Abstract
Here, we demonstrate that primed, single-stranded DNA (ssDNA) is sufficient for activation of the ATR-dependent checkpoint pathway in Xenopus egg extracts. Using this structure, we define the contribution of the 5′- and 3′-primer ends to Chk1 activation when replication is blocked and ongoing. In addition, we show that although ssDNA is not sufficient for checkpoint activation, the amount of ssDNA adjacent to the primer influences the level of Chk1 phosphorylation. These observations define the minimal DNA requirements for checkpoint activation and suggest that primed ssDNA represents a common checkpoint activating-structure formed following many types of damage.
The cellular response to DNA damage and replication stress is essential for the maintenance of genomic stability (Zhou and Elledge 2000; Melo and Toczyski 2002). The ATR (ATM and Rad3-related) kinase plays a central role in this pathway and responds to many types of genotoxic stress. Many of the effects of ATR are mediated by the downstream effector kinase Chk1, which is phosphorylated and activated by the ATR–ATRIP (ATR-interacting protein) complex (Zhou and Elledge 2000; Melo and Toczyski 2002). Activation of Chk1 also requires the function of several other proteins. Among these are the Rad9–Hus1–Rad1 (9–1–1) complex, a PCNA-related complex that is recruited to damaged chromatin, enhances ATR activation, and binds primer–template junctions in vitro (Ellison and Stillman 2003; Parrilla-Castellar et al. 2004; Majka et al. 2006b). Also important are TopBP1 and Claspin. TopBP1 activates the kinase activity of the ATR–ATRIP complex, and Claspin may both activate Chk1 and recruit it to ATR (Kumagai and Dunphy 2000; Lee et al. 2005; Kumagai et al. 2006).
Despite the growing knowledge of proteins involved in checkpoint processes, the precise structure responsible for checkpoint activation following DNA damage or replication inhibition is not known. Several studies suggest the lesions induced by ultraviolet radiation, methyl methanesulfonate, and cisplatin activate the checkpoint most efficiently in S phase (Lupardus et al. 2002; Stokes et al. 2002; Tercero et al. 2003; Ward et al. 2004; Marini et al. 2006). In these cases and upon treatment with the polymerase inhibitor aphidicolin it is thought that replication forks stall, generating a common checkpoint-activating intermediate through uncoupling of helicase and polymerase activities (Walter and Newport 2000; Pacek and Walter 2004; Byun et al. 2005; Cortez 2005). This process leads to accumulation of replication protein A (RPA)-coated single-stranded DNA (ssDNA), a structure sufficient to recruit the ATR–ATRIP complex and essential for checkpoint activation (Zou and Elledge 2003). However, several studies indicate that ssDNA is not sufficient and that additional replication is required for checkpoint activation and for loading of the 9–1–1 complex (Michael et al. 2000; Stokes et al. 2002; You et al. 2002; Byun et al. 2005). These observations suggest the checkpoint-activating structure is comprised of at least two parts, RPA-coated ssDNA and a primer–template junction.
Whether primed ssDNA alone is sufficient for checkpoint activation is unclear. In one study, primed, damaged ssDNA failed to induce any detectable Chk1 phosphorylation, leading to the suggestion that a replication fork is required (Stokes et al. 2002). However, simple DNA templates consisting of the annealed homopolymers (dA)70 and (dT)70 cause ATR-dependent phosphorylation of Chk1 in Xenopus egg extracts (Kumagai and Dunphy 2000). These homopolymers produce heterogeneous, oligomeric mixtures that form a variety of DNA structures, among which are blunt ends, cruciforms, and others that might mimic replication fork structures, making identification of the actual checkpoint-activating structure difficult. Here we report the identification of a well-defined checkpoint-activating structure lacking double-stranded ends, which we have used to determine the structural requirements for ATR-dependent checkpoint activation.
Results and Discussion
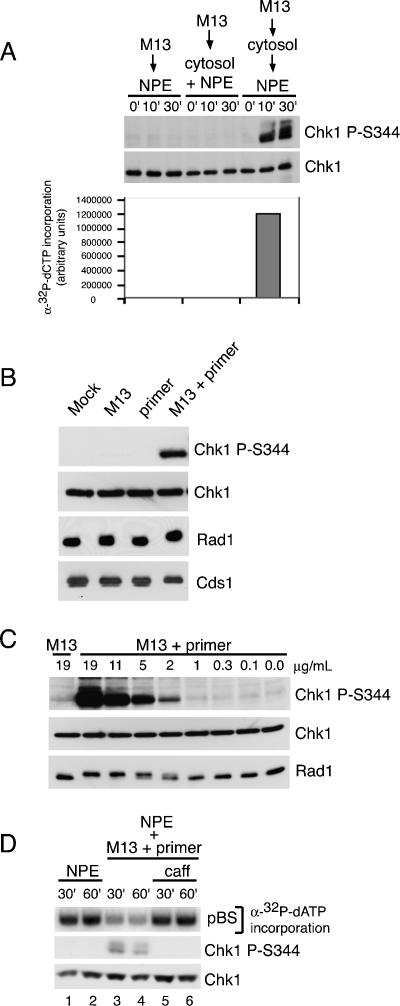
To investigate the ability of simple, well-defined DNA structures to activate the ATR-dependent checkpoint, we used Xenopus egg extracts, a cell-free system competent for DNA replication and checkpoint activation. First, we tested the ability of M13 ssDNA to induce the phosphorylation of Chk1 on S344, a site indicative of ATR activation (Liu et al. 2000). Two different preparations of extracts, cytosol and nucleoplasmic extract (NPE) (Walter et al. 1998), were used. Previous studies have led to conflicting results regarding the ability of M13 ssDNA to activate the checkpoint in NPE (Stokes et al. 2002; Shechter et al. 2004a). Following rigorous purification, we found that M13 ssDNA does not replicate when added to NPE or a mixture of NPE and cytosol, consistent with previous observations (Walter and Newport 2000; Stokes et al. 2002), and under these conditions, M13 ssDNA was unable to induce Chk1 phosphorylation (Fig. 1A). However, we have found that many sources of M13 ssDNA do replicate in NPE and induce the phosphorylation of Chk1 if not sufficiently purified prior to use (data not shown). These results indicate that ssDNA alone cannot induce Chk1 phosphorylation, and that in cases where checkpoint activation was induced by addition of M13 to NPE, some contaminant or a secondary structure formed by nicked M13 ssDNA was likely responsible.
Figure 1.
Primed M13 ssDNA induces Chk1 phosphorylation and checkpoint activation. (A) M13 ssDNA (30 μg/mL) was added to NPE, a 1:1 mixture of cytosol and NPE, or cytosol (30 min) and then NPE. Parallel samples were removed at the indicated times post-DNA addition for NPE and cytosol + NPE. For the sequential addition, times were post-NPE addition. Samples were either immunoblotted for phospho-Chk1 (S344) and Chk1 (top panels), or replication was monitored by measuring α32P-dCTP incorporation following gel electrophoresis (bottom). (B) Buffer, M13 ssDNA, the ssDNA primer (80-mer), or M13 preannealed to the primer (M13 + primer) was added to NPE. Samples were taken after 20 min and analyzed by immunoblotting after SDS-PAGE with antibodies for phospho-Chk1 (S344), Chk1, Rad1, and Cds1. (C) ssM13 DNA or M13 + primer was added to NPE at the indicated concentrations. Samples were analyzed as in B. (D) Double-stranded pBS (30 μg/mL) was incubated in cytosol for 30 min and added to an equal volume of NPE (lanes 1,2), NPE containing M13 + primer (20 min; 12.5 μg/mL; lanes 3,4), or NPE containing M13 + primer + caffeine (20 min; 12.5 μg/mL; caff, 4 mM; lanes 5,6). Parallel samples were removed 30 and 60 min post-addition of plasmid to NPE and analyzed for replication as in A (top panel) or immunoblotted for phospho-Chk1 (S344) and Chk1 (bottom panels). Final concentrations are shown.
In contrast, we observed robust Chk1 phosphorylation when purified M13 ssDNA was first incubated in cytosol, where replication can occur (Mechali and Harland 1982), and subsequently added to NPE (Fig. 1A). We were unable to detect significant Chk1 phosphorylation in cytosol alone, possibly due to a higher Chk1 phosphatase activity in cytosol (Kumagai and Dunphy 2000). Addition of geminin to the cytosol prior to addition of M13 ssDNA had no effect on Chk1 phosphorylation (Supplementary Fig. 1A). As geminin prevents initiation of replication on double-stranded DNA (McGarry and Kirschner 1998), this indicates that Chk1 phosphorylation is not due to rereplication of a double-stranded M13 template. We also found that DNA polymerase α (Pol α) was required for checkpoint activation (Supplementary Fig. 1B), consistent with previous studies (Michael et al. 2000; Byun et al. 2005). These results indicate that replication of M13 ssDNA is necessary for Chk1 phosphorylation.
Next, we asked if we could bypass the need for priming of ssDNA in cytosol by annealing an 80-nucleotide (nt) DNA primer to M13 ssDNA and adding this structure directly to NPE (Fig. 1B). This extract lacks the ability to synthesize new primers (Walter and Newport 2000; Stokes et al. 2002), although extension of existing primers can occur (Supplementary Fig. 2B; Stokes et al. 2002). Addition of primed M13 ssDNA directly to NPE induced robust Chk1 phosphorylation, whereas addition of M13 ssDNA or the primer alone did not (Fig. 1B). Although the Chk1 phosphorylation induced by addition of primed M13 ssDNA can be readily detected using a phospho-specific antibody, a significant change in Chk1 mobility is not observed (Supplementary Fig. 3). This likely accounts for the inability to observe Chk1 phosphorylation in previous studies using primed ssDNA, where the shift in total Chk1 was examined as an indicator of phosphorylation (Stokes et al. 2002). Importantly, similar results were observed using primers with a different sequence (data not shown).
ATR activation also leads to the phosphorylation of Rad1, decreasing its mobility (Lupardus and Cimprich 2006), and primed M13 ssDNA induced the phosphorylation of Rad1 (Fig. 1B). Approximately 1–2 ng of this structure per microliter of NPE was sufficient to induce Chk1 and Rad1 phosphorylation (Fig. 1C), an amount that corresponds to 10–30 primer–template junctions per somatic cell nucleus (see Supplemental Calculation). In addition, primed ssDNA induced a partial shift in the phosphorylation of Cds1/Chk2, a downstream substrate of ATM and ATR (Ahn et al. 2004), as monitored by a change in Cds1 mobility (Fig. 1B). Taken together, these data indicate that primed ssDNA can induce the phosphorylation of several targets of the ATR kinase.
We then asked whether Chk1 phosphorylation induced by replicating M13 ssDNA had functional consequences. ATR activation prevents firing of late origins (Machida et al. 2005). Thus, we assayed the effect of primed M13 ssDNA on replication of a double-stranded DNA plasmid. Because origin firing and replication of plasmid DNA requires sequential incubation in cytosol and NPE (Walter et al. 1998), we pretreated the NPE with primed M13 ssDNA and used this NPE to replicate a plasmid previously incubated in cytosol. We found primed M13 ssDNA inhibited replication of the double-stranded plasmid in trans (Fig. 1D, lanes 3,4). Consistent with ATR’s role in regulating late origin firing, this inhibition was reversed by the addition of caffeine, which inhibits the ATR kinase (Fig. 1D, lanes 5,6). Similar results were obtained with replicating M13 ssDNA (Supplementary Fig. 1C). These observations suggest that the checkpoint signal induced by primed M13 ssDNA has functional consequences and is sufficient to inhibit origin firing.
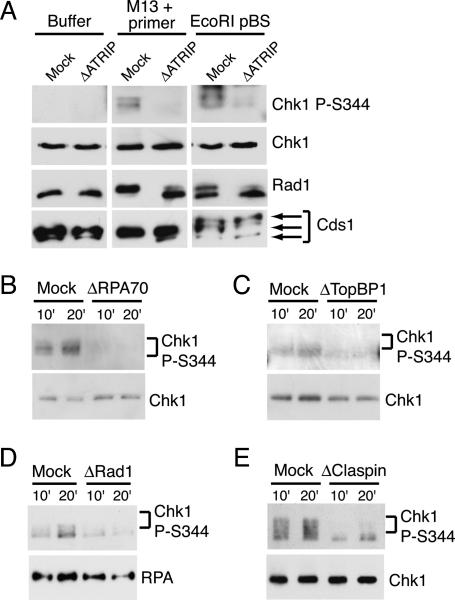
To determine if checkpoint activation induced by primed M13 ssDNA requires the ATR–ATRIP complex, we tested the effects of ATRIP depletion on Chk1, Rad1, and Cds1 phosphorylation. We found that phosphorylation of all three proteins induced by primed ssDNA was lost upon ATRIP depletion (Fig. 2A). An inhibitor of ATM, KU-55933 (Hickson et al. 2004), failed to block the Chk1, Rad1, or Cds1 phosphorylation induced by primed ssDNA (Supplementary Fig. 4). Chk1 phosphorylation induced by primed ssDNA was also lost in extracts depleted of RPA, TopBP1, Rad1, or Claspin (Fig. 2B–E), four proteins required for checkpoint activation induced by stalled replication forks (Melo and Toczyski 2002; Zou and Elledge 2003; Garcia et al. 2005). Similarly, Chk1 phosphorylation induced by replicating M13 ssDNA was lost upon depletion of these proteins (Supplementary Fig. 5). These observations indicate that phosphorylation of Chk1, Rad1, and Cds1 induced by primed ssDNA is mediated by ATR and not ATM. They further suggest that primed M13 ssDNA mimics the checkpoint-activating structure produced at stalled replication forks.
Figure 2.
Effect of checkpoint protein depletion on phosphorylation events induced by primed M13 ssDNA. (A) Buffer, M13 + primer, or EcoRI-digested pBS (0.2 μg/mL) was added to mock-depleted or ATRIP-depleted NPE, and samples were analyzed as in Figure 1B. (B–E) NPE was mock-depleted or depleted with antibodies to RPA70 (B), TopBP1 (C), Rad1 (D), or Claspin (E). Mock-depleted NPE or depleted NPE was then incubated with M13 + primer, and samples were analyzed as in Figure 1B. Chk1 (B,C,E) and RPA (D) levels were monitored as loading controls. The lower band in E represents cross-reacting residual IgG in the extract from depletion.
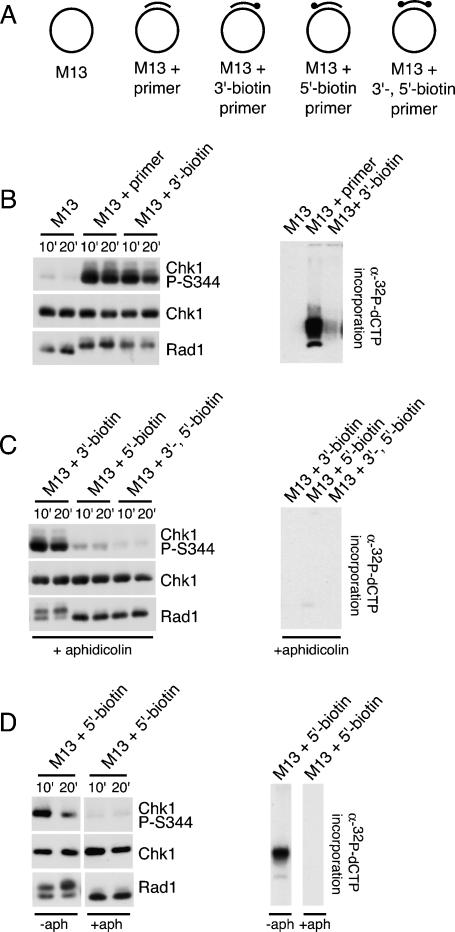
Primers annealed to M13 ssDNA are extended in NPE, raising the possibility that extension of the primer is required for checkpoint activation. To determine if primed M13 ssDNA is sufficient for phosphorylation of Chk1, we annealed M13 ssDNA to an 80-nt primer labeled at the 3′-end with biotin (Fig. 3A). We then added this structure to extracts after preincubation with streptavidin. Streptavidin had no effect on replication of the unmodified primer or its ability to induce Chk1 phosphorylation, but it prevented replication of the 3′-biotinylated structure (Fig. 3B, Supplementary Fig. 2). Importantly, this 3′-biotinylated structure induced the phosphorylation of Chk1 and Rad1 in the presence of streptavidin (Fig. 3B), strongly suggesting that primer extension is not required and that primed M13 ssDNA is sufficient to activate the checkpoint.
Figure 3.
Effect of modifying primer ends on Chk1 and Rad1 phosphorylation induced by primed M13 ssDNA. (A) Names and figures of structures used in B–D. Dot represents the biotinylated end of the primer. (B) M13, M13 + unmodified 80-mer, or M13 + 80-mer blocked at the 3′-end with biotin was incubated with streptavidin and added to NPE. Samples were taken and analyzed for phospho-Chk1 (S344), Chk1, or Rad1 by immunoblotting, or for replication as described in Figure 1A. (C) M13 ssDNA annealed to an 80-mer blocked at the 3′-end, 5′-end, or 3′- and 5′-ends with biotin were incubated with streptavidin, added to NPE in the presence of aphidicolin, and analyzed as described in B. (D) M13 ssDNA annealed to an 80-mer blocked at the 5′-end with biotin was incubated with streptavidin, added to NPE in the absence or presence of aphidicolin, and analyzed as described in B.
The ability of 3′-biotinylated primed M13 ssDNA to induce Chk1 phosphorylation suggests that a single free 5′-end is sufficient for checkpoint activation when replication is blocked, as at a stalled replication fork. To further test the role of the 5′ primer end in checkpoint activation, we prepared primers biotinylated at the 5′-end and both the 3′- and 5′-ends (Fig. 3A). The ability of these modified primed M13 structures to induce Chk1 and Rad1 phosphorylation was then monitored in the presence of a dose of aphidicolin sufficient to prevent replication of the 5′-biotinylated primer. The addition of aphidicolin models the scenario at a stalled replication fork, and also keeps the 5′-modified primer the same length as the 3′-modified and dual-modified primers. We found that modification of the 5′-end of the primer strongly reduced its ability to induce Chk1 and Rad1 phosphorylation under these conditions (Fig. 3C). Because the 5′-modified primer is more stable than the 3′-modified primer (Supplementary Fig. 6), the contribution of the free 5′-end to Chk1 phosphorylation is likely underestimated in this experiment. As expected, modification of both primer ends prevented essentially all Chk1 and Rad1 phosphorylation (Fig. 3C).
Next we asked if the 3′-end of a primer efficiently activates the checkpoint in its replicating state. In contrast to the results obtained in the presence of aphidicolin, we found that Chk1 and Rad1 phosphorylation were induced by M13 ssDNA annealed to the 5′-biotinylated primer in the absence of a replication block. It seems unlikely that the effect of replication on Chk1 phosphorylation in this experiment is due to the increased size of the primer, since primers of significantly smaller size are able to load the 9–1–1 complex and activate the ATR pathway in systems using purified proteins (Ellison and Stillman 2003; Majka et al. 2006a).
Taken together, these data suggest the free 5′-end, or some processed version of this end, is necessary for efficient Chk1 phosphorylation when replication is blocked as at a stalled replication fork. However, when replication is ongoing, the 3′-end of a primer is capable of inducing Chk1 phosphorylation. The contribution of the 5′-end in the absence of replication is consistent with studies using purified proteins, which demonstrate a preference for Rad17-dependent loading of the 9–1–1 clamp complex onto a 5′-end or gap (Ellison and Stillman 2003; Zou et al. 2003; Majka et al. 2006a). These data therefore extend the studies of checkpoint activation in purified systems by showing that while replication is not required, ongoing replication may alter the molecular requirements for checkpoint activation.
Both free 5′- and 3′-ends are found in many of the DNA structures formed following different types of DNA damage, including recombination intermediates, nucleotide excision repair (NER) intermediates and possibly stalled forks. Thus, in some cases two types of ends may contribute to activation. A free 5′-end is also found at resected double-strand breaks. It is interesting that in the case of a stalled replication fork, which in our system is modeled by the addition of aphidicolin, the 5′-end plays a more significant role in checkpoint activation. When a lesion is on the lagging strand, a free 5′-end would be found immediately downstream from the stalled polymerase and adjacent to the accumulated ssDNA. However, when the polymerase is stalled on the leading strand, this observation would suggest that signaling requires replication restart downstream from the stalled polymerase to generate a 5′-end or that primers on the lagging strand can contribute to signaling when the fork arrests. Indeed, there is evidence for replication restart on the leading strand in eukaryotes (Lopes et al. 2006).
The 5′-end of the structures formed at stalled replication forks would at least initially be composed of RNA, and previous studies indicate the primase activity of Pol α is required for checkpoint activation (Michael et al. 2000). These observations raise the question of whether an RNA primer is sufficient for checkpoint activation. We have tested the ability of an RNA primer annealed to M13 ssDNA to induce checkpoint activation and have been unable to detect any Chk1 or Rad1 phosphorylation. However, even large RNA primers are rapidly degraded in the extract (Supplementary Fig. 7). Thus, we cannot rule out the possibility that primer degradation prevents induction of Chk1 or Rad1 phosphorylation. In any case, our work clearly indicates the RNA portion of the primer is not required for activation of ATR and that a DNA primer is sufficient, consistent with the fact that many structures formed following different types of DNA damage would not contain RNA.
Activation of the ATR pathway is often accompanied by the accumulation of ssDNA (Shechter et al. 2004b; Cortez 2005). It is unclear if the additional ssDNA that accumulates during processes such as uncoupling or resection has any impact on the checkpoint response, and if small gaps, like those created during NER, are sufficient to induce ATR activation. We previously reported that the extent of uncoupling can influence the amount of Chk1 phosphorylation (Byun et al. 2005). In that study, however, we could not determine if the increased Chk1 phosphorylation observed with more extensive uncoupling was due to the additional ssDNA formed, or if additional primers were formed on this ssDNA and accounted for or contributed to the increased signaling.
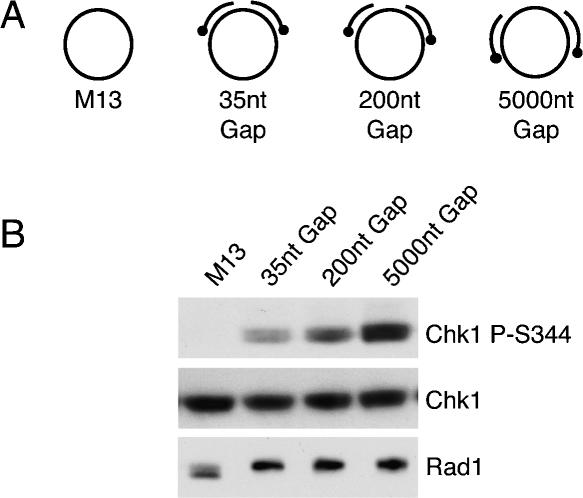
To determine the effect of ssDNA on Chk1 phosphorylation, we prepared a series of structures containing two primers, one biotinylated at the 5′-end and another at the 3′-end. These primers were annealed to M13 ssDNA to create ssDNA gaps of 35, 200, or 5000 nt (Fig. 4A). In order to keep the primers the same length and to maintain the ssDNA gap size, the structures were added to NPE in the presence of aphidicolin. While these structures had the same total amount of ssDNA, we found there were significant differences in the amount of Chk1 phosphorylation observed. The length of ssDNA between the free ends of the primers determined the amount of Chk1 phosphorylation observed, with larger gaps inducing higher levels of Chk1 phosphorylation (Fig. 4B). Rad1 phosphorylation, however, was unaffected by the size of the ssDNA gap. We also observed a similar effect of the gap size on Chk1 phosphorylation in the absence of aphidicolin. Thus, while ssDNA alone is unable to induce checkpoint activation, this result suggests the amount of ssDNA adjacent to the primer ends can modulate the strength of the signal generated in either the presence or absence of a replication block.
Figure 4.
Effect of ssDNA gap size on Chk1 phosphorylation. (A) Names and figures of structures used in B. Dot represents the biotinylated end of the primer. (B) The structures shown were incubated with streptavidin and then added to NPE in the presence of aphidicolin. Samples were analyzed as in Figure 3B.
These findings provide an explanation for why many types of DNA damage activate ATR more efficiently in S phase. For example, in the case of UV damage, the lesion can be repaired outside of S phase by NER, a process that generates a gap of ∼35 nt upon excision of the lesion (Huang et al. 1992). Our results directly demonstrate that a ssDNA gap of this size is sufficient to induce checkpoint activation, albeit weakly (Fig. 4B), and they suggest that outside of S phase, these intermediates may induce low levels of Chk1 phosphorylation, particularly when the dose of UV is high and a large number of small gaps are formed. Consistent with this idea, processing of UV lesions by NER is necessary for Chk1 phosphorylation in primary human fibroblasts during G0/G1 (Marini et al. 2006). However, because a larger region of ssDNA may be formed by helicase/polymerase uncoupling when a replication fork encounters the same lesion (Byun et al. 2005), more robust checkpoint activation would occur during S phase.
The ability to precisely define a minimal checkpoint-activating structure has allowed us to outline the structural components of DNA that are necessary to induce ATR-dependent checkpoint activation in Xenopus egg extracts. Our studies indicate that primed ssDNA mimics the checkpoint-activating structure produced at stalled replication forks and is sufficient to induce checkpoint signaling through ATR and Chk1. They also demonstrate the amount of ssDNA adjacent to the primer–template junction determines the ability of each primer to induce Chk1 phosphorylation, suggesting the effect of any given primer is influenced by its environment. Our primed ssDNA structure contains a large amount of ssDNA, and we have shown the limit of detectable Chk1 phosphorylation induced by this structure to be in the range of 10–30 primer–template junctions per somatic cell nucleus. When considered in the context of stalled replication forks where ssDNA accumulates, this number would represent only a small fraction of the total number of replication origins in eukaryotes (103–105) (Blow and Dutta 2005), suggesting that activation of the ATR checkpoint is very sensitive. During normal replication, however, primers are not associated with large regions of ssDNA. Thus, while multiple primer–template junctions may be present at any given time during replication, the sum of these primers may not reach the threshold necessary for significant checkpoint activation.
Importantly, primed ssDNA is formed following many types of DNA damage and may represent the common and minimal structure produced by damage that is responsible for ATR-mediated checkpoint activation. It seems likely that chromatin structure, the lesion itself, and other more complex protein–DNA structures, such as those found at a replication fork, may influence checkpoint activation in cells, and further studies to better define the contribution of these and other elements are necessary. Regardless, the ability to precisely define, quantitate, and manipulate a minimal checkpoint activating structure will facilitate future studies regarding the mechanism of checkpoint activation.
Materials and methods
Xenopus egg extracts
Membrane-free egg cytosol and NPE were prepared as described previously (Walter et al. 1998).
Chemicals, antibodies, and recombinant proteins
Antibodies used were from both our laboratory and several others. Recombinant geminin and Pol α-primase were produced as previously described (Stadlbauer et al. 1994; Lupardus et al. 2002). See the Supplemental Material for full details.
Immunodepletion experiments
Immunodepletions (three rounds, 1 h each) were carried out at 4°C with Protein A-Sepharose beads (Amersham Pharmacia). See the Supplemental Material for full details.
DNA structure preparation
M13mp18 ssDNA was purified by agarose gel electrophoresis. DNA oligonucleotides were synthesized and purified by Integrated DNA Technologies. The RNA oligonucleotide was generated by in vitro transcription. Quantitation and verification of structures were performed by Sybrgold staining after denaturing urea PAGE. See the Supplemental Material for full details.
DNA and RNA structure assays
M13 and M13 + primer structures were incubated in NPE in the absence or presence of aphidicolin. Samples were taken for Western blot analysis and to monitor replication. See the Supplemental Material for full details.
Acknowledgments
This work was supported by a 2003 Ford Foundation Predoctoral Minority Fellowship awarded to C.M., by a DOD Breast Cancer research grant (04-1-0311) to T.S.B. and C.V., and by grants to K.A.C. from the National Institutes of Health (GM62193) and the American Cancer Society (RSG-05-028-01-CCG). K.A.C. is a Leukemia and Lymphoma Society Scholar.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1522607
References
- Ahn J., Urist M., Prives C., Urist M., Prives C., Prives C. The Chk2 protein kinase. DNA Repair (Amst.) 2004;3:1039–1047. doi: 10.1016/j.dnarep.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Blow J.J., Dutta A., Dutta A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A., Pacek M., Yee M.C., Walter J.C., Cimprich K.A., Yee M.C., Walter J.C., Cimprich K.A., Walter J.C., Cimprich K.A., Cimprich K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes & Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D. Unwind and slow down: Checkpoint activation by helicase and polymerase uncoupling. Genes & Dev. 2005;19:1007–1012. doi: 10.1101/gad.1316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison V., Stillman B., Stillman B. Biochemical characterization of DNA damage checkpoint complexes: Clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:231–243. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V., Furuya K., Carr A.M., Furuya K., Carr A.M., Carr A.M. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst.) 2005;4:1127–1139. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C., Jackson S.P., Curtin N.J., Smith G.C., Curtin N.J., Smith G.C., Smith G.C. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Huang J.-C., Svoboda D.L., Reardon J.T., Sancar A., Svoboda D.L., Reardon J.T., Sancar A., Reardon J.T., Sancar A., Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl. Acad. Sci. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W.G., Dunphy W.G. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell. 2000;6:836–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Lee J., Yoo H.Y., Dunphy W.G., Lee J., Yoo H.Y., Dunphy W.G., Yoo H.Y., Dunphy W.G., Dunphy W.G. TopBP1 activates the ATR–ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lee J., Gold D.A., Shevchenko A., Shevchenko A., Dunphy W.G., Gold D.A., Shevchenko A., Shevchenko A., Dunphy W.G., Shevchenko A., Shevchenko A., Dunphy W.G., Shevchenko A., Dunphy W.G., Dunphy W.G. Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol. Biol. Cell. 2005;16:5269–5282. doi: 10.1091/mbc.E05-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Guntuku S., Cui X.S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Guntuku S., Cui X.S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Cui X.S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Carattini-Rivera S., DeMayo F., Bradley A., DeMayo F., Bradley A., Bradley A., et al. Chk1 is an essential kinase that is regulated by ATR and required for the G(2)/M DNA damage checkpoint. Genes & Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Fiorani M., Sogo J.M., Fiorani M., Sogo J.M., Sogo J.M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Lupardus P.J., Cimprich K.A., Cimprich K.A. Phosphorylation of Xenopus Rad1 and Hus1 defines a readout for ATR activation that is independent of Claspin and the Rad9 carboxy terminus. Mol. Biol. Cell. 2006;17:1559–1569. doi: 10.1091/mbc.E05-09-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupardus P.J., Byun T., Yee M.C., Hekmat-Nejad M., Cimprich K.A., Byun T., Yee M.C., Hekmat-Nejad M., Cimprich K.A., Yee M.C., Hekmat-Nejad M., Cimprich K.A., Hekmat-Nejad M., Cimprich K.A., Cimprich K.A. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes & Dev. 2002;16:2327–2332. doi: 10.1101/gad.1013502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y.J., Hamlin J.L., Dutta A., Hamlin J.L., Dutta A., Dutta A. Right place, right time, and only once: Replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Majka J., Binz S.K., Wold M.S., Burgers P.M., Binz S.K., Wold M.S., Burgers P.M., Wold M.S., Burgers P.M., Burgers P.M. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J. Biol. Chem. 2006a;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- Majka J., Niedziela-Majka A., Burgers P.M., Niedziela-Majka A., Burgers P.M., Burgers P.M. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell. 2006b;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F., Nardo T., Giannattasio M., Minuzzo M., Stefanini M., Plevani P., Falconi M.M., Nardo T., Giannattasio M., Minuzzo M., Stefanini M., Plevani P., Falconi M.M., Giannattasio M., Minuzzo M., Stefanini M., Plevani P., Falconi M.M., Minuzzo M., Stefanini M., Plevani P., Falconi M.M., Stefanini M., Plevani P., Falconi M.M., Plevani P., Falconi M.M., Falconi M.M. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc. Natl. Acad. Sci. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry T.J., Kirschner M.W., Kirschner M.W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Mechali M., Harland R.M., Harland R.M. DNA synthesis in a cell-free system from Xenopus eggs: Priming and elongation on single-stranded DNA in vitro. Cell. 1982;30:93–101. doi: 10.1016/0092-8674(82)90015-0. [DOI] [PubMed] [Google Scholar]
- Melo J., Toczyski D., Toczyski D. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- Michael W.M., Ott R., Fanning E., Newport J., Ott R., Fanning E., Newport J., Fanning E., Newport J., Newport J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- Pacek M., Walter J.C., Walter J.C. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilla-Castellar E.R., Arlander S.J., Karnitz L., Arlander S.J., Karnitz L., Karnitz L. Dial 9–1–1 for DNA damage: The Rad9–Hus1–Rad1 (9–1–1) clamp complex. DNA Repair (Amst.) 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Shechter D., Costanzo V., Gautier J., Costanzo V., Gautier J., Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 2004a;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- Shechter D., Costanzo V., Gautier J., Costanzo V., Gautier J., Gautier J. Regulation of DNA replication by ATR: Signaling in response to DNA intermediates. DNA Repair (Amst.) 2004b;3:901–908. doi: 10.1016/j.dnarep.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Stadlbauer F., Brueckner A., Rehfuess C., Eckerskorn C., Lottspeich F., Forster V., Tseng B.Y., Nasheuer H.P., Brueckner A., Rehfuess C., Eckerskorn C., Lottspeich F., Forster V., Tseng B.Y., Nasheuer H.P., Rehfuess C., Eckerskorn C., Lottspeich F., Forster V., Tseng B.Y., Nasheuer H.P., Eckerskorn C., Lottspeich F., Forster V., Tseng B.Y., Nasheuer H.P., Lottspeich F., Forster V., Tseng B.Y., Nasheuer H.P., Forster V., Tseng B.Y., Nasheuer H.P., Tseng B.Y., Nasheuer H.P., Nasheuer H.P. DNA replication in vitro by recombinant DNA-polymerase-α-primase. Eur. J. Biochem. 1994;222:781–793. doi: 10.1111/j.1432-1033.1994.tb18925.x. [DOI] [PubMed] [Google Scholar]
- Stokes M.P., Van Hatten R., Lindsay H.D., Michael W.M., Van Hatten R., Lindsay H.D., Michael W.M., Lindsay H.D., Michael W.M., Michael W.M. DNA replication is required for the checkpoint response to damaged DNA in Xenopus egg extracts. J. Cell Biol. 2002;158:863–872. doi: 10.1083/jcb.200204127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J.A., Longhese M.P., Diffley J.F., Longhese M.P., Diffley J.F., Diffley J.F. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- Walter J., Newport J., Newport J. Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45; RPA; and DNA polymerase α. Mol. Cell. 2000;3:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Walter J., Sun L., Newport J., Sun L., Newport J., Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Ward I.M., Minn K., Chen J., Minn K., Chen J., Chen J. UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. J. Biol. Chem. 2004;279:9677–9680. doi: 10.1074/jbc.C300554200. [DOI] [PubMed] [Google Scholar]
- You Z., Kong L., Newport J., Kong L., Newport J., Newport J. The role of single-stranded DNA and polymerase α in establishing the ATR, Hus1 DNA replication checkpoint. J. Biol. Chem. 2002;277:27088–27093. doi: 10.1074/jbc.M204120200. [DOI] [PubMed] [Google Scholar]
- Zhou B.B., Elledge S.J., Elledge S.J. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S.J., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L., Liu D., Elledge S.J., Liu D., Elledge S.J., Elledge S.J. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]