Abstract
The LIM domain-binding protein Ldb1 is an essential cofactor of LIM-homeodomain (LIM-HD) and LIM-only (LMO) proteins in development. The stoichiometry of Ldb1, LIM-HD, and LMO proteins is tightly controlled in the cell and is likely a critical determinant of their biological actions. Single-stranded DNA-binding proteins (SSBPs) were recently shown to interact with Ldb1 and are also important in developmental programs. We establish here that two mammalian SSBPs, SSBP2 and SSBP3, contribute to an erythroid DNA-binding complex that contains the transcription factors Tal1 and GATA-1, the LIM domain protein Lmo2, and Ldb1 and binds a bipartite E-box-GATA DNA sequence motif. In addition, SSBP2 was found to augment transcription of the Protein 4.2 (P4.2) gene, a direct target of the E-box-GATA-binding complex, in an Ldb1-dependent manner and to increase endogenous Ldb1 and Lmo2 protein levels, E-box-GATA DNA-binding activity, and P4.2 and β-globin expression in erythroid progenitors. Finally, SSBP2 was demonstrated to inhibit Ldb1 and Lmo2 interaction with the E3 ubiquitin ligase RLIM, prevent RLIM-mediated Ldb1 ubiquitination, and protect Ldb1 and Lmo2 from proteasomal degradation. These results define a novel biochemical function for SSBPs in regulating the abundance of LIM domain and LIM domain-binding proteins.
Keywords: Single-stranded DNA-binding proteins, LIM domain proteins, LIM domain-binding proteins, ubiquitination, transcription factors, erythroid differentiation
The LIM domain-binding protein Ldb1 was originally identified through its interaction with LIM-homeodomain (LIM-HD) and LIM-only (LMO) proteins (Agulnick et al. 1996; Jurata et al. 1996; Bach et al. 1997; Visvader et al. 1997), and these interactions are critical for Ldb1 function (Agulnick et al. 1996; Bach et al. 1997; Jurata and Gill 1997; Morcillo et al. 1997; Cassata et al. 2000; van Meyel et al. 2000; Thaler et al. 2002; Pueyo and Couso 2004; Nishioka et al. 2005). Ldb1 is required for normal development as evidenced by the severe defects in Ldb1-null mice, including the absence of primitive hematopoiesis (Mukhopadhyay et al. 2003). These and other studies have established that this LIM protein cofactor participates in diverse developmental programs (Matthews and Visvader 2003).
The relative abundance of Ldb1, LIM-HD, and LMO proteins is critical to formation of the multiprotein complexes to which they contribute and, ultimately, their biological actions (Fernández-Fúnez et al. 1998; Milán and Cohen 1999; van Meyel et al. 1999, 2000; Rincón-Limas et al. 2000). Proper stoichiometry of Drosophila Ldb1 (or Chip) and Apterous, a LIM-HD protein with which Chip interacts, is essential for the formation and function of this complex (van Meyel et al. 2000), and Ldb1 also contributes to DNA-binding complexes containing LIM-only proteins, GATA transcription factors, and basic helix–loop–helix (bHLH) protein heterodimers (Visvader et al. 1997; Wadman et al. 1997; Heitzler et al. 2003; Xu et al. 2003). While genetic studies have provided insight into Ldb1’s biological functions, little is known about how the levels of Ldb1 and its LIM protein interaction partners are controlled in cells or how assembly of Ldb1-containing transcriptional complexes is regulated.
The RING finger protein RLIM encoded by the RNF12 gene acts as an E3 ubiquitin ligase for Ldb1, LMO2, and histone deacetylase HDAC2, with the ubiquitinated forms of these proteins flagged for destruction in the 26S proteasome (Ostendorff et al. 2002; Hiratani et al. 2003; Krämer et al. 2003). Transient increases in the LIM-only protein LMO2 or LIM-HD protein Xlim-1 were shown to inhibit RLIM-mediated turnover of Ldb1, suggesting its LIM protein partners regulate Ldb1 levels in vivo (Ostendorff et al. 2002; Hiratani et al. 2003). However, LMO2 is also a substrate of RLIM (Ostendorff et al. 2002; Hiratani et al. 2003), dLMO protein expression in the Drosophila wing requires Chip rather than the reverse (Milán and Cohen 2000), and Xlim-1 levels do not correlate with those of Ldb1 (Hiratani et al. 2003). Thus, the specific contribution LIM domain proteins make to Ldb1 protein stability has been unclear.
CSDP, the founding member of a group of single-stranded DNA-binding proteins, was isolated through its single-stranded DNA-binding activity (Bayarsaihan and Lukens 1996), while the closely related single-stranded DNA-binding protein 2 (SSBP2) was discovered as a target of an unbalanced translocation in myeloid leukemia cells (Castro et al. 2002). The SSBPs have been widely studied as Ldb1 interaction partners, and there is considerable genetic evidence for their function in axis formation in Xenopus, wing development in Drosophila, and head morphogenesis in mice (Chen et al. 2002; van Meyel et al. 2003; Nishioka et al. 2005; Enkhmandakh et al. 2006). Both dSsdp and murine Ssbp3 have been shown to be essential for normal embryonic development (Chen et al. 2002; van Meyel et al. 2003; Nishioka et al. 2005; Enkhmandakh et al. 2006). While the physical and functional interactions between SSBPs and Ldb1 predict that these proteins contribute to common transcriptional complexes (Chen et al. 2002; van Meyel et al. 2003; Nishioka et al. 2005), the precise composition of such complexes, the target genes they regulate, and the actions of SSBPs in complex formation or function are not known.
We recently showed that the Protein 4.2 (P4.2) gene is a target of a DNA-binding complex in proerythroblasts that includes the transcription factors TAL1 (also known as SCL), E47, GATA-1, LMO2, and Ldb1 and determined that Ldb1 plays a positive role in erythroid gene expression and differentiation (Xu et al. 2003). We establish here that two SSBPs, Ssbp2 and Ssbp3, are associated with this DNA-binding complex and that Ssbp2 occupies and augments transcription from the P4.2 promoter in murine erythroid cells. Furthermore, SSBP2 was demonstrated to inhibit RLIM-mediated ubiquitination and proteasomal destruction of Ldb1 and inhibit Lmo2 turnover via a mechanism requiring Ldb1. These data define a novel biochemical function for the SSBPs that accounts for their Ldb1-dependent actions in development.
Results
Ssbp2 and Ssbp3 are integral components of an erythroid E-box-GATA DNA-binding complex
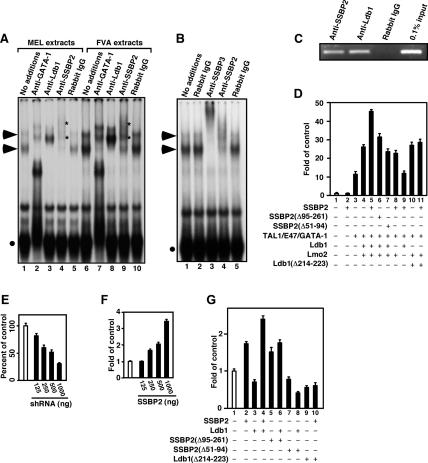
We and others have described a DNA-binding complex in erythroid cells that contains TAL1, GATA-1, Ldb1, and Lmo2 and binds an E-box-GATA sequence element (Wadman et al. 1997; Xu et al. 2003), and we established that the P4.2 gene promoter is one of its physiologic targets (Xu et al. 2003). To identify other members of this multiprotein complex, we undertook both candidate protein and protein purification approaches. Given the reports of SSBP interaction with Ldb1 (Chen et al. 2002; van Meyel et al. 2003), we investigated whether members of this class of proteins were components of the complex, initially through antibody supershift analysis. As described (Xu et al. 2003), incubation of extracts of both murine erythroleukemia (MEL) cells and splenic proerythroblasts elicited by the anemia-inducing stain of Friend virus (FVA cells) with a 32P-labeled E-box-GATA oligonucleotide corresponding to an E-box-GATA element in the P4.2 promoter led to formation of two highly retarded protein–DNA complexes (Fig. 1A, arrowheads). The more retarded, and less abundant, of these appears to contain two protein–DNA complexes linked in solution by Ldb1 and the faster a single protein–DNA complex (Xu et al. 2003). Both complexes were supershifted by incubating extracts with antibodies to two of its known components, GATA-1 and Ldb1 (Fig. 1A; Wadman et al. 1997; Xu et al. 2003). Furthermore, rabbit polyclonal antibody to Ssbp2 (asterisk in Fig. 1A [lanes 4,9], B [lane 4]) and to Ssbp3 (Fig. 1B, lane 3), but not normal rabbit IgG (Fig. 1A [lanes 5,10], Fig. 1B [lane 5]) also supershifted both complexes. The Ssbp2 and Ssbp3 antibodies had no effect, in contrast, on a less retarded protein–DNA complex (filled circles in Fig. 1A [lanes 4,9], B [lanes 3,4]) supershifted exclusively by antibody to GATA-1 (Fig. 1A, lanes 2,7). Additionally, mass spectrometry analysis of proteins that copurified with a tandem epitope (TAP)-tagged Tal1 protein in MEL cells (Y. Cai, Z. Xu, J. Xie, M.J. Koury, S.W. Hiebert, and S.J. Brandt, in prep.) revealed multiple peptides derived from both Ssbp2 and Ssbp3 (see Supplemental Material). Finally, chromatin immunoprecipitation (ChIP) analysis showed that antibodies to Ssbp2 and Ldb1, but not normal rabbit IgG, precipitated chromatin fragments from the P4.2 promoter (Fig. 1C), establishing that Ssbp2 occupies this promoter with Ldb1 in cells. These results demonstrate that Ssbp2 and Ssbp3 are components of the TAL1- and GATA-1-containing complex(es) in erythroid progenitors. While not excluding the possibility that individual complexes could contain only one of these two SSBPs, supershift analysis suggests that most complexes contain both Ssbp2 and Ssbp3.
Figure 1.
Ssbp2 and Ssbp3 are integral components of an erythroid E-box-GATA DNA-binding complex. (A) EMSA of nuclear extracts from MEL and FVA cells for E-box-GATA DNA-binding activity. Rabbit IgG or antibodies to GATA-1, Ldb1, or SSBP2 were used as indicated for supershift analysis. Complexes supershifted by antibodies to GATA-1 and Ldb1 are marked with solid arrowheads and complexes containing only GATA-1 are marked with filled circles. Complexes supershifted by SSBP2 antibody are marked to the right of the relevant lane with asterisks. (B) EMSA of nuclear extracts from MEL cells for E-box-GATA DNA-binding activity. Normal rabbit IgG or antibodies to SSBP3 and SSBP2 were used for supershift analysis and DNA-binding complexes marked as above. (C) ChIP analysis of SSBP2 and Ldb1 occupancy at the P4.2 promoter in differentiating MEL cells. (D) Transient transfection analysis with a plasmid (1.4 μg) containing 1.7 kb of the P4.2 promoter linked to a luciferase reporter gene in COS-7L cells transfected with (filled column) or without (unfilled column) indicated combinations of expression plasmids pcDNA3.1-Tal1 (100 ng), pcDNA3.1-E47 (3 ng), pcDNA3.1-GATA-1 (25 ng), pEFIRES-Ldb1 (40 ng), pEFIRES-Ldb1(Δ214–223) (40 ng), pEFIRES-Lmo2 (40 ng), pEFIRES-SSBP2 (200 ng), pEFIRES-SSBP2(Δ95–261) (200 ng), and pEFIRES-SSBP2(Δ51–94) (200 ng). The mean ± SEM luciferase activity relative to that with reporter alone from three independent experiments is plotted. Reporter analysis in DMSO-treated MEL cells transfected with (filled column) or without (unfilled column) indicated amounts of pSilencer-Ssbp2-B (E), pEFIRES-SSBP2 (F), and indicated combinations of pEFIRES-SSBP2 (500 ng), pEFIRES-SSBP2(Δ95–261) (500 ng), pEFIRES-SSBP2(Δ51–94) (500 ng), pEFIRES-Ldb1 (500 ng), and pEFIRES-Ldb1(Δ214–223) (500 ng) (G).
To evaluate the contribution of the SSBPs to P4.2 promoter function, a P4.2 promoter-luciferase reporter and expression vector for SSBP2 were transfected into COS-7L cells with different combinations of plasmids for the five previously described components of the complex. Expression of SSBP2 with TAL1, E47, GATA-1, LMO2, and Ldb1 effected a nearly 50-fold increase in reporter activity compared with the 25-fold induction observed in its absence (Fig. 1D, cf. lanes 5 and 4), representing an approximately twofold increase in activity specifically attributable to SSBP2. Transfection of SSBP2 alone, in contrast, had no effect on reporter activity (Fig. 1D, cf. lanes 2 and 1), indicating that SSBP2 must be recruited to DNA, by the E-box-GATA DNA-binding complex(es) at this promoter, to activate transcription. A deletion mutant, SSBP2(Δ95–261), lacking a major portion of SSBP2’s Gly–Pro-rich central domain was significantly impaired in transactivating activity (Fig. 1D, cf. lanes 6 and 5), while the SSBP2(Δ51–94) deletion mutant lacking half of the N-terminal LUFS domain and thus incapable of binding to Ldb1 (van Meyel et al. 2003) was completely inactive (Fig. 1D, cf. lanes 7 and 4). Likewise, no further increase in reporter activity was obtained with full-length SSBP2 in the absence of Ldb1 (Fig. 1D, cf. lanes 8 and 4), and SSBP2 had little to no effect on reporter activity when cotransfected with Ldb1(Δ214–223), a deletion mutant lacking part of the Ldb1/Chip conserved domain (LCCD) and impaired in SSBP2 interaction (Fig. 1D, cf. lanes 11 and 10; van Meyel et al. 2003). This Ldb1 mutant exhibited the same transactivating activity as wild-type Ldb1, in contrast, when transfected with the core components of this complex in the absence of SSBP2 (Fig. 1D, cf. lanes 10 and 4). Finally, when the Lmo2 expression plasmid was left out, reporter activity was reduced to the level observed with Tal1, E47, and GATA-1 (Fig. 1D, cf. lanes 9 and 3), presumably from the inability of the ternary complex to form in the absence of Lmo2 (Wadman et al. 1997; Xu et al. 2003).
To determine whether SSBP2 activated transcription in a more physiological cellular environment, expression plasmids for SSBP2 or a short-hairpin RNA (shRNA) targeted to Ssbp2 were transfected with the P4.2 promoter-luciferase reporter into differentiating MEL cells. Depletion of Ssbp2 by RNA interference (RNAi) decreased (Fig. 1E) while enforced expression of SSBP2 increased (Fig. 1F) reporter activity, both in a concentration-dependent manner. Finally, cotransfection of SSBP2 and Ldb1 into differentiating MEL cells converted overexpressed Ldb1 from an inhibitor to an activator of reporter activity (Fig. 1G, cf. lanes 4 and 3). This effect was attenuated when a truncation mutant of SSBP2 was used (Fig. 1G, cf. lanes 6 and 4) and abrogated with deletion of the LUFS domain of SSBP2 (Fig. 1G, cf. lanes 8 and 4) or LCCD of Ldb1 (Fig. 1G, cf. lanes 10 and 4). These results indicate that SSBP2 is an integral component of the E-box-GATA DNA-binding complex and that it promotes transcription from the P4.2 promoter in an Ldb1-dependent manner. Moreover, they suggest that SSBP2 levels in MEL cells could be limiting for complex activity.
SSBP2 positively regulates P4.2 and β-globin gene expression in erythroid progenitors
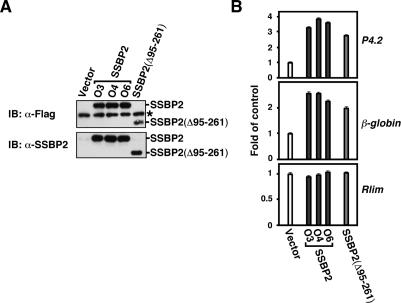
To examine whether SSBP2 regulates endogenous P4.2 gene expression in erythroid cells, Flag-tagged SSBP2 and the SSBP2(Δ95–261) mutant lacking the protein’s putative single-stranded DNA-binding domain were expressed in MEL cells. Western blot analysis of two polyclonal populations of transduced cells using antibodies to the Flag epitope and to SSBP2 showed that while transfected Flag-tagged SSBP2(Δ95–261) protein was readily detected (Fig. 2A), Flag-tagged full-length SSBP2 protein was not (data not shown). Because SSBP2 can potentially inhibit cellular proliferation (Liang et al. 2005), causing selection against overexpressing cells in a mixed population, oligoclonal lines of SSBP2-transduced MEL cells were derived and analyzed. As shown in Figure 2A, unambiguous expression of transduced SSBP2 protein was detected in three of these lines using antibodies to the Flag epitope and SSBP2.
Figure 2.
SSBP2 positively regulates P4.2 and β-globin gene expression in MEL cells. (A) Western blot analysis of Flag-tagged SSBP2 and SSBP2(Δ95–261) mutant in MEL whole cellular extracts with antibody to the Flag epitope (top) and nuclear extracts with antibody to amino acids 269–284 of SSBP2 (bottom). A cross-reacting protein recognized by Flag antibody is marked with an asterisk and served as loading control. (B) Quantitative RT–PCR analysis of P4.2, β-globin, and RLIM mRNAs in indicated MEL cell transductants. The mean ± SEM RNA level relative to vector control is plotted.
While vector-transduced cells were pale, pellets of cells transduced with SSBP2 and SSBP2(Δ95–261) appeared pink, even in the absence of an inducer of erythroid differentiation, prompting us to investigate the effect of these proteins on erythroid gene expression. Quantitative RT–PCR analysis showed that overexpression of these proteins similarly, and markedly, increased P4.2 and β-globin mRNA levels without affecting RLIM gene expression (Fig. 2B). These results suggest that SSBP2 is a positive regulator of gene transcription in erythroid progenitors.
Overexpression of SSBP2 increases assembly of the E-box-GATA DNA-binding complex in vitro and P4.2 promoter occupancy in vivo
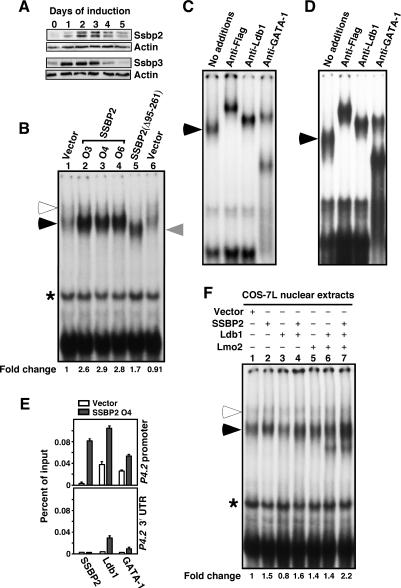
To elucidate the mechanism by which SSBPs regulate erythroid gene expression, we began by examining the expression pattern of Ssbp2 and Ssbp3 in MEL cells induced to differentiate with dimethyl sulfoxide (DMSO). Ssbp2 protein levels progressively increased over the first 2 d after addition of the inducer and then declined (Fig. 3A, top panel), with Ssbp3 protein levels showing a slightly earlier increase (Fig. 3A, bottom panel). As their abundance paralleled that of E-box-GATA DNA-binding activity during MEL-cell differentiation (Xu et al. 2003), we investigated whether the SSBPs had a role in complex formation or stability through use of nuclear extracts from MEL cells engineered to overexpress SSBP2. Overexpression of SSBP2 significantly increased the levels of the less retarded, and more abundant, of the two DNA-binding complexes (Fig. 3B, cf. lanes 2–4 and 1,6, solid arrowhead), and enforced expression of SSBP2(Δ95–261) did the same (Fig. 3B, cf. lanes 5 and 1,6, gray arrowhead). Supershift analysis confirmed the incorporation of Flag-tagged SSBP2 and Flag-tagged SSBP2(Δ95–261), as well as Ldb1 and GATA-1 proteins, into this complex (Fig. 3C,D). This E-box-GATA-binding complex had slightly greater mobility in SSBP2(Δ95–261)- than in SSBP2-transduced cells, due either to the smaller size of the SSBP2(Δ95–261) protein or the absence of proteins recruited by full-length SSBP2 but not SSBP2(Δ95–261). For reasons unclear at present, enforced expression of SSBP2 or SSBP2(Δ95–261) did not alter the more retarded of the two DNA-binding complexes (Fig. 3B, open arrowhead). In sum, overexpression of SSBP2 significantly stimulated the assembly of a DNA-binding complex to which this protein contributes.
Figure 3.
Overexpression of SSBP2 increases E-box-GATA DNA-binding activity and P4.2 promoter occupancy in MEL cells. (A) Western blot analysis of Ssbp2 (top) and Ssbp3 (bottom) protein expression in whole cellular extracts of differentiating MEL cells at times after DMSO addition. Actin was used as loading control. (B) EMSA of E-box-GATA DNA-binding activity in nuclear extracts from SSBP2-transduced MEL cells. The upper ternary complex is marked with an open arrowhead, and the lower complex, whose abundance was stimulated by SSBP2 overexpression, is marked with a solid arrowhead. The levels of DNA-binding activity for the lower complex were normalized to a nonspecific protein–DNA complex (asterisk). The fold change relative to vector control is shown below the autoradiographs. Antibody supershift analysis of nuclear extracts from oligoclonal population O4 transduced with full-length SSBP2 (C) or a polyclonal population transduced with SSBP2(Δ95–261) (D) using antibodies to Flag epitope, Ldb1, and GATA-1. DNA-binding complexes are marked as above. (E) Quantitative ChIP analysis of factor occupancy at the promoter or 3′ UTR of the P4.2 gene for vector-transduced (unfilled column) and SSBP2-transduced (filled column) MEL cells. The mean ± SEM occupancy as a percentage of input for SSBP2, Ldb1, and GATA-1 is plotted. (F) EMSA of E-box-GATA DNA-binding activity using nuclear extracts (5 μg) from COS-7L cells transfected with indicated combinations of pEFIRES-SSBP2, pEFIRES-Ldb1, and pEFIRES-Lmo2, assayed as mixed with nuclear extract (5 μg) from undifferentiated MEL cells. The levels of DNA-binding activity of the lower complex were normalized as above. The fold change relative to vector control is shown below the autoradiographs.
To investigate whether the increase in DNA-binding activity in SSBP2-transduced cells translated into increased occupancy of the P4.2 promoter, quantitative ChIP analysis was carried out. Overexpression of SSBP2 increased its loading on the P4.2 promoter (Fig. 3E, top panel), and, consistent with the increase in E-box-GATA DNA-binding activity detected by electrophoretic mobility shift analysis (EMSA) (Fig. 3B), also increased Ldb1 and GATA-1 recruitment (Fig. 3E, top panel). In contrast, these proteins did not occupy the 3′ untranslated region (3′ UTR) of this gene at significant levels, with or without SSBP2 overexpression (Fig. 3E, bottom panel).
Finally, the effect of SSBP2 on DNA-binding activity was determined in the presence and absence of Ldb1 and Lmo2. To that end, 5 μg of nuclear extracts of COS-7L cells transfected with different combinations of expression plasmids were mixed with 5 μg of nuclear extracts of undifferentiated MEL cells, and E-box-GATA DNA-binding activity was assayed. As shown in Figure 3F, addition of transfected SSBP2, either with or without Ldb1, modestly increased the abundance of the lower E-box-GATA DNA-binding complex (Fig. 3F, cf. lanes 2,4 and 1, solid arrowhead). The greatest increase in DNA-binding activity, however, was observed with balanced overexpression of SSBP2, Ldb1, and Lmo2 (Fig. 3F, cf. lanes 7 and 1). As in MEL cells transduced with SSBP2 (Fig. 3B), the amount of the upper complex did not change with provision of SSBP2 protein in vitro (Fig. 3F, lanes 2,4,7, open arrowhead). These results show that maximal assembly of this ternary complex required the simultaneous expression of SSBP2, Ldb1, and Lmo2 in cells.
Ssbp2 regulates the abundance of Ldb1 and Lmo2 proteins in erythroid progenitors
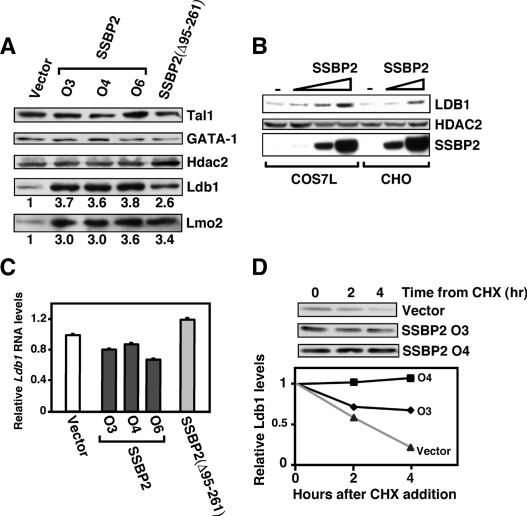
To gain insight into how SSBP2 stimulated complex assembly, the protein levels of the known components of the complex were determined. As shown in Figure 4A, oligoclonal populations overexpressing full-length SSBP2 and polyclonal populations expressing SSBP2(Δ95– 261) contained significantly greater amounts of Ldb1 and Lmo2 without any change in the abundance of Tal1, GATA-1, or an irrelevant protein, Hdac2. Transient overexpression of SSBP2 in both COS-7L and CHO cells also caused a concentration-dependent increase in endogenous Ldb1 levels, again without an effect on Hdac2 (Fig. 4B). Finally, quantitative RT–PCR analysis showed that Ldb1 mRNA was not altered in SSBP2- or SSBP2(Δ95– 261)-transduced MEL cells (Fig. 4C), while studies using the protein synthesis inhibitor cycloheximide (CHX) revealed a marked decrease in Ldb1 protein turnover in SSBP2-overexpressing cells (Fig. 4D). Specifically, the half-life of Ldb1 was ∼2 h in the vector control cells and >4 h in SSBP2-transduced cells (Fig. 4D). These data show that SSBP2 positively regulates Ldb1 and Lmo2 protein levels in differentiating erythroid progenitors.
Figure 4.
SSBP2 regulates the abundance of Ldb1 and Lmo2 proteins. (A) Western blot analysis of Tal1, GATA-1, Hdac2, Ldb1, and Lmo2 abundance in nuclear extracts from transduced MEL cells. The levels of Ldb1 and Lmo2 in SSBP2- or SSBP2(Δ95–261)-transduced cells relative to vector control are shown below the autoradiographs. (B) Western blot analysis of endogenous Ldb1 and Hdac2 expression and transduced Flag-tagged SSBP2 protein expression in COS-7L and CHO cells transiently transfected with increasing amounts of pEFIRES-SSBP2 (175 ng, 525 ng, and 1.6 μg for COS-7L cells; 125 ng and 1.0 μg for CHO cells). (C) Quantitative RT–PCR analysis of Ldb1 expression in indicated MEL cell transductants. The mean ± SEM RNA level relative to that in vector control is plotted. (D) Protein turnover analysis in SSBP2-transduced MEL cells treated with 100 μM CHX for the indicated times. (Top) Ldb1 expression was detected by Western blot analysis and quantified by densitometry of X-ray films. (Bottom) The amount of protein remaining is expressed as a function of time from addition of CHX.
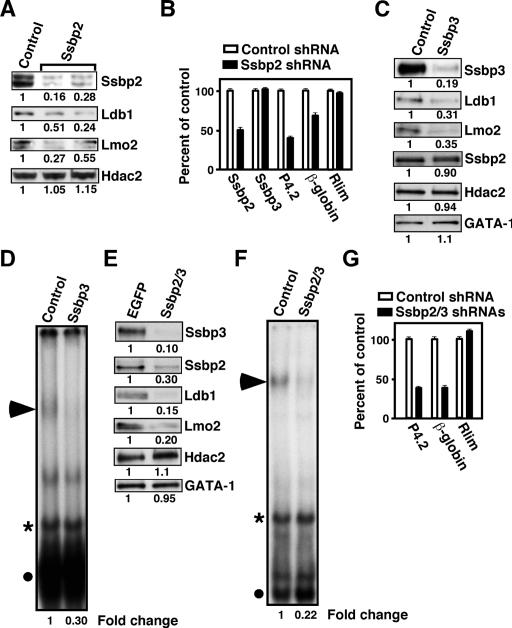
Depletion of Ssbp2 and Ssbp3 by RNAi reduces the abundance of Ldb1 and Lmo2 in erythroid progenitors
To determine whether Ssbp2 and Ssbp3 are required for Ldb1 and Lmo2 protein accumulation, shRNAs targeted to Ssbp2 and/or Ssbp3 were expressed in MEL cells. As shown in Figure 5A, knockdown of Ssbp2 expression in independent clonal populations of transduced MEL cells treated with 1.5% DMSO for 2 d to induce differentiation significantly reduced steady-state levels of Ldb1 and Lmo2 proteins without affecting Hdac2 levels (see the Supplemental Material for results with an additional clone). Likewise, transient expression of Ssbp2 shRNA in differentiating MEL cells significantly reduced Ssbp2, P4.2, and β-globin mRNA levels without affecting Ssbp3 or Rlim gene expression (Fig. 5B), and depletion of Ssbp3 by shRNA also markedly decreased steady-state levels of Ldb1 and Lmo2 (Fig. 5C). Furthermore, as shown in Figure 5D, the decline in Ssbp3 protein levels in shRNA-transduced cells was associated with a significant reduction in E-box-GATA DNA-binding activity (Fig. 5D, solid arrowhead) but not in a binding activity attributable to GATA-1 alone (Fig. 5D, filled circle). Finally, simultaneous depletion of Ssbp2 and Ssbp3 by specific shRNAs caused an even greater reduction in Ldb1 and Lmo2 abundance (Fig. 5E), E-box-GATA DNA-binding activity (Fig. 5F), and P4.2 and β-globin mRNA levels, again without affecting Rlim gene expression (Fig. 5G). Together, these studies indicate that at the concentrations prevailing in differentiating erythroid progenitors, Ssbp2 and Ssbp3 actively control the levels of Ldb1 and Lmo2 protein and E-box-GATA DNA-binding activity.
Figure 5.
Depletion of Ssbp2 and Ssbp3 reduces Ldb1 and Lmo2 abundance and E-box-GATA DNA-binding activity. (A) Western blot analysis of Ssbp2, Ldb1, Lmo2, and Hdac2 expression in whole cellular extracts from Ssbp2 shRNA-transduced MEL cells treated with 1.5% DMSO for 2 d. The levels of proteins in cells expressing Ssbp2 shRNA relative to those expressing a control shRNA are shown below the autoradiographs. (B) Quantitative RT–PCR analysis of Ssbp2, Ssbp3, P4.2, β-globin, and Rlim mRNAs in differentiating MEL cells transiently transfected with expression plasmid for Ssbp2 shRNA or control shRNA. (C) Western blot analysis of Ssbp3, Ldb1, Lmo2, Ssbp2, Hdac2, and GATA-1 expression in nuclear extracts from short-term puromycin-selected MEL cells transfected with expression plasmid for Ssbp3 shRNA or control shRNA. (D) EMSA of nuclear extracts prepared as above. The E-box-GATA DNA-binding complex is marked with a solid arrowhead. Its abundance was normalized to that of a nonspecific protein–DNA complex (asterisk). The fold change in E-box-GATA DNA-binding activity in cells expressing Ssbp3 shRNA relative to those expressing control shRNA is shown below the autoradiographs. (E) Western blot analysis of Ssbp3, Ssbp2, Ldb1, Lmo2, Hdac2, and GATA-1 abundance in nuclear extracts from short-term puromycin-selected MEL cells expressing shRNAs to both Ssbp2 and Ssbp3 or control (EGFP) shRNA. (F) EMSA of nuclear extracts prepared as above. Protein–DNA complexes are marked as in D. (G) Quantitative RT–PCR analysis of P4.2, β-globin, and Rlim mRNAs in differentiating MEL cells transfected with shRNA expression plasmids for Ssbp2 and Ssbp3 or a control sequence (EGFP).
SSBP2 prevents RLIM-mediated degradation of Ldb1, Lmo2, and LMO4 proteins
Since RLIM was reported to accelerate Ldb1 and Lmo2 destruction (Ostendorff et al. 2002; Hiratani et al. 2003), we investigated whether the SSBPs increased the abundance of these proteins through inhibition of RLIM action. To examine Ldb1 and Lmo2 protein levels simultaneously, their coding sequences were cloned in-frame with sequences encoding the V5 epitope, and V5-tagged Ldb1 and Lmo2 were expressed in CHO cells with RLIM and Flag-tagged SSBP2. The levels of Ldb1 and Lmo2 were then determined by Western blot analysis using an antibody to the V5 epitope tag.
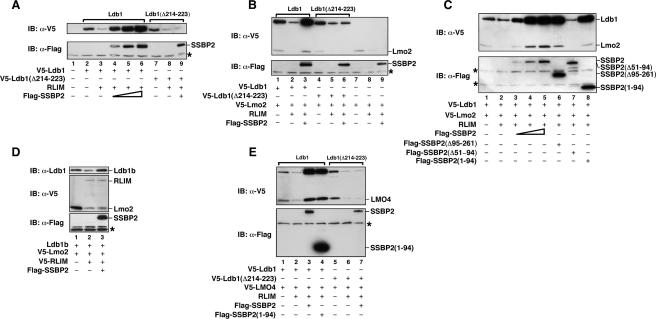
Confirming its reported role in Ldb1 degradation, RLIM expression markedly decreased Ldb1 protein levels (Fig. 6A, top panel, cf. lanes 3 and 2). Enforced expression of SSBP2 effected a concentration-dependent inhibition of RLIM-mediated Ldb1 degradation (Fig. 6A, top panel, cf. lanes 4–6 and 3) and increased Ldb1 expression to a level even greater than what was observed without RLIM transfection (Fig. 6A, top panel, cf. lanes 4–6 and 2), likely reflecting the actions of overexpressed SSBP2 on both transfected and endogenous RLIM (Ostendorff et al. 2002). While RLIM also promoted degradation of Ldb1(Δ214–223) (Fig. 6A, top panel, cf. lanes 8 and 7), SSBP2 did not protect this LCCD mutant (Fig. 6A, top panel, cf. lanes 9 and 8), suggesting that SSBP2 inhibition of RLIM-mediated degradation requires its direct interaction with Ldb1. In addition to Ldb1, SSBP2 also spared Lmo2 from RLIM-mediated destruction (Fig. 6B, top panel, cf. lanes 3 and 2). These actions of SSBP2 were not observed, however, when Ldb1(Δ214–223) was transfected in place of Ldb1 (Fig. 6B, top panel, cf. lanes 6 and 5) or Ldb1 was not transfected at all (Fig. 6B, top panel, cf. lanes 9 and 8). As shown earlier, overexpressed SSBP2 had no effect on the abundance of Hdac2, which is a substrate of RLIM but does not interact with Ldb1 (Fig. 4A,B). These results suggest that SSBP2 inhibition of RLIM-promoted destruction of Ldb1 and Lmo2 requires its physical interaction with Ldb1.
Figure 6.
SSBP2 prevents RLIM-mediated degradation of Ldb1 and Lmo2 proteins. (A) Western blot analysis of CHO cells transfected with indicated combinations of pTracer-EF/V5-Ldb1 (400 ng), pTracer-EF/V5-Ldb1(Δ214–223) (400 ng), pCMV6-XL4-RLIM (1.2 μg), and pEFIRES-SSBP2 (100, 200, or 400 ng). The total mass of DNA was adjusted to 2 μg with plasmid pCMV4 as needed. Whole cellular extracts were prepared 48 h after transfection and assayed for protein expression using a monoclonal antibody to the V5 epitope and polyclonal antibody to the Flag epitope. A cross-reacting protein recognized by the Flag antibody is marked with an asterisk and served as loading control. (B) Western blot analysis of CHO cells transfected with indicated combinations of pTracer-EF/V5-Ldb1 (300 ng), pTracer-EF/V5-Ldb1(Δ214–223) (300 ng), pTracer-EF/V5-Lmo2 (500 ng), pCMV6-XL4-RLIM (800 ng), and pEFIRES-SSBP2 (400 ng). V5- and Flag-tagged proteins were detected as above. (C) Western blot analysis of CHO cells transfected with indicated combinations of pTracer-EF/V5-Ldb1 (300 ng), pTracer-EF/V5-Lmo2 (500 ng), pCMV6-XL4-RLIM (800 ng), pEFIRES-SSBP2 (100, 200, or 400 ng), pEFIRES-SSBP2(Δ95–261) (400 ng), SSBP2(Δ51–94) (400 ng), and SSBP2(1–94) (400 ng). (D) Western blot analysis of CHO cells transfected as above using indicated combinations of pTracer-EF/V5-Ldb1b (300 ng), pTracer-EF/V5-Lmo2 (500 ng), pTracer-EF/V5-RLIM (800 ng), and pEFIRES-SSBP2 (400 ng). (E) Western blot analysis of CHO cells transfected as in C with pTracer-EF/V5-LMO4 instead of pTracer-EF/V5-Lmo2.
Next, the domains in SSBP2 important for its Lmo2 and Ldb1 protein-sparing effects were determined. While wild-type SSBP2 protected Ldb1 and Lmo2 from RLIM-mediated degradation in a concentration-dependent manner (Fig. 6C, top panel, cf. lanes 3–5 and 2), SSBP2(Δ51–94) had no effect on Ldb1 protein abundance (Fig. 6C, top panel, cf. lanes 7 and 2) at any level. In contrast, both SSBP2(Δ95–261) and SSBP2(1–94) protected Ldb1 from degradation in a manner similar to full-length SSBP2 (Fig. 6C, top panel, cf. lanes 6,8 and 2). These results demonstrate that its LUFS, or Ldb1 interaction, domain is necessary and sufficient for SSBP2 inhibition of RLIM-mediated Ldb1 and Lmo2 turnover.
To investigate whether SSBP2-mediated protection of Lmo2 from degradation requires Lmo2–Ldb1 interaction, we took advantage of a naturally occurring Ldb1 splice isoform, Ldb1b, that retains the protein’s dimerization domain and LCCD but lacks its LIM interaction domain (LID) (Tran et al. 2006). Similar to what was observed for wild-type Ldb1 (Fig. 6A–C), enforced RLIM expression decreased the abundance of Ldb1b (Fig. 6D, top panel, cf. lanes 2 and 1), and SSBP2 protected Ldb1b from RLIM-initiated degradation (Fig. 6D, top panel, cf. lanes 3 and 2). In contrast to full-length Ldb1, however, Ldb1b did not cooperate with SSBP2 in protecting Lmo2 from RLIM-promoted degradation (Fig. 6D, middle panel, cf. lanes 3 and 2). Finally, no effect of overexpressed SSBP2 was observed on RLIM protein levels (Fig. 6D, middle panel, cf. lanes 3 and 2), indicating that SSBP2 did not act by reducing RLIM abundance. These studies suggest that direct interaction with Ldb1 is essential for SSBP2 to protect this LIM-only protein from degradation.
The four LMO proteins, which are each important developmental regulators (de la Calle-Mustienes et al. 2003; Gering et al. 2003; Lee et al. 2005), share considerable sequence homology and the ability to interact with Ldb1, prompting us to investigate whether SSBP2 regulated the abundance of any LIM-only protein besides LMO2, and LMO4, the most divergent member of this protein family, was selected for analysis. Similar to the results with Lmo2 (Fig. 6B,C), RLIM expression decreased (Fig. 6E, top panel, cf. lanes 2 and 1) while enforced expression of SSBP2 or SSBP2(1–94) dramatically increased LMO4 protein levels (Fig. 6E, top panel, cf. lanes 3,4 and 1). Furthermore, this effect of SSBP2 was not observed with the Ldb1 LCCD deletion mutant (Fig. 6E, top panel, cf. lanes 7 and 6). These studies indicate that SSBP2 is capable of protecting multiple LMO proteins from RLIM-initiated degradation.
SSBP2 inhibits RLIM-mediated ubiquitination of Ldb1 by preventing RLIM interaction with Ldb1
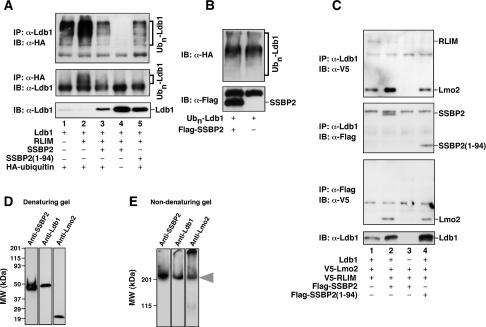
The E3 ubiquitin ligase RLIM catalyzes the ubiquitination of its substrates, targeting them for destruction in the proteasome (Ostendorff et al. 2002; Hiratani et al. 2003). We therefore examined whether SSBP2 inhibited RLIM-promoted ubiquitination of Ldb1. To that end, an expression plasmid for Ldb1 was transfected into CHO cells with plasmids encoding RLIM, SSBP2, and/or HA-tagged ubiquitin, and total cellular extracts of transfected cells were subjected to coimmunoprecipitation analysis. While ubiquitination of Ldb1 was detected in CHO cells not transfected with RLIM (Fig. 7A, top and middle panels, lane 1), likely reflecting endogenous ubiquitin ligase activity, ubiquitination was markedly increased in RLIM-transfected cells (Fig. 7A, top and middle panels, cf. lanes 2 and 1). In turn, expression of either SSBP2 or SSBP2(1–94) reduced RLIM-initiated Ldb1 ubiquitination to a level even lower than in the absence of transfected RLIM (Fig. 7A, top panel, cf. lanes 3,5 and 1), with inhibition of Ldb1 ubiquitination by SSBP2 correlating with higher steady-state levels of Ldb1 protein (Fig. 7A, bottom panel, cf. lanes 3,5 and 2). In control experiments, Ldb1 ubiquitination was not detected when HA-tagged ubiquitin was not transfected (Fig. 7A, top and middle panels, lane 4). These results establish that SSBP2 inhibits RLIM-mediated Ldb1 ubiquitination.
Figure 7.
SSBP2 inhibits RLIM-mediated Ldb1 ubiquitination and RLIM interaction with Ldb1. (A) Western blot analysis of CHO cells transfected with indicated combinations of pEFIRES-Ldb1 (400 ng), pCMV6-XL4-RLIM (1.0 μg), pEFIRES-SSBP2 (300 ng), pEFIRES-SSBP2(1–94) (300 ng), and CMV-HA-ubiquitin (300 ng). Whole cellular extracts prepared from MG132-treated transfectants were subjected to coimmunoprecipitation analysis as indicated. (B) Lack of deubiquitinating activity of SSBP2. Equivalent amounts of ubiquitinated Ldb1 immobilized on beads were incubated with or without purified Flag-SSBP2 protein. Ubiquitinated Ldb1 and Flag-SSBP2 retained on beads were then quantified by Western blot analysis. (C) Western blot analysis of CHO cells transfected as above with indicated combinations of pEFIRES-Ldb1 (500 ng), pTracer-EF/V5-Lmo2 (500 ng), pTracer-EF/V5-RLIM (500 ng), pEFIRES-SSBP2 (500 ng), and pEFIRES-SSBP2(1–94) (500 ng). Whole cellular extracts from MG132-treated transfectants were subjected to coimmunoprecipitation analysis as indicated. Western blot analysis of SSBP2, Ldb1, and Lmo2 in purified SSBP2 protein complexes was carried out following denaturing (D) and nondenaturing (E) polyacrylamide gel electrophoresis. CHO cells were transfected with expression plasmids for Ldb1, Lmo2, and Flag-tagged SSBP2, and Western blot analysis of SSBP2-associated proteins was carried out on purified complexes following elution with Flag peptide.
To exclude the possibility of SSBP2 having deubiquitinating activity (Amerik and Hochstrasser 2004), which could explain both the protein stability and ubiquitination results, purified SSBP2 protein was incubated with ubiquitinated Ldb1 under conditions optimal for deubiquitinating enzyme activity (Li et al. 2002). Although SSBP2 physically associated with ubiquitinated Ldb1 (Fig. 7B, bottom panel), it did not alter the extent of Ldb1 ubiquitination (Fig. 7B, top panel), indicating that SSBP2 lacks deubiquitinase function.
A member of the RING family of E3 ubiquitin ligases, RLIM must interact with its substrates to effect the transfer of ubiquitin from an E2 ubiquitin-conjugating enzyme (Pickart 2001). The affinity of Ldb1 for RLIM was therefore investigated in the presence and absence of SSBP2. Without transfected SSBP2, both RLIM and Lmo2 were immunoprecipitated by an antibody to Ldb1 (Fig. 7C, top panel, lane 1), indicating that an interaction existed between RLIM and Ldb1 in these cells. Enforced expression of SSBP2 or SSBP2(1–94) markedly decreased Ldb1’s association with RLIM without affecting its interaction with Lmo2 (Fig. 7C, top panel, cf. lanes 2,4 and 1). However, neither RLIM nor Lmo2 was immunoprecipitated by an antibody to Ldb1 when Ldb1 was not also transfected (Fig. 7C, top panel, lane 3), indicating the specificity of this coimmunoprecipitation analysis. Confirming published information, its LUFS domain was sufficient for SSBP2 interaction with Ldb1 (Fig. 7C, second panel, lanes 2,4).
Although these studies are consistent with SSBP2 inhibiting RLIM–Ldb1 interaction through direct binding to Ldb1, they would also be compatible with SSBP2 binding first to RLIM and preventing its interaction with Ldb1. To distinguish between these possibilities, antibody to the Flag epitope was used in coimmunoprecipitation analysis of SSBP2 and associated proteins. RLIM was not immunoprecipitated by Flag antibody in either the presence or absence of Ldb1 (Fig. 7C, the third panel from the top, lanes 2,4), demonstrating that SSBP2 did not act through direct interaction with RLIM. In contrast, Lmo2 was associated with SSBP2 and SSBP2(1–94) in the presence of Ldb1 (Fig. 7C, third panel, lanes 2,4) but not its absence (Fig. 7C, third panel, lane 3), supporting the notion that Ldb1 contacts SSBP2 and Lmo2 simultaneously through its separate SSBP- and LIM-interacting domains. The exclusion of RLIM from this trimolecular complex suggests, furthermore, that while capable of interacting with Ldb1 and Lmo2 in the absence of SSBP2, RLIM is not able to contact these proteins when they are complexed with SSBP2. These results suggest that SSBP2 binding prevents Ldb1 and LMO2 from encountering RLIM and in this way inhibits their ubiquitination.
To gain further evidence that SSBP2, Ldb1, and Lmo2 contribute to a common complex, an expression vector encoding Flag-tagged SSBP2 was transfected into CHO cells with plasmids for Ldb1 and Lmo2. SSBP2 and associated proteins were immunoprecipitated from transfected cells using antibody to the Flag epitope and then eluted from the agarose beads with Flag peptide. Western blot analysis of these purified SSBP2 protein complexes showed that SSBP2, Ldb1, and Lmo2 were each present (Fig. 7D), with SSBP2, Ldb1, and Lmo2 comigrating in a single high-molecular-weight complex when gels were run under nondenaturing conditions (Fig. 7E). These results demonstrate that SSBP2 forms a stable trimolecular complex with Ldb1 and Lmo2 in cells.
Discussion
Although the importance of SSBPs in development is well established (Chen et al. 2002; van Meyel et al. 2003; Nishioka et al. 2005; Enkhmandakh et al. 2006), the specific genes they regulate have not been determined. We show here that both Ssbp2 and Ssbp3 contribute to a multiprotein complex binding the proximal promoter of the P4.2 gene and that Ssbp2 augments P4.2 transcription and occupies this promoter with previously identified components of the complex. Given the direct interaction of SSBPs with Ldb1 (Chen et al. 2002; van Meyel et al. 2003) and the requirement of Ldb1 for multiple functions of LIM-HD, LMO, and even non-LIM proteins (Agulnick et al. 1996; Morcillo et al. 1997; van Meyel et al. 2000; Weihe et al. 2001; Nishioka et al. 2005), the SSBPs likely regulate a broader range of developmental processes than presently appreciated (Chen et al. 2002; van Meyel et al. 2003; Nishioka et al. 2005; Enkhmandakh et al. 2006).
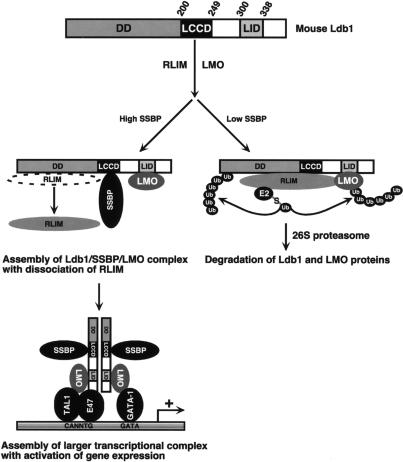
Genetic experiments have emphasized the Ldb1 dependence of SSBP action, particularly in developmental processes (Chen et al. 2002; van Meyel et al. 2003; Nishioka et al. 2005). Using the P4.2 E-box-GATA DNA-binding complex as a model, we confirmed the importance of this interaction, and the involvement of the LCCD and LUFS domain of Ldb1 and SSBP2, for Ldb1’s contribution to SSBP2 function. Given the evolutionary conservation of these two protein interaction domains (Chen et al. 2002; van Meyel et al. 2003) and the ability of every SSBP examined to interact with Ldb1 (Chen et al. 2002; van Meyel et al. 2003), all members of this protein family are likely able to protect all LDB isoforms from degradation. In support is the reduction in Ldb1 protein abundance observed in erythroid cells in which Ssbp2 and Ssbp3 expression was knocked down by RNA interference and in a mouse pituitary cell line in which Ssbp3 was similarly depleted (Y. Cai, Z. Xu, and S.J. Brandt, unpubl.). In addition to reducing Ldb1 turnover, SSBP2 also protected Lmo2 from RLIM-promoted degradation. While SSBP2 does not interact with LIM-only proteins directly, it was found to contribute to a complex containing both Lmo2 and Ldb1. That RLIM was excluded from this complex suggests that SSBP2 prevents this ubiquitin ligase from encountering its substrates. SSBP binding may mask Ldb1’s RLIM interaction domain or elicit a conformational change that achieves the same result, with SSBP2-bound Ldb1 likely acting similarly for LMO2. Such a mechanism (Fig. 8) is highly reminiscent of how TAF(II)31 enhances the stability of p53 (Buschmann et al. 2001).
Figure 8.
Model of SSBP regulation of Ldb1 and LMO protein stability and transcriptional complex assembly. Top shows schematic of mouse Ldb1 with dimerization domain (DD), Ldb1/Chip conserved domain (LCCD), and LIM interaction domain (LID) marked. The model predicts that when at sufficient levels, SSBPs interact with Ldb1 to promote formation of an SSBP/Ldb1/LMO trimolecular complex, with Ldb1 contacting SSBP and Lmo2 simultaneously through its separate LCCD and LID, respectively. This prevents their interaction with the ubiquitin ligase RLIM, RLIM-initiated ubiquitination, and proteasomal degradation. The SSBP/Ldb1/LMO complex could also help nucleate assembly of larger transcriptional complexes, including the E-box-GATA DNA-binding complex shown. When SSBPs are present at insufficient levels to interact with Ldb1, RLIM promotes the ubiquitination of Ldb1 and LIM-only proteins, targeting them for destruction in the 26S proteasome.
The LIM interaction domain of Ldb1, like its LCCD, has been highly conserved in evolution. This region mediates Ldb1 interaction with both LIM-HD and LIM-only proteins (Jurata et al. 1996), and the effect of SSBPs on LMO protein turnover probably extends to many if not most LIM proteins capable of binding Ldb1. If correct, this would explain the requirement of Drosophila Ldb1 (Chip) in both dLMO (Milán and Cohen 2000) and Apterous (Weihe et al. 2001) protein expression in the developing wing and the dose-sparing effect of Drosophila Ssdp and mouse Ssbp3 on enhancement of axis induction by Ldb1 and Xlim-1 in Xenopus embryos (Chen et al. 2002). Our studies predict, furthermore, that at comparable expression levels, dLMO would be stabilized by dSsdp to a greater extent than Apterous as a result of its greater affinity for Chip (Weihe et al. 2001).
Although SSBP2 might enhance the transactivating function of the E-box-GATA DNA-binding complex (Fig. 1D; Nishioka et al. 2005; Wu 2006), it also appeared to promote complex assembly (Fig. 3B,F). Ssbp2 and Ssbp3 concentrations may, in fact, be limiting for complex formation, a conclusion supported by the increase in E-box-GATA DNA-binding activity with SSBP2 overexpression and decrease with Ssbp3 depletion. While the increased abundance of Ldb1 and Lmo2 could itself increase DNA-binding activity, the SSBP/Ldb1/Lmo2 trimolecular complex may also help nucleate or stabilize the larger DNA-binding complex.
The results of supershift analysis showing that antibody to Ssbp2 largely and to Ssbp3 completely shifted the E-box-GATA-binding complexes suggest that both Ssbp2 and Ssbp3 are present in the majority of complexes. If the erythroid E-box-GATA DNA-binding complex contains an Ldb1 homodimer as proposed by Rabbitts and colleagues (Wadman et al. 1997), each Ldb1 molecule in this dimer could simultaneously contact one molecule of Ssbp2 or Ssbp3 and one molecule of Lmo2 through its separate LCCD and LIM interaction domain (Fig. 8). This predicts that the relative concentrations of the SSBP orthologs would dictate their incorporation. In support, proteomic and/or antibody supershift analyses of Tal1- and Ldb1-containing complexes in MEL cells, which express comparable levels of Ssbp2 and Ssbp3, identified both SSBPs (Goardon et al. 2006; Meier et al. 2006), whereas only Ssbp3 was detected in an Lhx2- and Ldb1-containing DNA-binding complex from a pituitary cell line that expresses considerably more Ssbp3 than Ssbp2 (Y. Cai, Z. Xu, and S.J. Brandt, unpubl.). Finally, while the N-terminal LUFS domains of the SSBPs are virtually identical, their internal amino acid sequences, including a potential activation domain in Ssbp3 (Wu 2006), diverge. It is therefore possible that certain SSBPs possess functions not shared with other family members, a notion supported by the finding that SSBP2, but not SSBP3, interacts with the adenoviral oncoprotein E1B55K (Fleisig et al. 2007).
From the results of both overexpression and knockdown studies, we propose that Ldb1 levels are regulated in cells by a dynamic equilibrium between the SSBPs acting to protect Ldb1 from ubiquitination and specific E3 ubiquitin ligases, including RLIM (Ostendorff et al. 2002; Hiratani et al. 2003) and possibly RNF6 (Hiratani et al. 2003) promoting Ldb1 ubiquitination. The reduction in Ldb1 levels in cells in which Ssbp2 and Ssbp3 were knocked down (see above) or the Ssbp2 gene had been knocked out (Y. Yang and L. Nagarajan, unpubl.) and the severe disruptions in head development in mice with reduced Ssbp3 expression owing to a transgene insertion (Nishioka et al. 2005) provide support for this notion and illustrate the consequences of unopposed LIM protein and Ldb1 ubiquitination.
Although our studies focused on cellular differentiation, several lines of evidence suggest they could also be relevant to tumorigenesis. First, misexpression of LMO genes is observed in multiple tumor types, including T-cell acute lymphoblastic leukemia, neuroblastoma, and carcinoma of the head and neck and breast (Valge-Archer et al. 1998; Visvader et al. 2001; Ferrando et al. 2002; Hacein-Bey-Abina et al. 2003; Mizunuma et al. 2003; Aoyama et al. 2005), and overexpression of LMO proteins in transgenic models has been shown to cause or contribute to tumor development (Larson et al. 1994; Chervinsky et al. 1999; Aoyama et al. 2005; Sum et al. 2005). Importantly, LMO4 and LDB1 protein levels are concordantly up-regulated in human oral cavity carcinomas (Mizunuma et al. 2003), and high-level expression of one or more SSBP genes was detected in a majority of samples in a small series of carcinomas of the head and neck (C. Chung and S.J. Brandt, unpubl.). In addition, clonal retroviral insertions predicted to increase transcription of these genes have been noted in all three Ssbp loci in lymphoid leukemias in certain inbred mouse strains (Mouse Retrovirus Tagged Cancer Gene Database; http://rtcgd.ncifcrf.gov). However, SSBP2 expression was also found to be decreased in human myeloid leukemias (Qian et al. 2002; Liang et al. 2005). We hypothesize that altered expression of SSBPs, either increased or decreased, would contribute to tumorigenesis by disrupting the normal stoichiometry of important LDB–LIM protein-containing complexes.
Materials and methods
Plasmid constructs
The P4.2 promoter-luciferase reporter plasmid pGL2-P4.2p1700-Luc and expression vectors pcDNA3.1-Tal1, pcDNA3.1-E47, pcDNA3.1-GATA-1, pEFIRES-Ldb1, pEFIRES-Lmo2, pFlag-CMV5-SSBP2, and CMV-HA-ubiquitin have been described (Xu et al. 2003; Liang et al. 2005). A cDNA for the Ldb1 splice isoform Ldb1b lacking the LIM interaction domain (Tran et al. 2006) was subcloned into pEFIRES-P. cDNAs encoding Flag-tagged human SSBP2 and its 94 N-terminal amino acids were amplified by PCR from pFlag-CMV5-SSBP2 and also subcloned into pEFIRES-P. pEFIRES-SSBP2(Δ51–94), pEFIRES-SSBP2(Δ95–261), and pEFIRES-Ldb1(Δ214–223) were constructed using PCR as described (Imai et al. 1991). Plasmid CMV6-XL4-RLIM was purchased from Origene. The full-length human LMO4 coding sequence was amplified using RNA from MCF-7 cells. cDNAs encoding RLIM, Ldb1, Ldb1(Δ214–223), and Lmo2 were obtained using PCR and subcloned into pTracer-EF/V5-His (Invitrogen). The correct sequences of their inserts and sizes of the proteins expressed from these plasmids were both verified. The small interfering RNA (siRNA) target sequences, shown in the Supplemental Material, were selected using siRNA Target Finder (http://www.ambion.com). Double-stranded oligonucleotides comprised of 19-base-pair (bp) sense and antisense sequences linked by a 9-bp loop were cloned into pSilencer 4.1-CMV-puro (Ambion) to construct the shRNA expression vectors pSilencer-Ssbp2-A, pSilencer-Ssbp2-B, and pSilencer-Ssbp2-D; pSilencer-Ssbp3-C; and pSilencer-EGFP. The pSilencer-CMV-puro-Control vector was also from Ambion.
Cell culture and generation of short-term transductants
MEL and COS-7L cells were handled as described (Xu et al. 2003). CHO cells were transfected using the FuGENE 6 reagent (Roche). For transient knockdown experiments, MEL cells were transfected with shRNA expression vectors targeted to Ssbp2, Ssbp3, EGFP, or a control sequence, transferred 15 h later to medium containing 6 μg/mL puromycin and 1.8% DMSO, and cultured for an additional 40 h. Viable cells enriched with Lymphoprep reagent (Axis-Shield) were used in the preparation of nuclear extracts or incubated in medium containing 1.8% DMSO for an additional 48 h before total cellular RNA was prepared.
Generation of stable transductants
pEFIRES-SSBP2, pEFIRES-SSBP2(Δ95–261), or parental vector was introduced into MEL cells, and transductants were selected with puromycin as described (Xu et al. 2003). Polyclonal populations of SSBP2(Δ95–261)- and vector-transduced MEL cells were used. Ten oligoclonal populations were derived for SSBP2-transduced MEL cells, and three lines—O3, O4, and O6—were used in the studies described. pSilencer-CMV-puro-Control or the combination of pSilencer-Ssbp2-A and pSilencer-Ssbp2-B was similarly handled. For Ssbp2 shRNA-transduced cells, nine monoclonal populations were analyzed for Ssbp2 abundance, and two lines were selected.
EMSA
Nuclear extracts from MEL and FVA cells were prepared, and EMSA of DNA-binding activity was carried out as described (Xu et al. 2003). Rabbit immunoglobulin and antibodies to Ldb1, GATA-1, and SSBP2 have also been described (Xu et al. 2003; Liang et al. 2005). Polyclonal antibody to the Flag epitope (F7425) was purchased from Sigma. Details on the antibody to SSBP3 will be published elsewhere.
Western blot and immunoprecipitation analyses
Western blot analysis was carried out as described (Xu et al. 2003). For immunoprecipitation, cells were extracted on ice in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM NaVO4, 0.5% NP-40, and 10% glycerol. Immunoprecipitated proteins were captured by Protein G-agarose and washed three times with 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 0.1% NP-40, and specific proteins were identified by Western blot analysis. In addition to the antibodies above, affinity-purified rabbit polyclonal antibody to Tal1 was prepared as described (Kallianpur et al. 1994). Rabbit antibodies to HDAC2 (sc-7899) and actin (sc-1615) were from Santa Cruz Biotechnology, mouse monoclonal antibody to the V5 epitope (catalog no. 46-0705) was from Invitrogen, sheep polyclonal antibody to Lmo2 (ab16132) was from Abcam, and monoclonal antibody to the HA epitope (MMS-101P) was from Covane.
Quantitative RT–PCR analysis
Total cellular RNA was prepared using RNeasy Midi reagents (Qiagen), and residual genomic DNA was removed by DNase I treatment (Ambion). One microgram of this RNA was then converted to cDNA with the iScript cDNA Synthesis product (Bio-Rad) and analyzed by real-time PCR using the iQ SYBR Green Supermix (Bio-Rad). Relative quantification of cDNAs used a standard curve generated by serial dilution of cDNAs, and levels of P4.2, β-globin, RLIM, and Ldb1 mRNAs were normalized to that of ribosomal S16. The sequences of the primers used in PCR analysis are given in the Supplemental Material.
ChIP analysis
ChIP analysis was performed with an EZ ChIP kit (Upstate Biotechnology), and immunoprecipitated DNA was analyzed by real-time PCR using the iQ SYBR Green Supermix (Bio-Rad). Levels of factor occupancy were determined by subtracting the values obtained with normal IgG from those with the indicated antibodies. The sequences of the primers used in the PCR analysis are given in the Supplemental Material.
Protein turnover analysis
Transduced MEL cells were treated with 100 μM CHX for the indicated times, and whole-cell extracts were subjected to Western blot analysis.
In vivo ubiquitination assay
CHO cells were transfected with the indicated combinations of expression plasmids for Ldb1, RLIM, SSBP2, and HA-ubiquitin. Beginning 43 h after transfection, the proteasomal inhibitor MG132 was added to the medium (25 μM), and the cells were incubated for an additional 5 h. Whole-cell extracts were then subjected to immunoprecipitation with antibodies to Ldb1 or HA epitope, and specific proteins were identified by Western blot analysis.
Deubiquitination assay
Flag-tagged SSBP2 protein was purified from SSBP2-transfected CHO cells using the FLAG M purification kit (Sigma). Ubiquitinated Ldb1 was immunoprecipitated from CHO cells that had been transfected with Ldb1, RLIM, and HA-ubiquitin and treated with MG132. Protein G beads loaded with ubiquitinated Ldb1 were washed three times with 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, and 0.1% NP-40, and twice with deubiquitination buffer (50 mM Tris-HCl at pH 8.0, 50 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol). Beads were incubated for 2 h at 37°C in deubiquitination buffer containing 1000 ng of purified Flag-SSBP2 protein or the same concentration of 3× Flag peptide as described (Li et al. 2002). After extensive washing, captured proteins were quantified by Western blot analysis with antibodies to the HA and Flag epitopes.
Autoradiographic analysis
Band intensities on photographic film were quantified with NIH Image software (version 1.62).
Acknowledgments
We thank Dr. Alan Agulnick for Ldb1 antibody, Dr. Long-Sheng Chang for the P4.2 promoter-reporter construct, Dr. Hannah Cheung for assistance in making the SSBP2(Δ95-261) construct, Dr. Mark Koury for FVA cells, and Dr. Akira Kato for the mouse Ldb1b cDNA. This work was supported in part by National Institutes of Health grants R01 HL49118 (to S.J.B.) and HL074449 (to L.N.), a Merit Review Award from the Department of Veterans Affairs (to S.J.B.), and an American Society of Hematology Fellow Scholar Award (to Z.X.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1528507
References
- Agulnick A.D., Taira M., Breen J.J., Tanaka T., Dawid I.B., Westphal H., Taira M., Breen J.J., Tanaka T., Dawid I.B., Westphal H., Breen J.J., Tanaka T., Dawid I.B., Westphal H., Tanaka T., Dawid I.B., Westphal H., Dawid I.B., Westphal H., Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature. 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- Amerik A.Y., Hochstrasser M., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Aoyama M., Ozaki T., Inuzuka H., Tomotsune D., Hirato J., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Ozaki T., Inuzuka H., Tomotsune D., Hirato J., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Inuzuka H., Tomotsune D., Hirato J., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Tomotsune D., Hirato J., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Hirato J., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Tokita H., Ohira M., Nakagawara A., Ohira M., Nakagawara A., Nakagawara A. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005;65:4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- Bach I., Carriere C., Ostendorff H.P., Andersen B., Rosenfeld M.G., Carriere C., Ostendorff H.P., Andersen B., Rosenfeld M.G., Ostendorff H.P., Andersen B., Rosenfeld M.G., Andersen B., Rosenfeld M.G., Rosenfeld M.G. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes & Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- Bayarsaihan D., Lukens L.N., Lukens L.N. Single-strand-DNA-binding factors specifically recognize the pyrimidine element in the chick α2(I) collagen gene promoter. Biochem. J. 1996;314:293–296. doi: 10.1042/bj3140293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann T., Lin Y., Aithmitti N., Fuchs S.Y., Lu H., Resnick-Silverman L., Manfredi J.J., Ronai Z., Wu X., Lin Y., Aithmitti N., Fuchs S.Y., Lu H., Resnick-Silverman L., Manfredi J.J., Ronai Z., Wu X., Aithmitti N., Fuchs S.Y., Lu H., Resnick-Silverman L., Manfredi J.J., Ronai Z., Wu X., Fuchs S.Y., Lu H., Resnick-Silverman L., Manfredi J.J., Ronai Z., Wu X., Lu H., Resnick-Silverman L., Manfredi J.J., Ronai Z., Wu X., Resnick-Silverman L., Manfredi J.J., Ronai Z., Wu X., Manfredi J.J., Ronai Z., Wu X., Ronai Z., Wu X., Wu X. Stabilization and activation of p53 by the coactivator protein TAFII31. J. Biol. Chem. 2001;276:13852–13857. doi: 10.1074/jbc.M007955200. [DOI] [PubMed] [Google Scholar]
- Cassata G., Rohrig S., Kuhn F., Hauri H.P., Baumeister R., Burglin T.R., Rohrig S., Kuhn F., Hauri H.P., Baumeister R., Burglin T.R., Kuhn F., Hauri H.P., Baumeister R., Burglin T.R., Hauri H.P., Baumeister R., Burglin T.R., Baumeister R., Burglin T.R., Burglin T.R. The Caenorhabditis elegans Ldb/NLI/Clim orthologue ldb-1 is required for neuronal function. Dev. Biol. 2000;226:45–56. doi: 10.1006/dbio.2000.9846. [DOI] [PubMed] [Google Scholar]
- Castro P., Liang H., Liang J.C., Nagarajan L., Liang H., Liang J.C., Nagarajan L., Liang J.C., Nagarajan L., Nagarajan L. A novel, evolutionarily conserved gene family with putative sequence-specific single-stranded DNA-binding activity. Genomics. 2002;80:78–85. doi: 10.1006/geno.2002.6805. [DOI] [PubMed] [Google Scholar]
- Chen L., Segal D., Hukriede N.A., Podtelejnikov A.V., Bayarsaihan D., Kennison J.A., Ogryzko V.V., Dawid I.B., Westphal H., Segal D., Hukriede N.A., Podtelejnikov A.V., Bayarsaihan D., Kennison J.A., Ogryzko V.V., Dawid I.B., Westphal H., Hukriede N.A., Podtelejnikov A.V., Bayarsaihan D., Kennison J.A., Ogryzko V.V., Dawid I.B., Westphal H., Podtelejnikov A.V., Bayarsaihan D., Kennison J.A., Ogryzko V.V., Dawid I.B., Westphal H., Bayarsaihan D., Kennison J.A., Ogryzko V.V., Dawid I.B., Westphal H., Kennison J.A., Ogryzko V.V., Dawid I.B., Westphal H., Ogryzko V.V., Dawid I.B., Westphal H., Dawid I.B., Westphal H., Westphal H. Ssdp proteins interact with the LIM-domain-binding protein Ldb1 to regulate development. Proc. Natl. Acad. Sci. 2002;99:14320–14325. doi: 10.1073/pnas.212532399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervinsky D.S., Zhao X.F., Lam D.H., Ellsworth M., Gross K.W., Aplan P.D., Zhao X.F., Lam D.H., Ellsworth M., Gross K.W., Aplan P.D., Lam D.H., Ellsworth M., Gross K.W., Aplan P.D., Ellsworth M., Gross K.W., Aplan P.D., Gross K.W., Aplan P.D., Aplan P.D. Disordered T-cell development and T-cell malignancies in SCL LMO1 double-transgenic mice: Parallels with E2A-deficient mice. Mol. Cell. Biol. 1999;19:5025–5035. doi: 10.1128/mcb.19.7.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Calle-Mustienes E., Lu Z., Cortés M., Andersen B., Modolell J., Gómez-Skarmeta J.L., Lu Z., Cortés M., Andersen B., Modolell J., Gómez-Skarmeta J.L., Cortés M., Andersen B., Modolell J., Gómez-Skarmeta J.L., Andersen B., Modolell J., Gómez-Skarmeta J.L., Modolell J., Gómez-Skarmeta J.L., Gómez-Skarmeta J.L. Xenopus Xlmo4 is a GATA cofactor during ventral mesoderm formation and regulates Ldb1 availability at the dorsal mesoderm and the neural plate. Dev. Biol. 2003;264:564–581. doi: 10.1016/j.ydbio.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Enkhmandakh B., Makeyev A.V., Bayarsaihan D., Makeyev A.V., Bayarsaihan D., Bayarsaihan D. The role of the proline-rich domain of Ssdp1 in the modular architecture of the vertebrate head organizer. Proc. Natl. Acad. Sci. 2006;103:11631–11636. doi: 10.1073/pnas.0605209103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Fúnez P., Lu C.H., Rincón-Limas D.E., Garcia-Bellido A., Botas J., Lu C.H., Rincón-Limas D.E., Garcia-Bellido A., Botas J., Rincón-Limas D.E., Garcia-Bellido A., Botas J., Garcia-Bellido A., Botas J., Botas J. The relative expression amounts of apterous and its co-factor dLdb/Chip are critical for dorso-ventral compartmentalization in the Drosophila wing. EMBO J. 1998;17:6846–6853. doi: 10.1093/emboj/17.23.6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando A.A., Neuberg D.S., Staunton J., Loh M.L., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., Neuberg D.S., Staunton J., Loh M.L., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., Staunton J., Loh M.L., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., Loh M.L., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., Pui C.H., Downing J.R., Gilliland D.G., Downing J.R., Gilliland D.G., Gilliland D.G., et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- Fleisig H.B., Orazio N.I., Liang H., Tyler A.F., Adams H.P., Weitzman M.D., Nagarajan L., Orazio N.I., Liang H., Tyler A.F., Adams H.P., Weitzman M.D., Nagarajan L., Liang H., Tyler A.F., Adams H.P., Weitzman M.D., Nagarajan L., Tyler A.F., Adams H.P., Weitzman M.D., Nagarajan L., Adams H.P., Weitzman M.D., Nagarajan L., Weitzman M.D., Nagarajan L., Nagarajan L. Oncogene. 2007. Adenoviral E1B55K oncoprotein sequesters candidate leukemia suppressor sequence-specific single-stranded DNA-binding protein 2 into aggresomes. [Epub February 19, 2007; DOI: 10.1038/sj.onc.1210281] [DOI] [PubMed] [Google Scholar]
- Gering M., Yamada Y., Rabbitts T.H., Patient R.K., Yamada Y., Rabbitts T.H., Patient R.K., Rabbitts T.H., Patient R.K., Patient R.K. Lmo2 and Scl/Tal1 convert non-axial mesoderm into haemangioblasts which differentiate into endothelial cells in the absence of Gata1. Development. 2003;130:6187–6199. doi: 10.1242/dev.00875. [DOI] [PubMed] [Google Scholar]
- Goardon N., Lambert J.A., Rodriguez P., Nissaire P., Herblot S., Thibault P., Dumenil D., Strouboulis J., Romeo P.H., Hoang T., Lambert J.A., Rodriguez P., Nissaire P., Herblot S., Thibault P., Dumenil D., Strouboulis J., Romeo P.H., Hoang T., Rodriguez P., Nissaire P., Herblot S., Thibault P., Dumenil D., Strouboulis J., Romeo P.H., Hoang T., Nissaire P., Herblot S., Thibault P., Dumenil D., Strouboulis J., Romeo P.H., Hoang T., Herblot S., Thibault P., Dumenil D., Strouboulis J., Romeo P.H., Hoang T., Thibault P., Dumenil D., Strouboulis J., Romeo P.H., Hoang T., Dumenil D., Strouboulis J., Romeo P.H., Hoang T., Strouboulis J., Romeo P.H., Hoang T., Romeo P.H., Hoang T., Hoang T. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25:357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E., Lim A., Osborne C.S., Pawliuk R., Morillon E., Osborne C.S., Pawliuk R., Morillon E., Pawliuk R., Morillon E., Morillon E., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Heitzler P., Vanolst L., Biryukova I., Ramain P., Vanolst L., Biryukova I., Ramain P., Biryukova I., Ramain P., Ramain P. Enhancer-promoter communication mediated by Chip during Pannier-driven proneural patterning is regulated by Osa. Genes & Dev. 2003;17:591–596. doi: 10.1101/gad.255703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I., Yamamoto N., Mochizuki T., Ohmori S.Y., Taira M., Yamamoto N., Mochizuki T., Ohmori S.Y., Taira M., Mochizuki T., Ohmori S.Y., Taira M., Ohmori S.Y., Taira M., Taira M. Selective degradation of excess Ldb1 by Rnf12/RLIM confers proper Ldb1 expression levels and Xlim-1/Ldb1 stoichiometry in Xenopus organizer functions. Development. 2003;130:4161–4175. doi: 10.1242/dev.00621. [DOI] [PubMed] [Google Scholar]
- Imai Y., Matsushima Y., Sugimura T., Terada M., Matsushima Y., Sugimura T., Terada M., Sugimura T., Terada M., Terada M. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 1991;19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata L.W., Gill G.N., Gill G.N. Functional analysis of the nuclear LIM domain interactor NLI. Mol. Cell. Biol. 1997;17:5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata L.W., Kenny D.A., Gill G.N., Kenny D.A., Gill G.N., Gill G.N. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc. Natl. Acad. Sci. 1996;93:11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallianpur A.R., Jordan J.E., Brandt S.J., Jordan J.E., Brandt S.J., Brandt S.J. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood. 1994;83:1200–1208. [PubMed] [Google Scholar]
- Krämer O.H., Zhu P., Ostendorff H.P., Golebiewski M., Tiefenbach J., Peters M.A., Brill B., Groner B., Bach I., Heinzel T., Zhu P., Ostendorff H.P., Golebiewski M., Tiefenbach J., Peters M.A., Brill B., Groner B., Bach I., Heinzel T., Ostendorff H.P., Golebiewski M., Tiefenbach J., Peters M.A., Brill B., Groner B., Bach I., Heinzel T., Golebiewski M., Tiefenbach J., Peters M.A., Brill B., Groner B., Bach I., Heinzel T., Tiefenbach J., Peters M.A., Brill B., Groner B., Bach I., Heinzel T., Peters M.A., Brill B., Groner B., Bach I., Heinzel T., Brill B., Groner B., Bach I., Heinzel T., Groner B., Bach I., Heinzel T., Bach I., Heinzel T., Heinzel T., et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson R.C., Fisch P., Larson T.A., Lavenir I., Langford T., King G., Rabbitts T.H., Fisch P., Larson T.A., Lavenir I., Langford T., King G., Rabbitts T.H., Larson T.A., Lavenir I., Langford T., King G., Rabbitts T.H., Lavenir I., Langford T., King G., Rabbitts T.H., Langford T., King G., Rabbitts T.H., King G., Rabbitts T.H., Rabbitts T.H. T cell tumours of disparate phenotype in mice transgenic for Rbtn-2. Oncogene. 1994;9:3675–3681. [PubMed] [Google Scholar]
- Lee S.K., Jurata L.W., Nowak R., Lettieri K., Kenny D.A., Pfaff S.L., Gill G.N., Jurata L.W., Nowak R., Lettieri K., Kenny D.A., Pfaff S.L., Gill G.N., Nowak R., Lettieri K., Kenny D.A., Pfaff S.L., Gill G.N., Lettieri K., Kenny D.A., Pfaff S.L., Gill G.N., Kenny D.A., Pfaff S.L., Gill G.N., Pfaff S.L., Gill G.N., Gill G.N. The LIM domain-only protein LMO4 is required for neural tube closure. Mol. Cell. Neurosci. 2005;28:205–214. doi: 10.1016/j.mcn.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Li M., Chen D., Shiloh A., Luo J., Nikolaev A.Y., Qin J., Gu W., Chen D., Shiloh A., Luo J., Nikolaev A.Y., Qin J., Gu W., Shiloh A., Luo J., Nikolaev A.Y., Qin J., Gu W., Luo J., Nikolaev A.Y., Qin J., Gu W., Nikolaev A.Y., Qin J., Gu W., Qin J., Gu W., Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Liang H., Samanta S., Nagarajan L., Samanta S., Nagarajan L., Nagarajan L. SSBP2, a candidate tumor suppressor gene, induces growth arrest and differentiation of myeloid leukemia cells. Oncogene. 2005;24:2625–2634. doi: 10.1038/sj.onc.1208167. [DOI] [PubMed] [Google Scholar]
- Matthews J.M., Visvader J.E., Visvader J.E. LIM-domain-binding protein 1: A multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 2003;4:1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier N., Krpic S., Rodriguez P., Strouboulis J., Monti M., Krijgsveld J., Gering M., Patient R., Hostert A., Grosveld F., Krpic S., Rodriguez P., Strouboulis J., Monti M., Krijgsveld J., Gering M., Patient R., Hostert A., Grosveld F., Rodriguez P., Strouboulis J., Monti M., Krijgsveld J., Gering M., Patient R., Hostert A., Grosveld F., Strouboulis J., Monti M., Krijgsveld J., Gering M., Patient R., Hostert A., Grosveld F., Monti M., Krijgsveld J., Gering M., Patient R., Hostert A., Grosveld F., Krijgsveld J., Gering M., Patient R., Hostert A., Grosveld F., Gering M., Patient R., Hostert A., Grosveld F., Patient R., Hostert A., Grosveld F., Hostert A., Grosveld F., Grosveld F. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133:4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- Milán M., Cohen S.M., Cohen S.M. Regulation of LIM homeodomain activity in vivo: A tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol. Cell. 1999;4:267–273. doi: 10.1016/s1097-2765(00)80374-3. [DOI] [PubMed] [Google Scholar]
- Milán M., Cohen S.M., Cohen S.M. Temporal regulation of apterous activity during development of the Drosophila wing. Development. 2000;127:3069–3078. doi: 10.1242/dev.127.14.3069. [DOI] [PubMed] [Google Scholar]
- Mizunuma H., Miyazawa J., Sanada K., Imai K., Miyazawa J., Sanada K., Imai K., Sanada K., Imai K., Imai K. The LIM-only protein, LMO4, and the LIM domain-binding protein, LDB1, expression in squamous cell carcinomas of the oral cavity. Br. J. Cancer. 2003;88:1543–1548. doi: 10.1038/sj.bjc.6600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo P., Rosen C., Baylies M.K., Dorsett D., Rosen C., Baylies M.K., Dorsett D., Baylies M.K., Dorsett D., Dorsett D. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes & Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay M., Teufel A., Yamashita T., Agulnick A.D., Chen L., Downs K.M., Schindler A., Grinberg A., Huang S.P., Dorward D., Teufel A., Yamashita T., Agulnick A.D., Chen L., Downs K.M., Schindler A., Grinberg A., Huang S.P., Dorward D., Yamashita T., Agulnick A.D., Chen L., Downs K.M., Schindler A., Grinberg A., Huang S.P., Dorward D., Agulnick A.D., Chen L., Downs K.M., Schindler A., Grinberg A., Huang S.P., Dorward D., Chen L., Downs K.M., Schindler A., Grinberg A., Huang S.P., Dorward D., Downs K.M., Schindler A., Grinberg A., Huang S.P., Dorward D., Schindler A., Grinberg A., Huang S.P., Dorward D., Grinberg A., Huang S.P., Dorward D., Huang S.P., Dorward D., Dorward D., et al. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development. 2003;130:495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- Nishioka N., Nagano S., Nakayama R., Kiyonari H., Ijiri T., Taniguchi K., Shawlot W., Hayashizaki Y., Westphal H., Behringer R.R., Nagano S., Nakayama R., Kiyonari H., Ijiri T., Taniguchi K., Shawlot W., Hayashizaki Y., Westphal H., Behringer R.R., Nakayama R., Kiyonari H., Ijiri T., Taniguchi K., Shawlot W., Hayashizaki Y., Westphal H., Behringer R.R., Kiyonari H., Ijiri T., Taniguchi K., Shawlot W., Hayashizaki Y., Westphal H., Behringer R.R., Ijiri T., Taniguchi K., Shawlot W., Hayashizaki Y., Westphal H., Behringer R.R., Taniguchi K., Shawlot W., Hayashizaki Y., Westphal H., Behringer R.R., Shawlot W., Hayashizaki Y., Westphal H., Behringer R.R., Hayashizaki Y., Westphal H., Behringer R.R., Westphal H., Behringer R.R., Behringer R.R., et al. Ssdp1 regulates head morphogenesis of mouse embryos by activating the Lim1–Ldb1 complex. Development. 2005;132:2535–2546. doi: 10.1242/dev.01844. [DOI] [PubMed] [Google Scholar]
- Ostendorff H.P., Peirano R.I., Peters M.A., Schluter A., Bossenz M., Scheffner M., Bach I., Peirano R.I., Peters M.A., Schluter A., Bossenz M., Scheffner M., Bach I., Peters M.A., Schluter A., Bossenz M., Scheffner M., Bach I., Schluter A., Bossenz M., Scheffner M., Bach I., Bossenz M., Scheffner M., Bach I., Scheffner M., Bach I., Bach I. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature. 2002;416:99–103. doi: 10.1038/416099a. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pueyo J.I., Couso J.P., Couso J.P. Chip-mediated partnerships of the homeodomain proteins Bar and Aristaless with the LIM-HOM proteins Apterous and Lim1 regulate distal leg development. Development. 2004;131:3107–3120. doi: 10.1242/dev.01161. [DOI] [PubMed] [Google Scholar]
- Qian Z., Fernald A.A., Godley L.A., Larson R.A., Le Beau M.M., Fernald A.A., Godley L.A., Larson R.A., Le Beau M.M., Godley L.A., Larson R.A., Le Beau M.M., Larson R.A., Le Beau M.M., Le Beau M.M. Expression profiling of CD34+ hematopoietic stem/progenitor cells reveals distinct subtypes of therapy-related acute myeloid leukemia. Proc. Natl. Acad. Sci. 2002;99:14925–14930. doi: 10.1073/pnas.222491799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón-Limas D.E., Lu C.H., Canal I., Botas J., Lu C.H., Canal I., Botas J., Canal I., Botas J., Botas J. The level of DLDB/CHIP controls the activity of the LIM homeodomain protein apterous: Evidence for a functional tetramer complex in vivo. EMBO J. 2000;19:2602–2614. doi: 10.1093/emboj/19.11.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sum E.Y., Segara D., Duscio B., Bath M.L., Field A.S., Sutherland R.L., Lindeman G.J., Visvader J.E., Segara D., Duscio B., Bath M.L., Field A.S., Sutherland R.L., Lindeman G.J., Visvader J.E., Duscio B., Bath M.L., Field A.S., Sutherland R.L., Lindeman G.J., Visvader J.E., Bath M.L., Field A.S., Sutherland R.L., Lindeman G.J., Visvader J.E., Field A.S., Sutherland R.L., Lindeman G.J., Visvader J.E., Sutherland R.L., Lindeman G.J., Visvader J.E., Lindeman G.J., Visvader J.E., Visvader J.E. Overexpression of LMO4 induces mammary hyperplasia, promotes cell invasion, and is a predictor of poor outcome in breast cancer. Proc. Natl. Acad. Sci. 2005;102:7659–7664. doi: 10.1073/pnas.0502990102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler J.P., Lee S.K., Jurata L.W., Gill G.N., Pfaff S.L., Lee S.K., Jurata L.W., Gill G.N., Pfaff S.L., Jurata L.W., Gill G.N., Pfaff S.L., Gill G.N., Pfaff S.L., Pfaff S.L. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein–protein interactions. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- Tran Y.H., Xu Z., Kato A., Mistry A.C., Goya Y., Taira M., Brandt S.J., Hirose S., Xu Z., Kato A., Mistry A.C., Goya Y., Taira M., Brandt S.J., Hirose S., Kato A., Mistry A.C., Goya Y., Taira M., Brandt S.J., Hirose S., Mistry A.C., Goya Y., Taira M., Brandt S.J., Hirose S., Goya Y., Taira M., Brandt S.J., Hirose S., Taira M., Brandt S.J., Hirose S., Brandt S.J., Hirose S., Hirose S. Spliced isoforms of LIM-domain-binding protein (CLIM/NLI/Ldb) lacking the LIM-interaction domain. J. Biochem. 2006;140:105–119. doi: 10.1093/jb/mvj134. [DOI] [PubMed] [Google Scholar]
- Valge-Archer V., Forster A., Rabbitts T.H., Forster A., Rabbitts T.H., Rabbitts T.H. The LMO1 AND LDB1 proteins interact in human T cell acute leukaemia with the chromosomal translocation t(11;14)(p15;q11) Oncogene. 1998;17:3199–3202. doi: 10.1038/sj.onc.1202353. [DOI] [PubMed] [Google Scholar]
- van Meyel D.J., O’Keefe D.D., Jurata L.W., Thor S., Gill G.N., Thomas J.B., O’Keefe D.D., Jurata L.W., Thor S., Gill G.N., Thomas J.B., Jurata L.W., Thor S., Gill G.N., Thomas J.B., Thor S., Gill G.N., Thomas J.B., Gill G.N., Thomas J.B., Thomas J.B. Chip and apterous physically interact to form a functional complex during Drosophila development. Mol. Cell. 1999;4:259–265. doi: 10.1016/s1097-2765(00)80373-1. [DOI] [PubMed] [Google Scholar]
- van Meyel D.J., O’Keefe D.D., Thor S., Jurata L.W., Gill G.N., Thomas J.B., O’Keefe D.D., Thor S., Jurata L.W., Gill G.N., Thomas J.B., Thor S., Jurata L.W., Gill G.N., Thomas J.B., Jurata L.W., Gill G.N., Thomas J.B., Gill G.N., Thomas J.B., Thomas J.B. Chip is an essential cofactor for apterous in the regulation of axon guidance in Drosophila. Development. 2000;127:1823–1831. doi: 10.1242/dev.127.9.1823. [DOI] [PubMed] [Google Scholar]
- van Meyel D.J., Thomas J.B., Agulnick A.D., Thomas J.B., Agulnick A.D., Agulnick A.D. Ssdp proteins bind to LIM-interacting co-factors and regulate the activity of LIM-homeodomain protein complexes in vivo. Development. 2003;130:1915–1925. doi: 10.1242/dev.00389. [DOI] [PubMed] [Google Scholar]
- Visvader J.E., Mao X., Fujiwara Y., Hahm K., Orkin S.H., Mao X., Fujiwara Y., Hahm K., Orkin S.H., Fujiwara Y., Hahm K., Orkin S.H., Hahm K., Orkin S.H., Orkin S.H. The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc. Natl. Acad. Sci. 1997;94:13707–13712. doi: 10.1073/pnas.94.25.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J.E., Venter D., Hahm K., Santamaria M., Sum E.Y., O’Reilly L., White D., Williams R., Armes J., Lindeman G.J., Venter D., Hahm K., Santamaria M., Sum E.Y., O’Reilly L., White D., Williams R., Armes J., Lindeman G.J., Hahm K., Santamaria M., Sum E.Y., O’Reilly L., White D., Williams R., Armes J., Lindeman G.J., Santamaria M., Sum E.Y., O’Reilly L., White D., Williams R., Armes J., Lindeman G.J., Sum E.Y., O’Reilly L., White D., Williams R., Armes J., Lindeman G.J., O’Reilly L., White D., Williams R., Armes J., Lindeman G.J., White D., Williams R., Armes J., Lindeman G.J., Williams R., Armes J., Lindeman G.J., Armes J., Lindeman G.J., Lindeman G.J. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc. Natl. Acad. Sci. 2001;98:14452–14457. doi: 10.1073/pnas.251547698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman I.A., Osada H., Grutz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H., Osada H., Grutz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H., Grutz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H., Westphal H., Forster A., Rabbitts T.H., Forster A., Rabbitts T.H., Rabbitts T.H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe U., Milán M., Cohen S.M., Milán M., Cohen S.M., Cohen S.M. Regulation of Apterous activity in Drosophila wing development. Development. 2001;128:4615–4622. doi: 10.1242/dev.128.22.4615. [DOI] [PubMed] [Google Scholar]
- Wu L. Structure and functional characterization of single-strand DNA binding protein SSDP1: Carboxyl-terminal of SSDP1 has transcription activity. Biochem. Biophys. Res. Commun. 2006;339:977–984. doi: 10.1016/j.bbrc.2005.11.098. [DOI] [PubMed] [Google Scholar]
- Xu Z., Huang S., Chang L.S., Agulnick A.D., Brandt S.J., Huang S., Chang L.S., Agulnick A.D., Brandt S.J., Chang L.S., Agulnick A.D., Brandt S.J., Agulnick A.D., Brandt S.J., Brandt S.J. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol. Cell. Biol. 2003;23:7585–7599. doi: 10.1128/MCB.23.21.7585-7599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]