Abstract
AUTO3DEM is an automation system designed to accelerate the computationally intensive process of three-dimensional structure determination from images of vitrified icosahedral virus particles. With minimal user input and intervention, AUTO3DEM manages the flow of data between the major image reconstruction programs, monitors the progress of the computations, and intelligently updates the input parameters as the resolution of the model is improved. It is designed to be used on any computer running the Linux or UNIX operating systems and can be run in parallel mode on multi-processor systems.
Keywords: 3D image reconstruction, Icosahedral virus, Origin and orientation determination, Electron cryo-microscopy, Automated 3D image reconstruction, Random model reconstruction, Traditional model reconstruction, Unbiased model reconstruction, Image processing software
1. Introduction
The fields of cryo-electron microscopy (cryoEM) and three-dimensional (3D) image reconstruction have grown dramatically in recent years and numerous macromolecular structures have been solved with these techniques at sub-nanometer resolutions (Jiang and Ludtke, 2005). These achievements are attributable not only to improvements in the computer hardware and transmission electron microscopes, but also advances in the software capabilities. In fact, software has played a major role in nearly every aspect of structure determination including image acquisition (Carragher, et al., 2000), particle identification (see Journal of Structural Biology, vol 145, 2004), image reconstruction (Frank, et al., 1996; Grigorieff, 1998; Ludtke, et al., 1999) and visualization (e.g. Pettersen, et al., 2004).
An overview of the steps required for a complete cryoEM structure study of virtually any macromolecule, including viruses, starting with sample preparation and culminating in interpretation of function, is shown in Figure 1. While each of the major steps can be quite demanding and time-consuming, structure determination, particularly for large viruses and/or high resolutions, can often become the rate limiting step. Image reconstruction procedures typically require tens to hundreds of iterations over the multiple programs responsible for origin and orientation refinement, model construction, and resolution determination. Additional complications arise because the optimal set of parameters for a given iteration may not necessarily be ideal for subsequent iterations. In addition, the structural biologist is faced with a substantial data management problem that requires tracking a large number of files containing digitized micrographs, boxed particle images, intermediate particle origins and orientations, 3D models, and Fourier Shell Correlation (FSC) values. Due to these complexities, a 3D image reconstruction project can end up requiring months, or even years, of hard work for an experienced researcher. These challenges are even more daunting for novice researchers and will become greater as more data are collected for higher-resolution reconstructions.
Fig 1.
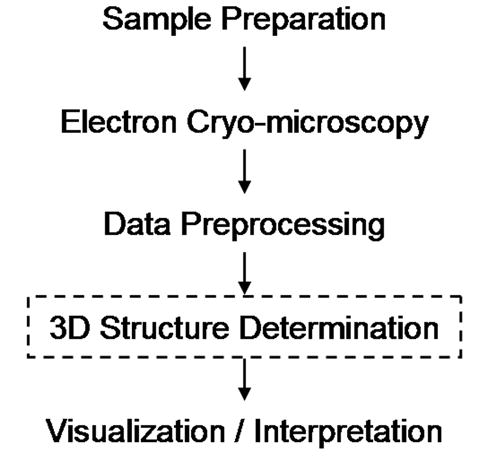
Primary steps involved in determining and analyzing the 3D structures of biological macromolecules. AUTO3DEM carries out all of the tasks required to produce the 3D reconstruction of icosahedral particles (boxed step).
Automating the image reconstruction process yields a number of important benefits. First, the time to solution can be greatly reduced. The output from one program can either be piped directly into the next program in the structure determination pipeline or parsed and used to set parameters for subsequent stages of the reconstruction, thereby eliminating the delay between executions of the programs. The user is relieved of many of the tedious tasks such as managing files, setting input parameters, extracting results from output files, and launching jobs. Not only does this free the researcher to focus on more important tasks, but it minimizes the likelihood of data entry errors. The importance of this second point cannot be overemphasized. Although the chance of making a typographical error at any one step may be fairly low, the cumulative probability of making at least one mistake over the course of a full image reconstruction project can be quite high. Another advantage of an automation system is that it helps novice users get up to speed quickly by taking advantage of the expert knowledge that has been built into the system. Finally, a well-designed automation framework provides a useful test bed for quickly evaluating new image reconstruction algorithms and procedures.
In the remainder of this paper, we describe AUTO3DEM, our solution for automating the process of structure determination for icosahedral viruses. We start with an overview of the capabilities of AUTO3DEM, and then follow with descriptions of both the numerically intensive image processing programs that are called and the calculations that are performed internally by AUTO3DEM in order to determine the course of the reconstruction. Finally, we present two case studies of structures that have been successfully solved using AUTO3DEM and briefly discuss our plans for future improvements.
2. Overview of AUTO3DEM capabilities
The main function of AUTO3DEM is to integrate the execution of the major image reconstruction programs and manage the flow of data from one application to the next. In our programming environment these include the programs PFTsearch, PO2R, P3DR, PCUT, and PSF, which are briefly described below (section 3). Here we provide an overview of the capabilities and modes of operation of AUTO3DEM.
AUTO3DEM is written in Perl and can be used on any machine running the UNIX or Linux operating system. The only hardware requirement is that the machine has sufficient memory to run the numerically intensive codes mentioned above. While it can be used on scalar systems, AUTO3DEM is designed to take full advantage of parallelism and can be run on an arbitrary number of processors. Software requirements are minimal and include an installation of Perl for the automation system itself and a standard Message Passing Interface (MPI) implementation for running the parallel codes.
AUTO3DEM was designed to perform three basic tasks: (i) traditional model (TM) reconstruction as described in Baker et al. (Baker, et al., 1999), (ii) unbiased model (UM) reconstruction (Grigorieff, 2000), and (iii) the generation of a starting model using the random model (RM) method (Yan, et al., 2006). The TM reconstruction method (Fig. 2) contains the 3D structure determination core (3DCore) that serves as the basis for the other two types of calculations. These three methods and the relations among them are discussed in subsequent sections (2.1–2.3).
Fig 2.
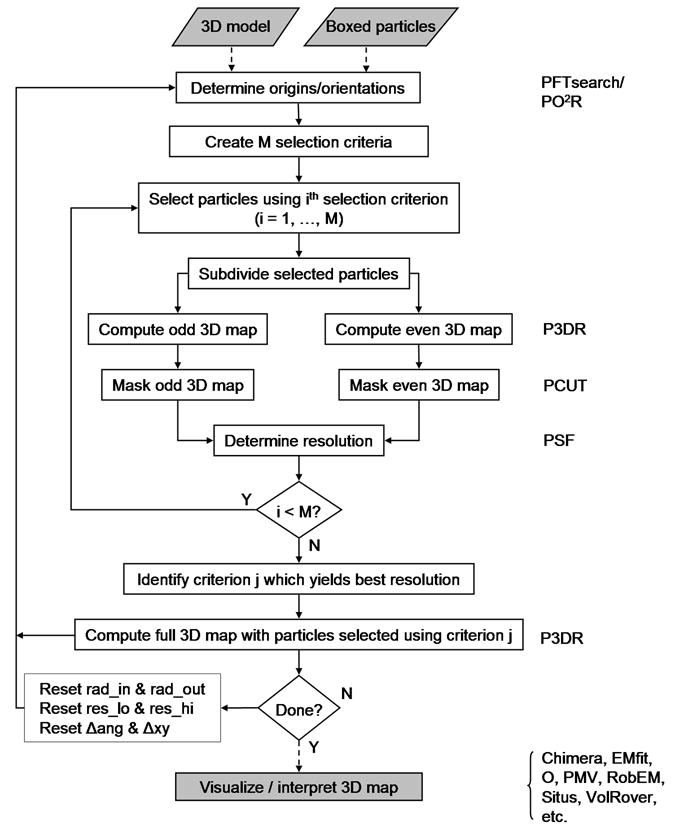
Flow chart of the traditional model (TM) reconstruction method. Procedures that comprise the core of 3D image reconstruction (3DCore) are identified in unshaded boxes. The programs needed at each computational step are listed at the right side of the figure. Dashed and solid lines indicate one-time and iterative operations, respectively. In our implementation, the programs P3DR, PO2R, and PFTsearch impose or assume icosahedral symmetry.
For both the TM and UM reconstructions, the determination of particle origins and orientations can be carried out in either SEARCH or REFINE mode. SEARCH mode is used in the early stages of a reconstruction when the particle orientations are not yet known to a high level of accuracy and the resolution of the model is still relatively low. REFINE mode is used in the later stages of the reconstruction when the goal is to reach the highest resolution that can be achieved given the quality of the available image data. The RM calculation, which is used only to generate a starting model for the conventional or unbiased model calculations, always uses SEARCH mode for determining origins and orientations.
2.1 Traditional model (TM) reconstruction method
The TM reconstruction method is based on execution of the 3DCore (Fig. 2). Input consists of a set of boxed particle images and an initial 3D model, together with a file that contains the values of the parameters dictating the overall flow control and execution of the image reconstruction programs. The usual procedure is to run the calculations in SEARCH mode using PFTsearch (Polar Fourier Transform search: Baker and Cheng, 1996) with the entire set of particle images until the resolution of the model fails to improve, and then shift to REFINE mode using PO2R (Parallel Origin and Orientation Refinement: Ji, et al., 2006).
The key difference between these two modes is the manner in which the orientations of the particles are determined. In SEARCH mode, each particle image is compared against a set of projections of the model evenly covering the asymmetric unit and the particle is assigned the orientation of the projection that best matches the image. REFINE mode also compares the particle image against a set of projections, but the orientations are chosen from a limited region of orientation space surrounding the most recent estimate for the orientation of the particle. The origins and orientations determined after running AUTO3DEM in SEARCH mode serve as input for the first iteration of REFINE mode. The latest set of origins and orientations determined from an iteration of REFINE mode serves as the starting point for the next iteration.
Although the subsequent model construction is generally performed using only a subset of the images, the origins and orientations of all images need to be updated. The reason for this strategy is that images are often chosen for inclusion in the model based upon the quality of their best match against projections of the model. The quality of this match can vary from one stage of the reconstruction to the next, and it is not until after the new origins and orientations have been determined that we can evaluate the suitability of the image for inclusion in the model.
Following origin and orientation determination, maps are constructed using one or more particle selection criteria. These criteria can be based, for example, on the orientations of the particles, defocus levels, or most commonly on the quality of the best match between the images and projections of the model. For the purpose of resolution estimation, the selected particle images are divided into two sets and a map is constructed for each set using the program P3DR (Parallel Three-Dimensional Reconstruction: Marinescu and Ji, 2003). The resulting maps are masked with the program PCUT (Ji and Marinescu, personal communication) to isolate the ordered portions of the maps. Fourier transforms of these masked maps are then calculated and compared by the program PSF (Parallel Structure Factor) in order to estimate the map resolution, where resolution is defined by the point where the FSC value first drops below a specified threshold (typically 0.5). The particle selection criterion that yields the highest resolution map is identified and the full map is constructed using all of the particle images that meet the selection criterion.
At this point, the progress of the reconstruction is evaluated. If the resolution had failed to improve over the course of a specified number of consecutive iterations, the target resolution had been reached, or the maximum number of iterations had been exceeded, AUTO3DEM terminates execution. Depending on the reason for termination, the user may have multiple courses of action. For example, if the resolution had reached a plateau when running in SEARCH mode, AUTO3DEM would typically be restarted in REFINE mode. If steady progress had been made, but the maximum number of iterations had been reached, AUTO3DEM might simply be restarted where the calculations had left off. If the execution terminated when running in REFINE mode and further progress had not been made, the user would need to consider manually changing AUTO3DEM input parameters, adding more particles, or simply ending the reconstruction process and concentrating on visualization and interpretation of the 3D data.
If none of the termination conditions are met, new values are calculated for the program parameters and the next iteration of the structure determination is initiated. Depending on the mode of operation, these updated parameters may include the resolution limits used in comparing the images to model projections, the inner and outer radii of the ordered region of the model, and the steps sizes required for origin and orientation determination.
2.2 Unbiased model (UM) reconstruction method
One shortcoming of the TM procedure is that it is subject to model bias. The particle images are split into two sets for the purpose of resolution determination and bias arises because the model used during origin and orientation refinement is reconstructed using the full set of images. The solution to this problem is to carry out a straightforward UM calculation (Fig. 3) in which the two reconstructions are generated independently.
Fig. 3.
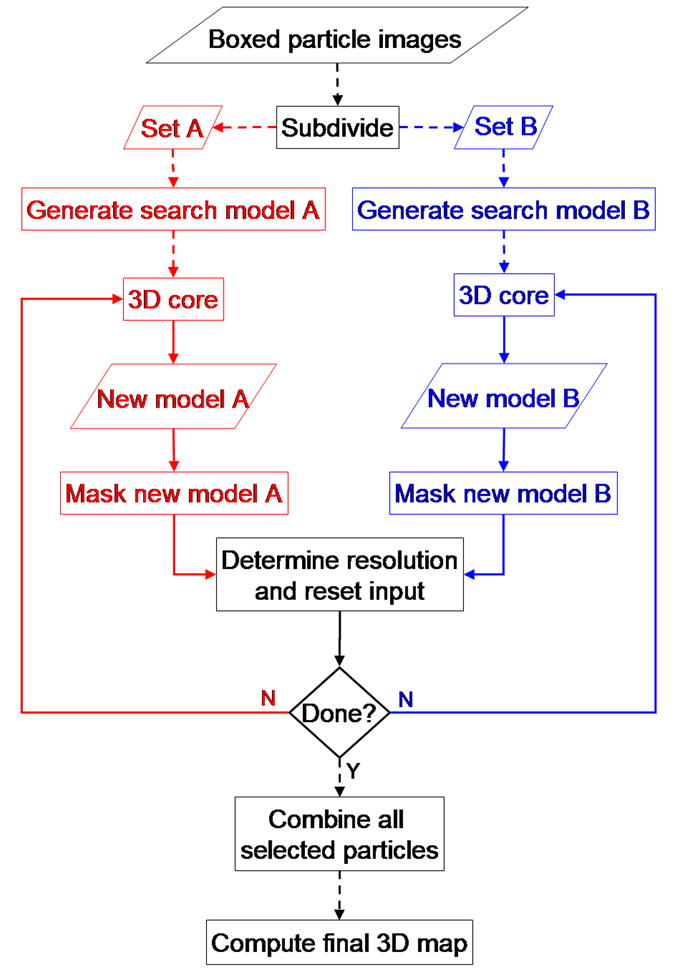
Flow chart of the unbiased model (UM) reconstruction method. The data set of boxed particle images is first subdivided into two subsets and each is processed in an independent branch following the 3DCore procedure illustrated in Fig. 2. Since each independent map is generated with an arbitrary handedness, for resolution determination the handedness of both maps, if necessary, must be fixed to be identical Dashed and solid lines indicate one-time and iterative operations, respectively.
At the outset of the reconstruction protocol, the particles are divided into two sets, A and B. One model is constructed from each set and the A and B models are then fed separately into the 3DCore (Fig. 2) described for the TM calculations. After each model has been independently refined to a specified level of resolution, the two models are compared to determine to what resolution they agree. The net effect is to use all of the particle images for resolution determination, but without contaminating one branch of the reconstruction with images from the other. Since each branch uses only one-half of the total number of particle images, the resolution determined from the FSC curve within each 3DCore would likely be lower than that obtained from the comparison of the A and B models. In a similar fashion to the conventional calculation, the unbiased model calculation is terminated when the maximum number of iterations is reached, a target resolution is achieved, or further progress fails to be made. Otherwise, the values of the program parameters based on the comparison of the two models are recalculated and control is returned to the 3DCore.
In order to eliminate bias in the model reconstruction, it is essential that two completely independent starting models be used. Even starting from a previously solved, lower-resolution model would introduce bias since the orientation and origin refinements of the particles in the two branches would be performed relative to the same starting model. The random model reconstruction method (section 2.3) avoids this problem and leads to a bias-free model.
2.3 Random model (RM) reconstruction method
As mentioned above (section 2.1), one of the inputs to the 3DCore is an initial model. Often, a model might already be available to use such as the reconstruction of a related virus, a lower resolution model of the same virus, a synthetic model constructed with the correct size and triangulation number, or a model based on structural data gleaned from the PDB. In other instances where no initial model exists, one needs to be constructed ab initio. AUTO3DEM can be used to perform a random model (RM) calculation (Fig. 4, Yan, et al., 2006) to generate a suitable model for further refinement.
Fig 4.
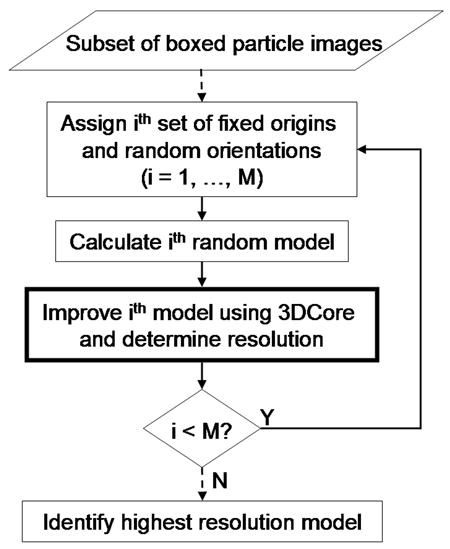
Flow chart of the random model (RM) reconstruction method. The RM calculations make use of the 3DCore (box with thick lines) shown in Fig. 2. Dashed and solid lines indicate one-time and iterative operations, respectively.
The RM reconstruction is performed using a small subset of particle images, typically numbering in the range of one or two hundred out of a much larger, full data set. The rationale for this is twofold. First, the goal at this point of the processing strategy is not to reconstruct a high-resolution model, but simply to produce a reasonable starting model that can be used as the input for conventional or unbiased model calculations. By restricting the number of images, the run time for the RM method is minimized. Second, experience shows that the use of too many images lowers the success of the RM calculations because the initial model approaches a nearly spherically symmetric structure, and one therefore that is less likely to generate reliable estimates of the particle origins and orientations.
In the RM calculations, an initial map is constructed from particles whose orientations are randomly assigned and origins are set at the center of the box window. This map, together with the corresponding set of particle images, is fed into the 3DCore and computations are repeated for a fixed number of iterations using SEARCH mode to improve the model. The process is repeated for multiple sets of initial random orientations and the resulting model with the highest resolution is chosen. Due to the small number of particles used, the FSC curve may be fairly noisy. For this reason, a different measure of resolution is used for the RM method in which the FSC is averaged over a range of spatial frequencies (typically 1/60-1/30 Å−1). Although we have no theoretical basis for determining whether or not a given set of random orientations will converge to a viable, low-resolution model, we find that 20–60% of the assignments do result in convergence (Yan, et al., 2006). The user can specify the number of random models to use, but our experience shows that choosing a total of ten models achieves a nice balance between run time and the quality of the suitable search model.
It is worth noting that running the RM calculation is equivalent to launching a series of TM calculations in SEARCH mode, each with a different set of random particle orientations. The advantage though of using the RM capabilities of AUTO3DEM is that it completely automates the process. AUTO3DEM generates the random orientations, launches the TM reconstruction, manages the input and output files, and interrogates the FSC curves to determine the best model.
3. Description of key programs
A full description of the image reconstruction programs used by AUTO3DEM is beyond the scope of this paper. Here we present a brief overview of each program at a level sufficient to understand the automation process.
The most numerically intensive step in the image reconstruction process is determination of the five parameters describing the orientations and origins of the particles. These are the three angles θ, ϕ, and ω that define the inclination of the view vector from one of the twofold axes (typically the z axis), the azimuthal angle relative to a line joining adjacent fivefold axes, and the rotation of the projected view relative to the x axis in the image, respectively; and x and y, the coordinates of the center of symmetry of the particle where all of the symmetry axes intersect (see Fig. 11 from Baker, et al., 1999). This determination can be performed either in SEARCH mode or REFINE mode, using the programs PFTsearch and PO2R, respectively.
PFTsearch is used in the early stages of the reconstruction when the particle origins and orientations are either unknown or only known to a relatively low level of accuracy and the model is still at low resolution. Orientations are derived in a two step process in which θ and |ϕ| are determined first, followed by ω and the sign of ϕ. The particle images are compared to projections of the model corresponding to values of θ and ϕ covering one-half of the asymmetric unit (ASU) of an icosahedron. At the typical angular spacing of 1°, this results in comparison against 370 projections. The key advantage of SEARCH mode is that the orientation space is evenly sampled and the particle can avoid being trapped in a local minimum and failing to find the true orientation. In theory, by reducing the angular spacing, SEARCH mode can be used to determine the particle orientations to an arbitrary level of accuracy. In practice, however, this is not done since the number of projections that each image needs to be compared against increases approximately as the square of the inverse of the angular spacing. For example, spacings of 0.5° and 0.25° result in 1430 and 5606 projections, respectively.
The particle origins and orientations are determined in REFINE mode using the program PO2R. Rather than searching the entire ASU, the particle images are compared against a set of projections in (θ, ϕ, ω) space centered about the current orientation obtained from PFTsearch or the previous iteration of the reconstruction using PO2R. The user can control both the size of the grid to be searched (i.e., the number of steps to be taken in each direction for each angle) and the magnitude of the angular spacing. Although the model projections used for one particle cannot in general be used for other particles, PO2R is much more efficient than PFTsearch for refining the particle orientations since the number of comparisons performed for each image can be kept fixed while the magnitude of the angular spacing is gradually reduced.
The 3D model is constructed from the particle images using the program P3DR. The discrete Fourier transform (DFT) is calculated for each 2D particle image and then interpolated into the 3D DFT of the model using the particle’s origins and orientations. After this process has been completed for all of the images, the real space electron density map is obtained by taking the inverse FT of the 3D DFT.
Two other external programs are required by AUTO3DEM. The ordered region of the virus is masked from the real space map using the program PCUT, thereby reducing the non-icosahedral ‘noise’ in the FT of the map. The key parameters specified by the user are the inner and outer radii that define a spherical annulus within which the ordered regions are concentrated. PCUT employs a Gaussian falloff at each boundary to avoid artifacts that would be caused by sharply cutting through a region of non-zero electron density. The FSC curve, and thus the resolution estimate, is determined by the program PSF. This program accepts as input the two masked maps and variables that define the range of spatial frequencies over which the correlation coefficients should be calculated.
4. AUTO3DEM description
An overview of AUTO3DEM and its capabilities, modes of operation, and a description of the underlying computationally intensive image reconstruction codes have been given. The following discussion provides more details of the workings of AUTO3DEM including the format of the user input, internal calculation and automatic updating of program parameters, particle selection, output and monitoring of results, user intervention, and restart capabilities.
4.1 User input
A single file is used to set the AUTO3DEM control parameters, the initial values of the input parameters for the image processing codes, and the names of the data file(s) containing the particle origins and orientations (Fig. 5). AUTO3DEM and program parameters are specified using a three-field record format, where the first field designates the program and the second and third fields form key-value pairs. The data records have two fields and are distinguished by the leading keyword ‘data’. Not all input parameters need to be assigned values, and in many instances reasonable default values are set automatically by AUTO3DEM.
Fig 5.
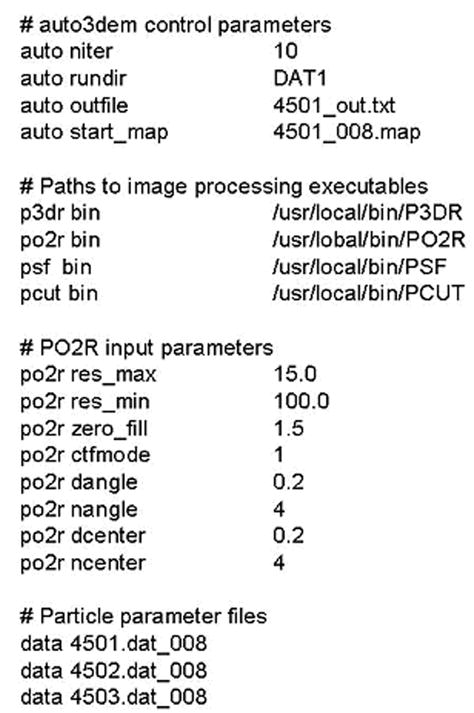
Short excerpt from AUTO3DEM input file illustrating the specification of AUTO3DEM control parameters, full paths to image processing programs, input for program PO2R, and data files. In all records except those listing data files, the first field specifies the hash used to store related data and the second and third fields form key value pairs within the hash.
Great care has been taken to maximize the flexibility of the format. Records can be listed in any order, extra white space (blank lines, leading and training spaces, additional spaces or tabs between fields) is ignored, and comments can be used throughout the file. Except for file names and string literals, all fields are case insensitive. Extensive error checking is done to make sure that valid program names and keys are specified.
4.2 Internal calculation of program parameters
Understanding and properly choosing the values for the input parameters is one of the most challenging hurdles in making effective use of image processing software. Sometimes values are chosen using empirical rules that become known only after extensive experience with the codes. In other instances, the optimal values can be derived either from the input data or from a previous step in the reconstruction. To help novice users quickly get up to speed and allow all users to work efficiently, AUTO3DEM sets reasonable default values and performs internal calculation of many of the parameters required by PO2R, P3DR, PFTsearch, PCUT, and PSF. A description of the key calculations is provided in the remainder of this section.
4.2.1 Inner and outer particle radii
The focus of most investigations is on the portion of the map corresponding to the ordered region of the viral structure. The interior of the virus, which typically contains disordered genomic material, and the region outside of the capsid contribute ‘noise’ to the reconstruction and should not be used in origin and orientation determination. In the early stages of the reconstruction process, where preliminary structural information is not available, it can be difficult to supply an accurate estimate for the inner and outer radii demarcating the ordered region of the particle. When running AUTO3DEM in SEARCH mode, the program PFTsearch generates the radially averaged correlation coefficient (CC) between projections of the model and raw particle images (Fig. 6). This coefficient drops to low values in disordered regions and can be used to derive values for the inner and outer radii. These are updated at the end of each iteration of the reconstruction process during SEARCH mode and then held fixed when REFINE mode is used.
Fig 6.
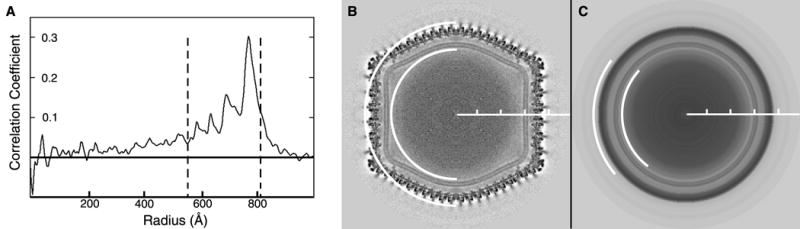
Determination of inner and outer particle radii by AUTO3DEM. (A) Plot of radially averaged CC between a 3D reconstruction of CIV and a subset of 84 particle images chosen from a full set of 6925 images. The summation over the radially averaged CC between the map and the particles is calculated over a sliding window whose default width is one eight of the box dimension. The boundaries of the window location that maximize the sum define the particle radii. The inner and outer boundaries are extended inwards or outwards, respectively, as long as the CC value does not drop below a threshold with a default value of one half of the maximum CC value. In this example with the CIV data, AUTO3DEM selected inner and outer radii of 565 and 805 Å, respectively (dashed lines). (B) Central section of CIV 3D map at a resolution of 13 Å viewed along a twofold axis. The box size is 2000 Å and the tick marks are 200 Å apart. White arcs define the inner and outer radii determined by AUTO3DEM (dashed lines in A). (C) Central section of spherically averaged CIV map. White arcs at 565 and 805 Å clearly illustrate that the radii selected by AUTO3DEM captures most of the highly ordered density and excludes disordered regions.
4.2.2 Low-pass and high-pass filters
The spatial frequency limits used during origin and orientation determination play a critical role in structure determination. The use of values outside of the optimal range can lead to reconstructions that are dominated by noise, overwhelmed by the low-frequency information describing the gross shape of the particle, or simply fail to reach the level of resolution that should be achievable given the available image data. AUTO3DEM sets the cutoff for the lower resolution (Reslo) to be equal to one-fifth of the size of the boxed particle image. The value of the upper resolution limit (Reshi) is determined to correspond to the spatial frequency at which the FSC curve drops below a specified threshold (default value of 0.3, but can be altered by the user) and is bounded by the Nyquist limit of double the pixel size.
The low-pass filter (1/Reshi) sets the upper bound of the spatial frequency spectrum to be used when comparing the image to projections of the model. However, the model itself and the FSC curve are calculated to a slightly higher spatial frequency limit. Rather than use a fixed offset in spatial frequency space to determine these higher resolutions, a fractional offset is employed to calculate the resolution of the map. The resolution of the map (Resmap) is calculated as follows: 1/(1/Reshi + δ1). To avoid sharp discontinuities, a Gaussian falloff is used in calculating the high-frequency portion of the map, with Resgauss given by 1/(1/Resmap + δ2). The constants δ1 and δ2 both have default values of 0.01, but these can be reset by the user. An upper resolution limit of 10 Å, for example, would result in a map calculated to a resolution of approximately 8.3 Å, with a Gaussian falloff starting at 9.1 Å.
4.2.3 Angular and spatial step sizes
When running AUTO3DEM in REFINE mode, the choice for the angular step size can significantly impact both the quality of the reconstruction and the time required for performing the calculation. Recall that REFINE mode carries out a local search of the (θ, ϕ, ω)-space surrounding the current orientation, rather than a global search over the entire ASU. To determine orientations to highest levels of accuracy, a region of the orientation space must be explored that is large enough to capture the true particle orientations, but small enough to avoid covering a range of (θ, ϕ, ω) larger than necessary. Ideally, the angular step size decreases as the resolution of the model improves. Experience has shown that an upper limit for the step size given by θ = (360 Reshi)/(2π d) works well, with Reshi defining the resolution limit from the last iteration and d, the box dimension, both measured in Angstroms. AUTO3DEM recalculates θ at the end of each iteration.
The value of Reshi can also be used to calculate the spatial step size used for the origin refinement. Consider a real-space model, f(x), and its corresponding transform, F(k). If the model is translated by a vector dr, then the new transform is simply the original transform multiplied by exp[2πidr·k]. Assuming a maximum acceptable phase residual of 45° (π/4) at the highest resolution |k|=1/Reshi, the inequality |dr| ≤ Reshi/8 is obtained. If we further assume that the contributions to |dr| from each component of the displacement, dx and dy, are equal, then dx becomes Reshi/16. For example, at a resolution of 20 Å, the step size would be equal to 1.77 Å.
4.3 Output, monitoring, and user intervention
AUTO3DEM generates distinctly named input and output files for each execution of the numerically intensive image reconstruction codes. This facilitates tracking of the input parameters that were used at each stage, troubleshooting the run to determine where the quality of the model deteriorated, or restarting in the event of hardware failure. The overall progress of AUTO3DEM is monitored in a master output file, which lists the jobs that were launched along with summary information such as the name of the executable, number of CPUs, and names of input and output files. The master file also lists the estimated resolution achieved at each iteration and summarizes changes made to the program input parameters. Before terminating, AUTO3DEM writes a new input file that can be used to restart the whole process.
AUTO3DEM is typically run without intervention, but users can manually reset parameters during the course of the reconstruction. To accomplish this, the user edits a separate parameter file that is interrogated after each of the iterations. If the timestamp of the file is more recent than the start of the last iteration, it is read and the contents are used to override the current values of the parameters. This feature can be used by highly experienced users to perform a limited degree of computational steering.
4.4 Particle selection
In most structure determination projects, not all of the particle images identified in the micrographs are used. Obviously bad images are usually eliminated at the onset of the project, but even images that pass initial screens will often be excluded at later stages of the reconstruction process for any of a number of reasons. For example, particles that show a preferred orientation in the micrograph may be left out during model construction in order to get a more representative sampling of views. More commonly, particles are rejected on the basis of some measure of the quality of fit between the image and the projection of the model that best matches the image.
The user can specify the ranges of allowed or prohibited values for the angles θ, ϕ, and ω. By default, particles of all orientations are considered. The user can also select particles on the basis of the quality metrics generated by PFTsearch or PO2R. Since the relative quality of the match between the image and model is generally what is considered, options are provided for selecting either a fraction of the best particles or the particles that have quality measures that are within a given number of standard deviations of the average quality. AUTO3DEM has the capability to generate and compare models using multiple selection criteria, thereby making it possible to determine which criterion yields a map with the highest estimated resolution.
5. Discussion
The AUTO3DEM process strategy has already been used by us and others to examine a variety of icosahedral viruses. Two examples of this are briefly described to illustrate how the procedure works with experimental data. In both examples, calculations were performed on a 44-node Linux PC cluster, where each node consisted of two 2.4 GHz Pentium 4 processors with a shared memory of 3 GB.
5.1.1 Chilo iridescent virus (CIV)
CIV is a large dsDNA virus (d = 1850 Å) of the family Iridoviridae that infects the rice stem borer insect. Images were recorded on an FEI/Philips CM300 FEG microscope at a nominal magnification of 33,000x with underfocus levels ranging from 0.8 to 3.0 μm. A total of 6925 particle images was extracted from 180 micrographs that were digitized with an effective pixel size of 4.24 Å. The starting map for the calculation was derived from a 26 Å resolution structure (Yan, et al., 2000). One cycle of refinement of the CIV data (4713 voxels) requires about 20 hours of wall clock time on 20 processors. Delays between executions of the programs significantly increased the amount of time required to complete this project. Eliminating these delays, thereby streamlining the structure determination process, was the initial motivation for the development of AUTO3DEM. The CIV structure was solved in conjunction with the development of AUTO3DEM and a resolution of 13 Å was ultimately achieved (Yan, et al., 2005).
5.1.2 Adeno-associated virus 1 (AAV1)
Adeno-associated viruses are small (d ~ 260 Å) ssDNA viruses belonging to the dependovirus genus of the family Parvoviridae. We studied the structure of AAV1, one of eleven known serotypes of AAV. Images were recorded on an FEI/Philips CM200 FEG microscope at 50,000x nominal magnification with defocus levels in the range of 1.1 to 5.6 μm. A total of 1700 particles was extracted from 11 micrographs, 1200 of which were used for constructing the final map of size 1313 voxels. Starting with a 15 Å resolution model, AUTO3DEM was run overnight (~12 hours; 25 cycles) without user intervention on 30 CPUs to reach a new resolution of 9.1 Å (Fig. 7). The benefits of using AUTODEM for relatively small data sets and map sizes were clearly demonstrated in this project. The run times for each cycle were relatively short (~1 hour), but without AUTO3DEM the time to solution would have been dominated by the delay between the submissions of individual jobs.
Fig 7.
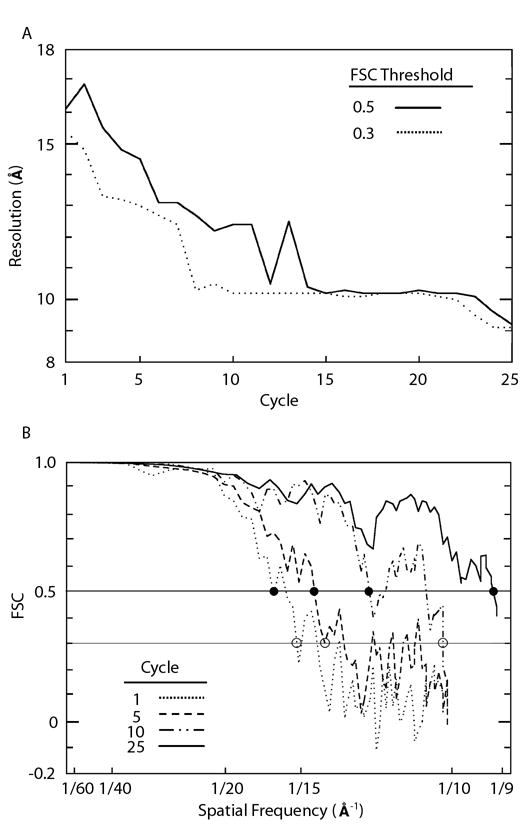
Progress of automated 3D image reconstruction for AAV1. A. Resolution at which the FSC curve drops below thresholds of 0.5 (solid line) and 0.3 (dotted line) as a function of cycle number. The resolution at which the FSC first drops below 0.5 is generally taken as the estimated resolution of the map, whereas the lower threshold is used to set the resolution limits used by programs PFTsearch, PO2R, and P3DR. The large fluctuations in the solid curve do not necessarily reflect variations in the quality of the map, but are an artifact of the noisy behavior of the FSC curve near the threshold. B. FSC curves at cycles one, five, ten, and twenty five. The horizontal lines indicate the FSC thresholds at 0.5 and 0.3. The solid and open disks show where the FSC curves first drop below 0.5 and 0.3 respectively.
5.2 Discussion and future enhancements to AUTO3DEM
AUTO3DEM has evolved into a reliable method for processing large data sets of icosahedral particle images, but remains a work in progress. Numerous enhancements will further expand its capabilities. For example, we plan to integrate the Leginon automated image acquisition system (Carragher, et al., 2000) with AUTO3DEM to allow users to carry out low-resolution image reconstructions on-line at the microscope. This would provide important feedback to the microscopist so that imaging conditions could be modified and optimized in real time. In addition, such immediate feedback would provide a basis for deciding whether a particular sample is appropriate for full scale data collection.
We also aim to extend the capabilities of AUTO3DEM so it can respond automatically in the event that the reconstruction procedure stalls at a particular resolution. It might prove advantageous to relax or tighten particle selection criteria, to identify and eliminate over-represented orientations that may dominate the reconstruction, or to modify the angular step sizes in a way that takes into account the individual histories of the particles over the course of the structure refinement.
Work is underway to further minimize the need for user input. Our goal is to calculate as many of the program parameters as possible so that the user would only be responsible for pre-processing the image data (e.g. boxing individual particles and determining defocus levels). Preliminary tests on a variety of experimental data indicate that AUTO3DEM can execute the RM and TM methods of image reconstruction in sequence without specifying anything other than the location of the image data.
Another area for improvement in AUTO3DEM is in the handling of the microscope contrast transfer function (CTF). At present, the defocus level is calculated only once for each micrograph and used unchanged for all of the images within the micrograph throughout the entire structure determination process. Work is underway for two enhancements to the calculation of the CTF. First, the defocus levels for the micrographs will be updated at each cycle of AUTO3DEM. A parallel program PCTFR (Parallel CTF Refinement: Ji and Marinescu, personal communication) already exists for this purpose and simply needs to be integrated into AUTO3DEM. Second, we are investigating techniques for evaluating the gradients in the defocus levels across a micrograph so that the CTF can be calculated individually for each particle image (Mindell and Grigorieff, 2003; van Heel, et al., 2000).
The use of AUTO3DEM is currently restricted to systems with icosahedral symmetry. This is due not to any inherent limitations of AUTO3DEM per se, but rather the underlying image processing code PFTsearch, which assumes the images are of particles with icosahedral symmetry. We plan to relax this constraint and extend the capabilities to other symmetries.
We will also be adding capabilities to AUTO3DEM that allow it to re-box particles in the digitized micrographs. During reconstruction, the origins of some particles in the images are found to be significantly different from those originally assigned by the particle identification software. While this can be indicative of poor particle images, often it is simply due to limitations of the image processing programs. Rather than discarding the images for which the particle origins deviate too far from the center of the box, the origins generated by PFTsearch or PO2R can be used as input to the particle identification software in an attempt to salvage the images.
Finally, it should be noted that the possibility always exists that an incorrect result may be obtained when using any automated image reconstruction system. The responsibility ultimately lies with the structural biologist to confirm the validity of the structure. The results should be consistent with biochemical and other relevant data and be confirmed whenever possible by independent means. Comparisons should also be made with the known structures of similar viruses or lower resolution maps of the same virus. In addition, if the CPU time required for the calculations is not prohibitive, it would be worthwhile to rerun the AUTO3DEM with slightly different initial conditions to test that the same structure emerges.
The source code for AUTO3DEM and the image reconstruction programs are available at http://cryoem.ucsd.edu/programs.shtm.
Acknowledgments
The authors thank W. Zhang, X. Zhang, J. Tang, Y. Ji, and D. Marinescu for valuable discussions that led to improvements in AUTO3DEM. This work was supported in part by grants R37 GM-033050 and R01 AI-055672 from the National Institutes of Health to TSB. RSS received partial support from the San Diego Supercomputer Center.
References
- Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, Pulokas J, Reilein A. Leginon: an automated system for acquisition of images from vitreous ice specimens. J Struct Biol. 2000;132:33–45. doi: 10.1006/jsbi.2000.4314. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. Resolution measurement in structures derived from single particles. Acta Crystallogr D Biol Crystallogr. 2000;56 ( Pt 10):1270–1277. doi: 10.1107/s0907444900009549. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22Å in ice. J Mol Biol. 1998;277:1033–1046. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]
- Ji Y, Marinescu DC, Zhang W, Zhang X, Yan X, Baker TS. A model-based parallel origin and orientation refinement algorithm for cryoTEM and its application to the study of virus structures. J Struct Biol. 2006;154:1–19. doi: 10.1016/j.jsb.2005.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Ludtke SJ. Electron cryomicroscopy of single particles at subnanometer resolution. Curr Opin Struct Biol. 2005;15:571–577. doi: 10.1016/j.sbi.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Marinescu DC, Ji Y. A computational framework for the 3D structure determination of viruses with unknown symmetry. J Parallel and Distrib Comput. 2003;63:738–758. [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, Patwardhan A. Single-particle electron cryo-microscopy: towards atomic resolution. Q Rev Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- Yan X, Chipman PR, Battisti AJ, Bergoin M, Rossmann MG, Baker TS. The Structure of the T=147 Iridovirus, CIV, at 13Å Resolution. Microsc Microanal. 2005;11(suppl 2):134–135. [Google Scholar]
- Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2006 doi: 10.1016/j.jsb.2006.07.013. (this issue), submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Olson NH, Van Etten JL, Bergoin M, Rossmann MG, Baker TS. Structure and assembly of large lipid-containing dsDNA viruses. Nat Struct Biol. 2000;7:101–103. doi: 10.1038/72360. [DOI] [PMC free article] [PubMed] [Google Scholar]


