product was detected by Western blot using an antiserum directed against an EspP C-terminal peptide. (D) Model of the topology of EspP*Δ1 derived from the data shown in (B) and (C). The distance of each TEV insertion from the passenger domain cleavage site is indicated. The passenger domain is shown in red.
These experiments provided a clear indication that passenger domain cleavage occurs inside the β barrel. We found that EspP*Δ1(TEV1), EspP*Δ1(TEV2) and EspP*Δ1(TEV3), which contain TEV sites 111, 49 and 35 residues upstream of the passenger domain cleavage site, respectively, were cleaved quantitatively by the TEV protease (Figure 2B). In contrast, EspP*Δ1(TEV4), EspP*Δ1(TEV5) and EspP*Δ1(TEV6), which contain TEV sites 13 and 4 residues upstream and 9 amino acids downstream of the passenger domain cleavage site, respectively, were completely resistant to cleavage. All of these sites are accessible to the protease when the protein is unfolded (R Ieva, unpublished data). Furthermore, proteinase K treatment of EspP*Δ1(TEV3) yielded a slightly smaller fragment than the one produced by TEV protease (Figure 2C). As this fragment was recognized on a Western blot by an antipeptide antiserum directed against the C-terminus of the β domain, proteinase K must have digested a larger segment of the passenger domain. Therefore, ∼15 amino acids downstream of the TEV3 site must be exposed outside the β barrel. Taken together, the data strongly suggest that the segment immediately surrounding the passenger domain cleavage site (∼20 residues upstream and ∼10 residues downstream) is embedded within the β barrel. Because in this location the cleavage site would very likely be inaccessible to exogenous proteases, the results raised the possibility that the passenger domain is processed autocatalytically.
Asp1120 plays an essential role in the cleavage of the EspP passenger domain
Given that the EspP β domain lacks the active site motifs found in classical serine proteases, aspartyl proteases and metalloproteases, lacks cysteines and lacks homology to the only known family of bacterial OM proteases (the ‘omptins', which include OmpT), the mechanism by which it might mediate an autoproteolytic reaction could not be predicted from its sequence. Nevertheless, proteases that lack canonical sequence motifs are not unprecedented. The GDSG motif found in the vast majority of serine proteases is absent from Tsp and the cytomegalovirus protease (Keiler and Sauer 1995; Chen et al, 1996), and a variety of aspartyl proteases including the omptins and presenilins lack the D(T/S)G consensus (Wolfe et al, 1999; Vandeputte-Rutten et al, 2001). Moreover, the observation that neither the rate nor the efficiency of EspP passenger domain cleavage was significantly affected by a broad range of serine, aspartyl- and metalloprotease inhibitors was consistent with the possibility that the β domain mediates a non-canonical proteolytic reaction (Supplementary Figure 2).
To test the hypothesis that the EspP β domain is directly involved in the cleavage of the passenger domain, we mutated each of the serine, aspartate and histidine residues in the β domain that are strictly conserved in the SPATE family (Figure 1). This mutagenesis strategy was based on the observation that the positions of key catalytic residues are generally conserved within families of proteases (Barrett et al, 2003). Serine and histidine residues were mutated to alanine and aspartate residues were mutated to asparagine. AD202 was transformed with pJH62 or pKMS3 (plasmids that encode espPΔ1 and espP*Δ1, respectively, under the control of the trc promoter) or a derivative of pJH62 containing a point mutation in the β domain. Cells were grown in LB, and the production of the plasmid-borne gene was induced by the addition of IPTG. The cleavage of the precursor form of the protein, in which the passenger and β domains are covalently linked (‘proEspPΔ1'; Figure 2A) into discrete passenger and β domain fragments, was monitored by Western blot using the C-terminal antipeptide antiserum.
Our analysis of the β domain mutants pinpointed a single residue that plays an essential role in passenger domain cleavage. Because the cleavage reaction is normally rapid (Skillman et al, 2005), only the free ∼30 kDa β domain was detected in cells that produced wild-type EspPΔ1 (Figure 3A, lane 1). In contrast, only the unprocessed ∼43 kDa pro form of the protein was observed in cells that produced EspP*Δ1 (Figure 3A, lane 2). The absence of proEspPΔ1 in cells that synthesized the serine and histidine mutants indicated that the mutations did not affect passenger domain processing and strongly suggest that the EspP β domain is neither a serine protease nor a metalloprotease (Figure 3A, lanes 3–14). Mutation of 2 of the 13 conserved aspartate residues, however, led to defects in passenger domain cleavage. One mutation, D1068N, led to the accumulation of a small amount of proEspPΔ1, but a second mutation, D1120N, completely abolished proEspPΔ1 processing (Figure 3A, lanes 18 and 21).
Figure 3.

Mutational analysis reveals a single β domain residue that is essential for EspP passenger domain cleavage. AD202 was transformed with a plasmid encoding EspPΔ1, EspP*Δ1 or the indicated EspPΔ1 mutant, and autotransporter synthesis was induced by the addition of IPTG. Passenger domain cleavage was then analyzed by Western blot using the C-terminal antipeptide antiserum. (A) Analysis of serine, histidine and aspartate mutations. (B) Analysis of Asp1120 mutations. (C) Analysis of glutamate mutations.
To rule out the possibility that the D1120N mutation blocks processing indirectly by altering protein folding or localization or translocation of the passenger domain across the OM, AD202 producing EspPΔ1, EspP*Δ1 or EspPΔ1(D1120N) was subjected to pulse-chase labeling. Each sample was divided in half, and one portion was treated with proteinase K to digest the passenger domain population that was exposed on the cell surface. EspP-containing polypeptides were then immunoprecipitated with the C-terminal antiserum and analyzed by SDS–PAGE. Consistent with the Western blot data, all of the wild-type proEspPΔ1 present in untreated cells was processed to discrete passenger and β domain fragments within 2 min, but none of the EspP*Δ1 or EspPΔ1(D1120N) precursor was processed even after 10 min (Figure 4, panels 1–3, lanes 1–4). Proteinase K treatment of pulse-labeled cells that produced EspP*Δ1 or EspPΔ1(D1120N), however, reduced virtually all of the proEspPΔ1 to a ∼33 kDa fragment (Figure 4, panels 1–3, lane 5). This fragment corresponds to a population of proEspPΔ1 whose passenger domain has been transported across the OM but not cleaved (Skillman et al, 2005). The data show that the passenger domain of both wild-type and mutant proteins is translocated across the OM rapidly, and imply that the D1120N mutation does not affect any step of protein biogenesis before passenger domain cleavage. Thus, Asp1120 plays a critical role in the release of the passenger domain from the cell surface. Interestingly, a topological model of the EspP β domain that generates a 12-stranded β barrel very similar to that of an autotransporter whose structure is known (Oomen et al, 2004) predicts that Asp1120 resides on the hydrophilic face of a β strand near the middle of the OM (Figure 5). Taken together with our TEV site mapping studies, this model predicts that Asp1120 projects into the middle of the β barrel in close proximity to the cleavage junction and therefore would be correctly positioned to be an active site residue.
Figure 4.
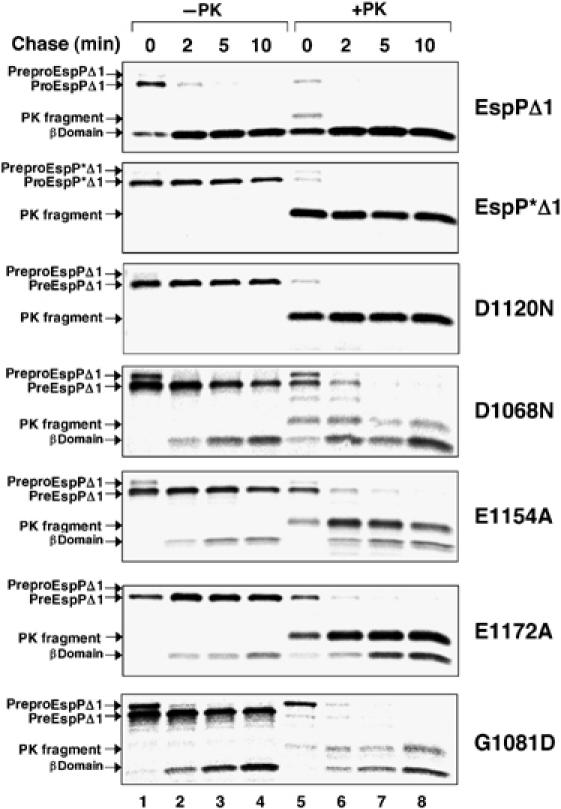
Effect of β domain mutations on the kinetics of EspPΔ1 translocation and cleavage. AD202 producing the indicated mutants was subjected to pulse-chase labeling after the addition of IPTG, and proteinase K was added to half of each sample. EspP-containing polypeptide was then immunoprecipitated with the C-terminal antipeptide antiserum. Lanes 1–4, no proteinase K added (−PK); lanes 5–8, proteinase K added (+PK).
Figure 5.
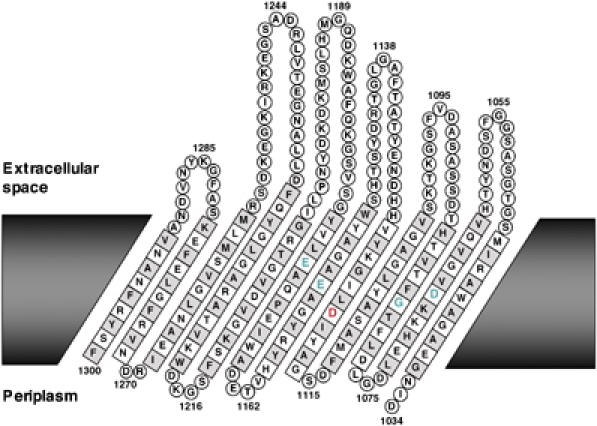
Predicted membrane topology of the EspP β domain. The topology of the EspP β barrel was predicted using PRED-TMMB (Bagos et al, 2004). The residues that comprise the hydrophobic face of each β strand are shown in grey. Residues that are essential for cleavage (Asp1120 and Asn1023) are shown in red and residues that play a secondary role in cleavage (Asp1068, Gly1080, Glu1154 and Glu1172) are shown in green. The first 10 residues of the β domain, which presumably loop into the barrel, have been removed for clarity.
We also changed Asp1120 to alanine, glutamine and glutamic acid to obtain further insight into its role in the cleavage reaction. None of the mutations affected protein localization or passenger domain translocation (Supplementary Figure 3), but two of the mutations, D1120A and D1120Q, completely abolished passenger domain cleavage (Figure 3B). In contrast, the D1120E mutation did not significantly affect proteolytic processing. These results show that the essential feature of Asp1120 is its acidic side chain.
Conserved residues that are predicted to reside near Asp1120 maximize the efficiency of passenger domain cleavage
Further efforts to define the mechanism of EspP passenger domain cleavage led to the identification of a class of residues that play a secondary role in the reaction. Because most aspartyl proteases (including the omptins) contain two active site aspartate residues (one of which acts as a base and the other as an acid), it seemed plausible that both Asp1120 and a second aspartate might be required for passenger domain cleavage. Our Western blot analysis, however, suggested a possible role for only one other aspartate, Asp1068, in the processing reaction. Pulse-chase experiments corroborated the conclusion that the D1068N mutation only partially impairs passenger domain cleavage. Although the mutation caused a considerable delay in processing, about half of the proEspPΔ1 was converted into discrete passenger and β domain fragments within 10 min (Figure 4, panel 4, lanes 1–4). The finding that all of the proEspPΔ1 was sensitive to proteinase K digestion by 2 min shows that the mutation primarily affects passenger domain release and not an earlier step in protein biogenesis (Figure 4, panel 4, lanes 5–8). Given the intermediate nature of the cleavage defect, however, Asp1068 cannot be an essential catalytic residue. Like Asp1120, Asp1068 is also predicted to reside near the middle of the membrane-spanning portion of the EspP β barrel (Figure 5).
Because some acidic proteases use glutamate as a catalytic residue, we also mutated all of the conserved glutamates in the β domain to alanine (Figure 1). AD202 that produced mutant versions of EspPΔ1 were grown in LB and passenger domain cleavage was assayed by Western blot as described above. None of the mutations abolished cleavage, but the E1154A and E1172A mutations partially impaired proEspPΔ1 processing (Figure 3C, lanes 4 and 6). Pulse-chase labeling of cells that produced EspPΔ1(E1154A) and EspPΔ1(E1172A) followed by proteinase K digestion revealed a substantial defect in passenger domain processing, but at most a slight delay in passenger domain translocation (Figure 4, panels 5 and 6). Interestingly, the E1154A and E1172A mutations not only produce essentially the same effect on the cleavage reaction as the D1068N mutation, but also modify residues that are predicted to neighbor Asp1120 in three-dimensional space (Figure 5).
Examination of a third mutation that alters a residue that cannot be catalytic provided a framework for rationalizing these results. The G1081D mutation was isolated in a separate mutational analysis of highly conserved β domain residues (KM Skillman, unpublished data). Pulse-chase experiments showed that the mutation causes a partial defect in passenger domain processing (Figure 4, panel 7). Like other residues that appear to influence cleavage efficiency, Gly1081 is predicted to reside inside the β barrel near Asp1120 (Figure 5). Taken together, the data suggest that the mutation of amino acids in the vicinity of the catalytic Asp1120 residue indirectly affects the cleavage reaction by improperly positioning the scissile N1023–N1024 bond or altering the environment inside the β barrel. It is especially easy to envision how replacement of Gly1081 with a much bulkier aspartate might displace the cleavage junction from Asp1120.
Because we could not identify a second acidic residue in the β domain that is essential for catalysis, we considered the possibility that passenger domain cleavage requires the contribution of catalytic residues from two EspP protomers. Indeed the active site residues of aspartic proteases are often contributed by two subunits of a dimeric enzyme (Barrett et al, 2003). Experiments involving the simultaneous production of catalytically active and inactive forms of EspPΔ1, however, effectively ruled out this scenario (Supplementary Figure 4).
Purified proEspPΔ1 undergoes self-cleavage in detergent solution
To further test the hypothesis that Asp1120 plays a catalytic role in an autoproteolytic reaction, we examined the ability of purified His-tagged proEspPΔ1 to mediate passenger domain cleavage. We could not obtain wild-type proEspPΔ1, however, because it is cleaved very rapidly in vivo (Figure 4) and because the residual precursor that was present in cell extracts was completely processed during purification. To overcome this problem, we used the slow-cleaving variant EspPΔ1(E1154A) to assay passenger domain cleavage in vitro. The mutant form of the protein was overproduced and purified in the presence of detergent under native conditions on an Ni-NTA column. Because the His tag is at the N-terminus of the protein, the cleaved passenger domain copurified with the unprocessed precursor. Upon incubation at 37°C, the ∼45 kDa proEspPΔ1(E1154A) band gradually disappeared, whereas increasing amounts of a ∼30 kDa band that corresponds to the EspP β barrel appeared (Figure 6A). Consistent with the observation that the cleavage of the EspPΔ1(E1154A) passenger domain is very inefficient in vivo, the processing of the purified mutant protein was slow in vitro. In contrast, when proEspPΔ1(D1120N) was purified and incubated at 37°C in parallel reactions, no precursor processing was observed (Figure 6B). These results provide strong additional evidence that Asp1120 plays a direct role in an autocatalytic cleavage reaction.
Figure 6.
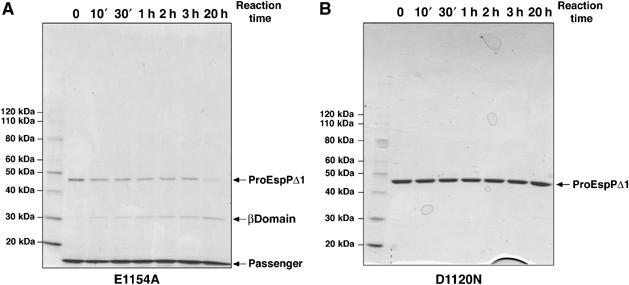
Purified EspPΔ1 undergoes self-cleavage in vitro. Purified 10His-EspPΔ1(E1154A) (A) or 10His-EspPΔ1(D1120N) (B) was incubated at 37°C. Aliquots were removed from each reaction at the indicated time points and analyzed by SDS–PAGE. Proteins were visualized by Coomassie blue staining.
The Asn1023 residue on the upstream side of the cleavage site is essential for EspP passenger domain processing
Although we could identify only one aspartate that is essential for passenger domain cleavage, proteases that contain a single acidic residue in the active site are not unprecedented. The cleavage of the major capsid protein of nodaviruses and tetraviruses into two fragments during virion maturation requires an acidic residue and an asparagine located in the ‘P1' position on the upstream side of the cleavage site (Schneemann et al, 1992; Zlotnick et al, 1994; Taylor et al, 2002; Taylor and Johnson, 2005). Remarkably, an asparagine (Asn1023) is strictly conserved at the P1 position of the SPATEs (Figure 1). In creating EspP*Δ1, we found that mutation of this residue and a second invariant asparagine at the P1' position on the downstream side of the cleavage site to serine abolishes cleavage. To dissect the significance of the P1 and P1′ residues in the cleavage reaction, we mutated each residue individually. AD202 was transformed with the mutant plasmids, and EspP biogenesis was monitored in pulse-chase experiments. We found that the N1023A, N1023D and N1023S mutations abolished passenger domain cleavage, but had no effect on earlier steps of protein biogenesis (as assessed by the susceptibility of the passenger domain to proteinase K digestion) (Figure 7A). Interestingly, a mutation that retains an amide group in the P1 position (N1023Q) only slowed the cleavage reaction (Figure 7A). In addition, mutation of the conserved residues Glu1021 and Phe1018 (P3 and P7, respectively) and insertion of the TEV5 site (which in effect mutates the P3–P10 residues) partially impaired the cleavage of the EspPΔ1 passenger domain but not its translocation across the OM (Figure 7B). Like the β domain mutations that produced intermediate effects, these mutations may hinder processing by misaligning the cleavage site relative to Asp1120. In contrast, mutation of either the P1' residue or the conserved P3' residue (Asn1026) had no effect on either passenger domain translocation or cleavage (Figure 7C). Taken together, the results suggest that Asp1120 and Asn1023 are analogous to the two residues that are essential for the maturation of nodavirus and tetravirus capsid proteins.
Figure 7.
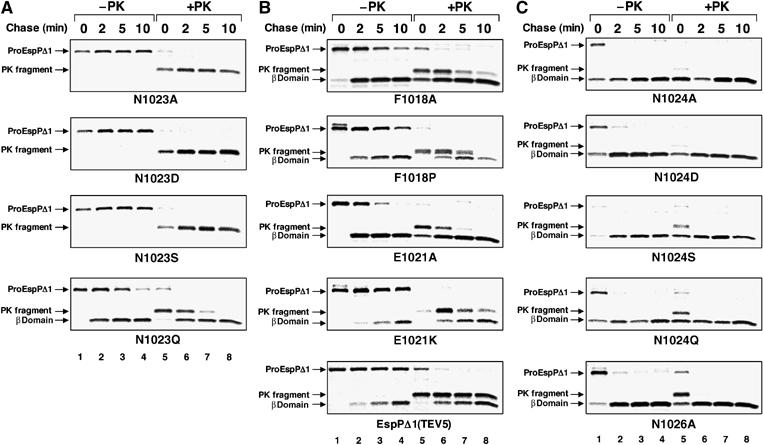
EspP passenger domain cleavage requires an amide at the P1 position. AD202 producing the indicated mutants was subjected to pulse-chase labeling after the addition of IPTG, and proteinase K was added to half of each sample. EspP-containing polypeptide was then immunoprecipitated with the C-terminal antipeptide antiserum. (A) Analysis of Asn1023 mutations. (B) Analysis of mutations near the C-terminus of the passenger domain. (C) Analysis of Asn1024 and Asn1026 mutations. Lanes 1–4, no proteinase K added (−PK); lanes 5–8, proteinase K added (+PK).
EspP passenger domain cleavage involves the formation of a succinimide intermediate
Based in part on an analysis of the active site by X-ray crystallography, it has been proposed that self-cleavage of nodavirus and tetravirus capsid proteins involves the formation of a hydrogen bond between the side chains of the catalytic asparagine and acidic residues that polarizes the amide nitrogen of the asparagine. The activated asparagine then performs a nucleophilic attack on its own main-chain carbonyl carbon and cleaves the polypeptide (Taylor et al, 2002; Taylor and Johnson, 2005). This reaction produces a cyclic succinimide intermediately at the C-terminus of the upstream fragment that is ultimately hydrolyzed to yield a mixture of asparagine and iso-asparagine.
To determine whether the EspP passenger domain might be released through this mechanism, we first purified the cleaved EspPΔ1 passenger domain and measured its mass by MS. Remarkably, the purified protein contained a mixture of two major species, one whose mass (15 027.2 Da) corresponded well to its theoretical mass (15 028.7 Da) and another that was 18 Da smaller (Supplementary Figure 5). We hypothesized that the smaller species contained a C-terminal succinimide, which is a dehydrated version of asparagine. To test this idea, we first digested the EspPΔ1 passenger domain with trypsin. In addition to identifying a peptide that has the predicted mass of the C-terminal tryptic fragment (806.4 Da) by MS, we also identified a peptide that was 18 Da smaller. Fragmentation of these two peptides by collisionally induced dissociation and analysis by tandem mass spectrometry (MS/MS) confirmed that the mass deficit in the smaller of the two EspPΔ1 passenger domain species was due to a C-terminal modification (most likely a dehydration) (Supplementary Figure 6). To perform an additional test of the cleavage mechanism, we first developed an LC method to separate synthetic peptides corresponding to the C-terminal tryptic fragment of the passenger domain (AFLNEVN) and a derivative containing a terminal iso-asparagine (AFLNEVN(iso)). When we used this method to analyze a tryptic digest of the purified EspPΔ1 passenger domain by LC/MS/MS, we identified fragments that eluted from the column at the same time and had the same mass (806.4 Da) as both synthetic peptides (Figure 8). The presence of a fragment corresponding to AFLNEVN(iso) is almost certainly due to the hydrolysis of a C-terminal succinimide. Taken together, these results provide compelling evidence that the EspP passenger domain is cleaved by the mechanism that has been proposed for the autocatalytic maturation of nodavirus and tetravirus capsids.
Figure 8.

A mixture of asparagine and iso-asparagine is present at the C-terminus of the EspPΔ1 passenger domain. Samples containing the indicated synthetic peptide(s) or a tryptic digest of the EspPΔ1 passenger domain were analyzed by LC/MS/MS as part of a series of runs shown from bottom to top. The elution of the synthetic peptides or C-terminal peptide of the EspPΔ1 passenger domain from the LC column is shown as the integrated intensity of the monoisotopic peak observed at 806.4 Da as a function of run time.
Distantly related autotransporters use the same passenger domain cleavage mechanism as EspP
Previous work has shown that the cleavage of the passenger domains of several members of the pertactin (Prn) family of autotransporters produced by B. pertussis occurs after an asparagine residue (Middendorf et al, 2005). While the β domains of these proteins are closely related to each other, they are only weakly related to the β domains of the SPATEs (Figure 1). Remarkably, examination of four of the six known members of this family has shown that proteins that have an asparagine at the P1 position (Prn, BrkA and TcfA) all undergo passenger domain processing, whereas Vag8, which has an aspartate at this position, has an uncleaved passenger domain (Finn and Amsbaugh, 1998). Moreover, despite the weak homology between the SPATE and Prn families, we found that the residues that are important for EspP cleavage are conserved in all of the Prn-like autotransporters except Vag8 and the uncharacterized Phg protein (Figure 1). These observations suggest that the passenger domains of the SPATEs and Prn-like proteins are cleaved by very similar mechanisms.
To gain insight into the processing of the B. pertussis autotransporters, we first cloned the genes encoding Prn (with an N-terminal HA tag) and BrkA (with a C-terminal His tag) into multicopy plasmids under the control of the trc promoter. These plasmids were introduced into an E. coli strain (BL21-CodonPlus(DE3)-RIL) that affords optimal translation of heterologous mRNAs. Cells were grown in LB and expression of the B. pertussis genes was induced with IPTG, and passenger domain cleavage was assayed by Western blot using anti-HA or anti-His antisera. As shown previously, the pro form of wild-type Prn (P.93) is rapidly processed into passenger domain (P.69) and β domain fragments (Charles et al, 1994; Figure 9A, lane 1). Mutation of the residue that corresponds to Asp1120 in EspP (Asp733) to asparagine abolished passenger domain cleavage (Figure 9A, lane 3). As in EspP, a glutamate is tolerated at this position, but cleavage is considerably less efficient (Figure 9A, lane 2). Mutation of the residue that corresponds to Asn1023 in EspP (Asn631) to either glutamine or alanine also abolished cleavage (Figure 9A, lanes 4 and 5). None of the mutations affected earlier steps in protein biogenesis as assessed by accessibility of the passenger domain to trypsin (Supplementary Figure 7A). Like Prn, wild-type BrkA was also cleaved to discrete passenger and β domains (Figure 9B, lane 1). Mutation of Asp833 (Asp1120 in EspP) to glutamate severely impaired processing; the free β domain could only be observed on overexposed blots (Figure 9B, lane 2 and data not shown). Mutation of Asp833 to asparagine or alteration of Asn731 (Asn1023 in EspP), however, completely abolished passenger domain cleavage but did not affect its translocation across the OM (Figure 9B, lanes 3–5 and Supplementary Figure 7B). Taken together, these results provide strong evidence that the Prn family of autotransporters is subject to autoproteolytic cleavage by an Asp–Asn dyad.
Figure 9.
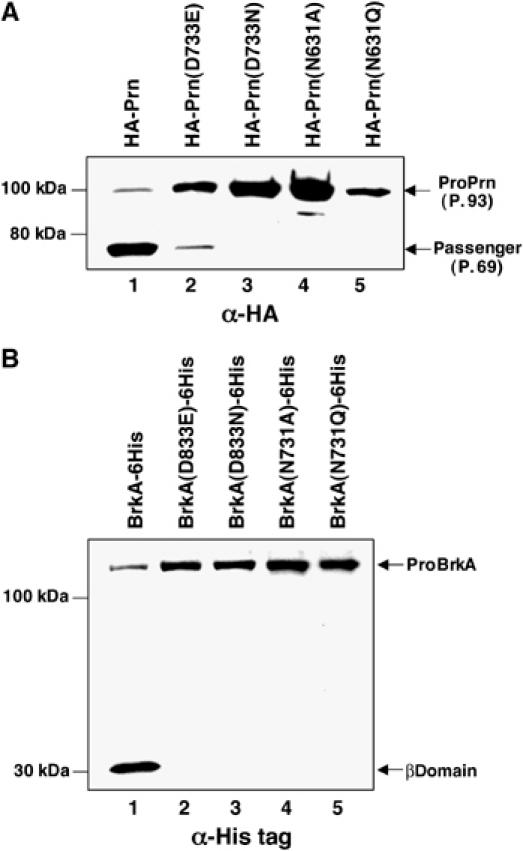
Residues that are required for EspP processing are also essential for the processing of B. pertussis autotransporters. BL21-CodonPlus(DE3)−RIL was transformed with a plasmid encoding HA-tagged Prn or the indicated mutant (A), or His-tagged BrkA or the indicated mutant (B) and grown in LB. Autotransporter synthesis was induced by incubating cultures with 100 μM IPTG for 2 h, and passenger domain cleavage was assayed by Western blot using anti-HA or anti-His tag antisera.
Discussion
In this report, we describe several lines of evidence that the EspP passenger domain is cleaved in an unusual autocatalytic reaction. Initial protease protection experiments strongly suggested that the cleavage site is sequestered inside the pore formed by the β domain when cleavage occurs. Although the mechanism by which the β domain might mediate passenger domain cleavage could not be predicted from the amino-acid sequence, we found that mutation of a single aspartate (Asp1120) to any non-acidic residue abolished processing. Mutation of several other conserved residues, all of which are likely to be close to Asp1120 in three-dimensional space, partially impaired cleavage. Interestingly, the properties of the cleavage site mirrored those of the β domain. Only a single residue (the P1 residue, Asn1023) was essential for processing, and a mutation that preserved the amide chemistry was tolerated. Likewise, the mutation of residues that are just upstream of (and therefore in close proximity to) Asn1023 produced intermediate cleavage defects. Although all of the mutagenesis was performed on EspPΔ1, the same results were obtained when the β domain and Asn1023 mutations were introduced into full-length EspP (Supplementary Figure 8). Finally, the observation that the passenger domain of purified EspPΔ1 is processed in vitro provided further evidence that the cleavage reaction is autocatalytic. Taken together, these data suggest a scenario in which Asp1120 and Asn1023 are catalytic residues that move into proximity as a consequence of a precise fitting of the polypeptide segment that surrounds the cleavage site into the β barrel. The conclusion that Asp1120 and Asn1023 play direct roles in catalysis was reinforced by the finding that they are both conserved and essential for cleavage in distantly related B. pertussis autotransporters. As the active site of omptins is located in the extracellular space, EspP represents the first example of an OM protein in which proteolysis occurs within the β barrel itself.
An analysis of the C-terminus of the cleaved EspPΔ1 passenger domain using LC and MS demonstrated that passenger domain processing proceeds through a mechanism that has been proposed for the autocatalytic maturation of structurally unrelated eukaryotic viral capsid proteins. The simplest explanation of the data is that Asp1120 activates the amide group of Asn1023 to mediate a nucleophilic attack on the polypeptide backbone. This reaction releases the passenger domain and produces a relatively stable C-terminal succinimide that is eventually hydrolyzed to yield a mixture of asparagine and iso-asparagine (Figure 10). Interestingly, as in EspPΔ1, the likely active site acidic residue in a tetravirus capsid protein can be either an aspartate or a glutamate (Taylor and Johnson, 2005). Although an asparagine to glutamine mutation is not tolerated in tetraviruses, the observation that the N1023Q mutation in EspP significantly slows (but does not abolish) cleavage is consistent with the proposed mechanism. Glutamine cyclizes less readily than asparagine because it has an extra CH2 group, but there is strong evidence that the formation of a glutarimide intermediate can promote a self-cleavage reaction (Amitai et al, 2004).
Figure 10.

Proposed mechanism of EspP self-cleavage. After translocation of the passenger domain across the OM, Asn1023 is positioned close to Asp1120. As a consequence of hydrogen bonding between the two side chains, the amide group of Asn1023 mediates a nucleophilic attack on the Asn1023–Asn1024 peptide bond (A). This reaction produces a succinimide intermediate at the C-terminus of the passenger domain (B), which is eventually hydrolyzed to yield a mixture of asparagine and iso-asparagine (C).
Despite these striking parallels, there also appear to be subtle differences between the autoproteolytic reactions catalyzed by viral proteins and autotransporters. The active site of the viral proteins all contain either a threonine or a tyrosine that appears to help position the amide group of the catalytic asparagine. Although we have mutated the conserved hydroxylated residues that are predicted to reside in the middle of the β barrel (T1079, Y1108, Y1118 and Y1157), none of the mutations affected passenger domain cleavage (N Dautin, unpublished data). In addition, the processing of EspP occurs in less than a minute, whereas the viral reactions are much slower. These disparities might reflect fundamental differences in the architecture of the two classes of proteins.
Our data suggest that autoproteolytic cleavage reactions involving an acid–asparagine dyad have evolved independently in at least two structurally distinct protein families. Although the mechanism by which cyclization of the asparagine at the cleavage site is catalyzed is unknown, it has been proposed that the same dyad might also be responsible for the autoproteolytic processing of FlhB, a polytopic inner membrane protein that regulates the ordered export of flagellar structural proteins in Salmonella (Ferris et al, 2005). Despite the fact that the viral and bacterial proteins are completely unrelated, in each case, proteolysis functions as a switch in a multistep self-assembly pathway. In both the biogenesis of autotransporters and the maturation of viral capsids, it is essential that cleavage be contingent on the formation of a specific intramolecular or intermolecular structure. A catalytic dyad may be particularly useful in this context because it requires only two key residues that can be kept apart and then brought together by a conformational change. Whereas in the case of RNA viruses formation of the active site requires a pH-induced conformational change in the assembled procapsid, in the case of autotransporters it requires precise positioning of the passenger domain inside the β barrel (which in turn may depend on integration of the β barrel into the OM and/or completion of passenger domain translocation). In addition, because asparagine cyclization is generally slow, the cleavage reaction can act as a timer. Indeed the composition of the bacterial flagellum may be dictated by the number of early substrate molecules that can be exported prior to the cleavage of FlhB.
Finally, the evidence that two autotransporter families whose β domains are only ∼15% identical are both cleaved by the same mechanism suggests that the Asp–Asn dyad evolved in an early common ancestor. Interestingly, a variety of uncharacterized autotransporters whose β domains are distantly related to the SPATE and Prn family β domains contain most of the residues that we implicated in the cleavage of the EspP passenger domain, and at least some of them might undergo the same autocatalytic processing reaction. Furthermore, the finding that the alteration of key residues differentially affects the processing of EspP, BrkA and Prn raises the possibility that naturally occurring mutations could be selected to alter the rate or extent of passenger domain cleavage. In this way, it would be possible to create passenger domains that function transiently at the cell surface and then subsequently in the extracellular environment.
Materials and methods
Reagents, bacterial strains and growth conditions
Peptides AFLNEVN and AFLNEVN(iso) were synthesized and HPLC-purified at the Facility for Biotechnology Resources, Center for Biologics Evaluation and Research (Food and Drug Administration). Rabbit antisera generated against C- and N-terminal peptides of EspP have been described (Szabady et al, 2005). Polyclonal antisera directed against the influenza hemagglutinin epitope HA.11 and pentahistidine were obtained from Covance and Qiagen, respectively. Purified recombinant TEV protease (hyperactive S219V mutant) was obtained from Susan Buchanan. The bacterial strains used in this study were AD202 (MC4100 ompT∷kan) (Akiyama and Ito, 1990), BL21 and BL21-CodonPlus(DE3)-RIL (Stratagene). Bacteria were grown at 37°C and media were supplemented with 100 μg/ml ampicillin, 50 μg/ml kanamycin or 30 μg/ml chloramphenicol as needed.
Plasmid construction
Plasmids pJH62, encoding espPΔ1, and pKMS3, encoding espP*Δ1, have been previously described (Skillman et al, 2005; Szabady et al, 2005). EspP mutants were constructed by overlap PCR (Ansaldi et al, 1996) using pJH62 as a template. To construct pKMS6 and pND2, a decahistidine tag was placed at the N-terminus of the EspP*Δ1 and EspPΔ1 passenger domains, respectively, by ligating the oligonucleotides Dir10HisLink and Rev10HisLink into the EagI site of pKMS3 and pJH62 (the sequence of all oligonucleotides is listed in Supplementary Table 1). PstI sites were then introduced at six locations in pKMS6 by overlap PCR using the oligonucleotides Pst1–6 and their complements. A 27 bp fragment was then inserted at each PstI site using the oligonucleotides TEV+ and TEV− to create plasmids p(TEV1)-p(TEV6). To make pTrc10HisEspPΔ1-E1154A and pTrc10HisEspPΔ1-D1120N, an ∼800 bp KpnI–HindIII fragment from the appropriate mutant versions of pJH62 was subcloned into pND2. As described in Supplementary data, brkA and prn were amplified using genomic DNA from B. pertussis strain Tomaha I as a template, cloned into pTrc99A (Pharmacia) and modified with epitope tags to construct pTrcBrkA-6His and pTrcHAPrn.
Purification of EspP*Δ1 derivatives containing TEV sites and TEV protease treatment
BL21 transformed with p(TEV1)-p(TEV6) was grown overnight in LB, washed and diluted into 250 ml medium at OD600=0.02. When the cultures reached OD600=0.2, synthesis of the EspP derivatives was induced by the addition of 10 μM IPTG (final concentration). After 2 h, the cells were pelleted and frozen on dry ice. Half of the cells were resuspended in 0.8 ml buffer A (50 mM K2HPO4 (pH 7.5), 200 mM NaCl, 20 mM imidazole, 1 mM MgCl2, 10 μg/ml DNAse I and 100 μg/ml 4-(2-aminoethyl) benzenesulfonyl fluoride) and lysed by sonication. The lysates were rotated for 30 min at 4°C after the addition of 5% Elugent and insoluble material was then removed by centrifugation (20 000 g, 4°C, 10 min). Clarified lysates were mixed with 20 μl of a 50% stock of Ni-NTA resin (Qiagen) and rotated for 30 min at 4°C. The resin was then pelleted (1000 g, 4°C, 10 s) and washed twice in buffer B (50 mM K2HPO4 (pH 7.5), 200 mM NaCl, 20 mM imidazole, 10% glycerol and 0.1% dodecylmaltoside). To elute the protein, the resin was pelleted and resuspended in 20 μl buffer C (buffer B containing 250 mM imidazole) four times. Proteolysis reactions (45 μl) were performed at 25°C in buffer D (buffer C with 1 mM DTT and 1 mM EDTA) and contained ∼2 μg of an EspP derivative and ∼7 μg/ml TEV protease.
Purification of ProEspPΔ1 and the cleaved EspPΔ1 passenger domain
To purify His-tagged proEspPΔ1, BL21 transformed with either pTrc10HisEspPΔ1(D1120N) or pTrc10HisEspPΔ1(E1154A) was grown in 500 ml LB. At OD550=0.5, expression of the plasmid-borne genes was induced by adding 10 μM IPTG. After 3 h, cells were pelleted and frozen. Thawed cells were resuspended in buffer A and disrupted by sonication at 4°C. To extract membrane proteins, 5% Elugent was added and the samples were agitated for 2 h at 4°C. After unbroken cells and cell debris were removed by centrifugation (3500 g, 4°C, 15 min), the cell extract was applied to an Ni-NTA column equilibrated with buffer C containing no imidazole. The column was washed with buffer C containing 20–50 mM imidazole and proEspPΔ1 was eluted with buffer C containing 300 mM imidazole. The His-tagged EspPΔ1 passenger domain was purified from BL21 transformed with pND2 as described in Supplementary data.
Analysis of autotransporter biogenesis in vivo
To analyze autotransporter biogenesis under steady-state conditions, cells were grown in LB to OD550=0.2–0.3. Unless otherwise noted, expression of plasmid-encoded genes was induced by the addition of 10 μM IPTG. After 30 min, aliquots of each culture were mixed with 10% cold trichloroacetic acid (TCA) to precipitate both cellular and secreted proteins. To examine the kinetics of EspP biogenesis, cells were grown in M9 containing 0.2% glucose and all the L-amino acids except methionine and cysteine. Overnight cultures were washed and diluted into fresh medium at OD550=0.02. When the cultures reach OD550=0.2, synthesis of plasmid-borne genes was induced for 30 min by the addition of 10 μM IPTG. Pulse-chase labeling was then conducted as described (Ulbrandt et al, 1997). Aliquots of radiolabeled cells were poured over ice and collected by centrifugation. Cell pellets were resuspended in PBS and divided into two portions. One half was precipitated by TCA and the other was treated with proteinase K (0.2 mg/ml) for 20 min on ice before TCA precipitation. TCA precipitates were then solubilized and immunoprecipitations were performed as described (Ulbrandt et al, 1997).
Gel electrophoresis and Western blotting
Unless otherwise noted, proteins were resolved by SDS–PAGE using 8–16% Tris–glycine minigels (Invitrogen). For Western blotting, horseradish peroxidase (HRP)-linked protein A (Amersham) or HRP-linked goat anti-mouse IgG (Sigma) was used together with the supersignal Pico chemiluminescence kit (Pierce) to detect antibody–antigen complexes.
LC/MS/MS
Synthetic peptides and a tryptic digest of the purified EspPΔ1 passenger domain were analyzed on a Nanoacquity HPLC system interfaced with a QTOF-2 MS/MS (Waters) as described in Supplementary data.
Supplementary Material
Supplementary Informaion
Supplementary Table 1
Supplementary Figures
Acknowledgments
We thank Janine Peterson and Kristen Skillman for help in constructing plasmids, Tod Merkel for providing B. pertussis and Susan Buchanan for providing helpful comments on the manuscript. This work was supported by the NIDDK Intramural Research Program.
References
- Akiyama Y, Ito K (1990) SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the OmpT protease in vitro. Biochem Biophys Res Commun 167: 711–715 [DOI] [PubMed] [Google Scholar]
- Amitai G, Dassa B, Pietrokovski S (2004) Protein splicing of inteins with atypical glutamine and aspartate C-terminal residues. J Biol Chem 279: 3121–3131 [DOI] [PubMed] [Google Scholar]
- Ansaldi M, Lepelletier M, Mejean V (1996) Site-specific mutagenesis by using an accurate recombinant polymerase chain reaction method. Anal Biochem 234: 110–111 [DOI] [PubMed] [Google Scholar]
- Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ (2004) PRED-TMBB: a web server for predicting the topology of β-barrel outer membrane proteins. Nucleic Acids Res 32: W400–W404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AJ, Rawlings ND, Woessner JF (2003) The Handbook of Proteolytic Enzymes, 2nd edn. Elsevier (Amsterdam): Academic Press [Google Scholar]
- Charles I, Fairweather N, Pickard D, Beesley J, Anderson R, Dougan G, Roberts M (1994) Expression of the Bordetella pertussis P.69 pertactin adhesin in Escherichia coli: fate of the carboxy-terminal domain. Microbiology 140: 3301–3308 [DOI] [PubMed] [Google Scholar]
- Chen P, Tsuge H, Almassy RJ, Gribdskov CL, Katoh S, Vanderpool DL, Margosiak SA, Pinko C, Matthews DA, Kan CC (1996) Structure of the human cytomegalovirus protease catalytic domain reveals a novel serine protease fold and catalytic triad. Cell 86: 835–843 [DOI] [PubMed] [Google Scholar]
- Dutta PR, Cappello R, Navarro-Garcia F, Nataro JP (2002) Functional comparison of serine protease autotransporters of Enterobacteriaceae. Infect Immun 70: 7105–7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egile C, d'Hauteville H, Parsot C, Sansonetti PJ (1997) SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol Microbiol 23: 1063–1073 [DOI] [PubMed] [Google Scholar]
- Eslava C, Navarro-Garcia F, Czeczulin JR, Henderson IR, Cravioto A, Nataro JP (1998) Pet, an autotransporter protein enterotoxin from enteroaggregative Escherichia coli. Infect Immun 66: 3155–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM (2005) FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem 280: 41236–41242 [DOI] [PubMed] [Google Scholar]
- Finn TM, Amsbaugh DF (1998) Vag8, a Bordetella pertussis bvg-regulated protein. Infect Immun 66: 3985–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP (1999) Characterization of Pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect Immun 67: 5587–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D (2004) Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev 68: 692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson DR, de la Morena ML, Stathopoulos C, St Geme JW III (1997) Structural determinants of processing and secretion of the Haemophilus influenzae Hap protein. Mol Microbiol 26: 505–518 [DOI] [PubMed] [Google Scholar]
- Keiler KC, Sauer RT (1995) Identification of active site residues of the Tsp protease. J Biol Chem 270: 28864–28868 [DOI] [PubMed] [Google Scholar]
- Middendorf B, Stübs D, Guiso N, Deppisch H, Gross R, Fuchs TM (2005) Phg, a novel member of the autotransporter family present in Bordetella species. Microbiol Res 160: 329–336 [DOI] [PubMed] [Google Scholar]
- Oomen CJ, van Ulsen P, van Gelder P, Feijen M, Tommassen J, Gros P (2004) Structure of the translocator domain of a bacterial autotransporter. EMBO J 23: 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SK, Dotson J, Allen KP, Fleckenstein JM (2004) Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect Immun 72: 1786–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlner J, Halter R, Beyreuther K, Meyer TF (1987) Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325: 458–462 [DOI] [PubMed] [Google Scholar]
- Schneemann A, Zhong W, Gallagher TM, Rueckert RR (1992) Maturation cleavage required for infectivity of a nodavirus. J Virol 66: 6728–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruto D, Adu-Bobie J, Scarselli M, Veggi D, Pizza M, Rappuoli R, Arico B (2003) Neisseria meningitidis App: a new adhesin with autocatalytic serine protease activity. Mol Microbiol 48: 323–334 [DOI] [PubMed] [Google Scholar]
- Shere KD, Sallustio S, Manessis A, D'Aversa TG, Goldberg MB (1997) Disruption of IcsP, the major Shigella protease that cleaves IcsA, accelerates actin-based motility. Mol Microbiol 25: 451–462 [DOI] [PubMed] [Google Scholar]
- Skillman KM, Barnard TJ, Peterson JH, Ghirlando R, Bernstein HD (2005) Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Mol Microbiol 58: 945–958 [DOI] [PubMed] [Google Scholar]
- Stein M, Kenny B, Stein MA, Finlay BB (1996) Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. Infect Immun 178: 6546–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabady RL, Peterson JH, Skillman KM, Bernstein HD (2005) An unusual signal peptide facilitates the late steps in the biogenesis of a bacterial autotransporter. Proc Natl Acad Sci USA 102: 221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Johnson JE (2005) Folding and particle assembly are disrupted by single-point mutations near the autocatalytic cleavage site of Nudaurelia capensis ω virus capsid protein. Protein Sci 14: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Krishna NK, Canady MA, Schneemann A, Johnson JE (2002) Large-scale, pH-dependent, quaternary structure changes in an RNA virus capsid are reversible in the absence of subunit autoproteolysis. J Virol 76: 9972–9980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DP, Wooldridge KG, Ala'Aldeen DA (2002) Autotransporter serine protease A of Neisseria meningitidis: an immunogenic, surface-exposed outer membrane, and secreted protein. Infect Immun 70: 4447–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrandt ND, Newitt JA, Bernstein HD (1997) The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88: 187–196 [DOI] [PubMed] [Google Scholar]
- van Ulsen P, van Alphen L, ten Hove J, Fransen F, van der Ley P, Tommassen J (2003) A Neisserial autotransporter NalP modulating the processing of other autotransporters. Mol Microbiol 50: 1017–1030 [DOI] [PubMed] [Google Scholar]
- Vandeputte-Rutten L, Kramer RA, Kroon J, Dekker N, Egmond MR, Gros P (2001) Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO J 20: 5033–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velarde JJ, Nataro JP (2004) Hydrophobic residues of the autotransporter EspP linker domain are important for outer membrane translocation of its passenger. J Biol Chem 279: 31495–31504 [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ (1999) Two transmembrane aspartates in presenilin-1 required for presinilin endoproteolysis and gamma-secretase activity. Nature 398: 513–517 [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Reddy VS, Dasgupta R, Schneemann A, Ray WJ Jr, Rueckert RR, Johnson JE (1994) Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single aspartic residue. J Biol Chem 269: 13680–13684 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Informaion
Supplementary Table 1
Supplementary Figures


