Abstract
Radioprotection of DNA from direct-type radiation damage by histones has been studied in model systems using complexes of positively charged polypeptides (PCPs) with DNA. PCPs bind to DNA via ionic interactions mimicking the mode of DNA-histone binding. Direct radiation damage to DNA in films of DNA-PCP complexes was quantified as unaltered base release, which correlates closely with DNA strand breaks. All types of PCPs tested protected DNA from radiation, with the maximum radioprotection being approximately 2.5-fold compared with non-complexed DNA. Conformational changes of the DNA induced by PCPs or repair of free radical damage on the DNA sugar moiety by PCPs are considered the most feasible mechanisms of radioprotection of DNA. The degree of radioprotection of DNA by polylysine (PL) increased dramatically on going from pure DNA to a molar ratio of PL monomer:DNA nucleotide ~1:2, while a further increase in the PL:DNA ratio did not offer more radioprotection. This concentration dependence is in agreement with the model of PCP binding to DNA that assumes preferential binding of positively charged side groups to DNA phosphates in the minor groove, so that the maximum occupancy of all minor-groove PCP binding sites is at a molar ratio of PCP:DNA = 1:2.
INTRODUCTION
Chromatin DNA is tightly packaged by forming a complex with histone and non-histone proteins. This DNA-protein complex has long been known to confer protection of DNA against radiation damage (1). Radiosensitivity of DNA, quantified as the yield of double-strand breaks (DSBs), was demonstrated to increase on going from intact cells to naked DNA, with the partially stripped chromatin being intermediate in this order (2, 3). Stepwise removal of DNA-bound proteins from the chromatin increases the amount of radiation-induced DSBs (4, 5), with removal of core histones having the largest impact on the radiosensitivity of DNA (5).
Positively charged polyamines (PCAs) such as spermine, putrescine (3, 6–8) or oligolysines (9), which mimic histones in the mode of DNA binding, also afford radiation protection for DNA in completely or partially dehistonized chromatin, and they do so in a concentration-dependent manner. No radioprotection of DNA by spermine, however, was observed for native chromatin, consistent with the role of histones as the major radioprotectors (3).
Two types of radiation damage to DNA are traditionally recognized: indirect-type damage, which is due to attack of DNA by radicals that are produced from ionizations in the surrounding medium, and direct-type damage, which is due to direct ionization of DNA or transfer of electrons or holes to DNA from its solvent shell. So far, radioprotection of DNA by chromatin proteins and PCAs has been attributed to protection from the indirect effect. The proposed mechanism of protection is believed to be due to polyamine-induced compaction and aggregation of the DNA (PICA effect) as well as by scavenging of radicals (primarily, the •OH) at the sites of DNA-protein binding (6, 9–11).
It appears likely that chromatin proteins and PCAs also protect DNA from direct-type damage (12). If one accepts that about 50% of the total radiation damage to DNA in chromatin is due to the direct effect (13), a number of observations are difficult to explain by the indirect effect alone. For example, it is difficult to explain why DNA in dehistonized chromatin is approximately 2.5 times more radiosensitive than in the native chromatin, even in the presence of the radical scavenger DMSO (14). While DMSO would suppress the indirect effect, it would not suppress direct-type damage. Furthermore, the direct effect plays a significant role in the formation of clustered lesions, such as DSBs and SSBs with proximal base damage (15). Given the critical role of clustered lesions in biological end points, it would seem likely that part of the radioprotection afforded to DNA by chromatin packaging stems to a significant extent from altering the degree of clustering, which must include direct damage processes. All together, these points support the hypothesis that radioprotection of DNA by chromatin or model compounds is only partially due to protection against the indirect effect; the direct effect is expected to play a significant role.
To date, there are very few definitive experiments that provide data on how the direct effect is modified when DNA is packaged in chromatin, but some progress has been made recently. Irradiated ‘‘dry’’ or hydrated chromatin and DNA-histone complexes have been investigated by low-temperature EPR spectroscopy (16, 17). Weiland and Hüttermann (17) observed the free radical transfer from protein to DNA in chromatin X-irradiated at 77 K. They measured an approximately twofold net increase in radical concentration in DNA packaged in dry chromatin compared to DNA alone, with an increased contribution of electron-gain centers (pyrimidine radical anions) and reduction in electron-loss centers (guanine radical cations) in DNA packaged in dry chromatin. These results suggest that there is an influx of electrons and efflux of holes from DNA packaged in dry chromatin. The underlying mechanisms and how these are affected by sample temperature remain poorly understood.
The present paper focuses on the mechanism of radio-protection of DNA from direct-type damage by PCP complexes using such PCPs as polylysine (PL), polyarginine (PR) and polylysinetyrosine (PLT). The DNA-PL, DNA-PR and DNA-PLT complexes were used as a model of DNA-histone complexes. As a method of quantifying direct-type damage, this work is restricted to measurement of free unaltered base release, which is known to correlate with DNA strand breaks (1, 18–21).
MATERIALS AND METHODS
Materials
Calf thymus DNA, poly-L-lysine hydrochloride with a weight-average molecular weight of 68,600 Da, poly-L-arginine hydrochloride with a weight-average molecular weight >70,000 Da, and a random 1:1 copolymer of L-lysine and tyrosine hydrobromide with a weight-average molecular weight of 128,000 Da, all from Sigma, were used as substrates. DNA bases used as reference compounds in HPLC experiments were from Sigma. Aqua Sil, used for plate surface silylation, was from Hampton Research. All other reagents and solvents were used as received from Sigma-Aldrich Chemical Co. or Fisher Scientific Co.
Film Preparation
PLT hydrobromide solutions were dialyzed against 100 mM NaCl for 48 h to exchange the bromide anion for chloride. PL, PR and PLT hydrochloride solutions were dialyzed against water for 48 h to reduce the amount of low-molecular-weight fragments and filtered. Concentrations of dialyzed polypeptide solutions were determined spectrophotometrically on a Cary 100 spectrophotometer (Varian) using extinction coefficients of 313 and 377 M−1 cm−1 at 225 nm and for PL and PR, respectively, and of 306 M−1 cm−1 at 230 nm for PLT. Solutions for film preparation were made by slowly adding DNA solution to the polypeptide solution, or vice versa depending on the desired moles amino acid:mole DNA nucleotide ratio in the resulting solution, at 70°C under constant stirring. These conditions ensure formation of stable soluble DNA-PCP complexes, which otherwise tend to precipitate. Solutions of DNA-PCP complexes show significant light scattering; for this reason we did not determine concentrations of DNA and PCP in the complexes based on individual absorptions of DNA and PCP. Instead, we assumed that the concentrations of each component of the complex are the same as the concentrations of DNA and PCP in the solutions used for complex preparation (taking dilutions into account).
The DNA-PR films were grown from a solution containing 0.83 mM DNA and 2.64 mM PR (a molar ratio of PR amino acid to DNA nucleotide of 3.2:1). The DNA-PLT films were grown from the solution containing 0.55 mM DNA and 4.7 mM PLT (a molar ratio of PLT amino acid to DNA phosphate of 8.5:1). The molecular weight of the PLT monomer was calculated as the average of the molecular weights of the Lys and Tyr monomers. The following concentrations of DNA and PL and the ratios of PL amino acid:DNA nucleotide in DNA-PL solutions were used for film preparation: (i) 1.9 mM DNA, 0.46 mM PL (PL:DNA = 0.24:1); (ii) 1.4 mM DNA, 0.8 mM PL (PL:DNA = 0.57:1); (iii) 1.6 mM DNA, 0.92 mM PL (PL:DNA = 0.59:1); (iv) 1.4 mM DNA, 0.93 mM PL (PL:DNA = 0.67:1); (v) 1 mM DNA, 0.77 mM PL (PL:DNA = 0.77: 1); (vi) 0.65 mM DNA, 2.25 mM PL (PL:DNA = 3.5:1); (vii) 0.73 mM DNA, 3.24 mM PL (PL:DNA = 4.4:1); (viii) 1 mM DNA, 10 mM PL (PL:DNA = 10:1). Variations in concentration of DNA and PCP solutions used for film preparations did not affect the properties of the films provided that the PCP:DNA ratio remained the same.
A total of 100 μM sodium azide was added to all resulting solutions to prevent decomposition from bacterial activity. The presence of sodium azide was demonstrated to have no effect on the results described in this paper.
DNA-PL, DNA-PR, DNA-PLT, DNA, PL, PR and PLT films were grown in silylated petri dishes or nine-well plates under dehydrating conditions (oversaturated NaOH, 5% relative humidity) by stepwise addition of the corresponding solutions. DNA, PL, PR and PLT films were prepared from corresponding solutions with concentrations of each substrate close to concentrations in the DNA-PL, DNA-PR or DNA-PLT complexes. Films were allowed to dry completely before adding the next portion. Typically, 3-ml aliquots were used for films grown on petri dishes (total volume 18–21 ml) and 200-μl aliquots for films grown on nine-well plates (total volume 800–1600 μl). DNA-PL (i, vi–viii), DNA-PR, DNA-PLT and DNA films are uniformly transparent, while films made of PCP solutions are turbid. DNA-PL films (ii–v) with PL:DNA ratios close to 1:1 show partially microcrystalline structure, which is likely indicative of the sample heterogeneity. Films grown in petri dishes were detached from the dishes and cut into pieces for further irradiation, while films grown in nine-well plates were irradiated directly in the plates. The films grown over saturated NaOH are referred to as dry films. Continuous vacuuming of dry DNA-PL films (over 48 h) did not lead to any weight change, so the dry DNA-PL, DNA-PR or DNA-PLT films were ascribed the lowest hydration level considered for DNA under these conditions (Γ = 2.5) (22, 23). It should be noted, however, that the exact lowest hydration level for the DNA-PCP films was not measured.
Film Hydration
The DNA-PL, DNA and PL films used for the hydration experiment were grown in petri dishes or in nine-well plates. Two types of the DNA-PL films were prepared: with PL:DNA ratios of 3.5:1 and 10:1. All films were grown under dehydrating conditions (saturated NaOH). The films grown in petri dishes were detached from the dishes and cut into pieces. The pieces of the films were attached to the interior of preweighed aluminum boxes using a small grease droplet. It has been found that the grease does not chemically interact with the films. The boxes were covered with preweighed aluminum lids and weighed to obtain the weight of the dry film. For film hydration, open boxes with films were then placed into chambers over various saturated salt solutions [100% relative humidity, deionized water; 92% relative humidity, saturated K2HPO4; 81% relative humidity, saturated (NH4)2SO4; 77% relative humidity, saturated oxalic acid; 66% relative humidity, saturated NaNO2]. Relative humidities were measured using a portable hygrometer (Sigma). The films were hydrated for at least 1 month, with the film weight measured periodically to ensure that the films were equilibrated with water vapor. The level of film hydration (Γ = number of water molecules per DNA nucleotide or per PL amino acid for the PL films) was calculated as the mean difference in the weight of the hydrated film and the starting dehydrated one, based on weighing at least three samples. Films grown in nine-well plates were hydrated directly in the plates placed into chambers over various saturated salt solutions. Since it is impossible to measure Γ directly for films in nine-well plates, these films were assumed to have the same level of hydration as the pieces of film grown in petri dishes prepared from the same solutions and placed into the same chambers as the films in nine-well plates.
It should be noted that we do not know how water is distributed between DNA and PL molecules; hence we do not know how many water molecules are associated specifically with DNA at a given hydration level. To estimate the amount of water attached specifically to the DNA moiety in the DNA-PL films, Γ′DNA-PL, we measured hydrations of the DNA and PL films for the same relative humidities as for the DNA-PL films. For the DNA films, ΓDNA values measured by Milano and Bernhard (24) were used in conjunction with the experimental data obtained in the present work. We assumed that at a given relative humidity, water in the DNA-PL films partitions between DNA and PL moieties in the same proportions as in the sum of hydration of the DNA films, ΓDNA, and of hydration of the PL films, 3.5 ΓPL (ΓPL is the amount of water bound per PL amino acid; this value is multiplied by 3.5 to account for the PL: DNA ratio of 3.5:1 in the DNA-PL films). Although this approximation is not quite accurate, especially at high hydration levels, it allowed us to give a rough estimate of Γ′DNA-PL using the formula
where ΓDNA-PL is a non-specific hydration of the DNA-PL films per DNA nucleotide measured experimentally.
Film Irradiation
For irradiation of films in nine-well plates, each well was sealed with aluminum foil. Film pieces were irradiated under air in covered boxes made of aluminum foil. The aluminum foil was used to maintain constant humidity of the films; additionally, it acted as an X-ray filter to attenuate low-energy X rays.
Films were irradiated at room temperature with X rays generated by a Phillips X-ray tube with a tungsten anode operated at 55 kV and 20 mA. The dose rates were measured spectrophotometrically using radiochromic films (Far West Technology, Inc.). For films irradiated in nine-well plates or in aluminum boxes, the dose rates were 92 kGy/h and 161 kGy/h, respectively.
Immediately after irradiation, 40 mM ammonium acetate solution, pH 6.8, containing about 10 μM uracil as an internal standard for the quantitative reverse-phase HPLC was added to the films. The DNA, PL, PR and PLT films dissolved easily in buffer solutions, while the films consisting of DNA-PCP complexes did not dissolve completely and formed suspensions. Solutions or suspensions were quantitatively transferred to Eppendorf tubes, incubated at 90°C during 5 min, and cooled down. Then 25 mM spermine solution was added for DNA precipitation (no spermine was added for PCP films), and solutions were vortexed and centrifuged. Supernatants were then used for product analysis by HPLC.
Product Analysis
DNA cleavage products (free unaltered DNA bases) were detected and quantified by reverse-phase HPLC (Waters Millipore Model 510, Waters Associates Model 440 Absorbance Detector) on a Luna C18 4.6 × 250-mm column (Phenomenex) equilibrated with 40 mM ammonium acetate. A linear acetonitrile gradient (1–9.6% over 20 min at a flow rate 1 ml) was applied to elute the products. The products were identified by comparison with authentic reference compounds by retention time and the ratio of peak intensities at 254 and 280 nm. Radiation yields of released DNA cleavage products were calculated from peak areas at 254 nm as described previously (25). The radiation chemical yield of total base release, Gt, was obtained as a sum of radiation chemical yields of individual base release unless otherwise specified.
RESULTS
Release of Unaltered DNA Bases from Dry DNA-PL, DNA-PR and DNA-PLT Films
The release of free unaltered DNA bases was monitored by reverse-phase HPLC. To measure the effect of PL, PR and PLT on DNA direct-type damage, the amount of unaltered DNA bases released from the DNA-PL, DNA-PR and DNA-PLT films were compared with the amount of unaltered DNA bases released from the X-irradiated DNA films (Fig. 1 and Table 1). No detectable amounts of products relevant to DNA chemistry were observed in control experiments with irradiated PL, PR and PLT films (results not shown). It can be seen that all three types of PCPs significantly protect DNA from direct radiation damage, as is evident from comparison of base release from irradiated DNA films with that from irradiated DNA-PCP films. Interestingly, all three types of PCPs show very similar levels of radioprotection of DNA, with PR being only a slightly less potent radioprotector. In a control experiment, heating solutions of irradiated DNA with PL did not significantly change the free base release (results not shown), indicating that radioprotection by PCPs most likely occurs during irradiation as opposed to postirradiation.
FIG. 1.
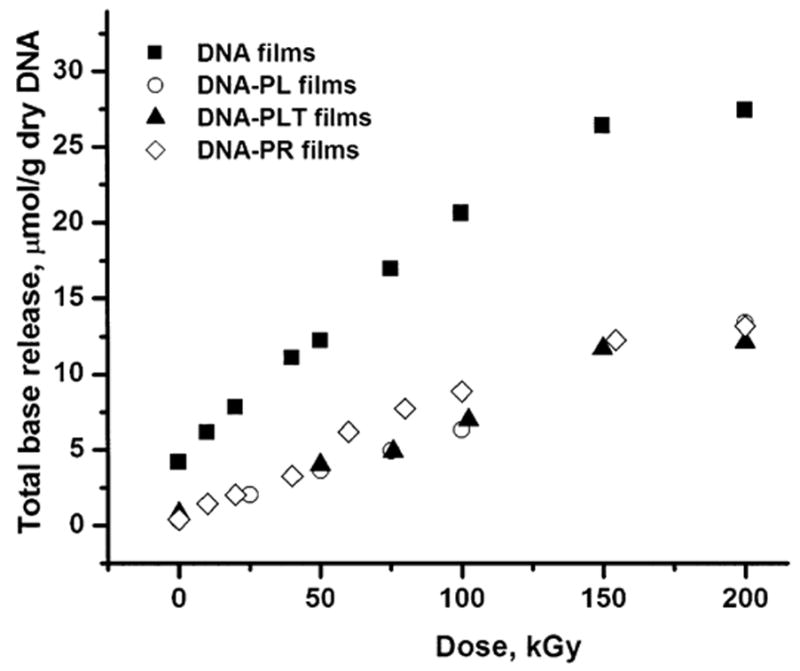
Dose dependence of total base release from dry (Γ = 2.5) DNA, DNA-PL (PL:DNA = 10:1), DNA-PR (PR:DNA = 3.2:1), and DNA-PLT (PLT:DNA = 8.5:1) films X-irradiated under aerobic conditions in nine-well plates. The films are labeled in the graph.
TABLE 1.
Radiation Chemical Yields of Unaltered Base Release (Gt) from Dry (Γ = DNA-PLT Films X-Irradiated in Air in Nine-Well Plates
| Film type | Release of cytosine (μmol J−1) | Release of guanine (μmol J−1) | Release of thymine (μmol J−1) | Release of adenine (μmol J−1) | Total base release (μmol J−1) |
|---|---|---|---|---|---|
| DNA | 0.041 (0.002)a | 0.037 (0.001) | 0.042 (0.002) | 0.042 (0.002) | 0.162 (0.007) |
| PL:DNA = 0.24:1 | 0.023 (0.002) | 0.018 (0.002) | 0.023 (0.001) | 0.030 (0.002) | 0.094 (0.006) |
| PL:DNA = 0.57:1 | 0.015 (0.001) | 0.011 (0.002) | 0.0142 (0.0006) | 0.0199 (0.0005) | 0.060 (0.004) |
| PL:DNA = 0.59:1 | 0.019 (0.003) | 0.021 (0.002) | 0.023 (0.003) | 0.029 (0.002) | 0.09 (0.01) |
| PL:DNA = 0.67:1 | 0.034 (0.002) | 0.027 (0.001) | 0.0302 (0.0007) | 0.038 (0.002) | 0.129 (0.006) |
| PL:DNA = 0.77:1 | 0.044 (0.004) | 0.037 (0.003) | 0.041 (0.003) | 0.050 (0.004) | 0.17 (0.01) |
| PL:DNA = 3.5:1 | 0.015 (0.002) | 0.014 (0.001) | 0.016 (0.001) | 0.0225 (0.0006) | 0.068 (0.005) |
| PL:DNA = 4.4:1 | 0.018 (0.003) | 0.011 (0.002) | 0.0149 (0.0004) | 0.023 (0.001) | 0.067 (0.006) |
| PL:DNA = 10:1 | 0.0178 (0.0003) | 0.016 (0.001) | 0.0151 (0.0007) | 0.016 (0.001) | 0.065 (0.003) |
| PR:DNA = 3.2:1 | 0.018 (0.0006) | 0.022 (0.001) | 0.022 (0.001) | 0.026 (0.002) | 0.088 (0.005) |
| PLT:DNA = 8.5:1 | NDb | NDb | 0.016 (0.001) | 0.019 (0.002) | 0.070 (0.006)c |
Mean ± 1 SD obtained by linear least-squares fit to the dose–response curves.
Not determined.
Cytosine and guanine were not quantified for the DNA-PLT films due to peak superimposition. Total base release was calculated as a doubled sum of thymine and adenine release.
A comparison of radiation chemical yields of total base release for the DNA and DNA-PL films (Table 1) shows that PL protects DNA from direct-type damage by approximately 2.5-fold, PLT by 2.3-fold, and PR by 1.8-fold. It is noteworthy that PLT in the DNA-PLT films gives essentially the same level of protection of DNA (Gt = 0.070 ± 0.006) as PL in the DNA-PL films (Gt = 0.065 ± 0.003) when both are in large excess (PL:DNA = 10:1 and PLT: DNA = 8.5:1), though only half of the residues in PLT carry a positive charge. A plot of total base release as a function of PL content is shown in Fig. 2. At low PL content, radioprotection of DNA increases with increasing the PL:DNA ratio, going from pure DNA films (Gt = 0.16) to PL:DNA = 0.24:1 (Gt = 0.094) to PL:DNA = 0.57:1 (Gt = 0.060). The dependence of on the PL:DNA ratio reaches a plateau above roughly 1:2, reaching its maximum radioprotection as measured by base release. However, it can be seen from Fig. 2 and Table 1 that at PL:DNA ratios between 0.5:1 and 1:1 there are large fluctuations in the total radiation yields of base release. We believe a likely cause of this is variations in the homogeneity in films formed with molar ratios ~1:1 due to formation of a mixture of complexed DNA and non-complexed DNA.
FIG. 2.
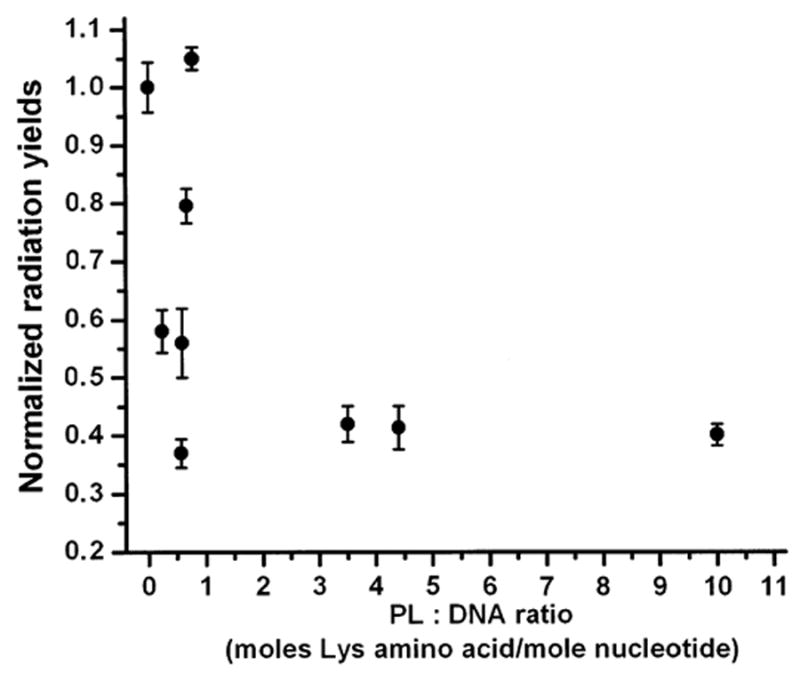
Dependence of radiation chemical yields of total base release from dry (Γ = 2.5) DNA and DNA-PL films X-irradiated under aerobic conditions in nine-well plates on the PL:DNA ratio. Radiation yields are normalized to the radiation yield of total base release from the DNA films.
Effect of the DNA-PL Film Hydration
To test the effect of film hydration on radioprotection of DNA by PL, the DNA, PL and DNA-PL films (PL:DNA = 3.5:1 or 10:1) were hydrated at different levels in chambers over various saturated salt solutions or, for the highest hydration level, over H2O. As expected, hydration levels for the DNA-PL films far exceeded those observed for the DNA films (22, 26) since water partitions between DNA and PL, with PL attaching more water than DNA due to its 3.5-fold or 10-fold molar excess. Table 2 and Fig. 3 show the dependence of base release on the hydration of DNA and DNA-PL films. For the DNA films, the dependence of the yields of total base release on the hydration level is generally consistent with the results obtained by Swarts et al. (22), with a decrease in base release on going from dry films to Γ ~ 15, followed by about a 60% increase in base release on further hydration [about a 50% increase in the yield of base release in DNA films on going from Γ ~ 15 to Γ = 33 was observed by Swarts et al. (22)]. The DNA-PL films respond differently. It can be seen from Fig. 3 that at low hydration levels, the yield of base release increases by about 30% on going from dry DNA-PL films (Γ = 2.5) to Γ = 10 for PL:DNA = 10:1 and by about 60% on going from dry DNA-PL films to Γ = 30 for PL:DNA = 3.5:1. Upon further hydration, the yield of base release is relatively constant. It should be noted that for the DNA-PL films, the values of Γ used in Fig. 3 and Table 2 show the amount of water distributed between DNA and PL rather than bound specifically to DNA. For this reason, the actual amount of water bound to DNA must be lower. The specific hydration of DNA in the DNA-PL films (PL:DNA = 3.5:1), Γ′DNA-PL, was estimated as described in the Materials and Methods. At low hydration levels, the dependence of non-specific hydration of the DNA-PL films, ΓDNA-PL, on relative humidity is described with fair accuracy as a sum of the dependences of ΓDNA and ΓPL as a function of relative humidity for the DNA and PL films, respectively (see Table 3). Based on these dependences, the plot of Gt as a function of Γ′DNA-PL in the DNA-PL films with PL:DNA = 3.5:1 is shown in the inset of Fig. 3.
TABLE 2.
Radiation Chemical Yields of Unaltered Base Release from DNA-PL Films Hydrated at Different Levels and X-Irradiated in Air in Nine-Well Plates
| DNA base release
|
||||||
|---|---|---|---|---|---|---|
| Film type | G | Release of cytosine (μmol J−1) | Release of guanine (μmol J−1) | Release of thymine (μmol J−1) | Release of adenine (μmol J−1) | Total base release (μmol J−1) |
| PL:DNA = 3.5:1 | 2.5 | 0.015 (0.002)a | 0.014 (0.001) | 0.016 (0.001) | 0.0225 (0.0006) | 0.068 (0.005) |
| 10 | 0.020 (0.001) | 0.015 (0.004) | 0.020 (0.001) | 0.025 (0.002) | 0.080 (0.008) | |
| 18 | 0.018 (0.003) | 0.013 (0.004) | 0.020 (0.001) | 0.030 (0.004) | 0.09 (0.01) | |
| 30 | 0.033 (0.004) | 0.014 (0.003) | 0.026 (0.002) | 0.040 (0.004) | 0.11 (0.01) | |
| 47 | 0.028 (0.004) | 0.013 (0.003) | 0.021 (0.003) | 0.032 (0.005) | 0.09 (0.02) | |
| PL:DNA = 10:1 | 2.5 | 0.013 (0.001) | 0.014 (0.001) | 0.011 (0.001) | 0.017 (0.002) | 0.055 (0.005) |
| 10 | 0.015 (0.002) | 0.018 (0.002) | 0.014 (0.002) | 0.024 (0.003) | 0.071 (0.009) | |
| 28 | 0.018 (0.002) | 0.019 (0.007) | 0.01 (0.01) | 0.031 (0.009) | 0.08 (0.03) | |
| 67 | 0.015 (0.005) | 0.015 (0.005) | 0.014 (0.005) | 0.018 (0.006) | 0.06 (0.02) | |
| 560 | 0.020 (0.002) | 0.012 (0.004) | 0.012 (0.004) | NDb | 0.06 (0.02)c | |
| DNA | 2.5 | 0.041 (0.002) | 0.037 (0.001) | 0.042 (0.002) | 0.042 (0.002) | 0.162 (0.007) |
| 7.5 | 0.031 (0.003) | 0.021 (0.002) | 0.033 (0.003) | 0.042 (0.003) | 0.13 (0.01) | |
| 10 | 0.030 (0.002) | 0.024 (0.002) | 0.032 (0.002) | 0.040 (0.003) | 0.126 (0.009) | |
| 12.5 | 0.0290 (0.0005) | 0.021 (0.002) | 0.031 (0.001) | 0.039 (0.002) | 0.120 (0.006) | |
| 22 | 0.040 (0.003) | 0.024 (0.002) | 0.041 (0.003) | 0.047 (0.003) | 0.15 (0.01) | |
| 33 | 0.055 (0.006) | 0.027 (0.003) | 0.043 (0.002) | 0.063 (0.007) | 0.19 (0.02) | |
Mean ± 1 SD obtained by linear least-squares fit to the dose–response curves.
Not determined.
The value for adenine from the column with Γ = 67 was used in calculating the total chemical yield.
FIG. 3.
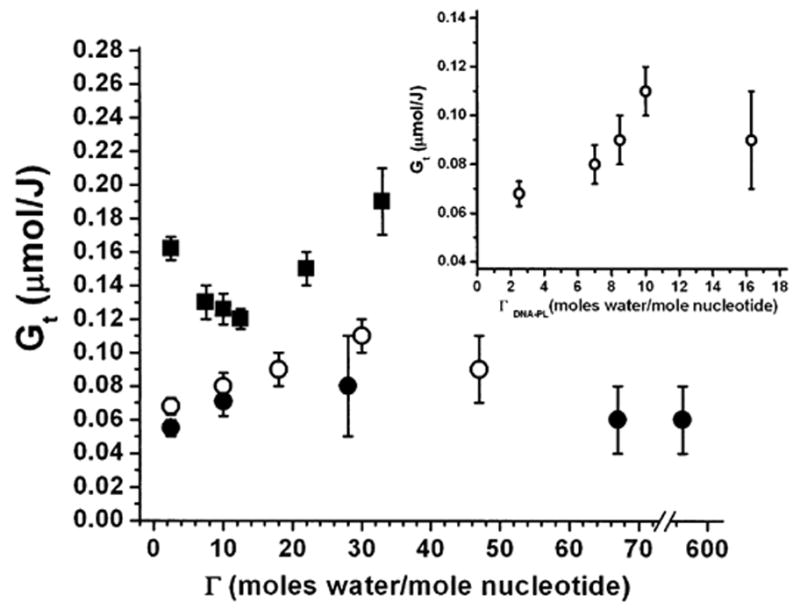
Dependence of radiation chemical yield of total base release (Gt) from DNA and DNA-PL films X-irradiated under aerobic conditions in nine-well plates upon film hydration. Squares, DNA films; open circles, PL-DNA films with PL:DNA = 3.5:1; closed circles, PL-DNA films with PL:DNA = 10:1. Inset shows the dependence of Gt on the estimated partial hydration of the DNA moiety, Γ′DNA-PL, in the DNA-PL films with PL:DNA = 3.5:1.
TABLE 3.
Hydration of DNA, PL and DNA-PL (PL:DNA = 3.5:1) Films
| Relative humidity (%) | ΓDNA DNA films | 3.5 ΓPLa PL films | ΓDNA-PLb DNA-PL films | ΓDNA + 3.5 ΓPL | Γ′DNA-PLc DNA-PL films |
|---|---|---|---|---|---|
| 66 | 7.5 | 3.2 | 10 | 10.7 | 7 |
| 77 | 10 | 11.2 | 18 | 21.2 | 8.5 |
| 81 | 12.5 | 25 | 30 | 37.5 | 10 |
| 92 | 22 | 41.4 | 47 | 63.4 | 16.3 |
Hydration of PL films, calculated per PL amino acid and multiplied by 3.5 to reflect the PL:DNA ratio of 3.5: 1 in the DNA-PL films.
Non-specific hydration of DNA-PL films, calculated per DNA nucleotide.
Estimated specific hydration of DNA only in the DNA-PL films, calculated as moles of water per DNA nucleotide attached to DNA only (see the Materials and Methods).
DISCUSSION
The yield of free unaltered bases released from DNA can serve as a marker of DNA damage. In a number of studies, the approach of HPLC-based quantification of free base release for determination of the radiation damage to DNA has been employed (21, 22, 25, 27, 28). It is now generally accepted that DNA strand scission stems almost exclusively from a chemically modified DNA sugar, which is produced via hydrogen abstraction at any of the five deoxyribose carbons (1, 19, 20). Sugar modification weakens the glycosidic bond, resulting in release of an unaltered free base. Eventually, a modified sugar results in a strand break. Some sugar alterations result in immediate or frank strand breaks without any additional treatment (1, 19, 29, 30); other types of sugar damage (so-called heat- or alkaline-labile lesions) require heat, alkaline or catalytic treatment to produce late strand breaks (19, 28, 31, 32). In rare cases, strand scission may be accompanied by release of a base attached to a sugar fragment (e.g. a base propenal) (19, 29). The latter process has been demonstrated to be a very minor one in irradiated DNA (33). Thus one can assume that, regardless of the type of radiation damage to DNA (direct or indirect), there is a strong correlation between the total yield of radiation-induced DNA strand breaks (immediate and late) and the total yield of free unaltered base release. Under conditions promoting complete decomposition of labile sugar lesions, the ratio of strand breaks and free base release should be close to 1:1, as demonstrated by Henle et al. (18) for DNA irradiated in aqueous solutions and then incubated at 37°C during 24 h. The postirradiation treatment employed in the present study (5 min at 90°C at pH 6.8) may not be sufficient to reveal all late strand breaks. In particular, decomposition of 2-deoxyribonolactones, which we proved in our earlier work (28) to be the major radiation-produced labile DNA lesions, requires a longer heat treatment in the presence of a catalyst. However, 2-deoxyribonolactones are abasic sites; their formation is preceded by release of a free unaltered base. The only known labile DNA sugar lesion that still contains a nucleobase is oligonucleotide 5′-aldehyde formed via the C5′ pathway of DNA sugar damage; the base release from this site requires longer heat and/or catalytic treatment (19, 34, 35). However, the contribution of the C5′ pathway to the radiation damage to DNA is, according to our unpublished data, quite low. Thus we can assume with fair accuracy that our post-irradiation treatment ensures practically complete release of free unaltered bases.
The release of altered bases from irradiated DNA was neglected in our study since this process appears to be very minor under our conditions. Only traces of released altered bases were observed in irradiated lyophilized DNA by Swarts et al. (22) and in the present work in irradiated DNA or DNA-PCP films.
Model of Interactions between DNA and PCPs
Polylysine protects DNA from direct radiation damage in a concentration-dependent manner (Fig. 2). The degree of radioprotection increases dramatically on going from pure DNA to PL:DNA ~1:2, while a further increase in the PL:DNA ratio does not offer more radioprotection. Thus one Lys group per base pair is enough for maximum radioprotection of DNA.
Polycations are believed to bind to the phosphate groups predominately in the DNA minor groove. This has been exemplified by spermine (36–38) and PCPs (39), in particular, PL, which appears to have modes of DNA binding similar to H1 linker histone (40, 41). The X-ray crystal structure of H1 linker histone (42) and circular dichroism studies of drug binding to DNA (43) indicate that positively charged groups of histones are inserted into the DNA minor groove. Assuming preferential binding of lysine amino groups to DNA phosphates in the minor groove, the maximum occupancy of all minor groove PL binding sites is at a molar ratio of PL:DNA = 1:2. This model of PL binding nicely fits the observation that PL at a molar ratio of PL: DNA ~1:2 offers the maximum radioprotection of DNA and the fact that a further increase in the PL:DNA ratio does not increase the degree of radioprotection. This suggests that only PL bound tightly to DNA affords radioprotection to DNA; excess PL forms a polypeptide matrix that plays no detectable role with respect to base release.
Based on the similar levels of radioprotection of DNA afforded by PL and PR (Fig. 1) and on the functional similarity of these two polypeptides (both are polymers of positively charged amino acids), it can be suggested that PR binds to DNA in the same fashion as PL. The same situation probably holds for the DNA-PLT complexes. Presumably the Lys groups bind tightly to the minor groove of DNA, while the Tyr groups form weaker hydrogen bonds with DNA and water. If this is the case, Tyr might play a smaller protective role than Lys. At the PLT:DNA ratio employed (8.5:1), the Lys groups in PLT can occupy all of the minor groove binding sites, which agrees with the similar radiation yields for DNA-PL and DNA-PLT films when the polyamino acids are in excess. All together, the above-mentioned model of Lys–DNA phosphate ionic interactions explains the finding that the maximum radioprotection of DNA is observed at a Lys:DNA nucleotide ratio as low as 1:2.
Mechanisms of Protection of DNA against Radiation Damage by PCPs
Four possible mechanisms of protection of DNA from direct radiation damage by PCPs in the DNA-PCP films are considered here. (1) PCP inhibits hole transfer from the solvent shell to DNA. (2) Holes formed on the DNA sugar-phosphate backbone transfer to PCP. (3) PCP repairs free-radical damage on the DNA sugar moiety by donating a hydrogen atom to a sugar free radical. (4) PCP maintains DNA in a more stable conformation, which is less susceptible to radiation damage. While mechanisms (1), (2) and (4) focus on events occurring shortly after DNA ionization, mechanism (3) occurs on a much slower time scale.
1. PCP inhibits hole transfer from the solvent shell to DNA
It has been demonstrated (20, 26, 44–46) that at low hydration levels (when water molecules forming the first hydration layer are very tightly bound to DNA) all holes on water are rapidly transferred to DNA. The efficiency of hole transfer from H2O+• is dictated by the proximity of the hole accepting moiety. Due to strong ionic interactions between negatively charged phosphate groups in DNA and positively charged Lys or Arg groups in PL or PR, the latter are likely located in proximity to the DNA backbone as well as to water molecules bound to DNA. Therefore, it seems likely that some portion of the holes transfer from H2O+• to PCP instead of DNA, thus reducing oxidative damage to DNA. Hole transfer from H2O+• to PCP is favorable because the PCP radical cations have a much lower oxidizing potential than H2O+•. Finally, the rate of irreversible deprotonation of PCP+•, yielding a neutral carbon-centered radical, should proceed at a competitive rate, which makes PCP a deep hole trap and hence further favors the process of hole transfer from H2O+• to PCP.
However, the experiments with the hydrated DNA and DNA-PL films do not support mechanism (1) playing a major role. The increase in DNA radiation damage in the DNA-PL films (PL:DNA = 3.5:1) in the 2.5 < Γ′DNA-PL < 10 range (see inset in Fig. 3) demonstrates that PL does not provide any essential degree of protection of DNA from the damage mediated by H2O+• formed from tightly bound water molecules. One can see from the inset in Fig. 3 that at Γ′DNA-PL about 10% of the radioprotective effect of PL practically disappears; at these values of Γ, both DNA and DNA-PL films show very similar yields of base release. At higher hydration levels, DNA damage in the DNA films increases by about 60% (see Fig. 3 and Table 2). DNA damage at high hydration levels is believed to be mediated by the hydroxyl radicals formed from outer loosely bound water molecules (20, 22, 26, 45, 46). Because no increase in DNA damage was observed in the DNA-PL films at high hydration levels, we argue that the hydroxyl radicals are efficiently scavenged by PL. This hypothesis is supported by the absence of base release in irradiated aqueous solutions of the DNA-PL complexes (results not shown).
Even without considering the film hydration data, a 2.5-fold radioprotection of DNA by PL cannot be explained solely by mechanism (1). The relative probability of initial ionization is assumed to be proportional to the electron density distribution and thereby to the number of electrons in a given moiety. There are 158.7 electrons per nucleotide, while at Γ = 2.5, water molecules + sodium give only 31 electrons. This means that the holes initially formed in the solvation layer (H2O + Na+) constitute only 20% of holes formed in DNA (20). Even if one assumes that all holes formed in the solvation layer are transferred to PL, only 20% protection of DNA against direct radiation damage could be provided. This estimation indicates the involvement of other mechanisms of radioprotection of DNA by PL.
2. Holes formed on the DNA sugar-phosphate backbone transfer to PCP
Possibility (2) requires that the rate of hole transfer from the DNA backbone to PCP is competitive with hole transfer to the DNA bases and irreversible deprotonation of the sugar (fixing the hole on the sugar). While we cannot rule out this possibility, it seems unlikely when one considers the relative oxidation potential and proximity of the DNA bases. Although the oxidation potentials of lysine and arginine are unknown, they certainly must be higher than that of adenine and guanine. Also, the distance between a hole located on the DNA backbone to the nearest purine base is not likely to be greater than the distance to the nearest Lys or Arg residue.
The experiment with the DNA-PLT films sheds some light on the possible involvement of mechanism (2). Thermodynamically tyrosine will compete with guanine as a hole trap. At pH 7, the redox potentials of tyrosine and guanosine are 0.93 V (47) and 1.29 V (48), respectively. The value for guanine in plasmid DNA is 1.39 V (49). Given that about half the residues of PLT are tyrosines, PLT should be an effective competitor for hole trapping. However, no increase in radioprotection of DNA was observed for the DNA-PLT films compared to the DNA-PL films (see Fig. 1 and Table 1). Of course, if the distance between the tyrosine residues and the DNA were large enough, this would explain the lack of change and consequently not discern against mechanism (2). While we have no direct measure of the steric arrangement between PLT and DNA, we do know that the DNA-PLT complex presents the same radioprotection profile as DNA-PL. And if our working hypothesis is correct with regard to the importance of the tightly bound fraction of PL (probably in the minor groove), then it follows that the tyrosine residues that are flanked by lysine residues in PLT must also lie in close proximity to the DNA. We argue, therefore, that mechanism (2) does not play a significant role in radioprotection of DNA by PCPs.
As noted in the Introduction, Weiland and Hüttermann (17), using low-temperature EPR spectroscopy, observed a slight decrease in guanine radical cations (oxidized guanine) and a large increase in one-electron reduced pyrimidines in dry chromatin compared to DNA, both X-irradiated at 77 K. While their results suggest that hole transfer may occur from DNA to chromatin protein, they do not provide information on hole transfer involving the DNA backbone. Likewise, our experiments do not provide information on the possibility of hole transfer between the DNA bases and PCP.
3. PCP repairs free radical damage on the DNA sugar moiety by donating a hydrogen atom to a sugar free radical
The efficiency of mechanism (3) depends on whether hydrogen transfer is energetically favorable. It is useful, therefore, to look at the difference in enthalpies of bond formation for the deoxyribose C-H bonds and the protein C-H, N-H or O-H bonds. While the energies of the C-H bonds in the aliphatic chain of the Lys or Arg side chains in PL or PR, respectively, are supposed to be rather high, the theoretically calculated energy of the α-C-H bond in a peptide with a Lys or Arg side chain is as low as ~340 kJ/mol (50). This is lower than the weakest of C-H deoxyribose bonds, C1′-H, which is ~376 kJ/mol according to the theoretical calculations of Miaskiewicz and Osman (51). Therefore, the process of transfer of a neutral free radical from deoxyribose to PL or PR, with the formation of a tertiary α-C-centered free radical, is expected to be thermodynamically favorable. Evidence for the formation of α-carbon radicals in chromatin at 77 K has been obtained by Weiland and Hüttermann (17), but it is not known to what degree, if any, the α-carbon radicals are formed by hole transfer from DNA. It should be noted that in the DNA-PLT films, possibility (3) does not exclude participation of Tyr in radioprotection of DNA. The theoretically calculated energy of the α-C-H bond in a peptide with a Tyr side chain is ~344 kJ/mol (50), which is only slightly higher than for Lys and still significantly lower than for the C-H bonds in deoxyribose. In terms of mechanism (3), therefore, the Tyr and Lys groups might be nearly equivalent in sugar-radical repair efficiency. This is consistent with PL and PLT giving the same level of radioprotection against base release from DNA.
4. PCP maintains DNA in a more stable conformation, which is less susceptible to radiation damage
This possibility stems from the finding, addressed in the section on mechanism (1), that there is a clear difference in the behavior of the irradiated DNA and DNA-PL films upon hydration (see Fig. 3 and Table 2). At low hydration levels, DNA films demonstrate a decrease in DNA damage, which was attributed by Swarts et al. (22) to DNA conformational changes from a pseudo-C conformation for a dry DNA to a B conformation for a hydrated DNA. It is well known that the DNA restructures upon progressive hydration (22, 52), although the information on how this restructuring affects radiation damage to DNA is very limited. According to Swarts et al. (22), DNA in the more flexible B form is less susceptible to radiation damage compared to the rigid pseudo-C form because of the stabilization of the base stacking interactions, which results in an increase in radical recombination reactions. It can be seen from Fig. 3 and Table 2 that, unlike the DNA films, the DNA-PL films with PL:DNA = 3.5:1 show an approximately 60% increase in DNA damage in approximately the same hydration range (when specific DNA hydration, Γ′DNA-PL, is calculated as moles of water attached to DNA only; see the inset in Fig. 3). At low hydration levels, the largest difference in radioprotection of DNA between the DNA and DNA-PL films is observed for Γ = 2.5. This could be a consequence of PL maintaining a B, or B-like, conformation even at the lowest hydration levels. Although the effect of polyamine binding on the DNA conformation is generally poorly understood, there are some indications that polyamines maintain DNA in the B form. Thus, in a Raman study by Deng et al. (53), it has been demonstrated that polyamine-bound genomic DNA maintains the native B-form conformation upon precipitation (condensation). In NMR studies of the complexes of polyamines with oriented fibers of DNA, it has been shown that at low hydration levels DNA in the complexes is in modified B conformations (54).
In conclusion, positively charged polypeptides complexed with DNA have been demonstrated to protect the DNA sugar-phosphate backbone against direct-type damage in dry DNA. This conclusion is based on a ~2.5-fold reduction in spontaneous unaltered base release from dry DNA-PL, DNA-PR and DNA-PLT films X-irradiated at room temperature. We hypothesize that the protection is attributed to the PCP-induced conformational changes of the DNA secondary structure and/or the partial repair of the deoxyribose radicals via hydrogen atom transfer from the α-C-H bonds of PCP. Thus the possibility is raised that protection of DNA against direct radiation damage in vivo, afforded by histones and other DNA-binding proteins, stems from repair by hydrogen atom transfer from the bound protein to deoxyribose radicals. Hole transfer from DNA to PCP or a competitive scavenging of holes generated in the primary solvation layer of DNA by PCP do not appear to contribute significantly to the radioprotection of DNAs by PCP. It was shown the while the degree of radio-protection of DNA by PL in moderately hydrated DNA (2.5 < Γ < 15) is lower than in dry DNA, it again increases upon further hydration (Γ > 15), most likely due to scavenging of hydroxyl radicals formed in the outer hydration layer by PL.
Acknowledgments
We thank David M. Close for useful discussions on this paper. The investigation was supported by PHS Grant 2-R01-CA32546, awarded by the National Cancer Institute, DHHS. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
References
- 1.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor & Francis; New York: 1987. [Google Scholar]
- 2.Ljungman M, Hanawalt PC. Efficient protection against oxidative DNA damage in chromatin. Mol Carcinog. 1992;5:264–269. doi: 10.1002/mc.2940050406. [DOI] [PubMed] [Google Scholar]
- 3.Chiu S, Oleinick NL. Radioprotection against the formation of DNA double-strand breaks in cellular DNA but not native cellular chromatin by the polyamine spermine. Radiat Res. 1997;148:188–192. [PubMed] [Google Scholar]
- 4.Sak A, Stuschke M, Wurm R, Budach V. Protection of DNA from radiation-induced double-strand breaks: Influence of replication and nuclear proteins. Int J Radiat Biol. 2000;76:749–756. doi: 10.1080/09553000050028896. [DOI] [PubMed] [Google Scholar]
- 5.Xue LY, Friedman LR, Oleinick NL, Chiu SM. Induction of DNA damage in gamma-irradiated nuclei stripped of nuclear protein classes: Differential modulation of double-strand break and DNA-protein crosslink formation. Int J Radiat Biol. 1994;66:11–21. doi: 10.1080/09553009414550901. [DOI] [PubMed] [Google Scholar]
- 6.Newton GL, Aguilera JA, Ward JF, Fahey RC. Effect of polyamine-induced compaction and aggregation of DNA on the formation of radiation-induced strand breaks: Quantitative models for cellular radiation damage. Radiat Res. 1997;148:272–284. [PubMed] [Google Scholar]
- 7.Warters RL, Newton GL, Olive PL, Fahey RC. Radio-protection of human cell nuclear DNA by polyamines: Radiosensitivity of chromatin is influenced by tightly bound spermine. Radiat Res. 1999;151:354–362. [PubMed] [Google Scholar]
- 8.Chiu S, Oleinick NL. Radioprotection of cellular chromatin by the polyamines spermine and putrescine: Preferential action against formation of DNA-protein crosslinks. Radiat Res. 1998;149:543–549. [PubMed] [Google Scholar]
- 9.Newton GL, Ly A, Tran NQ, Ward JF, Milligan JR. Radioprotection of plasmid DNA by oligolysines. Int J Radiat Biol. 2004;80:643–651. doi: 10.1080/09553000400005510. [DOI] [PubMed] [Google Scholar]
- 10.Newton GL, Aguilera JA, Ward JF, Fahey RC. Polyamine-induced compaction and aggregation of DNA—a major factor in radioprotection of chromatin under physiological conditions. Radiat Res. 1996;145:776–780. [PubMed] [Google Scholar]
- 11.Spotheim-Maurizot M, Ruiz S, Sabattier R, Charlier M. Radioprotection of DNA by polyamines. Int J Radiat Biol. 1995;68:571–577. doi: 10.1080/09553009514551561. [DOI] [PubMed] [Google Scholar]
- 12.Svoboda P, Harms-Ringdahl M. Influence of chromatin structure and radical scavengers on yields of radiation-induced 8-oxo-dG and DNA strand breaks in cellular model systems. Radiat Res. 2005;164:303–311. doi: 10.1667/rr3418.1. [DOI] [PubMed] [Google Scholar]
- 13.Krisch RE, Flick MB, Trumbore CN. Radiation chemical mechanisms of single- and double-strand break formation in irradiated SV40 DNA. Radiat Res. 1991;126:251–259. [PubMed] [Google Scholar]
- 14.Oleinick NL, Balasubramaniam U, Xue L, Chiu S. Nuclear structure and the microdistribution of radiation damage in DNA. Int J Radiat Biol. 1994;66:523–529. doi: 10.1080/09553009414551561. [DOI] [PubMed] [Google Scholar]
- 15.Goodhead DT, Nikjoo H. Clustered damage in DNA: Estimates from track-structure simulations. Radiat Res. 1997;148:485–486. [Google Scholar]
- 16.Faucitano A, Buttafava A, Marinotti F, Pedraly-Noy G. ESR study of the direct radiolysis of DNA, DNA-histones and DNA-intercalators complexes. Radiat Phys Chem. 1992;40:357–364. [Google Scholar]
- 17.Weiland B, Hüttermann J. Spin transfer from protein to DNA in x-irradiated ‘dry’ and hydrated chromatin: An electron spin resonance investigation of spectral components between 77 K and room temperature. Int J Radiat Biol. 2000;76:1075–1084. doi: 10.1080/09553000050111541. [DOI] [PubMed] [Google Scholar]
- 18.Henle ES, Roots R, Holley WR, Chatterjee A. DNA strand breakage is correlated with unaltered base release. Radiat Res. 1995;143:144–150. [PubMed] [Google Scholar]
- 19.Pogozelski WK, Tullius TD. Oxidative strand scission of nucleic acids: Routes initiated by hydrogen abstraction from the sugar moiety. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 20.Razskazovskiy Y, Debije MG, Bernhard WA. Direct radiation damage to crystalline DNA: What is the source of unaltered base release? Radiat Res. 2000;153:436–441. doi: 10.1667/0033-7587(2000)153[0436:drdtcd]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Debije MG, Razskazovskiy Y, Bernhard WA. The yield of strand breaks resulting from direct-type effects in crystalline DNA X-irradiated at 4 K and room temperature. J Am Chem Soc. 2001;123:2917–2918. doi: 10.1021/ja005790r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swarts SG, Sevilla MD, Becker D, Tokar CJ, Wheeler KT. Radiation-induced DNA damage as a function of hydration. I. Release of unaltered bases. Radiat Res. 1992;129:333–344. [PubMed] [Google Scholar]
- 23.Swarts SG, Becker D, Sevilla M, Wheeler KT. Radiation-induced DNA damage as function of hydration. II. Base damage from electron-loss centers. Radiat Res. 1996;145:304–314. [PubMed] [Google Scholar]
- 24.Milano MT, Bernhard WA. The influence of packing on free radical yields in solid-state DNA: Film compared to lyophilized frozen solution. Radiat Res. 1999;152:196–201. [PMC free article] [PubMed] [Google Scholar]
- 25.Razskazovskiy Y. Radiation-activated nuclease activity of o,o′-diphenyleneiodonium cations (DPI): A reductively initiated chain reaction involving the C1′ chemistry. Radiat Res. 2003;159:543–549. doi: 10.1667/0033-7587(2003)159[0543:ranaoo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 26.Milano MT, Bernhard WA. The effect of packing and conformation on free radical yields in films of variably hydrated DNA. Radiat Res. 1999;151:39–49. [PMC free article] [PubMed] [Google Scholar]
- 27.Razskazovskiy Y, Roginskaya M, Jacobs A, Sevilla MD. Reductively activated cleavage of DNA mediated by o,o′-diphenylenehalonium compounds. Radiat Res. 2000;154:319–325. doi: 10.1667/0033-7587(2000)154[0319:racodm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 28.Roginskaya M, Bernhard WA, Marion RT, Razskazovskiy Y. The release of 5-methylene-2-furanone from irradiated DNA catalyzed by cationic polyamines and divalent metal cations. Radiat Res. 2005;163:85–89. doi: 10.1667/rr3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breen A, Murphy J. Reactions of oxyl radicals with DNA. Free Radic Biol Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 30.Schulte-Frohlinde D. Radioprotectors and Anticarcinogens. Academic Press; New York: 1986. [Google Scholar]
- 31.Rydberg B. Radiation-induced heat-labile sites that convert into DNA double-strand breaks. Radiat Res. 2000;153:805–812. doi: 10.1667/0033-7587(2000)153[0805:rihlst]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Jones GDD, Boswell TV, Ward JF. Effects of postirradiation temperature on the yields of radiation-induced single- and double-strand breakage in SV40 DNA. Radiat Res. 1994;138:291–296. [PubMed] [Google Scholar]
- 33.Rashid R, Langfinger D, Wagner R, Schuchmann HP, von Sonntag C. Bleomycin versus OH-radical-induced malonaldehydic-product formation in DNA. Int J Radiat Biol. 1999;75:101–109. doi: 10.1080/095530099140852. [DOI] [PubMed] [Google Scholar]
- 34.Pratviel G, Pitie M, Bernadou J, Meunier B. Furfural as a marker of DNA cleavage by hydroxylation at the 5′-carbon of de-oxyribose. Angew Chem Int Ed Engl. 1991;30:702–704. [Google Scholar]
- 35.Pratviel G, Pitie M, Bernadou J, Meunier B. Mechanism of DNA cleavage by cationic manganese porphyrins: Hydroxylations at the 1′-carbon and 5′-carbon atoms of deoxyriboses as initial damages. Nucleic Acids Res. 1991;19:6283–6288. doi: 10.1093/nar/19.22.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korolev N, Lyubartsev AP, Laaksonen A, Nordenskiold L. On the competition between water, sodium ions, and spermine in binding to DNA: A molecular dynamics computer simulation study. Biophys J. 2002;82:2860–2875. doi: 10.1016/S0006-3495(02)75628-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sy D, Hugot S, Savoy C, Ruiz S, Charlier M, Spotheim-Maurizot M. Radioprotection of DNA by spermine: A molecular modelling approach. Int J Radiat Biol. 1999;75:953–961. doi: 10.1080/095530099139719. [DOI] [PubMed] [Google Scholar]
- 38.Schmid N, Behr JP. Location of spermine and other polyamines on DNA as revealed by photoaffinity cleavage with polyaminobenzenediazonium salts. Biochemistry. 1991;30:4357–4361. doi: 10.1021/bi00231a035. [DOI] [PubMed] [Google Scholar]
- 39.Sponar J, Votavova H. Selective binding of synthetic polypeptides to DNA of varying composition and sequence: Effect of minor groove binding drugs. J Biomol Struct Dyn. 1996;13:979–997. doi: 10.1080/07391102.1996.10508912. [DOI] [PubMed] [Google Scholar]
- 40.Hendrickson FM, Cole RD. Selectivity in the interaction of various DNA sequences with H1 histone. Biochemistry. 1994;33:2997–3006. doi: 10.1021/bi00176a032. [DOI] [PubMed] [Google Scholar]
- 41.Carotti D, Funiciello S, Lavia P, Caiafa P, Strom R. Different effects of histone H1 on de novo DNA methylation in vitro depend on both the DNA base composition and the DNA methyltransferase. Biochemistry. 1996;35:11660–11667. doi: 10.1021/bi9606051. [DOI] [PubMed] [Google Scholar]
- 42.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 43.Fodera R, Caneva R, Canzonetta C, Savino M. Chromatin accessibility to DNA minor groove ligands in vitro: Role of linker histones and amino-terminal domains of octamer histones. Boll Soc Ital Biol Sper. 2000;76:21–30. [PubMed] [Google Scholar]
- 44.Debije MG, Strickler MD, Bernhard WA. On the efficiency of hole and electron transfer from the hydration layer to DNA: An EPR study of crystalline DNA X-irradiated at 4 K. Radiat Res. 2000;154:163–170. doi: 10.1667/0033-7587(2000)154[0163:oteoha]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker D, La Vere T, Sevilla MD. ESR detection at 77 K of the hydroxyl radical in the hydration layer of gamma-irradiated DNA. Radiat Res. 1994;140:123–129. [PubMed] [Google Scholar]
- 46.LaVere T, Becker D, Sevilla MD. Yields of OH· in Γ-irradiated DNA as a function of DNA hydration: Hole transfer vs. OH· formation. Radiat Res. 1996;145:673–680. [PubMed] [Google Scholar]
- 47.DeFilippis MR, Murthy CP, Faraggi M, Klapper MH. Pulse radiolytic measurement of redox potentials: The tyrosine and tryptophan radicals. Biochemistry. 1989;28:4847–4853. doi: 10.1021/bi00437a049. [DOI] [PubMed] [Google Scholar]
- 48.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 49.Milligan JR, Aguilera JA, Ward JF. Redox equilibrium between guanyl radicals and thiocyanate influences base damage yields in gamma irradiated plasmid DNA. Estimation of the reduction potential of guanyl radicals in plasmid DNA in aqueous solution at physiological ionic strength. Int J Radiat Biol. 2001;77:1195–1205. doi: 10.1080/09553000110083988. [DOI] [PubMed] [Google Scholar]
- 50.Rauk A, Yu D, Taylor J, Shustov GV, Block DA, Armstrong DA. Effects of structure on alpha C-H bond enthalpies of amino acid residues: Relevance to H transfers in enzyme mechanisms and in protein oxidation. Biochemistry. 1999;38:9089–9096. doi: 10.1021/bi990249x. [DOI] [PubMed] [Google Scholar]
- 51.Miaskiewicz K, Osman R. Theoretical study on the deoxyribose radicals formed by hydrogen abstraction. J Am Chem Soc. 1994;116:232–238. [Google Scholar]
- 52.Brandes R, Rupprecht A, Kearns DR. Interaction of water with oriented DNA in the A- and B-form conformations. Biophys J. 1989;56:683–691. doi: 10.1016/S0006-3495(89)82715-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng H, Bloomfield VA, Benevides JM, Thomas GJJ. Structural basis of polyamine-DNA recognition: Spermidine and spermine interactions with genomic B-DNAs of different GC content probed by Raman spectroscopy. Nucleic Acids Res. 2000;28:3379–3385. doi: 10.1093/nar/28.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Dam L, Korolev N, Nordenskiold L. Polyamine-nucleic acid interactions and the effects on structure in oriented DNA fibers. Nucleic Acids Res. 2002;30:419–428. doi: 10.1093/nar/30.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]


