Summary
ClpXP, an ATP-dependent protease, degrades hundreds of different intracellular proteins. ClpX chooses substrates by binding peptide tags, typically displayed at the N or C terminus of the protein to be degraded. Here, we identify a ClpX mutant that displays a 300-fold change in substrate specificity, resulting in decreased degradation of ssrA-tagged substrates but improved degradation of proteins with other classes of degradation signals. The altered-specificity mutation occurs within “RKH” loops, which surround the entrance to the central pore of the ClpX hexamer and are highly conserved in the ClpX subfamily of AAA+ ATPases. These results support a major role for the RKH loops in substrate recognition and suggest that ClpX specificity represents an evolutionary compromise that has optimized degradation of multiple types of substrates rather than any single class.
Introduction
Enzymatic specificity is essential for biological function. For most enzymes, specificity depends upon the complementary interaction between a substrate and the enzyme active site. This simple paradigm, however, does not explain the specificity of energy-dependent proteases such as ClpXP, ClpAP, Lon, HslUV, FtsH, and the proteasome. The active sites that cleave peptide bonds in these enzymes are sequestered in an interior chamber and are only encountered after specific protein substrates have been recognized, unfolded, and translocated into this chamber by the AAA+ ATPase subunits of the protease (Sauer et al. 2004; Prakash and Matouschek, 2004). The ATPase active sites bind nucleotide but do not directly contact protein substrates. How these AAA+ enzymes, which function as ring hexamers, discriminate among potential target proteins is poorly understood. The identification of mutations that confer altered substrate specificity provides one approach to answering this question.
ClpX, the ATP-dependent unfoldase/translocase of ClpXP, recognizes specific protein substrates by binding to short exposed peptide tags. For example, ClpXP appears to degrade any protein with an ssrA tag at its C terminus (Gottesman et al., 1998; Kim et al., 2000; Singh et al., 2000; Burton et al., 2001; Flynn et al., 2001; 2003; Lee et al., 2001; Kenniston et al., 2003; 2004). The C-terminal residues of MuA transposase and the N-terminal residues of the λO protein can also target proteins for ClpXP degradation (Levchenko et al., 1997; Gonciarz-Swiatek et al., 1999). Indeed, five distinct classes of degradation signals for ClpXP have been identified in proteomic studies, including three distinct sequence motifs (N1, N2, and N3) found at the N terminus of natural substrates and two sequence motifs (C1 and C2) found at the C terminus (Flynn et al., 2003).
To enter the degradation chamber of ClpP, unfolded protein substrates must first pass through a central pore in the ClpX hexamer (Ortega et al., 2000). Several loops line this pore, including one with a GYVG sequence that is highly conserved in most proteolytic AAA+ ATPases. A mutation in the GYVG loop of Escherichia coli ClpX reduces binding to ssrA-tagged substrates without markedly affecting binding of other classes of substrates (Siddiqui et al., 2004). It is unlikely, however, that this loop alone forms a binding site for the ssrA tag because the ATPases of human mitochondrial ClpXP and bacterial HslUV contain identical GYVG motifs, but these proteases do not degrade ssrA-tagged substrates (Kang et al., 2002; Burton et al., 2005). Another ClpX pore loop contains an RKH sequence that is >99% conserved in the bacterial orthologues. This 15-residue RKH loop is completely absent in FtsH, is replaced by shorter loops of 3-4 residues in HslU and ClpA, and differs at many key residue positions in human ClpX. In a hexameric model based on the structure of the Helicobacter pylori ClpX subunit (Kim and Kim, 2003), the RKH loops surround the entry to the central pore (Fig. 1A) and thus are positioned to mediate initial interactions between ClpX and the degradation tag of a protein substrate.
Fig. 1.
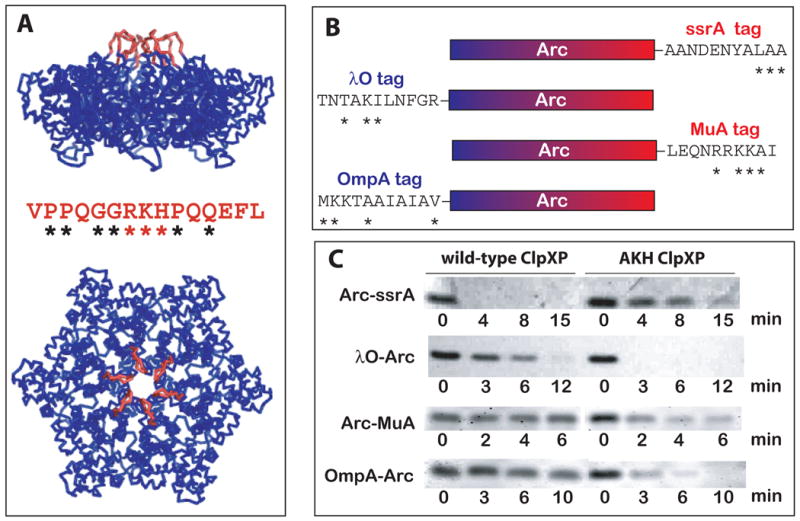
The RKH loops play roles in substrate specificity. (A) The RKH loops of the E. coli ClpX hexamer are colored red in a homology model based on the subunit structure of H. pylori ClpX (Kim and Kim, 2003). The conformations shown are hypothetical as these loops are disordered in the structure. The sequence of the E. coli RKH loop is shown (asterisks denote highly conserved residues in all bacterial ClpX orthologues). ClpP docks on the hexagonal face of ClpX opposite the RKH loops. (B) Schematic depiction of the substrates used in panel C. The N-terminal portion of Arc is blue and the C-terminal part is red. The sequence of the degradation tag of each substrate is shown. Asterisks mark residues known to be important for ClpX recognition and/or conserved in the C1 (ssrA), N1 (λO), C2 (MuA), and N2 (OmpA) classes of substrates (Flynn et al., 2001; 2003). (C) Arc-ssrA is degraded preferentially by wild-type ClpXP, whereas λO-Arc, Arc-MuA and OmpA-Arc are degraded preferentially by AKH ClpXP. Degradation of each substrate (5 μM) was performed for the times indicated using 0.3 μM ClpX6 and 0.8 μM ClpP14 and was assayed by SDS-PAGE.
In this paper, we explore the role of the RKH loops of ClpX in substrate selection and degradation. We find that a single mutation in this loop dramatically slows ClpXP degradation of substrates bearing ssrA tags (a C1 signal) but enhances degradation of substrates with three other classes of degradation tags. For example, comparison of the activities of the wild-type and the mutant enzymes reveals a 300-fold change in the second-order rate constants for degradation of an ssrA-tagged substrate and a λO-tagged substrate, caused almost completely by alterations in the strength of substrate binding. Unlike wild-type ClpX, the mutant enzyme does not recognize the C-terminal carboxylate of the ssrA-peptide tag. The mutant also displays higher maximal rates of degradation and ATP hydrolysis, indicating that the RKH loops play important roles in determining substrate specificity and in coordinating the catalytic and mechanical activities of the ClpXP machine.
Results
RKH loop mutations alter substrate specificity
To investigate the role of the RKH sequence, we constructed variants bearing the single substitutions R228A (AKH), K229A (RAH), and H230A (RKA) in His6-tagged E. coli ClpX. Following purification, we combined each of these mutants with ClpP, and assayed degradation of Arc repressor with a C-terminal ssrA tag (a C1 signal) or an N-terminal λO degradation tag (an N1 signal) (Fig. 1B). Compared to wild-type ClpXP, all three mutants degraded Arc-ssrA very poorly (data not shown). The RKA mutant also degraded λO-Arc poorly, but the AKH and RAH mutants degraded this substrate at a faster rate than wild type (data not shown). Because significant autodegradation of the His6-tagged mutant ClpX enzymes occurred in these experiments, we recloned the AKH mutation into an untagged background. The untagged AKH ClpX variant also showed poor ClpP-mediated degradation of Arc-ssrA but displayed enhanced degradation of λO-Arc relative to wild-type ClpX (Fig. 1C).
In principle, AKH ClpXP might prefer N-terminal degradation tags, whereas the wild-type enzyme prefers C-terminal tags. To test this possibility, we assayed degradation of Arc with a C-terminal MuA tag (a C2 signal) or an N-terminal OmpA tag (an N2 signal) (Fig. 1B). Both substrates were degraded faster by AKH ClpXP than by wild-type ClpXP (Fig. 1C), showing that the location of the degradation tag is not a major factor in the altered activity of the mutant enzyme. Thus, the AKH mutant catalyzes enhanced degradation of substrates with N1, N2, and C2 classes of degradation signals but shows reduced degradation of a substrate with a C1 degradation tag. These results demonstrate that the RKH loops of E. coli ClpX play a key role in determining substrate specificity.
Quantitative studies of degradation kinetics
For quantitative studies of specificity and degradation, we focused on the Arc-ssrA and λO-Arc substrates. Initial rates of degradation of 35S-labeled Arc-ssrA and λO-Arc by AKH ClpXP and the wild-type enzyme were determined at different substrate concentrations and fit to the Michaelis-Menten equation to determine kinetic parameters (Fig. 2; Table 1). The AKH mutation increased KM for degradation of Arc-ssrA from roughly 1 μM to greater than 100 μM. By contrast, this mutation decreased KM for degradation of λO-Arc from 13 to approximately 4 μM. SspB, an adaptor that helps ssrA-tagged substrates bind to ClpX (Levchenko et al., 2000), reduced K M for AKH degradation of Arc-ssrA to roughly 20 μM, but this value was still almost 40-fold higher than the corresponding value for wild-type ClpXP (Table 1). Thus, the AKH-loop mutation hinders recognition of one type of degradation signal but enhances recognition of another class of signal.
Fig. 2.

Kinetic properties of AKH and wild-type ClpXP. Michaelis-Menten plots of the variation of degradation rates as a function of the concentrations of the Arc-ssrA and λO-Arc substrates by wild-type ClpXP (left panel) and AKH ClpXP (right panel). Degradation was monitored by release of acid-soluble 35S-peptides using assay conditions listed in Fig. 1. The fits for wild-type ClpXP/λO-Arc and AKH ClpXP/Arc-ssrA included higher concentration data not shown on these graphs.
Table 1.
Kinetic parameters for degradation of substrates with ssrA or λO degradation tags by wild-type ClpXP and AKH ClpXP.
|
KM μM |
kdeg min−1 enz−1 |
kdeg/KM μM−1 min−1 enz−1 |
|
|---|---|---|---|
| wild-type ClpXP | |||
| Arc-ssrA | 1.2 ± 0.4 | 4.8 ± 0.5 | 3.9 ± 1.4 |
| Arc-ssrA + SspB | 0.5 ± 0.3 | 6.0 ± 0.2 | 11.0 ± 5.8 |
| λO-Arc | 13 ± 2.5 | 5.0 ± 1.1 | 0.38 ± 0.11 |
| λO-CM-titin | 41 ± 8.5 | 2.1 ± 0.2 | 0.052 ± 0.011 |
| CM-titin-ssrA | 2.4 ± 0.3 | 4.8 ± 0.5 | 2.0 ± 0.31 |
| AKH ClpXP | |||
| Arc-ssrA | 140 ± 11 | 13 ± 0.3 | 0.096 ± 0.0079 |
| Arc-ssrA + SspB | 20 ± 9 | 10 ± 1.5 | 0.52 ± 0.25 |
| λO-Arc | 4.4 ± 1.0 | 13 ± 0.1 | 2.9 ± 0.65 |
| λO-CM-titin | 11 ± 0.8 | 3.6 ± 0.2 | 0.31 ± 0.03 |
| CM-titin-ssrA | 80 ± 14 | 9.2 ± 0.8 | 0.11 ± 0.022 |
KM and kdeg are mean values from independent experiments (n=3, λO-titin/wild-type and Arc-ssrA/SspB/AKH; n=2, all others). The CM-titin substrate was denatured by carboxymethylation of cysteines (Kenniston et al., 2003). Errors were estimated as
AKH ClpXP degraded λO-Arc and Arc-ssrA at maximal rates (Vmax/[ClpX6]=kdeg) from 1.7 to 2.8-fold faster than observed for wild-type ClpXP (Fig. 2; Table 1). These increases in maximal degradation rates were accompanied by increases in the rate of ATP hydrolysis measured in the presence of ClpP and substrate (data not shown). For example, the AKH mutation caused a 2.5-fold increase in the maximal degradation rate of λO-Arc and a 2.1-fold increase in the ATP-hydrolysis rate measured in the presence of 70 μM substrate. These results suggest that the wild-type RKH loops help control the speed at which the ClpX machine functions by limiting the rate of ATP hydrolysis. These effects, however, must be long range and mediated through changes in protein structure, because any given RKH loop in the ClpX hexamer is 30 Å or more from the closest ATP-binding site (Kim and Kim, 2003).
The ratio of second-order rate constants for degradation (kdeg/KM) provides the best comparison of substrate specificities. By this measure, wild-type ClpXP preferentially degrades Arc-ssrA by a factor of roughly 10 compared to λO-Arc, whereas AKH ClpXP preferentially degrades λO-Arc by a factor of roughly 30. Thus, the AKH mutation changes substrate specificity by nearly 300-fold. This specificity change depends on the degradation signal but not the precise substrate; AKH ClpXP degraded a denatured titin variant with a λO tag better than the same protein with an ssrA tag, whereas wild-type ClpXP had the opposite preference (Table 1). For wild-type ClpXP, translocation of unfolded titin-ssrA is the rate-limiting step in degradation (Kenniston et al., 2005). Because AKH ClpXP degrades this substrate with an increased maximum velocity, it must translocate unfolded titin-ssrA faster than the wild-type enzyme. This increased rate of translocation is likely to be related to the enhanced ATPase activity of the mutant enzyme.
Loss of a specific contact in the ssrA tag
To determine whether AKH ClpXP still recognizes specific features of the ssrA tag, we assayed degradation of an Arc-ssrA variant in which the C-terminal Ala-Ala sequence, which is a strong determinant of recognition by wild-type ClpXP (Flynn et al., 2001), was changed to Gly-Gly. Although this mutant differs from Arc-ssrA by the absence of just two β-methyl groups, neither wild-type nor AKH ClpXP degraded the Gly-Gly substrate substantially over a 1-h time course (Fig. 3A). Because degradation of Arc-ssrA by AKH ClpXP is almost complete in 15 min (Fig. 1C), we conclude that the AKH mutant still recognizes some aspects of the Ala-Ala portion of the ssrA tag.
Fig. 3.
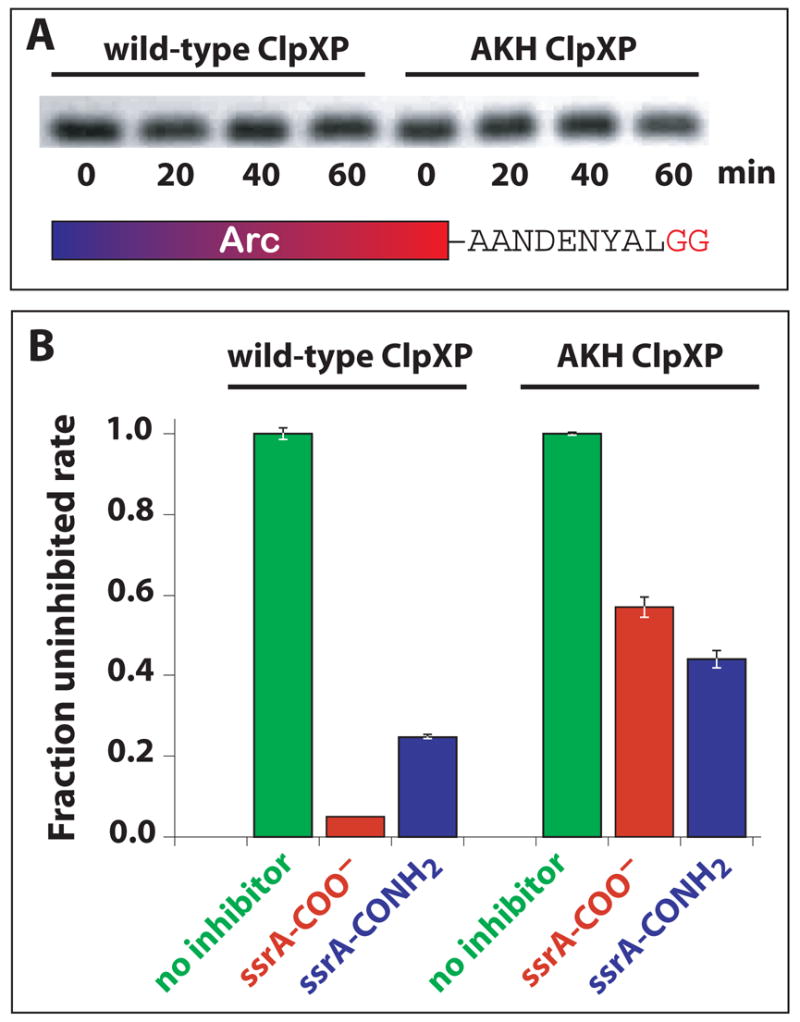
AKH ClpX recognizes the C-terminal Ala-Ala but not the C-terminal carboxylate of the ssrA tag. (A) Degradation of an Arc-ssrA variant with Gly-Gly replacing the C-terminal Ala-Ala was assayed using the conditions listed in Fig. 1. (B) Degradation of GFP-ssrA (1 μM) by wild-type ClpXP (0.3 μM ClpX6; 0.8 μM ClpP14) or AKH ClpXP (1 μM AKH ClpX6; 2 μM ClpP14) was assayed by changes in fluorescence without inhibitor or in the presence of 200 μM peptide inhibitor. The peptide sequences were NH2-CAANDENYALAA-COO− (ssrA-COO−) or NH2-CAANDENYALAA-CONH2 (ssrA-CONH2), where the underlined residues represent the ssrA-tag sequence.
The C-terminal carboxylate of the ssrA tag is another important binding determinant for wild-type ClpX (Kim et al., 2000; Flynn et al., 2001). We tested for ClpX recognition of this group by using an ssrA peptide and an otherwise identical peptide with a C-terminal–CONH2 to inhibit GFP-ssrA degradation by wild-type or AKH ClpXP. As expected (Kim et al., 2000; Flynn et al., 2001), the CONH2 peptide was less effective than the COO− peptide in inhibiting degradation by wild-type ClpXP (Fig. 3B). By contrast, the CONH2 peptide inhibited AKH ClpXP slightly better than the COO− peptide (Fig. 3B). This result indicates that the AKH mutant, in contrast to wild-type ClpX, does not interact favorably with the C-terminal carboxylate of the ssrA peptide.
Discussion
ClpX loops and substrate recognition
Our results show that the RKH loops, which form the entry to the central pore of the ClpX hexamer, play positive or negative roles in recognition of protein substrates with almost all classes of recognition signals. Unlike wild-type ClpX, the AKH mutant is defective in recognition of the negatively charged α-carboxylate of the ssrA-tag. The simplest interpretation of this result is that the R228 side chain of ClpX normally interacts favorably with this moiety. We note that the λO, MuA, and OmpA degradation tags all contain basic residues that are conserved in other members of related N1, N2, and C2 signals (Fig. 1B) and that each of these tags causes faster substrate degradation by AKH than by wild-type ClpXP. Because the R228A mutation reduces the positive charge of the RKH loop, it may enhance binding to the λO, MuA, and OmpA degradation tags by diminishing electrostatic repulsion. A role for the RKH loops in determining substrate specificity is also supported by phylogenetic comparisons. For example, human mitochondrial ClpX does not support degradation of λO or ssrA-tagged substrates (Kang et al., 2002) and has a loop sequence that differs significantly from the RKH loops of E. coli ClpX and other bacterial ClpX enzymes.
The ClpAP and FtsH proteases also degrade ssrA-tagged substrates (Gottesman et al., 1998; Herman et al., 1998), but the hexameric AAA+ ATPases of these enzymes do not contain RKH loops like those in ClpX. It is known, however, that ClpAP does not bind to the C-terminal residue or α-carboxylate of the ssrA tag and, indeed, recognizes this tag very differently than does ClpXP (Flynn et al., 2001). FtsH and ClpXP are also likely to interact with the ssrA tag in different manners (Herman et al., 1998). Hence, it is not surprising that the RKH loops of ClpX play critical roles in recognition of the ssrA tag, despite the fact that homologous loops are absent in other enzymes that recognize this degradation tag.
Previous studies established that a mutation in the GYVG loop of ClpX, which is positioned immediately below the RKH loop in the central pore of the hexamer, substantially reduced recognition of substrates with ssrA and related C1-motif tags but caused only modest decreases in degradation of substrates bearing other classes of degradation signals (Siddiqui et al., 2004). The RKH and GYVG pore loops may interact to form a binding site for the ssrA tag and related degradation signals, or could affect tag recognition indirectly. We favor direct binding, as ATP-hydrolysis dependent changes in pore-loop conformations could then begin to translocate the ssrA tag, resulting in unfolding of attached native proteins and ultimately in transport of the denatured polypeptide into ClpP (Kim et al., 2000; Kenniston et al., 2003; 2005; Sauer et al., 2004). A direct role of the RKH loops in substrate binding and in nucleotide-dependent conformational changes is also consistent with the enhanced ATP-hydrolysis and translocation/degradation phenotypes observed for AKH ClpXP. Direct pore binding would also explain the finding that only one ssrA-tag peptide binds to a ClpX hexamer (Piszczek et al., 2005), as binding of a single ssrA tag in the pore would sterically block the binding of other tag molecules even though symmetry in the hexamer would generate multiple potential tag binding sites.
Substrate-recognition tradeoffs
Our results indicate that the RKH loops of wild-type ClpX play important roles in its ability to recognize protein substrates with different classes of degradation signals. At low substrate concentrations, AKH ClpXP is more active than wild-type ClpXP in degrading proteins with N1, N2, and C2 signals but is less active in degrading proteins with C1 signals. Thus, the substrate specificity of wild-type ClpX appears to represent an evolutionary compromise in which the ability to degrade all types of substrates at reasonable rates was selected rather than optimal degradation of N1, N2 or C2 substrates. Clearly, enhancing recognition/degradation of these latter ClpX substrates is possible, but comes at the cost of reduced recognition/degradation of the C1 class of substrates, which include proteins tagged by the ssrA quality-control system (Keiler et al., 1996; Gottesman et al., 1998; Flynn et al., 2003). Such compromises may be common for enzymes that have a large number of protein substrates. Indeed, mutants of the GroEL chaperonin selected for the ability to enhance folding of a single substrate were found to have diminished renaturation activities for other substrate proteins (Wang et al., 2002). Like ClpXP, other energy-dependent proteases and molecular chaperones probably interact with hundreds or even thousands of different cellular substrates (Flynn et al., 2003; Kerner et al., 2005). Enzymes that interact with diverse substrates are likely to show suboptimal activity for any particular substrate, and thus mutations that enhance activity selectively will probably be far more common than would be expected for monogamous enzymes.
Experimental Procedures
Degradation and ATPase assays as well as overexpression and purification of ClpX, ClpX mutants, ClpP, and substrates followed published protocols (Kim et al., 2000; Flynn et al., 2001; 2003; Siddiqui et al., 2004; Kenniston et al., 2003; 2005). The His6-tagged AKH, RAH, and RKA were purified by Ni++-NTA chromatography and gel filtration on a Pharmacia Sephacryl 16/60 S-300 column. Untagged AKH ClpX was purified by sequential chromatography steps using a Pharmacia 16/10 phenyl sepharose column, a Sephacryl 16/60 S-300 column, and a 16/10 Q sepharose column. Arc substrates contained Arc residues 1–53 followed by H6KNQHE. The sequences of the ssrA, λO, MuA, and OmpA degradation tags are shown in Fig. 1B. Titin substrates contained the I27 domain of human titin, a His6 tag, either a C-terminal ssrA tag or an N-terminal λO tag, and were denatured by carboxymethylation (Kenniston et al., 2003). The NH2-CAANDENYALAA-COO− and NH2-CAANDENYALAA-CONH2 peptides (Kim et al., 2000) were synthesized by the MIT Biopolymers Facility. Degradation assays were performed at 30 °C in a buffer containing 25 mM Hepes (pH 7.6), 100 mM KCl, 10 mM MgCl2, and 10% glycerol with an ATP regeneration system containing 4 mM ATP, 16 mM creatine phosphate, and 0.32 mg/mL creatine phosphokinase.
Acknowledgments
We thank K. McGinness for providing the GG variant of Arc-ssrA and A. Martin for helpful discussions. Supported by NIH-grants AI-16892 and GM-49224. T.A.B. is an employee of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Burton RE, Baker TA, Sauer RT. Nucleotide-dependent substrate recognition by the AAA+ HslUV protease. Nat Struct Mol Biol. 2005;12:245–251. doi: 10.1038/nsmb898. [DOI] [PubMed] [Google Scholar]
- Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci USA. 2001;11:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Gonciarz-Swiatek M, Wawrzynow A, Um SJ, Learn BA, McMacken R, Kelley WL, Georgopoulos C, Sliekers O, Zylicz M. Recognition, targeting, and hydrolysis of the λ O replication protein by the ClpP/ClpX protease. J Biol Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman C, Thevenet D, Bouloc P, Walker GC, D'Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Ortega J, Singh SK, Wang N, Huang NN, Steven AC, Maurizi MR. Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J Biol Chem. 2002;277:21095–21102. doi: 10.1074/jbc.M201642200. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PRH, Sauer RT. Role of a peptide-tagging system in degradation of proteins translated from damaged mRNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of a AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Burton RE, Siddiqui SM, Baker TA, Sauer RT. Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J Struct Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc Natl Acad Sci USA. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, Hartl FU. Cell . Vol. 122. 2005. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli; pp. 209–220. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J Biol Chem. 2003;278:50664–50670. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Yamauchi M, Baker TA. ClpX and MuB interact with overlapping regions of Mu transposase: implications for control of the transposition pathway. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fueled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Ortega J, Singh SK, Ishikawa T, Maurizi MR, Steven AC. Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol Cell. 2000;6:1515–1521. doi: 10.1016/s1097-2765(00)00148-9. [DOI] [PubMed] [Google Scholar]
- Piszczek G, Rozycki J, Singh SK, Ginsburg A, Maurizi MR. The molecular chaperone, ClpA, has a single high affinity peptide binding site per hexamer. J Biol Chem. 2005;280:12221–12230. doi: 10.1074/jbc.M411733200. [DOI] [PubMed] [Google Scholar]
- Prakash S, Matouschek A. Protein unfolding in the cell. Trends Biochem Sci. 2004;29:593–600. doi: 10.1016/j.tibs.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui SM, Sauer RT, Baker TA. Role of the protein-processing pore of ClpX, a AAA+ ATPase, in recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc Natl Acad Sci USA. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Herman C, Tipton KA, Gross CA, Weissman JS. Directed evolution of substrate-optimized GroEL/S chaperonins. Cell. 2002;111:1027–1039. doi: 10.1016/s0092-8674(02)01198-4. [DOI] [PubMed] [Google Scholar]


