Abstract
In humans, there are at least seven Sir2-like proteins (SirT1–7) with diverse functions, including the regulation of chromatin structure, and metabolism. SirT3 levels have been shown to correlate with extended life span, to localize to the mitochondria, and to be highly expressed in brown adipose tissue. In humans, SirT3 exists in two forms, a full-length protein of ∼44 kDa and a processed polypeptide lacking 142 amino acids at its N terminus. We found that SirT3 not only localizes to the mitochondria, but also to the nucleus under normal cell growth conditions. Both the full-length and processed forms of SirT3 target H4-K16 for deacetylation in vitro and can deacetylate H4-K16 in vivo when recruited to a gene. Using a highly specific antibody against the N terminus of SirT3, we found that SirT3 is transported from the nucleus to the mitochondria upon cellular stress. This includes DNA damage induced by Etoposide and UV-irradiation, as well as overexpression of SirT3 itself.
Keywords: Histone deacetylation, chromatin, mitochondria, SirT3
SirT3 is a member of the Sir2 family of NAD+-dependent protein deacetylases (Frye 1999). This family of proteins is implicated in chromatin structure, transcriptional silencing, and aging in organisms ranging from yeast to humans. It has been proposed that human sirtuins generally have nonhistone substrates. Yet the SirT proteins (especially SirT1, SirT2, and SirT3) are highly related to the yeast Sir2 protein, a dedicated histone deacetylase with specificity for Lys 16 of histone H4 (H4-K16), a residue important in attaining a repressed chromatin state in yeast upon its deacetylation (Suka et al. 2002; Shogren-Knaak et al. 2006). Recent speculations suggest a function for SirT1 (and other SirTs) in caloric restriction and life span (Haigis and Guarente 2006). SirT1 initially was found to deacetylate the tumor suppressor p53 (Luo et al. 2001; Vaziri et al. 2001; Langley et al. 2002), as well as other transcription factors (Brunet et al. 2004; Giannakou and Partridge 2004; Haigis and Guarente 2006). However, SirT1 was later found to function in the formation of facultative heterochromatin through its deacetylation of core histones, specifically acetyl-Lys 16 of histone H4 (H4-K16ac) (Vaquero et al. 2004). SirT1 also interacts with histone H1 and deacetylates acetyl-Lys 26 (Vaquero et al. 2004). The ability of sirtuins to deacetylate H4-K16ac is not restricted to SirT1, as SirT2 has been shown to deacetylate H4-K16ac during mitosis (Vaquero et al. 2006).
SirT3 is the only Sirtuin with an apparent direct link to extended life span in humans. Mutations in an enhancer region of the SirT3 gene that potentially up-regulate its expression were found at a high frequency in long-lived individuals, suggesting that high expression of SirT3 can be an important marker in life extension (Bellizzi et al. 2005). Overexpression of murine SirT3 also has been shown to increase the expression of genes involved in mitochondrial biogenesis and metabolism in brown fat cells, linking SirT3 concentration to the regulation of nuclear gene expression (Shi et al. 2005). However, SirT3 has been reported to localize exclusively to the mitochondria, and it has been speculated that this localization may have important implications in SirT3 function in life span (Onyango et al. 2002; Schwer et al. 2002). It has also been reported that once SirT3 enters the mitochondria the protein becomes processed at the N terminus, resulting in the activation of its enzymatic activity (Schwer et al. 2002). Finally, studies demonstrated that SirT3 is able to deacetylate and activate the enzyme acetyl-CoA synthetase 2 within the mitochondria, showing an important role of SirT3 at this organelle (Hallows et al. 2006; Schwer et al. 2006). The previous studies used overexpressed SirT3 and did not address whether or not the endogenous SirT3 also resides exclusively in the mitochondria.
Using highly specific antibodies against the N terminus of SirT3 and hence specific to the full-length human protein, we show that under normal cellular conditions, full-length SirT3 resides in the nucleus. Full-length SirT3 does, indeed, localize to the mitochondria; however, we show that such translocation occurs upon stress caused by either environmental conditions or by the massive overexpression of SirT3 itself. We further observe that SirT3 completely exits the nucleus upon these conditions in a manner sensitive to Leptomycin B (LMB). In characterizing the biochemical activity of SirT3, we show that both the full-length and processed forms of SirT3 have a strong NAD+-dependent histone deacetylation activity in vitro with specificity for acetyl-Lys 16 of histone H4 and to a lesser extent acetyl-Lys 9 of histone H3. We further show that SirT3 is an in vivo histone deacetylase (HDAC), which can repress transcription of a gene when recruited to its promoter. Our work complements studies that revealed a mitochondrial function for SirT3 by showing that SirT3 also localizes to the nucleus and deacetylates acetyl-Lys 9 and acetyl-Lys 16 of histones H3 and H4, respectively.
Results
Full-length SirT3 is a nuclear protein
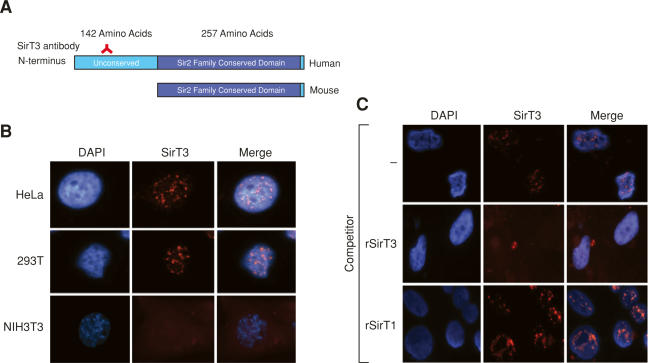
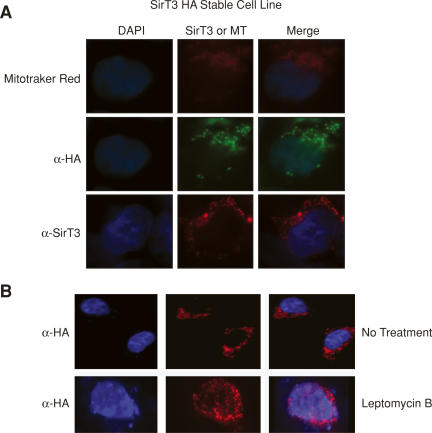
In order to study SirT3 function in vivo, we initially sought to generate a highly specific antibody. We used the N terminus of human SirT3 (hSirT3) as it does not share homology with other Sirtuins and it is specific to the human protein, as the mouse protein lacks this domain (Fig. 1A). After affinity purification of the antibodies, we examined SirT3 localization in several cell lines (Fig. 1A). Using immunofluorescence studies, we observed that the SirT3 antibodies recognize a protein that resides exclusively in the nucleus and is excluded from the dark DAPI regions (Fig. 1B). Since the murine SirT3 is devoid of the N-terminal region, no immunofluorescence signal was observed in the mouse cell line NIH3T3, demonstrating specificity of the antibodies. Moreover, in competition experiments only recombinant SirT3 competed away the signal, further verifying the specificity of the antibody (Fig. 1C). Since the antibody is generated against the N terminus, it is not possible to probe for the localization of endogenous SirT3 that has been processed. Using a highly specific antibody generated against the C terminus of SirT3 (Schwer et al. 2002), we demonstrated using Western blot analysis the presence of full-length and processed forms of SirT3 in the nucleus (Fig. 6B [below], lane 1). However, and in agreement with previous studies, we also observed that a large population of the N-terminal-processed SirT3 is localized to the mitochondria (Fig. 6B, below). Therefore, from these observations, we conclude that SirT3 is localized in the nucleus and mitochondrion.
Figure 1.
Full-length SirT3 is nuclear. (A) Schematic representation of full-length human and mouse SirT3. (B) Immunofluorescence analyses of the cell lines indicated using antibody specific to the N terminus of human SirT3. DAP1 was used to visualize the nuclei. (C) HeLa cells analyzed as in B but in the presence of the competitors indicated.
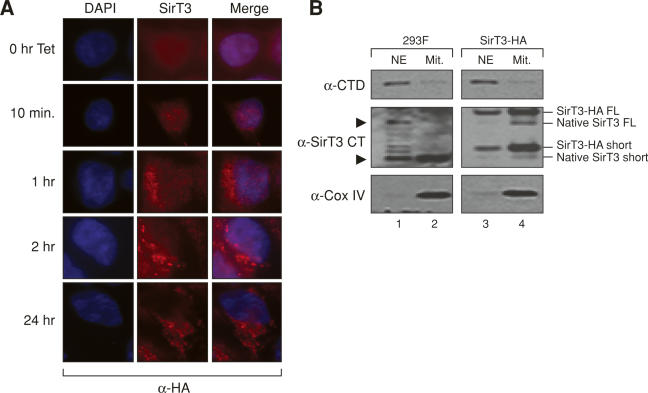
Figure 6.
Full-length endogenous SirT3 is expelled from the nucleus upon SirT3 overexpression. (A) Immunofluoresence experiment showing the localization of HA-tagged SirT3 as a function of induction time with tetracycline in a stable cell line, using antibody specific to the HA tag. (B) Western blot using antibody specific to the SirT3 C terminus and nuclear or mitochondrial fractions derived from either 293F cells or from a 293F stable cell line expressing tetracycline-inducible HA-tagged SirT3 after treatment with tetracycline overnight. Arrows on the right indicate both tagged and endogenous SirT3. Large arrows on the left indicate the sizes of endogenous SirT3. The antibodies used for Western analysis are indicated on the left.
SirT3 has NAD+-dependent histone deacetylase activity
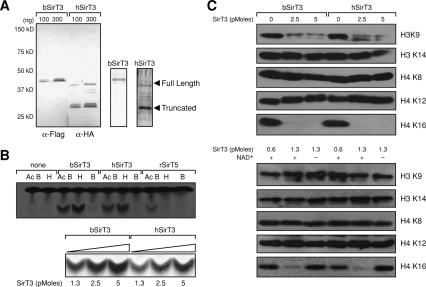
In order to understand the nuclear function of SirT3, we examined its enzymatic activity and substrate specificity. Only the processed SirT3 is considered biochemically active (Schwer et al. 2002). Yet given our findings that full-length SirT3 exists in the nucleus, we re-examined whether both forms of SirT3 have detectable NAD+-dependent activity. Recombinant SirT3 expressed in Escherichia coli generated exclusively full-length protein, but it was highly aggregated and exhibited very weak enzymatic activity; therefore, we generated a baculovirus SirT3 (bSirT3), which contained both a histidine and a Flag tag at the N terminus. After double affinity purification, only full-length bSirT3 was observed (Fig. 2A). We compared the bSirT3 to SirT3 purified from human cells (hSirT3) after transfection of a SirT3 expression plasmid containing an HA-tag at the C terminus. After HA affinity purification, >90% of the hSirT3 was in the processed form (Fig. 2A). Both SirT3 preparations displayed strong NAD+ -exchange activity (Vaquero et al. 2004) to either acetylated BSA or hyperacetylated core histones (Fig. 2B, top panel). As expected, unacetylated BSA as a substrate was inactive. Most importantly, SirT5, which localizes exclusively to the mitochondria (Haigis and Guarente 2006), displayed NAD+-exchange activity with specificity for acetylated BSA but not to hyperacetylated core histones. These observations collectively demonstrate that the histone-dependent exchange activity observed with SirT3 is specific.
Figure 2.
Full-length and truncated SirT3 exhibit similar HDAC activity and substrate specificity. (A, left side) Western blot of affinity-purified SirT3 from baculovirus-derived or human cells as indicated. (Right side) Coomassie blue stain of purified bSirT3 shows that only full-length SirT3 is detectable. (B, top) Nicotinamide exchange reaction in the presence of full-length SirT3 from baculovirus (bSirT3), truncated SirT3 from human cells (hSirT3), or recombinant SirT5 (rSirT5) as indicated. Substrates were acetylated BSA (Ac.B) histones derived from cells treated with histone deacetylase inhibitors (H) and BSA (B). (Bottom) Titration of bSirT3 and hSirT3 with histones as substrate. (C) Comparison of bSirT3 and hSirT3 in histone deacetylation reactions analyzed by Western blot using hyperacetylated histone substrates and with respect to specificity and NAD+ requirement. Antibodies against specific acetylated lysine residues of particular histones are indicated.
We next compared the SirT3 preparations in a histone deacetylation assay using hyperacetylated core histones purified from TSA and sodium butyrate-treated HeLa cells as substrate, coupled with Western blot analyses using antibodies against specific histone acetylated residues. In the absence of SirT3, deacetylation of histones was not observed (Fig. 2C, top panel). However, the addition of equivalent amounts of nicotinamide exchange units of both forms of SirT3 (full-length and processed forms shown in Fig. 2B, bottom panel) revealed a strong specificity for deacetylation of H3-K9ac and H4-K16ac in each case (Fig. 2C, top panel). We further analyzed the specificity and NAD+ dependence of baculovirus-derived, full-length SirT3 and human-derived, processed SirT3 by titrating down the enzymes, in the presence and absence of NAD+ (Fig. 2C, bottom panel). In each case, SirT3 exhibits apparently a higher specificity for H4-K16ac, and this HDAC activity is NAD+-dependent. As well, full-length SirT3 containing a Flag tag at the N terminus and isolated from human cells using anti-Flag affinity purification exhibited equivalent HDAC activity and specificity (data not shown), demonstrating that its expression in mammalian cells has no effect on its activity.
SirT3 deacetylates histones in vivo
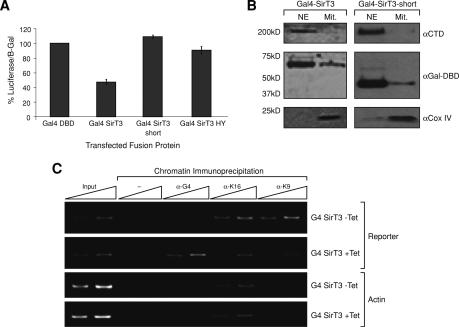
The results described above indicate that SirT3 is localized to the nucleus and that it deacetylates histone H4-K16ac and H3-K9ac in vitro. To analyze whether the above observations are biologically relevant, we analyzed whether SirT3 can deacetylate histones in vivo. We first tested if RNA interference (RNAi)-mediated reduction of cellular SirT3 would affect the overall levels of acetylated H3-K9 and H4-K16; these results were negative (data not shown). We then speculated that SirT3-mediated deacetylation of histone residues may be gene-specific. To test whether SirT3 can repress transcription in a gene-specific manner and in a deacetylation-dependent manner, we transfected a luciferase reporter directed by the TK promoter with Gal4 DNA-binding sites and Gal4-SirT3 expression plasmids into 293F cells. Recruitment of wild-type full-length Gal4-SirT3 to the luciferase reporter led to its repression (Fig. 3A). Transfection of a SirT3 mutant protein with a single substitution in the catalytic domain was ineffectual, demonstrating that the repression is dependent on SirT3 enzymatic activity. Since the N terminus of SirT3 may have an important role in the nucleus, we tested whether a truncated Gal4-SirT3 could repress transcription. Surprisingly, the N terminus was required for repression (Fig. 3A). Gal4-SirT3 fusion proteins were expressed and remained almost exclusively nuclear as shown by fractionation of cells transfected with full-length and processed forms of SirT3 (Fig. 3B). We next tested a cell line in which the full-length Gal4-SirT3 is stably integrated and its expression is regulated by tetracycline. Recruitment of Gal4-SirT3 to an integrated luciferase reporter was analyzed by chromatin immunoprecipitation (ChIP) experiments (Fig. 3C). After tetracycline induction, Gal4-SirT3 was present at the luciferase reporter gene as expected. Importantly, the levels of acetylated H4-K16 and H3-K9 were clearly reduced, relative to the uninduced case. Of note, tetracycline treatment was without effect at the actin promoter with respect to the levels of acetylated H4-K16 (Fig. 3C).
Figure 3.
SirT3 directed to a promoter mediates gene repression and HDAC activity toward H4-K16ac. (A) Results of luciferase assays performed using a stable cell line containing an integrated luciferase reporter under the TK promoter with GAL4 DNA-binding sites. Transient expression of the GAL4 fusion proteins indicated was performed. (B) Western blots of extracts fractionated into nuclear and mitochondrial fractions after transfection with expression vectors for the indicated protein. (C) ChIP experiments performed with a stable cell line containing both an integrated luciferase reporter and an integrated tetracycline-inducible GAL4-SirT3 fusion protein. The experiment was performed in the presence or absence of tetracycline as indicated and with the specific antibodies shown at the top. Primers targeted to the luciferase reporter as well as the actin promoter were used to assay the immunoprecipitations.
Overexpression of SirT3 leads to its relocalization to the mitochondria
We next sought to rationalize previous results showing the exclusive localization of SirT3 to the mitochondria. These previous studies used conditions under which SirT3 was overexpressed. Toward this end, we generated a stable cell line that constitutively expresses SirT3 containing an HA tag at its C terminus. Consistent with the current literature, when SirT3 was overexpressed, the protein localized to the mitochondria as evident with anti-HA antibodies (Fig. 4A). Interestingly, when SirT3 antibodies that recognize the N terminus of either endogenous or overexpressed protein were used for immunofluorescence, we no longer observed any signal in the nucleus and found SirT3 only in the mitochondria. This result led us to postulate that prolonged overexpression of SirT3 created an environment that provoked nuclear SirT3 to vacate the nucleus. It also is evidence of a mechanism whereby SirT3 function in the nucleus can be rapidly and negatively controlled through its potential physical exclusion from this site.
Figure 4.
Constitutively overexpressed SirT3 is actively transported to the mitochondria. (A) Immunofluoresence experiment performed with a stable cell line that constitutively expresses SirT3 with an HA tag using either HA-specific or SirT3 N-terminal-specific antibodies and Mitotracker. (B) Immunofluorescence studies of 293F cells transfected with SirT-HA and then treated with LMB as indicated. Cells were stained with antibodies against the HA tag.
Hst2, the closest homolog of SirT2, has recently been shown to contain an NES and its cytoplasmic localization to be sensitive to LMB (Wilson et al. 2006). LMB inhibits CRM1 (chromosomal region maintenance 1), a protein required for nuclear export of proteins containing a nuclear export signal (NES). Examination of the primary sequence of SirT3 did not reveal an NES; however, it is possible that SirT3 export is mediated through an interaction with a protein containing an NES. We tested the possibility that LMB would inhibit SirT3 export from the nucleus upon its overexpression. After transfection of SirT3-HA, cells were treated with LMB followed by immunofluorescence. Since transfection of SirT3 leads to constitutive expression of SirT3, we reasoned that treatment with LMB would inhibit newly expressed protein from leaving the nucleus. Following transfection of SirT3 under normal conditions, we did not observe any cells containing nuclear SirT3; however, upon treatment with LMB, we observed many cells that contained a nuclear SirT3 signal (Fig. 4B). This result suggests that SirT3 is actively transported from the nucleus to the mitochondria by a pathway dependent on CRM1.
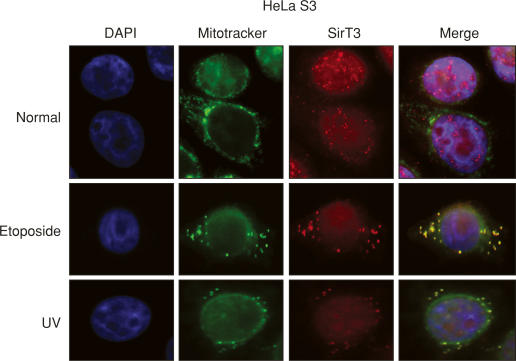
Full-length SirT3 resides in the nucleus and translocates to the mitochondria upon cellular stress
In light of the results presented above as well as the proposal that the main function of sirtuins might be to promote survival and stress resistance in times of adversity (Guarente and Picard 2005), we hypothesized that SirT3 localization to the mitochondria may be biologically relevant during cellular stress—a condition attained by its overexpression. Toward this end, we analyzed SirT3 localization under other conditions that lead to cellular stress: UV-irradiation or treatment with Etoposide, an inhibitor of topoisomerase II that causes DNA damage. In agreement with the results presented above, untreated cells displayed most of the endogenous full-length SirT3 in the nucleus (Fig. 5). However, upon UV or Etoposide treatment, a large population of the endogenous SirT3 was now localized to the mitochondria, as detected using the antibodies directed against its N terminus recognizing full-length human protein (Fig. 5). Since the SirT3 antibodies used in the last two experiments (Figs. 4, 5) are directed toward the N terminus of SirT3 and give rise to a strong mitochondrial signal under the conditions used (cellular stress induced by SirT3 overexpression, UV irradiation, or Etoposide treatment), we surmised that the full-length SirT3 protein is transported into the mitochondria after stress (see below).
Figure 5.
SirT3 is expelled from the nucleus upon cellular stress. Immunofluorescence experiment performed in HeLa S3 cells as a function of UV or Etoposide treatment using antibodies specific to the N terminus of hSirT3 and Mitotracker.
Newly synthesized SirT3 is processed in the nucleus
To analyze the fate of SirT3 upon its overexpression, we used a stable cell line encoding a tetracycline-inducible SirT3 harboring an HA-tag at its C terminus. We examined SirT3 localization as a function of time after tetracycline induction using anti-HA antibodies in immunofluorescence experiments (Fig. 6). Early after induction (10 min), SirT3 localized primarily to the nucleus, and at subsequent time points, SirT3 starts to accumulate in the mitochondria (Fig. 6A). After 1 h of induction, a large fraction of SirT3 localized to the mitochondria, but detectable amounts in the nucleus were also observed. At 2 h of SirT3 induction, most of the protein was localized to the mitochondria (Fig. 6A). This result is consistent with the observation shown above demonstrating that prolonged overexpression of SirT3 results in its complete translocation to the mitochondria. Similar results were observed when cells were fractionated into nuclear and mitochondrial fractions and SirT3’s localization analyzed by Western blots (Fig. 6B). Normal uninduced 293 cells displayed endogenous SirT3 in the nucleus and mitochondria as detected using an antibody against the C terminus of the SirT3 protein. The presence of SirT3 in the nucleus was not due to mitochondrial contamination in the nuclear fraction, as Western blots using antibodies against the mitochondria-specific protein Cox IV demonstrate its absence in the nuclear fraction (Fig. 6B, lanes 1,2). Consistent with the immunofluorescence studies presented above, after induction of SirT3-HA, most of the nontagged, endogenous SirT3 quickly localized to the mitochondria (Fig. 6B, lane 4). The C-terminal HA-tagged SirT3 protein can still be detected in the nuclear fraction as this represents the newly synthesized protein before it translocates to the mitochondria. That SirT3 is still detected in the nucleus by Western analysis and not by immunofluorescence may be due to the differences in the two methods. The normalization of the protein loaded for Western blot may not accurately represent the physiological ratio that is present in the fixed cells used for immunofluorescence. These observations further stress the presence of full-length SirT3 in the nucleus. Moreover, they suggest a path by which newly synthesized SirT3 translocates to the nucleus and then to the mitochondria.
Interestingly, both the truncated and full-length forms of SirT3 were present in the nuclear fraction, while only the truncated form was detected in the mitochondria (Fig. 6B, lanes 1,2). The most likely explanation for the presence of truncated SirT3 in the nucleus is that SirT3 becomes cleaved at this site. Overexpression causes the cell to become stressed (and oversaturated with SirT3), and this signals the full-length SirT3 to translocate to the mitochondria.
Discussion
In this study, we analyzed SirT3, a protein related to the yeast Sir2 (Frye 1999), which is involved in the formation of repressive chromatin structures through its ability to deacetylate histone H4-K16ac (Imai et al. 2000; Landry et al. 2000). In agreement with previous results (Onyango et al. 2002; Schwer et al. 2002), we found that SirT3 exists in two different forms, a full-length protein and a protein in which the N-terminal 142 amino acids are removed. However, in contrast to previous studies demonstrating that SirT3 localizes exclusively to the mitochondria, we found that the full-length SirT3 actually resides completely in the nucleus. It is intriguing that conditions that foster cellular stress such as those that induce apoptosis and even those caused by overloading the cell with ectopic expression of SirT3 result in the expulsion of full-length SirT3 from the nucleus. This suggests that the nuclear function of SirT3 can be completely and quickly halted and, in keeping with this, that the enzyme has an extremely important and time-sensitive role in the nucleus. Since a large amount of processed SirT3 is localized to the mitochondria, it certainly performs important roles there. However, we posit that this role is not as sensitive as the nuclear one in that most of the cellular processed SirT3 resides in the mitochondria already. Thus the small increase of SirT3 in the mitochondria from the pool of nuclear SirT3 would not be expected to lead to a strong effect on its concentration there. On the other hand, the exclusion of 100% of the full-length SirT3 from the nucleus would create a dramatic change in concentration at this site.
The nuclear full-length SirT3 protein is enzymatically active and deacetylates histones H3-K9ac and H4-K16ac both in vivo and in vitro. Previous reports indicated that only the processed SirT3 is enzymatically active (Schwer et al. 2002). In this case, SirT3 was generated by in vitro translation using rabbit reticulocyte extract and was tested for activity by using a histone H4 peptide that was chemically labeled in vitro. Our results were obtained using endogenous core histones that were acetylated in vivo. That the previous studies used a chemically labeled H4 peptide as opposed to using core histones isolated from cells may have in part led to the results obtained. Another possible explanation of the difference observed could be due to expression of SirT3 in reticulocyte extracts. We found that the bacterially expressed SirT3 protein tends to aggregate, prompting us to generate a baculovirus vector for SirT3 expression. The possibility exists that the SirT3 protein used in previous studies was misfolded, thereby inhibiting its enzymatic activity, and that treatment with peptidase removes the N terminus and this putative block.
Our studies also indicate that SirT3 is processed in the nucleus. This finding is consistent with studies of the mouse SirT3 (Shi et al. 2005), which lacks the N terminus and localizes to the mitochondria. This is in contrast to previous studies of the human SirT3 that reported a requirement for the N terminus in its localization to the mitochondria based on immunofluorescence studies and using a SirT3-GFP fusion protein (Schwer et al. 2002). Rather than the suggested possibility that mouse and human SirT3 proteins use two different pathways to localize to the mitochondria (Shi et al. 2005), it is more likely that both proteins are transported to the mitochondria by the same process. Should this be the case, the mouse protein would initially go to the nucleus and then travel to the mitochondria, while the human protein goes to the nucleus, where it is cleaved, and then the processed protein travels to the mitochondria via the same pathway. Furthermore, a possible explanation as to why truncated SirT3-GFP lost its localization to the mitochondria (Schwer et al. 2002) is that the N terminus is required for the human SirT3 to correctly localize to the nucleus before it can be transported to the mitochondria.
Our results also show that the N terminus of human SirT3 is required for transcription repression, further suggesting a regulatory role in the nucleus for this domain. It is possible that in mice, a murine adapter protein performs a similar function as the human SirT3 N terminus. The N terminus of human SirT3 might mediate protein recruitment and substrate specificity in the nucleus as compared with the mitochondria. For example, the N terminus might facilitate SirT3 activity in histone deacetylation in vivo through the recruitment of chromatin remodeling factors or an intrinsic chromatin-related activity. It is also possible that other HDACs are recruited to the gene and that the SirT3 activity is required for deacetylation of another transcription factor required for full repression.
Since Sirtuins, like Sir2, are dependent on NAD+ for enzymatic activity (Imai et al. 2000; Landry et al. 2000), SirT3-mediated gene repression is probably regulated by changes in metabolism and NAD+/NADH levels during normal cell conditions, and, as such, SirT3 functions in the mitochondria to deacetylate acetyl-CoA synthetase 2, leading to metabolic changes. Furthermore, given that upon cellular stress SirT3 completely leaves the nucleus, this might cause the rapid activation of nuclear genes involved in mitochondrial function. Hence, SirT3 may autoregulate itself between the nucleus and the mitochondria.
From diverse studies (Vaquero et al. 2004, 2006; Haigis and Guarente 2006), it is clear that the sirtuin family of proteins uses NAD+ for at least two functions, histone/protein deacetylation and protein ADP-ribosylation (Haigis and Guarente 2006). Three members, SirT3, SirT4, and SirT5, have been directly implicated in mitochondrial activity through their localization to the mitochondria and in the case of SirT1 through its ability to deacetylate and thereby enhance the activity of the nuclear coactivator PGC-1α, which regulates the expression of proteins involved in oxidative phosphorylation and mitochondrial biogenesis in skeletal muscle and brown adipose tissue (Koo and Montminy 2006; Lagouge et al. 2006). That sirtuins use NAD+ as a cofactor for enzymatic activity prompted speculations that these proteins, including ySir2, may be involved in the response to caloric restriction and in prolonging life span (Haigis and Guarente 2006). This speculation gained credibility when resveratrol, a naturally existing compound enriched in red grapes and thus in red wine, was found to specifically stimulate SirT1 activity. Further precise biochemical studies uncovered that the resveratrol effect was actually specific for one among several types of assays that measure SirT1-mediated protein deacetylation, and thus disputed the resveratrol effects both in mammals (Borra et al. 2005) and in yeast (Kaeberlein et al. 2005). However, recent studies performed in mice suggest that resveratrol does function in regulating caloric restriction by stimulating the activity of SirT1, which, in turn, deacetylates PGC-1α and stimulates the expression of nuclear genes involved in mitochondrial function and biogenesis (Lagouge et al. 2006). Certainly, the possible role of resveratrol in caloric restriction and in prolonging life span requires further studies. However, despite the existence of contradictory data, it is surprising and certainly encouraging that independent studies uncovered that increasing SirT1 activity through resveratrol or SirT3 expression through enhancer mutations is associated with prolonged life span. Our results predict that increased SirT3 expression would result in its expulsion from the nucleus and translocation to the mitochondria. Perhaps future experiments should concentrate on understanding how the SirT3 nuclear function together with its mitochondrial function participate in the processes of aging, metabolism, mitochondrial biogenesis, and thermogenesis (Bellizzi et al. 2005; Shi et al. 2005). Based on the studies described herein, it appears that drugs targeted toward the regulation of SirT3 activity and/or its expression as well as to the protease responsible for its cleavage may have important consequences in a cell’s response to stress and its life span.
Materials and methods
Plasmids and antibodies
All constructs were generated using standard PCR-based cloning strategy, and the identities of individual clones were verified through DNA sequencing using an ABI prism DNA sequencer. SirT5 was cloned into pET30a (Novagen), SirT3-HA was cloned into pcDNA4/TO, and baculovirus SirT3 was cloned into pAcHLT-B. Site-directed mutagenesis was performed to change histidine 247 of SirT3 to tyrosine to generate SirT3 HY. N-terminal SirT3 antibodies were generated in rabbits using the N-terminal peptide sequence (H2N-VGPFQACGCRLVLGGRD DVSAGLRGSHGARGEPLDPARPLQRPPRPEVPR-COOH). SirT3 C-terminal antibodies were a gift from Eric Verdin and are described in Schwer et al. (2002). The antibodies used in HDAC assays and ChIP were previously described (Vaquero et al. 2004). Anti-HA antibodies and HA peptide were purchased from Sigma. Anti-H3 acK9 antibodies were purchased from Abcam. Mitotracker was purchased from Molecular Probes.
Preparation of nuclear- and mitochondrial-enriched extracts
To generate fractions enriched in nuclear and mitochondrial proteins, 5.0 × 106 cells were harvested and washed in PBS. Cells were resuspended in 0.2 mL of buffer MgRSB (10 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl at pH 7.5, 0.1 mM PMSF, 1 mM DTT) and incubated on ice for 10 min. Cells were dounced to yield free nuclei, and 0.034 mL of sucrose buffer (2 M sucrose, 35 mM EDTA, 50 mM Tris-HCl at pH 7.5) was immediately added. Free nuclei were collected by centrifugation at 800 rcf for 10 min. Mitochondria were collected by centrifugation at 10,000 rcf for 10 min. Nuclei were resuspended in buffer C (20 mM Tris-HCl at pH 7.9, 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1 mM PMSF, 0.5 mM DTT) and incubated on ice for 20 min. To generate nuclear extract, nuclei were spun down at 20,000 rcf, and the supernatant was collected. Mitochondria were resuspended in buffer C to be used in Western blots.
Immunofluorescence
Mitotracker Red and Green were incubated with cells at a concentration of 0.1 nM and 0.4 nM, and the cells were allowed to grow for 45 min prior to fixation. For Etoposide treatment, cells were incubated in media containing 50 μM Etoposide for 4 h. For UV treatment, media was removed from the cells that were then irradiated with 40,000 μJ/cm2 UV followed by the addition of fresh media. The cells were allowed to grow for 4 h after UV treatment. Immunofluorescence was performed as described (Vaquero et al. 2006). For LMB experiments, 293F cells were transfected on coverslips with a vector expressing SirT3-HA overnight. The following day, cells were treated with 5 ng/mL LMB for 120 min, and immunofluorescence was performed as described.
In vitro enzymatic assays and cell-based assays
All assays were as previously described (Vaquero et al. 2004, 2006).
Stable cell lines
Cells harboring a Gal4-inducible luciferase reporter were generated by transfecting 293F TREX cells (Invitrogen) with a plasmid containing the luciferase gene downstream from Gal4-binding sites described previously (Vaquero et al. 2004) and selected for G418 resistance. After selection for luciferase expression, cells were transfected with pcDNA4/TO Gal4-SirT3 and were selected for G418 and Zeocin resistance. Inducible SirT3 stable cells were generated by transfection of pcDNA4/TO SirT3-HA into 293F TREX cells and selected for resistance.
Acknowledgments
We thank Dr. Eric Verdin and Dr. Bjoern Schwer for α-SirT3 C-terminal antibodies. We also thank Dr. Lynne Vales for comments on the manuscript and members of the Reinberg laboratory for helpful discussions. This work was supported by grants from NIH GM64844 (to D.R.) and the HHMI (to D.R.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1527307
References
- Bellizzi D., Rose G., Cavalcante P., Covello G., Dato S., De Rango F., Greco V., Maggiolini M., Feraco E., Mari V., Rose G., Cavalcante P., Covello G., Dato S., De Rango F., Greco V., Maggiolini M., Feraco E., Mari V., Cavalcante P., Covello G., Dato S., De Rango F., Greco V., Maggiolini M., Feraco E., Mari V., Covello G., Dato S., De Rango F., Greco V., Maggiolini M., Feraco E., Mari V., Dato S., De Rango F., Greco V., Maggiolini M., Feraco E., Mari V., De Rango F., Greco V., Maggiolini M., Feraco E., Mari V., Greco V., Maggiolini M., Feraco E., Mari V., Maggiolini M., Feraco E., Mari V., Feraco E., Mari V., Mari V., et al. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85:258–263. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Borra M.T., Smith B.C., Denu J.M., Smith B.C., Denu J.M., Denu J.M. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Ross S.E., Mostoslavsky R., Cohen H.Y., Mostoslavsky R., Cohen H.Y., Cohen H.Y., et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Frye R.A. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- Giannakou M.E., Partridge L., Partridge L. The interaction between FOXO and SIRT1: Tipping the balance towards survival. Trends Cell Biol. 2004;14:408–412. doi: 10.1016/j.tcb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Guarente L., Picard F., Picard F. Calorie restriction—The SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Haigis M.C., Guarente L.P., Guarente L.P. Mammalian sirtuins—Emerging roles in physiology, aging, and calorie restriction. Genes & Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Hallows W.C., Lee S., Denu J.M., Lee S., Denu J.M., Denu J.M. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong C.M., Kaeberlein M., Guarente L., Armstrong C.M., Kaeberlein M., Guarente L., Kaeberlein M., Guarente L., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McDonagh T., Heltweg B., Hixon J., Westman E.A., Caldwell S.D., Napper A., Curtis R., DiStefano P.S., Fields S., McDonagh T., Heltweg B., Hixon J., Westman E.A., Caldwell S.D., Napper A., Curtis R., DiStefano P.S., Fields S., Heltweg B., Hixon J., Westman E.A., Caldwell S.D., Napper A., Curtis R., DiStefano P.S., Fields S., Hixon J., Westman E.A., Caldwell S.D., Napper A., Curtis R., DiStefano P.S., Fields S., Westman E.A., Caldwell S.D., Napper A., Curtis R., DiStefano P.S., Fields S., Caldwell S.D., Napper A., Curtis R., DiStefano P.S., Fields S., Napper A., Curtis R., DiStefano P.S., Fields S., Curtis R., DiStefano P.S., Fields S., DiStefano P.S., Fields S., Fields S., et al. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- Koo S.H., Montminy M., Montminy M. In vino veritas: A tale of two sirt1s? Cell. 2006;127:1091–1093. doi: 10.1016/j.cell.2006.11.034. [DOI] [PubMed] [Google Scholar]
- Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Messadeq N., Milne J., Lambert P., Elliott P., Milne J., Lambert P., Elliott P., Lambert P., Elliott P., Elliott P., et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Landry J., Sutton A., Tafrov S.T., Heller R.C., Stebbins J., Pillus L., Sternglanz R., Sutton A., Tafrov S.T., Heller R.C., Stebbins J., Pillus L., Sternglanz R., Tafrov S.T., Heller R.C., Stebbins J., Pillus L., Sternglanz R., Heller R.C., Stebbins J., Pillus L., Sternglanz R., Stebbins J., Pillus L., Sternglanz R., Pillus L., Sternglanz R., Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley E., Pearson M., Faretta M., Bauer U.M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T., Pearson M., Faretta M., Bauer U.M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T., Faretta M., Bauer U.M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T., Bauer U.M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T., Minucci S., Pelicci P.G., Kouzarides T., Pelicci P.G., Kouzarides T., Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W., Chen D., Su F., Shiloh A., Guarente L., Gu W., Su F., Shiloh A., Guarente L., Gu W., Shiloh A., Guarente L., Gu W., Guarente L., Gu W., Gu W. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Onyango P., Celic I., McCaffery J.M., Boeke J.D., Feinberg A.P., Celic I., McCaffery J.M., Boeke J.D., Feinberg A.P., McCaffery J.M., Boeke J.D., Feinberg A.P., Boeke J.D., Feinberg A.P., Feinberg A.P. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., North B.J., Frye R.A., Ott M., Verdin E., North B.J., Frye R.A., Ott M., Verdin E., Frye R.A., Ott M., Verdin E., Ott M., Verdin E., Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Bunkenborg J., Verdin R.O., Andersen J.S., Verdin E., Bunkenborg J., Verdin R.O., Andersen J.S., Verdin E., Verdin R.O., Andersen J.S., Verdin E., Andersen J.S., Verdin E., Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T., Wang F., Stieren E., Tong Q., Wang F., Stieren E., Tong Q., Stieren E., Tong Q., Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J. Biol. Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M., Ishii H., Sun J.M., Pazin M.J., Davie J.R., Peterson C.L., Ishii H., Sun J.M., Pazin M.J., Davie J.R., Peterson C.L., Sun J.M., Pazin M.J., Davie J.R., Peterson C.L., Pazin M.J., Davie J.R., Peterson C.L., Davie J.R., Peterson C.L., Peterson C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Suka N., Luo K., Grunstein M., Luo K., Grunstein M., Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D., Erdjument-Bromage H., Tempst P., Reinberg D., Tempst P., Reinberg D., Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Vaquero A., Scher M.B., Lee D.H., Sutton A., Cheng H.L., Alt F.W., Serrano L., Sternglanz R., Reinberg D., Scher M.B., Lee D.H., Sutton A., Cheng H.L., Alt F.W., Serrano L., Sternglanz R., Reinberg D., Lee D.H., Sutton A., Cheng H.L., Alt F.W., Serrano L., Sternglanz R., Reinberg D., Sutton A., Cheng H.L., Alt F.W., Serrano L., Sternglanz R., Reinberg D., Cheng H.L., Alt F.W., Serrano L., Sternglanz R., Reinberg D., Alt F.W., Serrano L., Sternglanz R., Reinberg D., Serrano L., Sternglanz R., Reinberg D., Sternglanz R., Reinberg D., Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes & Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A., Pandita T.K., Guarente L., Weinberg R.A., Guarente L., Weinberg R.A., Weinberg R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wilson J.M., Le V.Q., Zimmerman C., Marmorstein R., Pillus L., Le V.Q., Zimmerman C., Marmorstein R., Pillus L., Zimmerman C., Marmorstein R., Pillus L., Marmorstein R., Pillus L., Pillus L. Nuclear export modulates the cytoplasmic Sir2 homologue Hst2. EMBO Rep. 2006;7:1247–1251. doi: 10.1038/sj.embor.7400829. [DOI] [PMC free article] [PubMed] [Google Scholar]