Abstract
‘Gene' has become a vague and ill-defined concept. To set the stage for mathematical analysis of gene storage and expression, we return to the original concept of the gene as a function encoded in the genome, basis of genetic analysis, that is a polypeptide or other functional product. The additional information needed to express a gene is contained within each mRNA as an ensemble of signals, added to or superimposed onto the coding sequence. To designate this programme, we introduce the term ‘genon'. Individual genons are contained in the pre-mRNA forming a pre-genon. A genomic domain contains a proto-genon, with the signals of transcription activation in addition to the pre-genon in the transcripts. Some contain several mRNAs and hence genons, to be singled out by RNA processing and differential splicing. The programme in the genon in cis is implemented by corresponding factors of protein or RNA nature contained in the transgenon of the cell or organism. The gene, the cis programme contained in the individual domain and transcript, and the trans programme of factors, can be analysed by information theory.
Keywords: gene, genon, gene expression, regulation, information theory
Introduction
What is a ‘gene'? Surprisingly, in the world of biology and genetics there is no longer a straightforward answer (cf. Pearson, 2006). For instance, Snyder and Gerstein (2003) define a gene as ‘a complete chromosomal segment responsible for making a functional product' and then discusses five criteria for identifying genes in the DNA sequence of a genome. The most common feeling is that it should be a piece of nucleic acid. At the onset of molecular biology (Benzer, 1959), the significance of the term gene was clear: it was the unit of function identified by genetic methods, as colours of flowers, the shape of a wing, number and shape of bacterial colonies on a Petri dish. This analysis had nothing to do with DNA nor RNA but functions exclusively. According to current insight in molecular biology, the only meaningful conception of a gene is the one of a functional and not of a hereditary unit (see for example Brosius, 2006).
The concept of the cistron (contiguous genomic elements acting in cis, essentially the protein coding sequence) introduced by Benzer (Benzer, 1959, 1961; Benzer and Champe, 1961) and extended by Jacob and Monod (1961) related the gene to an un-interrupted piece of DNA, able to complement a function in a cis/trans test. The equation function = gene = polypeptide = continuous piece of DNA=cistron seemed acceptable in first approximation. However, when several genes were found to constitute an ‘Operon' (Jacob and Monod, 1961), representing a programme of gene expression, other problems arose with the introduction of the notion of regulatory genes; for instance, the gene coding for the lac-repressor protein. The latter has to attach to the operator, a DNA sequence placed in cis upstream of the genes in the operon. The operator—is it (part of) a ‘gene'? The lac function is based on operator action, thus it is related to the phenotype; but the lac repressor gene is not part of the cistrons controlled by the operator.
With the advent of eukaryotic molecular biology, the problem of defining the gene became even more complicated. In eukaryotes, the tight physical complex linking transcription and translation in bacteria does not exist; the polyribosomes are removed from the DNA, which is stored away in the nucleus. As a consequence, the dimensions of space and time entered gene expression (see Figure 1, inset A; and The Cascade of Regulation (Scherrer and Marcaud, 1968)) and new types of controls had to be considered, in particular at the level of the, by now, autonomous messenger RNA (mRNA). There is an untranslated region (UTR) of about 50–250 nt (Gray and Hentze, 1994; Hess and Duncan, 1996) preceding the coding sequence in the mRNA, and at the end of the mRNA chain the 3′-side UTR which, surprisingly, in some genes (for example, the Prion mRNA) grew to become longer than the coding sequence. Being contiguous and in cis, upstream and downstream of the coding sequence, such a construct of nature did not fit the original concept of the gene.
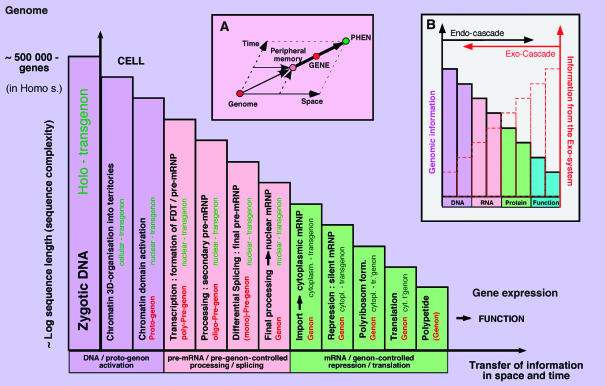
Figure 1.
The Cascade of Regulation (Scherrer, 1967, 1980): The information content of the zygotic genome is gradually reduced to that expressed in a differentiated cell. In Homo sapiens an estimated 500 000 polypeptide-genes are reduced to a few hundred in gradual steps; as few as 3 genes may account for up to 90% protein output, as is the case in red blood cells. The Holo-Cascade (not shown) includes additional steps, leading upstream from the information content of an entire species to that of populations and individuals, and downstream from the polypeptide to the assembled protein including all post-translational modifications (Scherrer, 1980). Under the direction of the holo-genon and holo-transgenon, the genomic information is reduced by DNA rearrangements to that of an individual cell, and then by individual steps to the expression of an individual function, as shown here and outlined in the text. These may include the following (1–2) chromatin modification and activation (protogenon-dependent); (3) transcription and formation of pre-mRNP (pre-genon); (4–6) gradual processing and splicing (pre-genon); (7) export and formation of cytoplasmic mRNP (genon); (8–9) activation (de-repression) of mRNP (genon); (10) translation of mRNA (genon) followed by peptide formation (genon has expired) and gene expression. (A) The spatial transfer of a genomic transcript to RNA processing centres (Iarovaia et al, 2001) and the nuclear periphery and (specific sites in) the cytoplasm (De Conto et al, 1999) induces a delay and, hence, a vector in time. Processing and transport steps may be temporarily interrupted, and lead to considerable delay of expression (up to 30 years in the case of histone mRNA in human oocytes), and the constitution of ‘peripheral memories' (Scherrer, 1974, 1980), from where the gene will be created and/or expressed, eventually. These may be in the form of unspliced (fragmented genes) or finally spliced pre-mRNPs, cytoplasmic mRNPs or miRNA complexes. (B) The genomic information being gradually reduced along the Cascade, information from the external space of the cell and organism plays a gradually increasing function. Highest at the periphery, concerning for example all cell-surface receptor functions, some external instructions may reach the genomic DNA to bring about a physiologic change. This can be conceived as an ‘exo-cascade' as proposed here, ‘infiltrating' the ‘endo-cascade' of gene expression.
Another problem arose with the observation that mRNA was able to form mRNA–protein (mRNP) complexes. It was found that specific proteins recognise and attach to specific sequence motifs along the mRNA chain, and not only in the UTRs, but right inside the coding sequence, as could be proven early on for globin mRNAs (Dubochet et al, 1973). This indicated that, superimposed onto the coding sequence, there must exist protein-binding sites most likely subject to a particular code of interaction. ‘Free' mRNPs, found in vivo outside the translation machinery of the polyribosomes, are not translatable in vitro, unless most of the mRNP proteins are removed (Civelli et al, 1980); these proteins seem hence to relate to repression. RNPs may also form higher order complexes (cf. review in Dreyfuss et al, 2002), assembled by interaction of (pre-)mRNA with proteins and protein complexes (De Conto et al, 1999), or cellular structures, such as, the cytoskeleton (Singer, 1992); they constitute part of the backbone of the nuclear matrix (De Conto et al, 2000; Razin et al, 2004; Ioudinkova et al, 2005).
A fatal blow to the original gene concept came with the observation of giant precursor RNA and their processing (Perry, 1962; Scherrer and Darnell, 1962; Georgiev et al, 1963; Scherrer et al, 1963; Perry et al, 1964) (review in (Scherrer, 2003)). After the experimental observation of pre-mRNA (Scherrer et al, 1970), the discovery of ‘splicing' (Berget et al, 1977; Chow et al, 1977) implied the fragmentation of the coding sequence at the genomic level: in most cases, only fragments and not intact genes are stored in DNA. According to the original genetic definition of the gene, and the cistron concept, this indicated that, each time it needs to be expressed, the gene has to be created from its parts encoded in the DNA (Figure 1, inset A; cf. disc. in Scherrer, 1980, 1989; Brosius and Gould, 1992). Interestingly, the occurrence of differential splicing and, as a consequence, the fact that the same DNA domain can contain the information for different genetically identifiable functions, indicated clear separation of the gene as a function from its genomic counterpart in the form of DNA, transmitted from generation to generation. Accordingly, these two matters might also be separated conceptually and in terminology.
The discovery of ‘polycistronic' giant RNA (Scherrer et al, 1966) (review in Scherrer, 2003) and the formulation of the pre-mRNA concept (Scherrer et al, 1966; Scherrer and Marcaud, 1968) made it particularly evident that it is impossible to concentrate into a single term all types of information involved in the expression of a single genetic function. To lead out of the actual confusion we suggest to break up the process of gene expression into its basic mechanistic, and thus logical units, namely gene function on the one hand and the mechanisms of storage and expression on the other. In this process, one is led to propose new concepts and terms that give precise definitions to comprehend gene expression in terms of Molecular Biology, and make it possible to analyse gene storage and expression in terms of information processing. Our intention is to present here a functional and information theoretic analysis of gene storage and expression.
Gene definition, expression and regulation
The basic system of information involved in gene expression, and the one most easily defined, is the coding sequence contained within the mRNA and its counterpart after translation, the nascent polypeptide. This leads back to the original definition of the gene, the unit of genetic function and analysis as used by Mendel and Morgan. It implies the polypeptide chain as the underling basic unit of function, or its equivalent, the uninterrupted nucleic acid stretch of the coding sequence, the ‘cistron' of Benzer (1959). This nucleic acid stretch emerges at the level of the mRNA in eukaryotes, and in most cases is not present at the DNA level as an uninterrupted sequence. In the discussion below, this should be the unique and exclusive definition and meaning of the term ‘gene'.
Attached to such a gene is the ‘history' of its ‘creation' from pieces in the genome before its expression; in other words, along with the transcript comes a programme that secures the formation of the mRNA and its expression in time and space. This programme will be conceptualised as the genon. Within this programme, two kinds of elements of control may be distinguished: (1) the cis-acting signals, which form (oligo-)sequence motifs contained in the same strand of DNA or RNA as the fragments of the coding sequence, and (2) the trans-acting factors which act on the signals placed in cis. Both participate in the programme that secures the generation of the gene, in the cellular space and in time, through the many steps of gene expression.
The genon concept concerns gene expression at large. Some genetic information, however, is only indirectly related to gene expression, like the 3D organisation of DNA and chromatin (see the Unified Matrix Hypothesis (Scherrer, 1989), and the recent observation of ‘3D-gene regulation' (Spilianakis et al, 2005)). Furthermore, epigenetic mechanisms of gene expression and transmission modify the genon in cis and its precursors at DNA level; the genon is, thus, flexible and not a rigid programme. Quite in general, we consider here only regulation directly related to gene expression, leaving out other types of signalling and metabolic controls. These points will be detailed in a more extensive analysis of gene expression and the genon concept (Scherrer and Jost, submitted to Th Biosci).
Types of information related to gene expression and regulation
Before developing the genon concept, we describe the types of information involved in gene expression, as well as the nature of the products. Gene expression results in the synthesis of products that may be either protein or RNA in nature; these products may carry out a given structural or enzymatic function, or may control the gene expression pathway in a mechanistic or regulative manner. Accordingly, we should make a distinction between protein genes (P-genes) and RNA genes (R-genes) on the one side, and between structural genes (s-genes) and controlling genes (c-genes). Combined, we have sP-genes and sR-genes as well as cP-genes and cR-genes.
The P-gene is the equivalent of the triplet-based coding sequence in the mRNA
The protein-coding sequence is the equivalent of the gene in the mRNA, being defined by genetic analysis carried out at the level of the phenotype. The outcome of this analysis constitutes the genotype as the ensemble of defined functions, which may be transmitted by heredity. Such physiological functions are based on the expression of an ensemble of unit functions. The unit function, subject to mutation, is carried by the polypeptide in its nascent form. The actual function is exerted in general by a quaternary protein or RNP complex, which may integrate several identical and/or different proteins, possibly modified chemically, as well as low-molecular-weight cofactors of organic or inorganic chemical nature.
The unit of a coding sequence is the triplet of nucleotides which, according to the genetic code, directs during translation of an mRNA the choice of a given anticodon carried by a given tRNA. Owing to the degeneracy of the code, incorporation of an identical amino acid may be directed by different triplets. As a consequence, according to the triplet chosen for a given amino acid, at the level of the mRNA a different nucleotide sequence is formed. The resulting different secondary structure of the nucleic acid may be ‘recognised' by proteins interacting with the mRNAs, or by small interfering RNAs (siRNAs)/micro RNAs (miRNAs) controlling RNA interference (RNAi).
Structural protein genes (sP-genes)
By definition, structural protein genes contribute to cellular structure and function either directly or via enzymatic activities. They may constitute the building blocks of the nuclear and plasmatic membranes, the endoplasmic reticulum, the nuclear matrix and the cytoskeleton. As enzymes they govern the intermediary metabolism as well as protein, RNA or lipid biosynthesis and degradation. The proteins acting as the mechanistic and enzymatic carriers of the system of protein biosynthesis do not discriminate among specific types of DNA, pre-mRNA or mRNA. Among them are, for example, the RNA polymerases, the non gene-specific splicing factors, the nonspecific transport factors such as ‘exportin' (Rodriguez et al, 2004; Kutay and Guttinger, 2005) or NLS (nuclear localisation signal) binding to RNA sequences (Rodriguez et al, 2004), the translation initiation and elongation factors, the proteins binding the CAP motif on the 5′-start or the poly(A) tail of the mRNA.
Regulatory protein genes (cP-genes)
Regulatory protein genes control gene expression from transcription to translation; they may function as repressors or activators of transcription, or act at post-transcriptional levels by interaction with pre-mRNA and mRNA. Four sets of such regulatory nucleic acid binding proteins (NABPs) can be distinguished: (1) the transcription factors of non-histone type as well as the histone- and DNA-modulating factors, which control local remodelling of chromatin, regulating access of the transcription machinery to DNA, (2) The nuclear pre-mRNA binding proteins (pre-mRNPs or hnRNPs), which interact, in specific sets, with given types of pre-mRNA, in statu nascendi as well as at the level of the nuclear matrix; there are several hundreds of relatively acidic proteins bound in part by hydrophobic bonds, and relatively fewer (some dozens) basic proteins bound by ionic interaction (the ‘histone-type' of pre-mRNP proteins) (Maundrell et al, 1979). (3) The cytoplasmic proteins binding in variable sets the non-translated mRNAs (‘silent' or (ribosome-)‘free' mRNPs); these proteins bind in general by hydrophobic interaction, they act positively in guiding the cytodistribution of mRNA, and negatively as cytoplasmic repressors (Maundrell et al, 1979). (4) The prosome particles (also known to constitute the core of the 26S proteasomes), a population of protein complexes built of 2 × 14 subunits in variable composition (Schmid et al, 1984), which bind on one side to chromatin, pre-mRNA and cytoplasmic repressed mRNA (review in Scherrer and Bey, 1994), and on the other to the nuclear matrix (De Conto et al, 2000; Ioudinkova et al, 2005) and the cytoskeleton (Arcangeletti et al, 2000).
Among these cP-gene products, two types can be distinguished: (1) those that act on specific individual genons in mRNAs, thus regulating the expression of specific genes, and (2) those that control the expression of whole sets of genes or gene families. Among the latter are, for instance, some types of transcription and translation factors.
Structural RNA genes (sR-genes)
The most important member of this class of RNA is the ribosomal RNA (rRNA), which serves as the scaffold of ribosomal subunits by organising the sequential alignment of ribosomal proteins (Scheer and Hock, 1999; Tschochner and Hurt, 2003). In addition, ‘rRNA is a ribozyme' (Steitz and Moore, 2003).
The metabolic precursor of rRNAs (Perry, 1962; Scherrer and Darnell, 1962; Scherrer et al, 1963) in the small (16S and 18S rRNA, respectively in prokaryotes and eukaryotes) and large (23S/28S rRNA) ribosomal subunits is the nascent pre-rRNA (45S in eukaryotes), which functions in a similar way to align not only the proteins ending up in the final ribosome, but also other proteins that, in eukaryotic cells, never leave the nucle(ol)us. The latter proteins play a structural role in ribosome biosynthesis and the nucleolar dynamic architecture; pre-ribosomes form the fibrillar centre of the nucleolus, whereas the final ribosomal subunits constitute its granular zone (Scheer and Benavente, 1990).
Regulatory RNA genes (cR-genes)
Among the regulatory RNAs operating in gene expression we have to distinguish those that act on many types of (pre-)mRNA without individual selection, from those that selectively recognise and control individual types of mRNA and pre-mRNA in a sequence-specific manner. The latter allow for strict recognition and control of individual gene expression, whereas the former RNA may discriminate among classes but not between individual mRNAs.
Non-discriminating cR-genes
The most straightforward example of such RNA are the tRNAs, which select individual triplets in the coding sequence. Concentration and availability of specific types of tRNA corresponding to types of (degenerate) triplets, or the many chemically modified tRNA types may influence and coordinate the expression of classes of mRNA. Similar limited regulatory function is exerted by the U-type RNAs involved in splicing (Valadkhan, 2005; Will and Luhrmann, 2005) and the snoRNAs (small nucleolar RNA), often encoded within introns of pre-mRNA (Filipowicz and Pogacic, 2002), which guide RNA modification (Dennis and Omer, 2005).
Discriminating cR-genes: siRNA and miRNA
The advent of RNAi marked the unexpected discovery of natural antisense transcripts, which silence sequence-specific mRNA (see review in Sontheimer, 2005). Although still controversial, it is not excluded that this type of post-transcriptional regulation may occur also at pre-mRNA level in the nucleus (Matzke and Birchler, 2005). The basic mechanism of RNAi is the synthesis of, by an RNA-dependent RNA polymerase and an RNA replicase, double-stranded RNA copies of target RNAs, in particular mRNA. From such double-stranded RNA, several hundred basepair long, short 21–25 nt-long fragments are cut out within the RISC RNA–protein complex. Such siRNA induces destruction of the target mRNA after sequence-specific hybridisation, whereas the miRNAs, often encoded in introns, silence temporarily a given mRNA. SiRNA and miRNA form distinct siRISC and miRISC complexes (for a recent review see Sontheimer and Carthew, 2005).
Basic principles and development of the genon concept
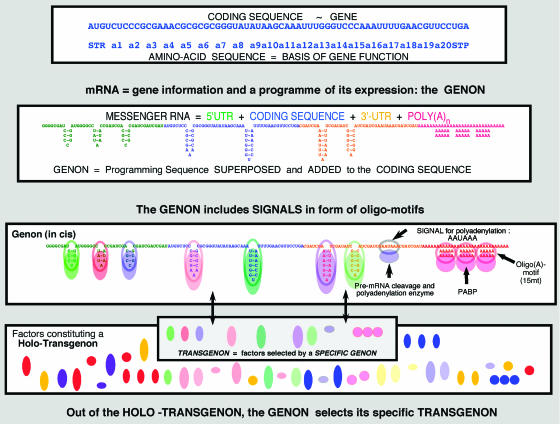
The process of gene expression entails many steps within ‘the Cascade of Regulation' (Scherrer and Marcaud, 1968; Scherrer, 1967, 1974, 1980), which reduce the genomic information to that of a gene in a stepwise manner. These include chromatin modification and activation, transcription and RNP formation, processing and transport of the pre-mRNA, formation and export of the mRNA to the cytoplasm, activation (or de-repression) of mRNA and, finally, translation (see Figure 1; Scherrer and Jost, submitted to Th Biosci). The cis programme guiding this process is unique to each distinct mRNA and polypeptide to be formed, although the same signals, in distinct combinations, may be used on the expression pathways of similar or different genes. To express this fact, we suggest the term ‘genon' (contraction of ‘Gene' and ‘operon') for the cis-acting programme, associated with a specific gene at mRNA level but encoded originally in the DNA. The ensemble of trans-acting factors bearing on a genon constitutes the ‘transgenon' of an mRNA or an ensemble of mRNA in a cell compartment, a cell or organism. Figure 2 shows the basic propositions of the genon concept.
Figure 2.
Coding sequence, gene, genon and transgenon: the amino-acid sequence of a polypeptide represents the gene, as the basis of a function; its equivalent at RNA level is the coding sequence which is inserted into the mRNA and framed by the 5′-end and 3′-end UTRs. In the latter and superimposed onto the coding sequence is the genon, a programme in cis of sequence oligomotifs, eventual binding sites for regulatory proteins (or si/miRNAs—not shown). The holo-Transgenon of a given cell is constituted by all these factors, which eventually will recognise the oligomotifs (empty coloured circles) in the genon in cis. A subset of factors (filled circles) interacting with a specific mRNA constitute the latter's Transgenon (PABP: poly(A)-binding protein). If the gene is a functional RNA, the same formalism applies.
Genons and transgenons are flexible programmes and may be modified without touching the DNA sequence. In cis, the holo-genon is modified when somatic or heritable epigenetic modifications occur, for example by DNA methylation. In somatic cells, the transgenon is constantly adapted by addition and elimination of factors of genomic or environmental origin and there are heritable protein and RNA factors involved in genetic and epigenetic regulation (for recent reviews see Delaval and Feil, 2004; Peaston and Whitelaw, 2006)).
The genon acting in cis
As defined above, the genon represents a regulatory programme superimposed and attached to a given coding sequence. It is materialised in cis by the ensemble of signals within the mRNA primary and secondary structure that control the expression of the coding sequence contained. These signals (henceforth referred to as ‘oligomotifs') are either superimposed onto the coding sequence or materialise within the mRNA sequence of the 5′- and 3′-side UTR; the mRNA sequence carrying a given programme is, therefore, longer than the coding sequence to which it is attached. In this manner, a specific genon in cis is defined for every gene (Figure 2). Implementation of the genon-programme in cis is carried out in trans by NABPs on the one side, and by interfering small RNAs (siRNAs, miRNAs) on the other; altogether, these factors provide the transgenon the programme in trans (see below), corresponding to a given genon (respectively mRNA).
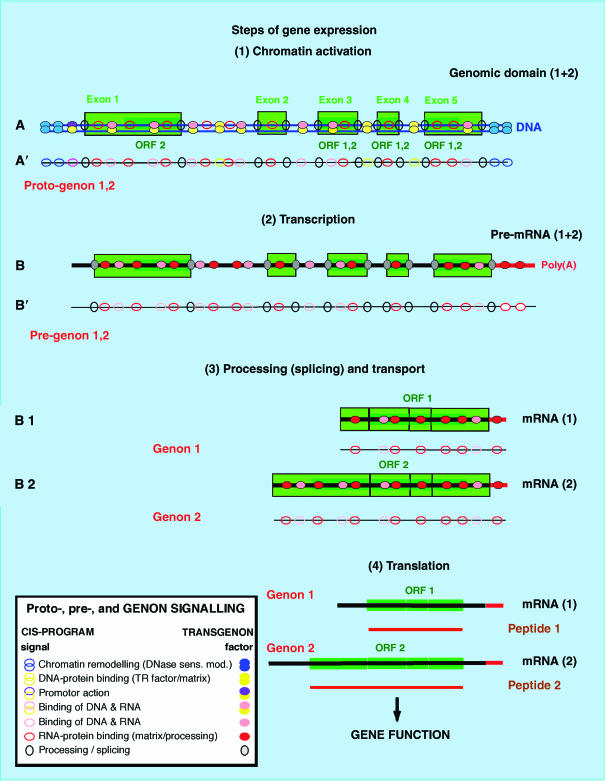
A polycistronic pre-mRNA and/or a full domain transcript (FDT) (Broders et al, 1990) might thus carry a ‘Pre-genon', controlling in cis one or several coding sequences. It may be polycistronic (several (fragmented) coding sequences in a row) or polygenic, containing the fragments of several genes to be crated by differential splicing. ‘Proto-genon' designates the signals of a DNA domain including a specific pre-genon and, in addition, the signals for chromatin modification and transcriptional activation. Each mRNA produced by alternative splicing would thus carry a genon as the remaining elements of its pre-genon. Eventually it will form a distinct (mono-)genon in the mRNA, including all cis-acting signals. At the genomic level, the term ‘holo-genon' designates the sum of all (proto-)genons. Figure 3 shows this process from DNA to mRNA expression.
Figure 3.
From DNA to mRNA expression: proto-genon, pre-genon and genon: (1) the genomic domain (line A) with exons (light green) and fragments of coding sequences (dark green) as well as intra-genic and inter-genic (not shown) DNA, contains instructions for modelling and activation of chromatin; this constitutes the proto-genon (A′). (2) From these, a pre-mRNA (B) or a FDT with its pre-genon (B′) may spring off. The latter may contain gene fragments subject to differential splicing; shown is the case of a pre-mRNA containing the two ORFs 1 and 2. (3) After processing and splicing, the two mRNAs are created with their respective genons and, thereafter, the two gene equivalents, the mRNAs (1) and (2). (4) After translation, peptides 1 and 2 secure two functions. If the gene is a functional RNA, the the same formalism applies. Inset: To the proto-genon, pre-genon and genon signals (oligomotifs) carrying distinct instructions for specific steps of processing and gene expression (left), correspond factors from the transgenon (right), in active or inactive states, which may (or not) implement the corresponding control.
The concept of the genon relates to the cis programme directly, and only indirectly to the transgenon, the system of trans-acting factors. Indeed, each trans-acting factor of protein or RNA nature is the result of a gene and its own genon. Implicit in the genon concept is the fact that there are at least as many genes and genons as distinct open reading frames (ORFs) encoded in the genome. Accordingly, the 36 000 or so genomic domains identified within the human genome project (cf. Venter et al, 2001; for more recent estimates, see Pennisi, 2003) would encode about 500 000 genes producing as many polypeptides. The genomic domains emerging, possibly, from sequence data correspond—by order of magnitude—to the highest molecular weight transcripts (FDTs) in eukaryots and to the DNA in loops of lampbrush chromosomes, or in the chromosome bands of polytene chromosomes of diptera. These were identified as units of transcription, and in sciaridae as units of local DNA amplification, and cytogenetically as units of meiotic recombination (cf. discussion in Scherrer and Marcaud, 1968; Scherrer, 1980, 1989; Scherrer and Jost, submitted to Th Biosci).
The genon and its precursors act at the transcriptional and post-transcriptional levels and lose their function with mRNA translation and subsequent degradation. Therefore, we will not consider here the downstream programmes governing gene expression post-translationally, or the catabolic aspect of protein homeostasis. However, most obviously, gene expression implies control of amounts as well as types of gene products and, therefore, RNA and protein degradation as well as biogenesis. This is achievable only by interplay and coordination of protein biosynthesis and degradation, as conceivable for example, within the prosome–proteasome system (Scherrer and Bey, 1994).
The transgenon
The genon in cis as outlined above is materialised by the ensemble of factor binding sites (‘oligomotifs') within an individual mRNA sequence. These sites are recognised by protein or RNA factors supplied by the programme in trans. These are available—or not—within the holo-transgenon of a given cell, nucleus or cytoplasm. We exclude here all mechanisms directly related to constitutive and basic protein biosynthesis within the frame of the genetic code, such as the ribosome and the basic tRNA machinery.
Regulation of transcription, and hence of programmes of differentiation and physiological change, occurs largely under the influence of cell-external factors, constituting some kind of Exo-cascade (see Figure 1, inset B), which act either directly or via the transgenon.
The genon of an mRNA is, thus, plunged into the pool of trans-acting factors recognised by the receptor oligomotifs in cis, ‘fishing out' its specific transgenon. The presence of these factors is thus crucial for execution of the expression programme encoded in the genon in cis. Being automatically picked-up by the oligomotifs in cis, these factors have a discriminative regulatory function. Their presence or absence controls the implementation of the cis programme; furthermore they may be present in active or inactive state. As proteins are capable of integrating many types of input, small molecular weight agents may influence factor–signal interactions either directly or as allosteric effectors.
Nucleic acid-binding proteins as carriers of the transgenon
A major part of the transgenon is constituted by NABPs, which are produced from cP-genes by the normal mechanisms of gene expression and regulation by protein biosynthesis. All types of RNA in the cell are covered by proteins (1:3 ratio in mRNPs). In the case of mRNA, it was shown by electron microscopy (Dubochet et al, 1973) that proteins are aligned along the entire length of the RNA molecules (and it is thus likely also for pre-mRNA), protecting specific oligomotifs from degradation by RNase (Goldenberg et al, 1979). Only in rare cases the oligonucleotide sequence that binds a given regulatory protein is known; in addition to the poly(A)-binding protein (PABP), one might mention, for example, the IRE-BP (iron response element binding protein), a protein that binds an oligomotif in the 5′-side or 3′-side UTR of the mRNAs for ferritin and transferrin, respectively (Thomson et al, 1999).
These observations indicate that there must be a ‘code' governing the interaction of a limited number of NABPs in chromatin and mRNPs, which, in general, are specifically DNA- or RNA-binding proteins. However, relatively new data have confirmed the earlier observation that a given protein may bind both, DNA and RNA. This was originally observed for the large T-antigen of SV40 and polyoma virus (Darlix et al, 1984) and more recently confirmed for a series of DNA-binding MAR proteins, long identified as hnRNP proteins (von Kries et al, 1994).
Nucleic acids as carriers of the transgenon within RNAi
As mentioned above, RNAi represents another mechanism of post-transcriptional regulation acting on pre-mRNA and mRNA interacting with the genon in cis. Because this topic is presently undergoing rapid development, we should defer this discussion to our forthcoming more extensive analysis (Scherrer and Jost, submitted to Th Biosci).
Mathematical analysis of genetic information and gene expression
General considerations
We will go beyond the classical application of information theory to molecular biology by defining a mathematical framework that distinguishes the mere coding information from process and product information contained in the genon and thus naturally includes gene expression and regulation.
Thus, we will approach here the mathematical analysis of the genon, that is the programme governing the expression of a gene. The analysis will start with the programme in cis, that is, the ensemble of genon-related signals encoded in DNA and RNA, and then be extended to trans-programme that is, the—rather heterogeneous—ensemble of factors, provided either by the genome or the environment of cell and organism, which interact with the signals in cis. The decision-making processes may be analysed in a different manner, according to whether the hologenon (all proto-genons encoded in a genome) of an organism or specific cell is considered, or the genon of an individual mRNA. Analysing gene expression by starting at the DNA level, the first decision-making process logically concerns the ‘immersion' of a holo-genon in cis into the population of factors provided by the transgenon of a given cell, as occurring during the dispersion of sperm DNA (devoid of any attached protein) into the ooplasm of an egg after fertilisation. It is simpler, however, to start from the other end, and first consider the immersion of a genon, as present in a given mRNA, into the cytoplasm of a given specialised cell. In a given cytoplasm, there are only about 500–1000 RNA-binding protein factors to which a given mRNA may be exposed. This approach reduces the selection process to the interaction of such a population of factors with the dozens or so signals in a given genon. In a first step, we consider a theoretically maximal trans programme, assuming that all possible trans-factors within a given genome were available to an mRNA; the decision process in this case would be reduced to the ‘fishing-out' by a given mRNA of the factors corresponding to its genon in cis (see Figure 2). The constitution of the trans programme for a specific cell might be analysed in a further step.
Information theoretic aspects of gene storage and expression
Information theory, as developed by Shannon, is concerned with the reduction in uncertainty obtained by receiving a particular message m drawn from some ensemble W when, before actually receiving it, one only knows the probabilities pm of the messages m (which satisfy the normalization ∑m∈w pm=1). These probabilities may be constructed as relative frequencies obtained from counting the messages received in the past. The information gained or, equivalently, the uncertainty reduced by receiving the actual message then is quantified by the entropy
The simplest nontrivial situation arises when we have only two messages, each of them occurring with equal probability 1/2. In that case, I=1(bit). Messages with probability=0 do not contribute, because 0 log 0=0, and when one message m is certain to occur, that is pm=1, then we gain no information (1 log 1=0) by receiving it because we knew it already before.
In molecular biology as well as in other applications of information theory, the messages are often sequences S=(i1, i2, i3, …, im) composed of symbols i drawn from some alphabet A. Each symbol again occurs with some probability pi, and when the symbols in the sequence S are chosen independent of each other, the probability of that sequence is
As such, information theory is a formal tool, and for applying it, we need to specify the ensemble of messages or the alphabet of symbols. In molecular biology, the symbols are either nucleotides or amino acids. The former applies to DNA sequences, which are composed of four nucleotides, A, C, G and T. When each of them occurs with relative frequency pi (i=A,C,G,T), each position contributes an information of
In particular, when all pi=1/4, this information is 2 (bits). For other values of the pi (still satisfying the normalization ∑i=A,C,G,Tpi=1), Inuc is smaller. When all positions in a sequence of a length N are independent, the sequence information is Iseq=N Inuc. Sequence correlations, however, will decrease that information.
The other type of sequences are polypeptides, which are composed of amino acids. There exist 20 different amino acids, and we denote the relative frequency of an amino acid α by pα. Thus, the average information required for specifying an amino acid is
When all these frequencies are equal, Iaa=log2 20. Otherwise, the value of Iaa is again smaller.
Owing to the degeneracy of the genetic code which leads to redundant coding for amino acids, the information needed to specify an amino acid is smaller than the one contained in a triplet (log 20<log 64=6).
Therefore, when we pass from a coding sequence in the DNA to the polypeptide that it codes, we see a reduction of information.
This is the standard application of information theory to molecular biology, but for applying the concept of the genon, we need to turn things around. Instead of the backward perspective where one starts with a polypeptide and quantifies the uncertainty about the coding sequence in the DNA or RNA, we start with a genomic domain including a variety of gene fragments and look at the ensemble of functional products derived from it (a collection of polypeptides or functional RNAs, grouped according to type). As the case of functional RNA is similar, for simplicity we here only consider the situation where those functional products are polypeptides. One such coding domain can be transcribed and translated several times, and during the expression process, mechanisms like alternative splicing or translational frameshift may even lead to chemically different polypeptides containing amino acids derived from a single coding sequence. Thus, we have a single coding domain S at DNA or RNA level, but an ensemble of products x derived from it. Let the relative frequency of x in that ensemble be qx. Therefore, the uncertainty of the result of the expression of S is given by
The important point we want to make here is that this information I(S) is not contained in the sequence S, but is rather provided by the (proto-, pre-)genon that accompanies it on the expression pathway and controls in which polypeptide it will end up. Therefore,
A finer analysis evaluates the information contributed by the different steps in the regulation of the expression process. At each step, we have certain binding sites in cis, and we have certain trans factors that bind or could possibly bind there. At the transcription and RNA level, we see mechanisms of transcription activation, RNA processing, promotion, enhancement or repression. Most of them are affected by nucleotides outside the coding sequence itself, but also the coding region can provide specific protein-binding sites. Each of these yields information about the ensemble of products derived from the coding region that is not contained in the nucleotide sequence information obeying the genetic code. More precisely, the information that counts here is not about the identity of a nucleotide or an amino acid derived from it, but about the relative frequency of the transcription and generation of a particular type of coding sequence. This then contributes to the determination of the types and numbers of functional products derived from the DNA coding region under consideration. Whereas the selection of proteins that can bind at those regulatory sites is determined by the chemical identity of their nucleotides and therefore represents a contribution from the genon in cis, the availability or the relative frequency of different proteins competing for a given binding site counts as a contribution from the transgenon. At a later stage, the mechanism of alternative splicing selects between different compositions of the final functional product. Again, that selection yields a reduction of uncertainty, that is, an information because before the splicing process, we do not know which alternative will be chosen. And that information is provided by the genon because the number of possibilities cannot be directly read from the chemical composition of individual nucleotides without knowing the context. In other words, it is a process and not a product information. In a similar manner, we can look at all the different steps in the cascade of regulation. Whereas nucleotide identities specify a final amino-acid product, on the expression pathway, here the important information is which proteins or RNAs (in the case of RNAi) bind to a given mRNA and in which manner they affect the expression process. Again, this is part of the information carried by the genon. As before, the presence or absence of those regulatory proteins or RNAs has to be counted as information provided by the transgenon.
With our information theoretic analysis, we can also compare different DNA segments S and [Smacr], where the smaller segment S is contained in the longer one [Smacr]. Here, from the backward point of view of product information, [Smacr] contains more information when it codes for a longer polypeptide than S. From our forward analysis of process information, however, the genon information in [Smacr] is higher when the end products that can be derived from it are fewer, that is, more specific than the ones derived from the shorter segment S.
As argued, equation (6) expresses the information provided by the genon, but perhaps not contained in S. Of course, the genon is at least partly superposed on S. But what, then, is the information contained in S itself about the end product derived from it? It turns out that in the present context, this is somewhat more arbitrary to specify. The question means to what extent the possibilities about abstractly possible polypeptides are reduced when we know the sequence S. In order to make this precise and quantifiable, we need to select an ensemble W of polypeptides in which S could possibly be represented. Here, S is conceived as any coding sequence, that is, one whose nucleotide composition is unknown. By determining that composition, we then gain information. But at this point, we are not interested in that chemical composition, but in the identity of the possible end products derived from it. Therefore, determining the nucleotide composition contributes only indirectly to the information desired here. Thus, we need to specify the ensemble of possible end products. This could be the ensemble of all biochemically possible polypeptides—an astronomical number—or the ones that can be derived from the genome in question—about 500 000 in the case of the human genome— or those that actually are made in the given cell—perhaps only several hundred. Of course, there exist other possible choices. In any case, within such an ensemble, each polypeptide x has a relative frequency px and the uncertainty about x then is
When only one single such x can be derived from S, then that is the information contained in S, because this is the amount of uncertainty deduced about the end product by knowing S. Our point, however, is that S does not completely specify that end product, but rather the additional information Igenon from equation (6) is needed. Therefore, the information provided by S is only
According to the cascade principle, the choice of final products is divided into multiple steps. Along these steps, the cisgenon is reduced by RNA processing and the transgenon modified according to cell compartment and physiological context. The relative information from cis and trans can be expressed in similar terms for each of these steps. The segmentation of this process facilitates individual selection of products as there is less uncertainty as the number of possibilities is reduced in each step.
Conclusion
In conclusion, we have provided an information theoretic analysis of the information provided by coding sequences and genons. We have distinguished the sequence information, determined by the nucleotide or amino-acid frequencies and the sequence correlations, the process information contributed by the genon according to the types and numbers of end products that can be derived from a given sequence, and the product information, expressing how much the number of possibilities for a polypeptide is reduced when we know the sequence. The difference between the numbers of possibilities when one does not know or knows the sequence again is the contribution of the genon. This should facilitate a deeper formal understanding of the respective contributions of coding domains and genons. The condition ‘sine qua non' to carry out this analysis is to assign a restrictive meaning to the term ‘gene' and separate this information from the process information of the genon.
Acknowledgments
We thank our colleagues who contributed by discussion over the years to evolution of the ideas presented here, and in particular Manfred Eigen and the participants of the Klosters Winter Seminar. We thank Kinsey Maundrell for checking an early version for language, and a referee as well as the MSB editors for insightful comments. This work was supported by the French CNRS, the Universities Paris 6 and 7, and by bioMérieux SA. The first author thanks the Max Planck Institute for Mathematics in the Sciences for its hospitality and good working conditions.
The authors wish to dedicate this paper to the memory of Ulrike Martyn, godchild of one of us (KS) who suddenly died on 8 Nov. 2005, away from two little children and having just finished her thesis in developmental biology (Martyn et al, 2006).
References
- Arcangeletti C, De Conto F, Sütterlin R, Pinardi F, Missorini S, Géraud G, Aebi U, Chezzi C, Scherrer K (2000) Specific types of prosomes distribute differentially between intermediate and actin filaments in epithelial, fibroblastic and muscle cells. Eur J Cell Biol 79: 423–437 [DOI] [PubMed] [Google Scholar]
- Benzer S (1959) On the topology of the genetic fine structure. Proc Natl Acad Sci USA 45: 1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S (1961) On the topography of the genetic fine structure. Proc Natl Acad Sci USA 47: 403–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S, Champe S (1961) Ambivalent rII mutants of phage T4. Proc Natl Acad Sci USA 47: 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S, Moore C, Sharp P (1977) Spliced segments at the 5′terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci USA 74: 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broders F, Zahraoui A, Scherrer K (1990) The chicken a-globin gene domain is transcribed into a 17-kilobase polycistronic RNA. Proc Natl Acad Sci USA 87: 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J (2006) The Definition of Gene Evolves—Blurring the Line between Coding and Non-coding. History and Epistemology of Molecular Biology and Beyond: Problems and Perspectives. Max Planck Institute, MPI for the History of Science. Vol. preprint 310
- Brosius J, Gould S (1992) On ‘genomenclature': a comprehensive (and respectful) taxonomy for pseudogenes and other ‘junk DNA'. Proc Natl Acad Sci USA 89: 10706–10710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L, Gelinas R, Brocker T, Roberts R (1977) An amazing sequence arrangeement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12: 1–8 [DOI] [PubMed] [Google Scholar]
- Civelli O, Vincent A, Maundrell K, Buri JF, Scherrer K (1980) The translational repression of globin mRNA in free cytoplasmic ribonucleoprotein complexes. Eur J Biochem 107: 577–585 [DOI] [PubMed] [Google Scholar]
- Darlix J, Khandjian E, Weil R (1984) Nature and origin of the RNA associated with simian virus 40 large tumor antigen. Proc Natl Acad Sci USA 81: 5425–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Conto F, Pilotti E, Razin SV, Ferraglia F, Graud G, Arcangeletti C, Scherrer K (2000) In mouse myoblasts the nuclear prosomes are associated with the nuclear matrix and accumulate preferentially in the peri-nucleolar areas. J Cell Sci 113: 2399–2407 [DOI] [PubMed] [Google Scholar]
- De Conto F, Razin S, Geraud G, Arcangeletti C, Scherrer K (1999) In the nucleus and cytoplasm of chicken erythroleukemic cells, prosomes containing the p23K subunit are found in centers of globin (pre-)mRNA processing and accumulation. Exp Cell Res 250: 569–575 [DOI] [PubMed] [Google Scholar]
- Delaval K, Feil R (2004) Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev 14: 188–195 [DOI] [PubMed] [Google Scholar]
- Dennis P, Omer A (2005) Small non-coding RNAs in Archaea. Rev Curr Opin Microbiol 8: 685–694 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim V, Kataoka N (2002) Messenger-RNA-binding proteins and the messages they carry. Rev Nat Rev Mol Cell Biol 3: 195–205 [DOI] [PubMed] [Google Scholar]
- Dubochet J, Morel C, Lebleu B, Herzberg M (1973) Structure of globin mRNA and mRNA–protein particles: use of dark-field electron microscopy. Eur J Biochem 36: 465–472 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Pogacic V (2002) Biogenesis of small nucleolar ribonucleoproteins. Rev Curr Opin Cell Biol 14: 319–327 [DOI] [PubMed] [Google Scholar]
- Georgiev G, Samarina O, Lerman M, Smirnov M, Svertzov A (1963) Biosynthesis of messenger and ribosomal ribonucleic acids in the nucleochromosamal apparatus of animal cells. Nature 200: 1291–1294 [DOI] [PubMed] [Google Scholar]
- Goldenberg S, Vincent A, Scherrer K (1979) Evidence for the protection of specific RNA sequences in globin messenger ribonucleoprotein particles. Nucleic Acids Res 6: 2787–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N, Hentze M (1994) Regulation of protein synthesis by mRNA structure. Mol Biol Rep 19: 195–200 [DOI] [PubMed] [Google Scholar]
- Hess M, Duncan R (1996) Sequence and structure determinants of Drosophila Hsp70 mRNA translation: 5′UTR secondary structure specifically inhibits heat shock protein mRNA translation. Nucleic Acids Res 24: 2441–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iarovaia O, Razin SV, Linares-Cruz G, Sjakste N, Scherrer K (2001) In chicken leuklemia cells globin genes are fully transcribed but their RNAs are retained in the perinucleolar area. Exp Cell Res 270: 159–165 [DOI] [PubMed] [Google Scholar]
- Ioudinkova E, Razin S, Borunova V, De Conto F, Rynditch A, Scherrer K (2005) RNA-dependent nuclear matrix contains a 33 kb globin full domain transcript as well as prosomes but no 26S proteasomes. J Cell Biochem 94: 445–457 [DOI] [PubMed] [Google Scholar]
- Jacob F, Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3: 318–356 [DOI] [PubMed] [Google Scholar]
- Kutay U, Guttinger S (2005) Leucine-rich nuclear-export signals: born to be weak. Rev Trends Cell Biol 15: 121–124 [DOI] [PubMed] [Google Scholar]
- Martyn U, Weigel D, Dreyer C (2006) In vitro culture of embryos of the guppy, Poecilia reticulata. Dev Dyn 235: 617–622 [DOI] [PubMed] [Google Scholar]
- Matzke M, Birchler J (2005) RNAi-mediated pathways in the nucleus. Rev Nat Rev Genet 6: 24–35 [DOI] [PubMed] [Google Scholar]
- Maundrell K, Maxwell ES, Civelli O, Vincent A, Goldberg S, Buri J-F, Imaizumi-Scherrer M-T, Scherrer K (1979) Messenger RNP complexes in avian erythroblasts: carriers of post-transciptional regulation? Mol Biol Rep 5: 43–51 [DOI] [PubMed] [Google Scholar]
- Pearson H (2006) What is a gene? Nature 441: 399–401 [DOI] [PubMed] [Google Scholar]
- Peaston A, Whitelaw E (2006) Epigenetics and phenotypic variation in mammals. Mamm Genome 17: 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E (2003) A low number wins the genesweep pool. Science 300: 1484. [DOI] [PubMed] [Google Scholar]
- Perry R, Srinivasan P, Kelley D (1964) Hybridization of rapidly labeled nuclear ribonucleic acids. Science 145: 504–507 [DOI] [PubMed] [Google Scholar]
- Perry RP (1962) The cellular sites of synthesis of ribosomal and 4S RNA. Proc Natl Acad Sci USA 48: 2179–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin S, Borunova V, Rynditch A, Ioudinkova E, Smalko V, Scherrer K (2004) The 33 kb transcript of the chicken alpha-globin gene domain is part of the nuclear matrix. J Cell Biochem 92: 445–457 [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Dargemont C, Stutz F (2004) Nuclear export of RNA. Biol Cell 96: 639–655 [DOI] [PubMed] [Google Scholar]
- Scheer U, Benavente R (1990) Functional and dynamic aspects of the mammalian nucleolus. BioEssays 12: 14–21 [DOI] [PubMed] [Google Scholar]
- Scheer U, Hock R (1999) Structure and function of the nucleolus. Curr Opin Cell Biol 11: 385–390 [DOI] [PubMed] [Google Scholar]
- Scherrer K (1967) Pattern of messenger RNA in animal cells and the concept of ‘cascade regulation'. In International Symposium on Biochemistry of Ribosomes and Messenger-RNA (1967), Lindigkeit R, Langen P, Richter J (eds) Vol. 1968 (1), pp 259–277. Akademie Verlag (Berlin), Castle Reinhardsbrunn [Google Scholar]
- Scherrer K (1974) Control of gene expression in animal cells: the cascade regulation hypothesis revisited. Adv Exp Med Biol 44: 161–219 [DOI] [PubMed] [Google Scholar]
- Scherrer K (1980) Cascade regulation: a model of integrative control of gene expression in eukaryotic cells and organisms. In Eukaryotic Gene Regulation, Vol. 1, pp 57–129. Boca Raton, FL: CRC Press Inc. [Google Scholar]
- Scherrer K (1989) A unified matrix hypothesis of DNA-directed morphogenesis, protodynamism and growth control. Biosci Rep 9: 157–188 [DOI] [PubMed] [Google Scholar]
- Scherrer K (2003) Historical review: the discovery of ‘giant' RNA and of RNA processing: 40 years of enigma. Trends Biochem Sci 28: 566–571 [DOI] [PubMed] [Google Scholar]
- Scherrer K, Bey F (1994) The prosomes (multicatalytic proteinase—proteasomes) and their relation to the untranslated messenger ribonucleoproteins, the cytoskeleton and cell differentiation. Progr Nucleic Acids Res Mol Biol 49: 1–64 [DOI] [PubMed] [Google Scholar]
- Scherrer K, Darnell JE (1962) Sedimentation characteristics of rapidly labelled RNA from HeLa cells. Biochem Biophys Res Comm 7: 486–490 [DOI] [PubMed] [Google Scholar]
- Scherrer K, Latham H, Darnell JE (1963) Demonstration of an unstable RNA and of a precursor to ribosomal RNA in HeLa cells. Proc Natl Acad Sci USA 49: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer K, Marcaud L (1968) Messeger RNA in avian erythroblasts at the transcriptional and translational levels and the problem of regulation in animal cells. J Cell Physiol 72: 181–212 [DOI] [PubMed] [Google Scholar]
- Scherrer K, Marcaud L, Zajdela F, London IM, Gros F (1966) Patterns of RNA metabolism in a differentiated cell: a rapidly labeled, unstable 60S RNA with messenger properties in duck erythroblasts. Proc Natl Acad Sci USA 56: 1571–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer K, Spohr G, Granboulan N, Morel C, Grosclaude J, Chezzi C (1970) Nuclear and cytoplasmic messenger-like RNA and their relation to the active messenger RNA in polyribosomes of HeLa cells. In Cold Spring Harbor Symposium 1970, Vol. 35, pp 539–554. Cold Spring Harbor Laboratory [Google Scholar]
- Schmid HP, Akhayat O, Martins de SA C, Puvion F, Koehler K, Scherrer K (1984) The Prosome: a ubiquitous morphologically distinct RNP particle associated with repressed mRNPs and containing specific ScRNA and a characteristic set of proteins. EMBO J 3: 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer RH (1992) The cytoskeleton and mRNA localization. Curr Opin Cell Biol 4: 15–19 [DOI] [PubMed] [Google Scholar]
- Snyder M, Gerstein M (2003) Genomics. Defining genes in the genomics era. Science 300: 258–260 [DOI] [PubMed] [Google Scholar]
- Sontheimer E (2005) Assembly and function of RNA silencing complexes. Rev Nat Rev Mol Cell Biol 6: 127–138 [DOI] [PubMed] [Google Scholar]
- Sontheimer E, Carthew R (2005) Silence from within: endogenous siRNAs and miRNAs. Rev Cell 122: 9–12 [DOI] [PubMed] [Google Scholar]
- Spilianakis C, Lalioti M, Town T, Lee G, Flavell R (2005) Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645 [DOI] [PubMed] [Google Scholar]
- Steitz T, Moore P (2003) RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Rev Trends Biochem Sci 28: 411–418 [DOI] [PubMed] [Google Scholar]
- Thomson A, Rogers J, Leedman P (1999) Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Rev Int J Biochem Cell Biol 31: 1139–1152 [DOI] [PubMed] [Google Scholar]
- Tschochner H, Hurt E (2003) Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Valadkhan S (2005) snRNAs as the catalysts of pre-mRNA splicing. Rev Curr Opin Chem Biol 9: 603–608 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C,Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X (2001) The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- von Kries J, Buck F, Stratling W (1994) Chicken MAR binding protein p120 is identical to human heterogeneous nuclear ribonucleoprotein (hnRNP) U. Nucleic Acids Res 22: 1215–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C, Luhrmann R (2005) Splicing of a rare class of introns by the U12-dependent spliceosome. Rev Biol Chem 386: 713–724 [DOI] [PubMed] [Google Scholar]