Abstract
Collagen XVII/BP180 is a transmembrane constituent of the epidermal anchoring complex. To study the role of its non-collagenous linker domain, NC16A, in protein assembly and stability, we analyzed the following recombinant proteins: the collagen XVII extracellular domain with or without NC16A, and a pair of truncated proteins comprising the COL15-NC15 stretch expressed with or without NC16A. All four proteins were found to exist as stable collagen triple helices; however, the two missing NC16A exhibited melting temperatures significantly lower than their NC16A-containing counterparts. Protein refolding experiments revealed that the rate of triple helix assembly of the collagen model peptide GPP10 is greatly increased by the addition of an upstream NC16A domain. In summary, the NC16A linker domain of collagen XVII exhibits a positive effect on both the rate of assembly and the stability of the adjoining collagen structure.
Keywords: BP180, transmembrane collagen, protein stability, protein folding, nucleation site
1. Introduction
Collagen XVII (also known as BP180 or BPAG2) is a major transmembrane component of the epithelial anchoring complex [1,2], a structure that functions in maintaining the adhesion of cells in the basal layer of stratified epithelia to the underlying basement membrane. The importance of collagen XVII in maintaining tissue architecture is underscored by genetic and acquired skin diseases in which its loss or lack of function leads to blister formation at the level of the dermal-epidermal junction (reviewed in [3]).
Biochemical studies from several groups have established that the collagen XVII protein exists as a transmembrane homo-trimer, with an N-terminal globular head region comprising the intracellular domain, and a long, highly extended, semi-rigid extracellular domain corresponding to a series of collagen triple helices (Gly-Xaa-Yaa tripeptide repeats) separated by non-collagenous interruptions (see Fig. 1) [4].
Fig. 1. Schematic representation of recombinant proteins of collagen XVII.
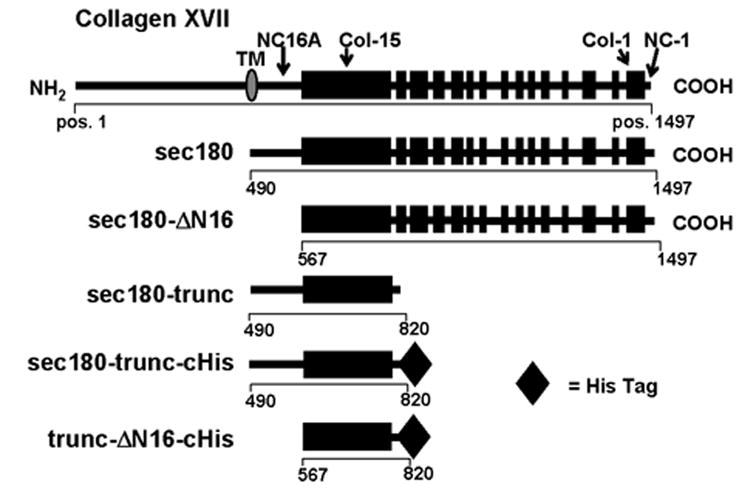
The extracellular domain of type XVII collagen (top diagram) consists of 15 collagenous domains (solid boxes) and 16 non-collagenous domains (horizontal lines). Also shown are schematic representations of the recombinant proteins used in this study. The corresponding AA positions are indicated for the termini of each protein (from GenBank accession NM_130778). The lengths (in AA) of sec180, sec180-ΔN16, sec180-trunc and trunc-ΔN16 are 1008, 931, 331 and 254, respectively. The His-tag, when present, adds 13 AA to the length of the protein. TM, transmembrane domain; pos, position.
It has been hypothesized that collagen XVII and other members of the membrane-associated collagen sub-family undergo triple helix assembly in the N-terminal (membrane-proximal) to C-terminal direction [5], rather than in the C-terminal to N-terminal direction as is typical for the “classical” collagens. Therefore, the membrane-proximal non-collagenous linker region of collagen XVII (designated NC16A; see Fig. 1), has been identified as a likely candidate for the site of association and initiation of triple helix assembly.
Supporting this idea is the fact that NC16A contains a predicted α-helical coiled-coil structure [4], as do the linker regions of the other members of the membrane-associated collagen sub-family [6]. Also it has been demonstrated that the carboxy-terminal region of collagen XVII is not required for triple helix formation [7]. More recently, Franzke and co-workers [8] reported that deletion of NC16A from full-length collagen XVII decreases its thermal stability. A disadvantage of using a transmembrane form of the protein for such an analysis is that a detergent is required for solubilization. In the present study we further probed the effects of NC16A on the stability and refolding of collagen XVII using soluble recombinant forms of the ectodomain of this protein.
2. Materials and Methods
2.1. Generation of recombinant forms of the collagen XVII ectodomain
The DNA vector pCEP4-sec180, which is based on the pCEP4 episomal expression vector (Invitrogen, Carlsbad, CA) and which encodes a secreted form of the collagen XVII ectodomain, was described previously [4,9]. pCEP4-sec180 was used as the template for PCR amplification of the segments of collagen XVII represented in Figure 1.
For use in the refolding experiments, a collagen model protein, consisting of 10 GPP triplets (GPP10), with an N-terminal NC16A moiety, was produced as a GST fusion protein using the pGEX-2T bacterial expression system (Pharmacia Biotech, Piscataway, NJ).
2.2. Peptide synthesis
A GPP10 synthetic peptide (sGPP10) was synthesized using Fmoc chemistry on an Applied Biosystems 432A Synergy peptide synthesizer (Applied Biosystems, Foster City, CA). Peptide purity was confirmed by MALDI-TOF (matrix-assisted laser desorption time-of-flight) mass spectrometry.
2.3. Immunodetection
Three rabbit antisera reacting with distinct epitopes on the extracellular region of collagen XVII were used in this study. Rabbit sera R594 and R136 react with sites in NC16A and near the C-terminus of collagen XVII, as previously described [9]. Rabbit antiserum, R5151, a generous gift from Dr. Kim Yancey, recognizes sites within the C-terminal two-thirds of the collagen XVII ectodomain.
Immunoblotting was carried out as previously described [9]. When the His-tag was needed as a means of detection, a mouse anti-penta-His antibody (Qiagen, Valencia, CA) was used.
2.4. Concentration by immunoprecipitation or by nickel-NTA affinity-purification
Immunoprecipitation was performed as described previously [9] to detect small amounts of the metabolically radiolabeled recombinant proteins in column fractions. Alternatively, when the His-tag was needed as a means of concentration, the samples were incubated for 2 h with Ni-NTA agarose beads (Qiagen, Valencia, CA).
2.5. Gel filtration chromatography
The recombinant forms of collagen XVII were analyzed by gel filtration chromatography using a Superose-6 column (Amersham, Piscataway, NJ), as previously reported [9].
2.6. Calculations
The frictional ratio (f/fo) was obtained using the following equations [4]:
| (1) |
| (2) |
where N is Avogadro’s number (6.022 1023 mol −1), η is the viscosity of water at 20 °C (1 x 10−3 kg m −1 s −1), ν¨ is the partial specific volume, ρ is the density of water at 20 °C (0.998 g/mL), rs is the Stoke's radius, and s20,ω is the sedimentation coefficient. The axial ratio, P, was calculated from the frictional ratio by fitting the protein as a cylinder using the equation:
| (3) |
2.7. Glycerol gradient sedimentation
The sedimentation properties of the collagen XVII recombinant proteins were analyzed by glycerol gradient sedimentation (linear gradient, 10% to 30% v/v), as previously described [4].
2.8. Chemical cross-linking
The various truncated forms of collagen XVII were subjected to chemical cross-link analysis using DSS (disuccinimidyl suberate; Pierce, Rockford, IL), as previously described [9].
2.9. Sensitivity to proteolysis
The thermal stability of the recombinant proteins was determined as previously described [9]. When the affinity purification was done by Ni-NTA purification, the reaction was performed on the 0.5 M imidazole eluate. The results were quantified by densitometry and the data points were fitted to sigmoidal curves using Sigma Plot (Jandel Scientific). The melting temperature, i.e., the temperature at which 50% of the proteins are unfolded, was determined based on the xo value from the resulting sigmoidal equation.
The rate of triple helix assembly of NC16A-GPP10 was calculated by monitoring the recovery of resistance to trypsin digestion. After denaturation at 70 °C for 20 min, 10 μl aliquots were incubated at 4 °C for varying lengths of time (from 5 min to 8 h). At the end of each incubation, duplicate samples received 10 μl of trypsin (100 μg/ml; Sigma, St. Louis, MO) and were incubated at 4 °C for 5 min. The reaction was stopped by adding 10 μl of soybean trypsin inhibitor (500 μg/ml; Sigma, St. Louis, MO). After incubation, the samples were analyzed by SDS-PAGE and immunoblotting. The kinetic data were fitted to exponential curves using Sigma Plot (Jandel Scientific) software package. The initial rate of assembly is the calculated slope of the tangent to the curve at time zero.
2.10. Circular dichroism
To monitor the rate of triple helix assembly, the synthetic GPP10 (sGPP10) was denatured by heating for 20 min at 70 °C, and then quickly loaded into a thermostatted 1 mm path-length quartz cell at 4 °C [10,11]. CD spectra of sGPP10 were acquired on a Jasco J715 spectropolarimeter. Kinetics of refolding were determined by monitoring the temporal change in ellipticity at a fixed wavelength of 228 nm. The ellipticity measurement was converted into the fraction of protein in the folded state following the equation:
where θobs is the observed ellipticity and θmax and θmin are the ellipticities of the folded and unfolded proteins, respectively, acquired from melting curves. The initial rate of assembly was calculated as described above.
Note about the refolding experiments
Due to technical considerations, different methods were employed to monitor the refolding of sGPP10 and NC16A-GPP10. There are two well-characterized methods for monitoring the assembly of collagens – CD spectropolarimetry and measuring resistance to proteolysis. While CD was ideal for following the folding of sGPP10, it was unsuitable for analyzing NC16A-GPP10 because the collagen-specific portion of this peptide’s CD spectrum was obliterated by its non-collagenous moiety. The refolding of NC16A-GPP10 was readily monitored using the trypsinization approach; however, this method could not be used for analyzing sGPP10, since this peptide is not trypsin sensitive (no Arg or Lys) and no antibodies are available for its detection in a Western blot.
3. Results
3.1. Analysis of the conformation of the recombinant collagen XVII fragments
A series of recombinant forms of collagen XVII (shown schematically in Fig. 1) were expressed in the human kidney cell line, 293-EBNA (see Figure S1). To facilitate detection and purification, one protein, trunc-ΔN16-cHis, was engineered with a C-terminal His-tag. The protein partner for trunc-ΔN16-cHis, sec180-trunc, was prepared both with and without a C-terminal His-tag.
Chemical cross-link analysis (Figure 2) revealed homo-trimeric conformations for sec180 (panel A, lanes 1–3), sec180-ΔN16 (panel A, lanes 4–6) and sec180-trunc (panel B, lanes 1–3), as well as for sec180-trunc-cHis (data not shown), indicating that the presence of the His-tag did not affect the ability of the protein to trimerize. Significantly, the truncated protein lacking NC16A, trunc-ΔN16-cHis, assayed at the same subunit concentration as those used for the other four proteins, exhibited no cross-linked products (Fig. 2B, lanes 4–6). However, at a 35-fold higher subunit concentration, the homo-dimeric and homo-trimeric forms of trunc-ΔN16-cHis were observed (Fig. 2B, lanes 7–9), albeit with a low trimer-to-monomer ratio.
Fig. 2. Cross-link analysis of collagen XVII recombinant proteins.
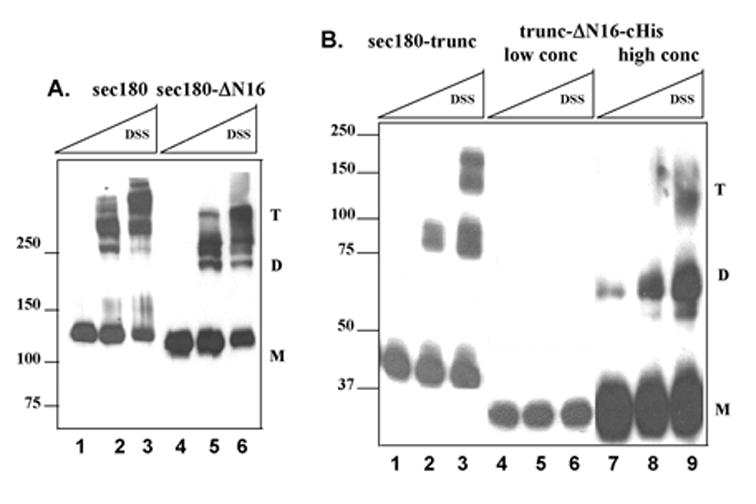
The collagen XVII truncation mutants were subjected to chemical cross-linking using disuccinimidyl suberate (DSS). Panel A, This figure is a Western blot of sec180 and sec180-ΔN16 treated with no DSS (lanes 1 and 4) or with DSS at 0.1 mM (lanes 2 and 5) or 0.5 mM (lanes 3 and 6). Panel B, This is a Western blot of sec180-trunc and trunc-ΔN16-cHis at low and high concentrations treated with no DSS (lanes 1, 4 and 7) or with DSS at 0.1 mM (lanes 2, 5 and 8) or 0.5 mM (lanes 3, 6 and 9). In both panels A & B, the positions of the monomer, dimer and trimer forms are indicated by "M", "D", and "T", respectively. The positions of the molecular weight markers are shown to the left of the blots.
Gel filtration analysis of the collagen XVII recombinant proteins provided additional insight into the multimeric nature of these proteins. As examples, the chromatographic profiles of sec180 and sec180-trunc are shown in Figure S2. For each of these two recombinant proteins, two distinct peaks were identified -- the first one, with the higher Stoke’s radius, corresponds to the trimeric form and the second one to the monomeric form of the protein. The gel filtration results for all of the recombinant proteins are shown in Table I. Interestingly, trunc-ΔN16-cHis exhibited only a monomeric peak, presumably due to its very low trimer-to-monomer ratio, as shown by cross-linking.
Table 1.
Structural parameters of collagen XVII recombinant proteins
| Average Stoke’s radius (nm) | ||||
|---|---|---|---|---|
| Recombinant proteins | Trimer | Monomer | sedimentation coefficient | Axial ratio (P) |
| sec180 | 13.6 +/− 0.4 | 9.1 +/− 0.6 | 5.3 +/− 0.2 | 63 |
| sec180- N16 | 14.3 | 9.0 | 5.5 +/− 0.4 | 81 |
| sec180-trunc | 10.3 +/− 0.07 | 5.1 +/− 0.4 | 2.6 | 79 |
| sec180-trunc-cHis | 11.2 | 5.3 | ||
| trunc- N16-cHis | undetectable | 4.5 +/− 0.3 | ||
The Stoke’s radius of each protein was determined from gel filtration chromatographic data (see Figure 4), and the sedimentation coefficient was obtained from glycerol gradient sedimentation profiles. The axial ratio was calculated as described in the text. Each result represents the average of 2 to 5 runs. The standard deviation is given only when at least 3 runs were performed.
The axial ratio P, the ratio of a protein’s length to width, can be deduced from the observed Stoke’s radius (Fig. S2 and Table I), sedimentation coefficient (data summarized in Table I) and molecular mass (see equations in Materials and Methods). The molecular mass of each trimer was calculated from the known amino acid sequence: sec180, 302.4 kDa; sec180- N16, 275.4 kDa; and sec180-trunc, 102.1 kDa. Using equation 1 and 2 to obtain the frictional ratio for each protein, and modeling them as cylinders (equation 3), the axial ratios of sec180, sec180- N16 and sec180-trunc are 63, 81 and 79, respectively (Table I). For comparison, collagen I has an axial ratio of 200 with a subunit length of about 1000 amino acids [12].
3.2. Thermal stability of the recombinant collagen XVII fragments
The denaturation of the triple helical regions of the various collagen XVII recombinant proteins was measured at various temperatures by monitoring the degradation of the protein by trypsin. Representative results are shown in Figure 3A, and a summary of the thermal denaturation results is presented in Figure 3B. Sec180-trunc and sec180-trunc-cHis had very similar Tm’s of 42 and 43 °C respectively; however, their counterpart lacking NC16A, trunc-ΔN16-cHis, had a significantly lower Tm (36 °C; p = 0.022), and its melting curve showed a more gradual transitional phase that extended from 25 °C to about 45 °C. The Tm’s of sec180 (46 °C) and sec180-ΔN16 (41 °C) were also shown to be significantly different (p = 0.007). When the two proteins lacking NC16A (sec180-ΔN16 and trunc-ΔN16-cHis) were compared, the longer of the two, sec180-ΔN16, was shown to have a significantly higher Tm (p = 0.037).
Fig. 3. Thermal denaturation analysis of collagen XVII recombinant proteins.
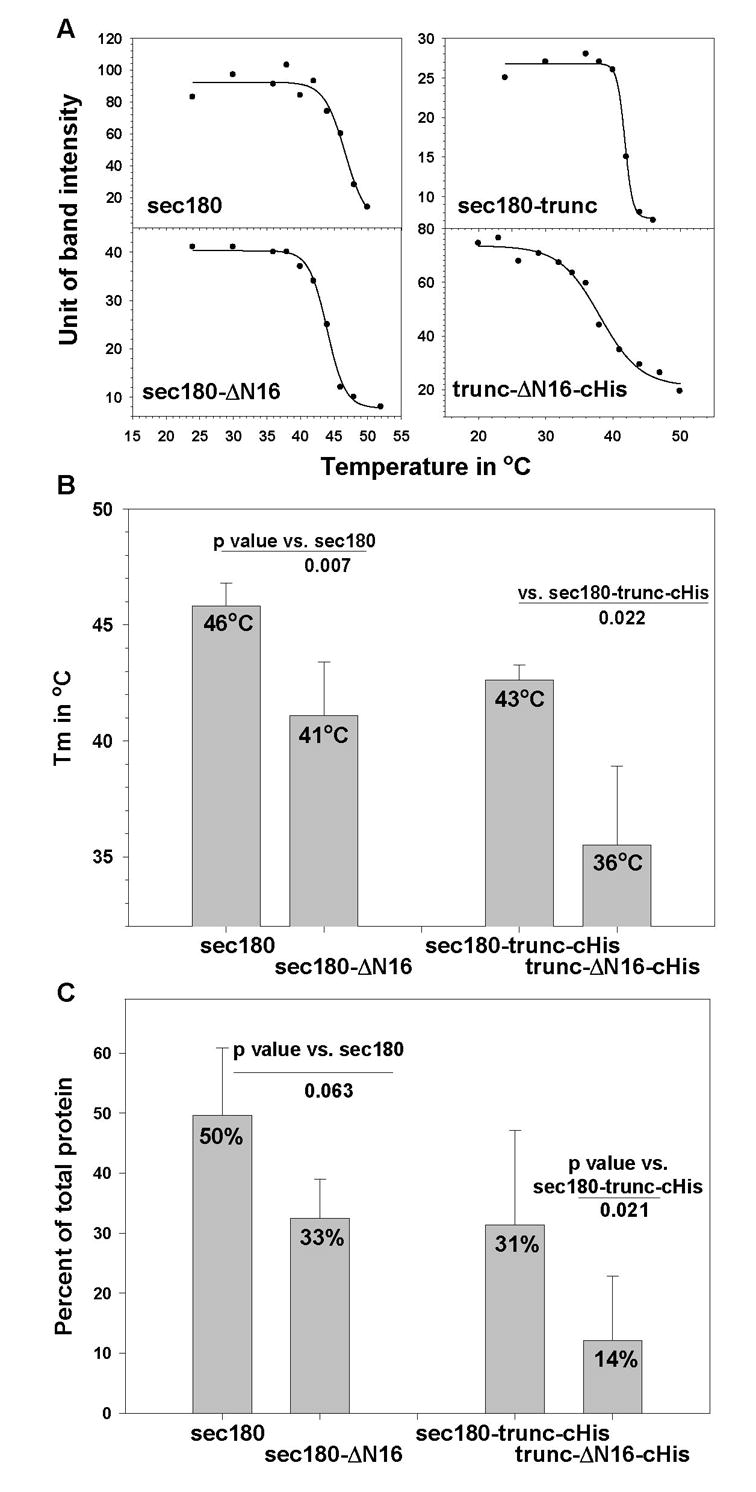
Panel A, This panel shows representative thermal denaturation curves that were generated by monitoring trypsin resistance. Panel B. The bar graph displays the mean melting temperature (Tm; 4–6 experiments) and standard deviation obtained for each protein. Panel C. Each recombinant protein was incubated with or without trypsin at 4 °C. Shown in the bar graph are the mean percentages of protein resistant to trypsin (4–8 digestions) and the corresponding standard deviations.
For each recombinant protein, we measured the percent of protein that is resistant to trypsin digestion at 4 °C. This provides information about the relative amounts of properly folded protein at steady state. As shown in Figure 3C, sec180-trunc-cHis exhibited a higher fractional level of trypsin-resistant material when compared with its counterpart that lacks the NC16A domain, trunc-ΔN16-cHis (p = 0.021).
3.3. Refolding of model proteins
NC16A-GPP10 and the sGPP10 were used in a series of experiments to compare their initial rates of in vitro triple helix assembly either by quantifying the level of trypsin-resistant material for NC16A-GPP10 or by following the ellipticity by circular dichroism at 228 nm for sGPP10. Figure 4A shows the refolding results obtained for NC16A-GPP10 at 40 μM and sGPP10 at 200 μM. Technical problems prevented us from measuring refolding of the two proteins at the same concentration. Forty μM was the maximum concentration that we were able to achieve for NC16A-GPP10, and refolding of sGPP10 at 40 μM was too slow to obtain an accurate initial rate. To circumvent this problem, we determined the initial rate of refolding of sGPP10 at 40 μM by extrapolation using a linear regression of the logarithm of the initial refolding rate of sGPP10 versus the logarithm of the peptide concentration (Figure 4B). Based on this analysis, the rates of refolding of sGPP10 and NC16A-GPP10 (both at 40 μM) were 2.3 μM/hour and 61.7 μM/hour, respectively, a 27-fold difference. The initial rate of sGPP10 at 40 μM obtained by extrapolation is based on the assumption that the reaction order does not change over the concentration range of 40–200 μM. This is a reasonable assumption since Boudko et al. showed that this was the case for a GPP10 peptide [10]. In addition, any change in reaction order that were to occur over this concentration range would result in a reaction rate lower than that obtained by the linear extrapolation and would increase the difference in initial rates exhibited by NC16A-GPP10 and sGPP10.
Fig. 4. Refolding of model proteins.
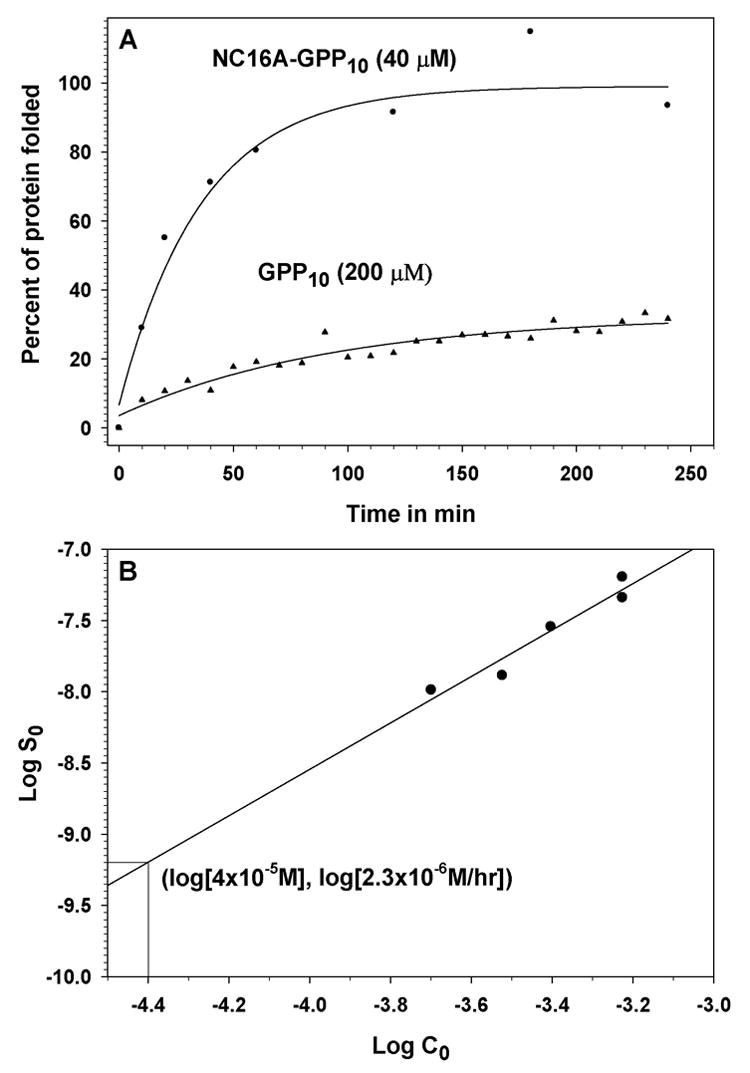
Panel A, This plot shows refolding curves for the model protein NC16A-GPP10 (closed circles) and the synthetic collagen peptide sGPP10 (closed triangles). Panel B, This plot shows the logarithm of initial rate (in M/sec) of triple helix formation of sGPP10 as a function of the logarithm of total chain concentration (in M).
4. Discussion
This study was designed to test the hypothesis that the NC16A domain of collagen XVII contains elements that influence the stability and rate of assembly of the adjacent triple helical domain. The approach involved structurally comparing, in a pair-wise fashion, recombinant forms of collagen XVII, with or without NC16A.
Our results demonstrated that the presence or absence of the NC16A domain had no major impact on the global conformation of the protein. The larger protein pair exhibited highly similar Stoke’s radii, axial ratios and cross-linking results, consistent with a semi-rigid rod-like structure. A complete structural comparison of the shorter collagen XVII protein pair was not possible, since the truncated protein without NC16A was expressed largely as a monomer. While the trimeric form of this protein was detectable by cross-linking only at high subunit concentrations, and was undetectable by gel filtration, it did yield a typical collagen spectrum by circular dichroism (data not shown). The dramatic differences in the trimer-to-monomer ratios for the two truncated proteins, revealed in both the protein structure and thermal denaturation analyses, indicated that the presence of NC16A leads to an increase in the steady state level of the correctly assembled protein form. This phenomenon is likely due to a difference in protein stability, although a difference in the rate of protein folding could also be a contributing factor.
The thermal denaturation experiments revealed that all four proteins were expressed in two distinguishable forms – one that is highly trypsin resistant (presumed to be folded correctly) and the other that is sensitive to trypsinization at 4 °C (presumed to be misfolded). For both pairs of collagen XVII proteins, the presence of NC16A resulted in a statistically significant increase in collagen triple helix stability (quantified by their melting temperatures), though this difference was more pronounced in the shorter protein pair. It is noteworthy that the denaturation of the truncated protein lacking NC16A occurred over a broad temperature range, suggestive of structural heterogeneity. This means that some of the trimers that do assemble might well be misaligned. These findings are consistent with previous data [8] and support the hypothesis that the NC16A domain facilitates chain association, forming a nucleus for triple helix assembly [10].
One key point from these studies is that two recombinant forms of collagen XVII – one missing the N-terminal NC16A and the other missing the C-terminal portion – are capable of folding into a stable conformation that resembles the native structure. Therefore, neither end of the ectodomain contains a site that is essential for proper folding. It must be stressed, however, that this does not exclude the terminal regions as potential sites of nucleation. Rather, the proper folding of these recombinant proteins is likely due to self-nucleation occurring at sites with sufficient numbers of GXP repeats, as shown for synthetic collagen peptides [11].
To address the question of whether NC16A contains a nucleation site, defined as a site that accelerates the folding of a protein, we measured the rate of triple helix assembly for the two collagen model peptides, sGPP10 and NC16A-GPP10. Our analysis revealed that the NC16A-containing peptide refolded at a rate 27 times higher than that of its counterpart, sGPP10. Based on these data, we can conclude that NC16A has a positive effect on one, or both, of the processes associated with triple helix assembly, i.e., subunit association and helix propagation [10]. At the low subunit concentrations used in this study it is very likely that the rate limiting step was subunit association and not helix propagation [10]. To distinguish between these possibilities an in-depth kinetic analysis of the effects of NC16A on triple helix formation will be carried out.
In addition, our structural studies revealed important clues about the overall shape of collagen XVII. For example, the N-terminal one-third of the collagen XVII ectodomain (consisting of NC16A, col-15 and NC15) accounted for 76% of the Stoke’s radius of the entire ectodomain (sec180) and exhibited an axial ratio very similar to that of a collagen triple helix of comparable size. In contrast, while sec180 and collagen I are very similar in terms of amino acid count, the axial ratio of sec180 is significantly lower. These findings strongly support a model in which the N-terminal one-third of the collagen XVII ectodomain is highly extended and rigid, while the remainder is highly flexible and might have a stable conformation in which this tail is folded back on itself (see Fig. S3). Such a model has previously been proposed based on ultrastructural data [3,13].
In conclusion, we have presented a systematic analysis of the structure of the membrane-proximal portion of the collagen XVII ectodomain. We have shown that the presence of the NC16A domain of collagen XVII confers a statistically significant increase in both the stability and the rate of assembly of the adjacent collagen triple helix.
Supplementary Material
The abbreviations used are
- AA
amino acid
- CD
circular dichroism
- Col
collagen
- NC
non-collagen
- DMSO
dimethyl sulfoxide
- DSS
disuccinimidyl suberate
- ECL
enhanced chemiluminescence
- HRP
horseradish peroxidase
- TM
melting temperature
Footnotes
This work was supported by grants from the National Institutes of Health (AR048901 to F.V.d.B.; AR040410 to G.J.G.).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Diaz LA, Ratrie H, 3rd, Saunders WS, Futamura S, Squiquera HL, Anhalt GJ, Giudice GJ. Isolation of a human epidermal cDNA corresponding to the 180-kD autoantigen recognized by bullous pemphigoid and herpes gestationis sera. Immunolocalization of this protein to the hemidesmosome. J Clin Invest. 1990;86:1088–94. doi: 10.1172/JCI114812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giudice GJ, Emery DJ, Diaz LA. Cloning and primary structural analysis of the bullous pemphigoid autoantigen BP180. J Invest Dermatol. 1992;99:243–50. doi: 10.1111/1523-1747.ep12616580. [DOI] [PubMed] [Google Scholar]
- 3.Van den Bergh F, Giudice GJ. BP180 (type XVII collagen) and its role in cutaneous biology and disease. Adv Dermatol. 2003;19:37–71. [PubMed] [Google Scholar]
- 4.Balding SD, Diaz LA, Giudice GJ. A recombinant form of the human BP180 ectodomain forms a collagen-like homotrimeric complex. Biochemistry. 1997;36:8821–30. doi: 10.1021/bi970675n. [DOI] [PubMed] [Google Scholar]
- 5.Pihlajaniemi T, Rehn M. Two new collagen subgroups: membrane-associated collagens and types XV and XVII. Prog Nucleic Acid Res Mol Biol. 1995;50:225–62. doi: 10.1016/s0079-6603(08)60816-8. [DOI] [PubMed] [Google Scholar]
- 6.Snellman A, Tu H, Vaisanen T, Kvist AP, Huhtala P, Pihlajaniemi T. A short sequence in the N-terminal region is required for the trimerization of type XIII collagen and is conserved in other collagenous transmembrane proteins. EMBO J. 2000;19:5051–9. doi: 10.1093/emboj/19.19.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Areida SK, Reinhardt DP, Muller PK, Fietzek PP, Kowitz J, Marinkovich MP, Notbohm H. Properties of the collagen type XVII ectodomain. Evidence for n- to c-terminal triple helix folding. J Biol Chem. 2001;276:1594–601. doi: 10.1074/jbc.M008709200. [DOI] [PubMed] [Google Scholar]
- 8.Franzke CW, Tasanen K, Borradori L, Huotari V, Bruckner-Tuderman L. Shedding of collagen XVII/BP180: structural motifs influence cleavage from cell surface. J Biol Chem. 2004;279:24521–9. doi: 10.1074/jbc.M308835200. [DOI] [PubMed] [Google Scholar]
- 9.Fu CL, Giudice GJ, Van den Bergh F. Protein structural analysis of BP180 mutant isoforms linked to non-Herlitz junctional epidermolysis bullosa. J Invest Dermatol. 2006;126:232–4. doi: 10.1038/sj.jid.5700024. [DOI] [PubMed] [Google Scholar]
- 10.Boudko S, Frank S, Kammerer RA, Stetefeld J, Schulthess T, Landwehr R, Lustig A, Bachinger HP, Engel J. Nucleation and propagation of the collagen triple helix in single-chain and trimerized peptides: transition from third to first order kinetics. J Mol Biol. 2002;317:459–70. doi: 10.1006/jmbi.2002.5439. [DOI] [PubMed] [Google Scholar]
- 11.Buevich AV, Silva T, Brodsky B, Baum J. Transformation of the mechanism of triple-helix peptide folding in the absence of a C-terminal nucleation domain and its implications for mutations in collagen disorders. J Biol Chem. 2004;279:46890–5. doi: 10.1074/jbc.M407061200. [DOI] [PubMed] [Google Scholar]
- 12.Wallace DG. The role of extrahelical peptides in stabilization of collagen fibrils. Biopolymers. 1990;30:889–97. doi: 10.1002/bip.360300904. [DOI] [PubMed] [Google Scholar]
- 13.Nonaka S, Ishiko A, Masunaga T, Akiyama M, Owaribe K, Shimizu H, Nishikawa T. The extracellular domain of BPAG2 has a loop structure in the carboxy terminal flexible tail in vivo. J Invest Dermatol. 2000;115:889–92. doi: 10.1046/j.1523-1747.2000.00136.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


