Abstract
We studied expression of the 5-lipoxygenase (5-LO) pathway in normal human skin. In situ hybridization revealed a 5-LO mRNA-containing epidermal cell (EC) population that was predominantly located in the midportion of the spinous layer, in outer hair root sheaths, and in the epithelial compartment of sebaceous glands. Examination of skin specimens by immunohistochemistry and of primary ECs by flow cytometry mapped the 5-LO protein exclusively to Langerhans cells (LCs). The LC 5-LO protein was largely found in the nuclear matrix, in nuclear envelopes, and perinuclear regions as indicated by in situ confocal laser scan microscopy. Reverse transcription–PCR and immunoblot analyses of purified primary EC populations further indicated that LCs are major 5-LO expressing cells. Enriched primary LCs were also found to contain 5-LO activating protein (FLAP), leukotriene (LT) C4 synthase, and LTA4 hydrolase. By contrast, 5-LO, FLAP, and LTC4 synthase were undetectable or largely reduced, but LTA4 hydrolase transcripts and protein were identified in ECs depleted of LCs. These data show that naïve LCs are major, and possibly the sole, 5-LO pathway expressing cells in the normal human epidermis.
Keywords: dendritic cells, 5-lipoxygenase pathway, arachidonic acid cascade
Products of the 5-lipoxygenase (5-LO; arachidonate:oxygen 5-oxidoreductase, EC 1.13.11.34) pathway, including leukotriene (LT) B4 and the cysteinyl LTs, are considered potent mediators of allergic and hypersensitivity reactions and may also act as physiological autocrine/paracrine signaling molecules (1). Several white blood cell lineages have been reported to express the 5-LO gene (1), whereas its occurrence in extrahematopoietic tissues has received less attention. We reported that the human keratinocyte cell line, HaCaT, can be induced to up-regulate the 5-LO gene when maintained for prolonged time periods in culture medium supplemented with fetal calf serum (2). Other experiments indicated that normal human epidermal keratinocytes can also be induced to express 5-LO under similar in vitro conditions (2). These data raised the possibility that skin keratinocytes in vivo express the 5-LO gene during their normal differentiation program or that it is induced in response to cytokines generated in diseased skin. To address these issues we decided to focus attention on normal human skin and to initially study epidermis, the outermost skin compartment (3). Epidermis is a highly organized stratified squamous epithelium consisting of keratinocytes at different stages of differentiation as its major cellular constituent. Moreover, epidermis harbors several numerically minor cell populations including melanocytes, Langerhans cells (LCs), Merkel cells, and occasionally white blood cells (4, 5). In the report detailed below we used in situ hybridization (ISH), immunohistochemistry, flow cytometry, and cell purification methods to identify the lineage(s) of 5-LO positive cells in the normal unperturbed epidermis. We found that the physiological keratinocyte differentiation pathway is not associated with up-regulation of the 5-LO gene but that LCs are the major, and possibly sole, 5-LO expressing cells in the normal human epidermis.
MATERIALS AND METHODS
Materials.
The materials used were as follows: [35S]CTP (1,000 Ci/mmol; Amersham; 1 Ci = 37 GBq); avian myeloblastosis virus (AMV) reverse transcriptase (Boehringer Mannheim); DNA stain Hoechst 33258 (Sigma); Cy2 (green) and Cy3 (red) fluorochrome-conjugated secondary antisera (Biotrend, Rockland, ME, and Dianova, Hamburg); rabbit or mouse antisera against human antigens: collagen IV (kindly provided by J. M. Foidart, Liege) and collagen VII mAb (kindly provided by I. Leigh, London); a mixture of mAbs against lamin A, B1, B2, and C (Progen, Heidelberg); melanocyte-associated protein, MEL5, mAb (Signet Laboratories, Dedham, MA); vimentin (Monosan, Uden, The Netherlands); keratin (Sigma); and CD1a and HLA-DR (Immunotech-Coulter, Hamburg). All other reagents were from Sigma if not otherwise noted.
Isolation of Epidermal Cell (EC) Populations and Flow Cytometry.
ECs were prepared and subjected to repeated immunobead adsorption by using dynabeads M-280 sheep anti-mouse IgG (Dynal, Oslo) and CD1a as reported (6). Analyses were performed with a FACScan (Lysis II software, 1024 channel mode; Becton Dickinson) on saponin-permeabilized ECs as reported (6). CD1a+ and CD1a− EC populations were gated manually and the percentage of 5-LO+ cells was determined by using 5-LO antiserum 1550 that had been prepared as described under Immunohistochemistry. Collagen IV antiserum served as control and a F(ab′)2 donkey anti-rabbit IgG fluorescein isothiocyanate (Dianova) was used as the secondary antiserum.
ISH and Reverse Transcription–PCR (RT-PCR).
Human 5-LO cDNA (nucleotides 1–2496) was subcloned into pBluescript II KS vector (Stratagene) and 35S-labeled cRNA antisense and sense fragment lengths were adjusted to 250 nucleotides by using standard techniques as reported (7–9). RT-PCR analyses were performed as described (2). Details of tissue preparation, hybridization conditions, probe specificities, primer sequences, PCR parameters, and competitive PCR are available on request.
Immunohistochemistry and Confocal Laser Scan Microscopy (CLSM).
Three polyclonal rabbit anti 5-LO antisera, LO 32, 1550 (directed against native purified recombinant human 5-LO), and 1551 (directed against SDS-denatured purified recombinant human 5-LO), were used with similar results. The 1550 and 1551 antisera were affinity-purified on recombinant 5-LO bound to nitrocellulose paper by using established techniques. To reduce unspecific binding, all antisera were preadsorbed on cytoskeletal proteins prepared from human epidermis as described (10). Skin specimens were embedded in Tissue TeK (Miles) and snap frozen in liquid nitrogen. Cryosections (4–7 μm) were transferred onto slides and air dried. For optimal results 5-LO antisera required 12- to 18-h incubations. Affinity-purified fluorochrome-conjugated secondary antisera were donkey anti-mouse IgG F(ab′)2 (Dianova), goat anti-mouse IgG, goat or donkey anti-rabbit IgG F(ab′)2, or intact IgG (Biotrend). For the generation of electronical microscopical images (Fig. 2I) a Zeiss Axiophot I microscope equipped with a charge-coupled device (CCD) camera (Photometrics, Tuscon, AZ) was used. CLSM was performed on paraformaldehyde-fixed skin cryosections by using a Leica DM IRBE microscope (Leica) equipped with a scanner head Leica TCS NT and argon-crypton laser. 5-LO protein was visualized with LO32 antiserum (or 1550, data not shown), DNA with propidium iodide, and nuclear envelopes with lamin antisera as described under Materials. Details of fixation, staining, antibody dilutions, software, and recording are available on request.
Figure 2.
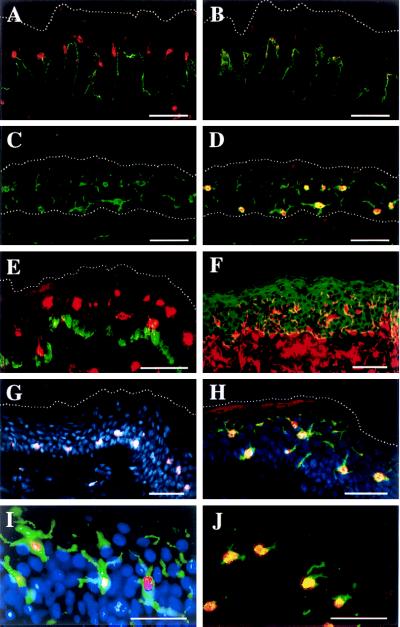
Immunofluorescence histochemistry localizes 5-LO protein in naïve LCs of normal epidermis. Sample designation: female upper arm, 79 years (C, D, and G–J); male back, 68 years (E); female breast, 29 years (F); female breast, 23 years (A and B). Upper dotted lines (A–E, G, and H) delineate the outer border of stratum corneum. Lower dotted lines (C and D) delineate the border between dermis and epidermis. (A) 5-LO (LO 32 Cy3, red) and collagen VII (Cy2, green). (B) Same as A but antiserum preadsorbed with recombinant 5-LO as described. (C) CD1a Cy2. (D) Same as C plus 5-LO 32, Cy3. (E) 5-LO 1550, Cy3 plus melanin-associated protein, MEL5, Cy2. (F) Vimentin, Texas red plus keratin, fluorescein isothiocyanate green. (G) 5-LO, 1550, Cy3 plus Hoechst 33258, blue. (H) 5-LO LO 32, Cy3 plus CD1a, Cy2 plus Hoechst 33258 blue. (I) Same as H but CCD camera-recorded as described. (J) 5-LO LO 32, Cy3 plus HLA DR, Cy2. (Bars = 50 μm.)
Immunoblot.
Analyses were performed by using polyclonal rabbit antisera against 5-LO, 5-LO activating protein (FLAP), and LTA4 hydrolase as described (2) and under Immunohistochemistry. Triton X-100 (1%) soluble extracts (35 μg protein) or leukocyte membranes (5 μg protein) were separated by 7.5% (5-LO, LTA4 hydrolase) or 15% (FLAP) SDS/PAGE, transferred to Hybond-C Super membranes, followed by enhanced chemiluminescence (Amersham, Braunschweig). Dilutions of antisera were as follows: 5-LO, 1:2,000; FLAP, 1:500; and LTA4 hydrolase, 1:2,000.
RESULTS
mRNA Expression of 5-LO Pathway Constituents in Normal Epidermis.
Epidermis and dermis from normal human skin specimens (obtained from different body sites of both sexes and ages ranging from 30 to 76 years) were separated and studied by RT-PCR for the presence of transcripts of 5-LO, FLAP, LTA4 hydrolase, and LTC4 synthase. PCR products of all four 5-LO pathway constituents were found in both skin compartments. The amounts of 5-LO transcripts in total epidermis RNA, as determined by competitive RT-PCR, were significantly lower than those previously obtained in RNA from cultured differentiated HaCaT keratinocytes or buffy coat RNA (ref. 2; see below).
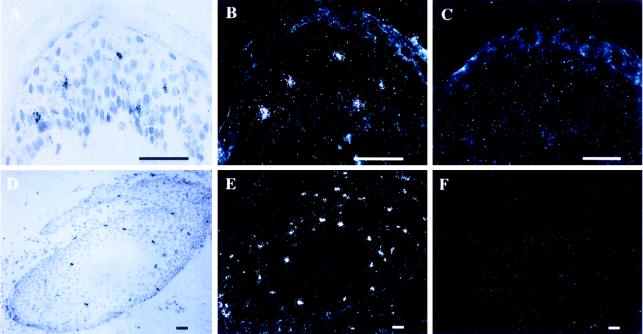
To identify individual 5-LO mRNA positive ECs, ISH analyses were performed. Most ECs, particularly the majority of keratinocytes including the differentiated keratinocytes of the granular layer, did not yield specific hybridization signals (Fig. 1 A–C). However, cells located predominantly in the midportion of the spinous layer and, to a lesser extent, in the basal layer, as well as few cells in the dermis were strongly positive (Fig. 1 A–C; additional data not shown). 5-LO mRNA positive cells accounted for only 1–2% of the total EC population and were uniformly located in all specimens examined. When epidermal appendages were inspected, a substantial number of 5-LO ISH positive cells were noted in the hair outer root sheaths (Fig. 1 D–F) and in the cellular compartment of sebaceous glands (data not shown). These findings suggested that distinct suprabasal ECs within the midportion of the spinous layer, a substantial number of hair outer root sheath cells, and cells in sebaceous glands express significant amounts of 5-LO transcripts.
Figure 1.
ISH of 5-LO mRNA in human skin. (A) Epidermis: 5-LO antisense bright field. (B) Epidermis 5-LO antisense. (C) Epidermis 5-LO sense. (D) Hair root antisense bright field. (E) Hair root antisense. (F) Hair root 5-LO sense. (Bar = 45 μm.)
Indirect Immunofluorescence of Normal Human Skin.
Initial attempts to identify 5-LO protein-expressing ECs in situ by using several polyclonal rabbit 5-LO antisera and control antisera yielded divergent staining patterns in the epidermis but not in the dermis (data not shown). Thus, rabbit 5-LO antisera and several unrelated rabbit antisera bound to granular or basal layer keratinocytes or produced diffuse patterns of cytoplasmic epidermal fluorescence. Similar phenomena have been found by others to be caused by unspecific binding of rabbit preimmune or immune antisera to cytoskeletal and other proteins (11–13). To avoid unspecific binding both 5-LO and control antisera were preadsorbed on Triton X-100 insoluble extracts of normal human epidermis (10). In addition, two polyclonal rabbit anti 5-LO antisera, 1550 and 1551, were affinity purified. Apparent unspecific binding was largely eliminated in immunohistochemistry and reexamination of human skin specimens by using preadsorbed 5-LO antisera revealed that most ECs did not yield positive fluorescence signals in any keratinocyte layer (Fig. 2A). However, analogous to the distribution pattern of positive 5-LO ISH signals (Fig. 1), ECs located predominantly and regularly in the midportion of the spinous layer, distinct hair outer root sheath cells, and scattered dermal cells showed positive fluorescence signals (Fig. 2A; additional data not shown). When control rabbit IgG was used or primary antisera were omitted, no fluorescence signals were recorded (data not shown). Preincubation of 5-LO antisera with recombinant 5-LO protein at 2 μg/100 μl largely abolished fluorescence positivity in immunohistochemistry and abolished a 78-kDa band in epidermis immunoblots performed on Triton X-100 soluble protein, indicating that binding was specific for 5-LO or 5-LO-related epitopes in situ (Fig. 2B; additional data not shown).
The scattered and regular distribution pattern of putative 5-LO positive cells in the spinous layer was suggestive of LCs that can be readily identified by their dendritic morphology and CD1a positivity in normal human epidermis (4, 5). Double immunofluorescence microscopy (Cy2 green fluorochrome for CD1a antigen; Cy3 red fluorochrome for 5-LO antigen; colocalization yields yellow) suggested localization of 5-LO and CD1a antigens in individual suprabasal cells with a dendritic morphology (Fig. 2 C and D). By contrast, a melanocyte-specific antibody, MEL5, largely separated 5-LO+ cells from melanocytes (Fig. 2E). That the majority of presumptive 5-LO+ skin cells are located in the epidermis can be appreciated in specimens treated with both vimentin antiserum (stains melanocytes and LCs in the epidermis and endothelial cells, fibroblasts, and hematopoietic cells in the dermis) and keratin antiserum (stains all epidermal keratinocytes) (Fig. 2F). However, it is noteworthy that 5-LO positive and CD1a/5-LO double positive cells were also found in the dermis indicating that 5-LO expression in dendritic cells is not restricted to epidermal LCs (lower part of Fig. 2E; additional data not shown). 5-LO positivity appeared to largely reside in the nucleus in double stains of 5-LO antisera and DNA binding dye Hoechst 33258 (Fig. 2G), and in triple stains of 5-LO, CD1a, and Hoechst 33258 (Fig. 2 H and I). Although we have inspected many 5-LO+ cells in >200 sections, strong cytoplasmic positivity was never observed in conventional immunohistochemistry (see below under CLSM). Although all 5-LO+ nuclei were associated with peripheral CD1a positivity (Fig. 2D), not all CD1a+ structures were associated with 5-LO+ nuclei (Fig. 2 D, H, and I). Because most of LCs’ CD1a occupies the cell surface (4), some LCs’ dendrites were visualized with no associated localizable 5-LO+ nucleus (Fig. 2 D, H, and I). Thus, though these data are compatible with the possibility that all LCs are 5-LO+, we cannot firmly establish this notion due to the limitations of histochemistry in thin tissue sections (see Flow Cytometry below). For better localization of separate signals within single cells triple fluorescence signals were recorded with a CCD camera. Colocalization of nuclear 5-LO, cell surface CD1a, and nuclear Hoechst 33258 was apparent in individual epidermal dendritic cells, whereas presumptive surrounding keratinocytes stained exclusively for Hoechst 33258 (Fig. 2I). HLA-DR antisera, which have been used previously to localize LCs in the skin (4), also localized to 5-LO+ cells (Fig. 2J).
When taken together, these data suggested that the majority of 5-LO protein-containing ECs are LCs, and that the 5-LO protein is located in or close to the nucleus.
Flow Cytometry Analyses of EC Suspensions.
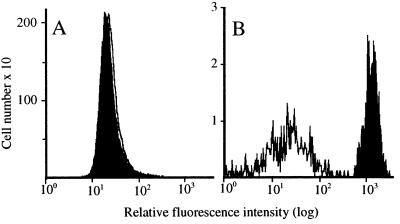
Two color flow cytometric analyses were performed on two freshly prepared EC suspensions from independent normal donors. As shown by histogram analysis, no 5-LO+ ECs were detected in the CD1a− population (a total of 49,309 cells was analyzed in the experiment shown in Fig. 3). Thus an unrelated rabbit IgG (against collagen IV) yielded fluorescence intensity that was as low as that obtained with the anti 5-LO antiserum (Fig. 3A). Also, the collagen antiserum yielded a low signal in the CD1a+ EC population (Fig. 3B, open histogram). By contrast, 5-LO fluorescence intensity in CD1a+ ECs (a total of 532 CD1a+ cells was analyzed in the experiment shown) was high (Fig. 3B, closed histograms). This result gives a ratio of approximately 59 for 5-LO versus collagen signals in the CD1a+ ECs. These data provided evidence against the presence of 5-LO+ cells in the CD1a− EC population and indicated that the majority of 5-LO positivity in freshly prepared EC suspensions resides in the CD1a+ EC subpopulation.
Figure 3.
Flow cytometry maps 5-LO protein to CD1a+ ECs. ECs were prepared and flow cytometry analyses performed as described under Methods. (A) CD1a− cells: collagen IV (open), 5-LO 1550 (filled); (B) CD1a+ cells: collagen IV (open), 5-LO 1550 (filled).
CLSM Localizes 5-LO Protein in Distinct Pools of Naïve LCs.
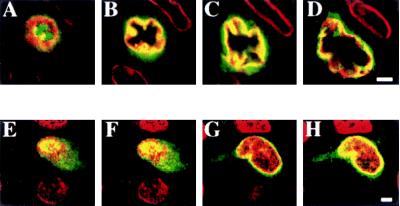
To localize 5-LO protein within CD1a+ cells in situ, CLSM was applied. Analyses of 20 cells in several independent specimens indicated that fluorescence intensity was strongest in nuclear envelopes (Fig. 4 B–D, G, and H) and adjacent cytoplasmic areas (Fig. 4 B and C), significant within the nucleus (Fig. 4 E–H), and notably weaker and variable in the dendrites (Fig. 4 G and H). Identical results were obtained with two independent anti-5-LO antisera, LO 32 and 1550. Why not all dendrites stained 5-LO+ is unclear but may be caused by detectability problems because dendrite-associated positivity was found to be relatively low when compared with that in the nucleus or to areas adjacent to the nucleus. Double staining with 5-LO antiserum and lamin antisera (to delineate the nuclear envelope) revealed that 5-LO fluorescence colocalized with lamin positivity (Fig. 4 B–D). However, it appeared to extend beyond lamin positivity in areas adjacent to both the inner and the outer nuclear envelopes (Fig. 4 B–D). Double staining of 5-LO with propidium iodide to visualize DNA showed that 5-LO positivity also colocalized with DNA (compare Fig. 4 A and E). Again, areas that extend beyond DNA were found positive (Fig. 4H). These data indicated that 5-LO protein is present in distinct cellular pools: (i) in nuclear envelopes (Fig. 4 B–D, G, and H) and within the nucleus (Fig. 4 A, E, and F); (ii) in areas adjacent to the cytoplasmic face of the outer nuclear envelope (Fig. 4 B–D, G, and H); and (iii) in the dendrites (Fig. 4 G and H). These findings suggested that the distribution of 5-LO protein of naïve epidermal LCs differs from that of resting human blood leukocytes and that it resembled that of Ca2+ ionophore-stimulated human blood leukocytes (14). Immunoelectron microscopy will be required to more precisely localize 5-LO protein to specific sites within these pools.
Figure 4.
CLSM reveals distinct intracellular pools of 5-LO protein in naïve LCs. Epidermis was fixed and prepared as described under Methods. (A–D) Colocalization of lamin (Cy3) and 5-LO, LO32, antisera (Cy2). (E–H) Colocalization of DNA binding dye propidium iodide (red) and 5-LO, LO 32 Cy2. (A–D) Eight planes with distances of 0.9 μm each were recorded; planes 2, 4, 6, and 8 are shown. (E–H) Eleven planes with distances of 1 μm each were recorded; planes 2, 5, 8, and 11 are shown. (Bars = 2 μm.)
Analyses of Purified EC Populations.
To obtain further evidence for the hypothesis that LCs express 5-LO mRNA and protein, LCs were purified. For this purpose we prepared single cell suspensions of total ECs, and CD1a immunobead adsorption was used to obtain highly purified LC populations (>90%) and ECs that were depleted of LCs (ECs − LCs) (6). RNA was prepared from these preparations (and from human buffy coat as control) and subjected to RT-PCR analyses for transcripts of all 5-LO pathway constituents, a panel of EC and hematopoietic cell markers, and two housekeeping genes, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin. RT-PCR analyses for CD1a confirmed the enrichment of cells expressing CD1a in the purified LC fractions from two donors (Table 1). The EC − LC fraction was largely depleted of LCs. Further RT-PCR analyses for marker enzymes showed that the purified LCs were practically devoid of mast cells (tryptase) and natural killer cells, neutrophils, and macrophages (CD16). Furthermore, keratin 10 PCR products were abundant in the EC − LC preparation but scarce in the enriched LC preparation. Small amounts of tryptase transcripts, FLAP, and CD1a transcripts in EC − LC preparations could reflect a minor mast cell and LC contamination (Table 1).
Table 1.
RT-PCR analyses of LCs, ECs − LCs, and human leukocyte buffy coat
| PCR product | LCs | ECs − LCs | Buffy coat |
|---|---|---|---|
| 5-LO | +++ | − | +++ |
| FLAP | +++ | + | +++ |
| LTA4 hydrolase | +++ | +++ | +++ |
| LTC4 synthase | ++ | − | + |
| CD1a | +++ | + | ++ |
| Tryptase | − | + | ++ |
| CD16 | − | − | +++ |
| Keratin 10 | + | +++ | − |
For preparation of ECs and RNA, and RT-PCR, see Materials and Methods. +++, Detectable at 28 cycles; ++, detectable at 34 cycles; +, detectable at 40 cycles; −, not detectable at 40 cycles. Markers: CD1a, LCs; tryptase, mast cells; CD16, natural killer cells, neutrophils, and macrophages; keratin 10, keratinocytes.
RT-PCR analyses of LCs revealed significant amounts of 5-LO transcripts. The relative amount of 5-LO PCR product in LCs exceeded that in ECs − LCs by at least two orders of magnitude, as judged by competitive RT-PCR by using external standard 5-LO and GAPDH cDNAs. Additional calculations yielded a 5.9-fold abundance of GAPDH relative to 5-LO transcripts in purified LCs. Also, the level of 5-LO transcripts in purified LCs was found to be similar or higher than found for 55-day differentiated HaCaT keratinocytes (ref. 2; additional data not shown). These data were consistent with the ISH and immunohistochemical analyses and supported the conclusion that practically all 5-LO transcripts in normal human epidermis reside in LCs and that LCs contain high levels of 5-LO transcripts (2).
Although the sensitivity of our FLAP ISH analyses (probably caused by a shorter cRNA probe when compared with that of 5-LO) was insufficient to detect FLAP ISH signals in normal epidermis in situ, RT-PCR analyses indicated the presence of FLAP transcripts in LCs (Table 1). Indeed, LCs contained similar or higher amounts of FLAP transcripts when compared with 5-LO transcripts as judged by competitive RT-PCR analyses (data not shown). Only small amounts of FLAP transcripts were found in ECs − LCs. In contrast, high levels of LTA4 hydrolase transcripts were found in both LCs and in ECs − LCs (Table 1; see also immunoblots in Fig. 5). Results from RT-PCR analyses also showed that LCs contained LTC4 synthase (15) transcripts. However, further work should determine the EC lineage that expresses LTC4 synthase transcripts, quantify their abundance and determine whether the enzyme is expressed at the protein and enzyme activity levels. When taken together, our data suggested significant 5-LO, FLAP, and LTA4 hydrolase transcript levels in LCs and indicated that LTA4 hydrolase transcripts are present also in ECs other than LCs.
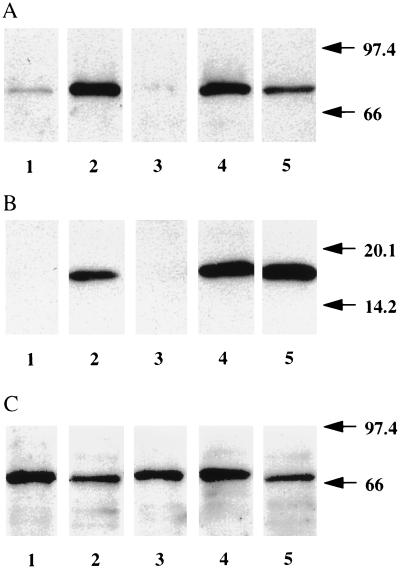
Figure 5.
Immunoblots of 5-LO, FLAP, and LTA4 hydrolase proteins in ECs, LCs, and EC − LCs. (A) 5-LO. Lanes: 1, ECs; 2, LCs; 3, ECs − LCs; 4, differentiated HaCaT keratinocytes; 5, RBL1 cells. (B) FLAP. Lanes: 1, ECs; 2, LCs; 3, ECs − LCs; 4, leukocyte membrane; 5, RBL1 cells. (C) LTA4 hydrolase. Lanes: 1, ECs; 2, LCs; 3, ECs − LCs; 4, differentiated HaCaT keratinocytes; 5, RBL1 cells.
Immunoblot Analyses of ECs and Purified LCs.
Total cellular Triton X-100 soluble extracts were used because it was found that these gave better results in SDS/PAGE followed by immunoblot for 5-LO and FLAP when compared with samples obtained by solubilizing the entire cells in SDS sample loading buffer. It was ascertained in independent experiments performed with HaCaT keratinocytes and RBL1 cells that Triton X-100 largely solubilized 5-LO and FLAP. 5-LO immunoblot analyses showed that 5-LO protein was detectable in Triton X-100 extracts of total EC suspensions (Fig. 5). However, purified LCs contained considerably higher amounts of 5-LO protein when compared with EC suspensions. Moreover, analogous to 5-LO transcripts, 5-LO protein was undetectable in ECs − LCs (Fig. 5). The apparent amounts of 5-LO protein in purified LCs appeared to be similar or higher when compared with levels previously found in cultured differentiated HaCaT keratinocytes or RBL1 cells (ref. 2; Fig. 5). By using densitometric scanning of specified aliquots of recombinant 5-LO protein that had been applied to the same gels, we estimated that LCs and ECs contain approximately 4,521 ng/mg and 296 ng 5-LO protein per mg Triton X-100 soluble protein, respectively (Fig. 5A; additional data not shown). When judged from standard curves generated by densitometric scanning of recombinant 5-LO protein applied to identical immunoblots this amounts to a 15-fold enrichment of 5-LO protein in LCs when compared with total EC suspensions. A similar cell lineage distribution pattern was found in immunoblots of FLAP protein, although the sensitivity of the FLAP immunoblot was not sufficient to detect the protein in Triton X-100 extracts of total EC suspensions (Fig. 5B). No attempts were made to purify FLAP protein from total epidermis. Moreover, FLAP protein was undetectable in ECs − LCs consistent with the low abundance of FLAP transcripts in this cell fraction (Table 1). In sharp contrast to 5-LO and FLAP proteins, similar levels of LTA4 hydrolase protein were found in all three EC preparations (Fig. 5). Indeed, in two experiments, performed with purified cell preparations of two donors, LTA4 hydrolase protein appeared to be expressed at somewhat higher levels in ECs − LCs when compared with LCs. Given the relative abundance of LCs and keratinocytes in the epidermis (approximately 1–2% LCs versus approximately 98% keratinocytes), the majority of total epidermal LTA4 hydrolase gene product appeared to reside in cells other than LCs. When taken together, these data supported the conclusion that LCs are the major 5-LO, FLAP, and possibly LTC4 synthase expressing cell lineage in normal epidermis and that, in addition to LCs, other ECs express LTA4 hydrolase.
DISCUSSION
We have identified LCs as the major, and most likely sole, 5-LO pathway expressing cell lineage of normal human epidermis. This conclusion is based on ISH, immunohistochemical, flow cytometry, CLSM, immunoblot, and RT-PCR analyses of epidermis tissue in situ, EC suspensions, and freshly isolated purified primary ECs. Although no information on activated LCs has been available until now, 5-LO protein of naïve LCs is located in distinct intracellular pools with preference for the nuclear envelopes and the adjacent cytoplasmic area. Competitive RT-PCR analyses of 5-LO transcripts and densitometric determination of immunoblots of 5-LO protein indicate that expression levels of the 5-LO gene in naïve LCs are similar or higher to that of differentiated HaCaT keratinocytes or RBL1 cells. Our results also strongly indicate that LCs express significant levels of FLAP and LTA4 hydrolase transcripts and protein. However, unlike 5-LO, FLAP, and possibly LTC4 synthase, LTA4 hydrolase is expressed in ECs − LCs at levels that are similar to those in LCs. Although our RT-PCR analyses are compatible with the possibility that LCs express LTC4 synthase, further work is required to more firmly demonstrate this assumption. Finally, our results indicate that the normal epidermal keratinocyte differentiation program is not associated with up-regulation of the 5-LO pathway in vivo (2).
LCs are specialized members of the family of dendritic cells in squamous stratified epithelia and comprise all of the accessory cell activity in normal human epidermis (16). They are regarded as outposts and sentinels of the skin immune system and are crucially involved in the initiation and propagation of immune responses directed against epicutaneous antigens of various origins including pollen, bacteria, viruses, fungi, and tumor cells (16). The accepted paradigm has been that LCs and other dendritic cells directly activate T lymphocytes after sampling and processing of antigen and subsequent migration through afferent lymph vessels into regional lymph nodes, the site where antigen presentation is likely to occur (16). More recently, Dubois et al. (17) have shown that dendritic cells can also directly activate B lymphocytes. Thus LCs initiate, govern, and maintain primary and secondary immune responses. Though a trace leukocyte population, dendritic cells are widely distributed in vivo at portals of entry of foreign antigen, in all solid organs, and in lymphoid tissues (16). However, phenotypes of different LCs and of other dendritic cell family members are distinct in different tissues, and so far expression of 5-LO has been demonstrated only for epidermal LCs. It will be a challenge for future studies to determine potential roles of 5-LO in the biology of LCs and, possibly, other dendritic cells.
Acknowledgments
R.S. and H.-J.S. contributed equally to this work. We thank Dr. J. Evans (Merck Frosst Center for Therapeutic Research, Quebec) for kind gifts of 5-LO, FLAP, and LTA4 hydrolase antisera. Antisera 1550 and 1551 were prepared by Drs. Y.-Y. Zhang, T. Hammarberg, and H. Okamoto. The technical assistance of I. Tomiç, and support in CLSM by Dr. S. Dietzel, Center for Molecular Biology Heidelberg, are gratefully acknowledged. This work was supported by the Deutsche Forschungsgemeinschaft (Ha 1083/8-2; SFB 217/D4), the Swedish Medical Council (03X-217), the Vårdal Foundation (A95 067), the German Bundesministerium für Forschung und Technologie (01GB 9401/6), the European Union (BMH4-CT96-0229), and a grant of the Stiftung VERUM.
ABBREVIATIONS
- CLSM
confocal laser scan microscopy
- EC
epidermal cell
- FLAP
5-LO activating protein
- ISH
in situ hybridization
- LC
Langerhans cell
- LT
leukotriene
- 5-LO
5-lipoxygenase
- RT-PCR
reverse transcription–PCR
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- CCD
charge-coupled device
References
- 1.Samuelsson B, Dahlén S-E, Lindgren J A, Rouzer C A, Serhan C N. Science. 1987;237:1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- 2.Janßen-Timmen U, Vickers P, Wittig U, Lehmann W D, Stark H-J, Fusenig N E, Rosenbach T, Rådmark O, Samuelsson B, Habenicht A J R. Proc Natl Acad Sci USA. 1995;92:6966–6970. doi: 10.1073/pnas.92.15.6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs E. Mol Biol Cell. 1997;8:189–203. doi: 10.1091/mbc.8.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holbrook K A. In: Physiology, Biochemistry and Molecular Biology of the Skin. Goldsmith L A, editor. Vol. 1. New York: Oxford Univ. Press; 1991. pp. 63–110. [Google Scholar]
- 5.Murphy G F. In: Dermatopathology: A Practical Guide to Common Disorders. Murphy G F, editor. Philadelphia: Saunders; 1995. pp. 3–27. [Google Scholar]
- 6.Wollenberg A, Kraft S, Hanau D, Bieber T. J Invest Dermatol. 1996;106:446–453. doi: 10.1111/1523-1747.ep12343596. [DOI] [PubMed] [Google Scholar]
- 7.Chomzcynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 9.Bosch F X, Leube R E, Achtstätter T, Moll R, Franke W W. J Cell Biol. 1988;106:1635–1648. doi: 10.1083/jcb.106.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stark H-J, Breitkreutz D, Limat A, Bowden P, Fusenig N E. Differentiation. 1987;35:236–248. doi: 10.1111/j.1432-0436.1987.tb00174.x. [DOI] [PubMed] [Google Scholar]
- 11.Osborn M, Franke W W, Weber K. Proc Natl Acad Sci USA. 1977;74:2490–2494. doi: 10.1073/pnas.74.6.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D, editors. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 632–633. [Google Scholar]
- 13.Bérubé B, Coputu L, Lefièvre L, Bégin S, Dupont H, Sullivan R. Anal Biochem. 1994;217:331–333. doi: 10.1006/abio.1994.1128. [DOI] [PubMed] [Google Scholar]
- 14.Woods J W, Evans J F, Ethier D, Scott S, Vickers P J, Hearn L, Heibein J A, Charleson S, Singer I I. J Exp Med. 1993;178:1935–1946. doi: 10.1084/jem.178.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam B K, Penrose J F, Freeman G J, Austen K F. Proc Natl Acad Sci USA. 1994;91:7663–7667. doi: 10.1073/pnas.91.16.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinman R M. Annu Rev Immunol. 1991;9:271–296. doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- 17.Dubois B, Vanbervliet B, Fayette J, Massacrier C, Van Kooten C, Brière, Banchereau J, Caux C. J Exp Med. 1997;185:941–951. doi: 10.1084/jem.185.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]