Abstract
Mapping protein–protein interactions is an invaluable tool for understanding protein function. Here, we report the first large-scale study of protein–protein interactions in human cells using a mass spectrometry-based approach. The study maps protein interactions for 338 bait proteins that were selected based on known or suspected disease and functional associations. Large-scale immunoprecipitation of Flag-tagged versions of these proteins followed by LC-ESI-MS/MS analysis resulted in the identification of 24 540 potential protein interactions. False positives and redundant hits were filtered out using empirical criteria and a calculated interaction confidence score, producing a data set of 6463 interactions between 2235 distinct proteins. This data set was further cross-validated using previously published and predicted human protein interactions. In-depth mining of the data set shows that it represents a valuable source of novel protein–protein interactions with relevance to human diseases. In addition, via our preliminary analysis, we report many novel protein interactions and pathway associations.
Keywords: human interactome, IP-HTMS, protein–protein interaction
Introduction
Biomolecular interactions play a critical role in the vast majority of cellular processes. Understanding the roles and consequences of protein interactions is fundamental for the development of systems biology as well as the development of novel therapeutics. Our current knowledge of biomolecular interactions in terms of cataloging interactions and understanding their biophysical properties is still very limited and is hindered by the limitations (primarily throughput and reproducibility) of existing technologies. Different techniques for mapping protein interactions, such as the yeast two-hybrid approach (Y2H) (Chien et al, 1991) and the LUMIER approach (Barrios-Rodiles et al, 2005), are available, and address the question of whether two proteins interact in a pairwise fashion. We have developed a high-throughput platform combining immunoprecipitation and high-throughput mass spectrometry (IP-HTMS) to rapidly identify potentially novel protein interactions for a bait protein of interest. We (Ho et al, 2002) and others (Gavin et al, 2002) previously used this approach to map protein–protein interactions in yeast, creating invaluable data sets for yeast biology and extrapolation into mammalian biology. We have since extended this approach to the high-throughput mapping of protein–protein interactions in humans and refined the computational processing with new methodology to assign a confidence score to each interaction. Mapping protein interactions in human cells has its own set of challenges owing to the number of potentially expressed genes, the number of different cell types and the numbers of internal and external factors that impact the cellular system. Although a complete mapping of the human interactome is still beyond current capabilities, more focused studies are possible. For example, application of IP-HTMS on a smaller scale was used to study the human TNF-alpha/NF-kappa B signal transduction pathway (Bouwmeester et al, 2004). On a more global scale, the Y2H system has recently been applied to study pairwise human interactions (Rual et al, 2005; Stelzl et al, 2005). Here, we report the first large-scale application of IP-HTMS to the mapping of protein–protein interactions in human cells using 338 human bait proteins of significant biomedical interest. The complete data set is provided as a table of bait–prey pairs with associated confidence values (Supplementary Table II) and in PSI-MI (Hermjakob et al, 2004) format from the Intact database (www.ebi.ac.uk/intact), accession EBI-1059370.
Results and discussion
Bait selection and analytical processing
An initial set of 407 human bait proteins was selected based on known or implied disease associations and functional annotation. These proteins are implicated in a diverse set of biological processes and pathways. The most well-represented biological process categories among the set of baits are protein modification, cell cycle, transcription and signal transduction, reflecting the choice of bait proteins that are fundamental to essential cellular processes. Many of the baits also have known disease associations, the most well represented being breast cancer, colon cancer, diabetes and obesity, reflecting our objective to target important human diseases. Approximately 10% of the baits selected were hypothetical or poorly annotated proteins, chosen in some cases for their homology to proteins with disease or functional associations of interest. The data set reported here maps interactions for 338 of the initial set of bait proteins. A complete listing of the bait proteins and a representative biological process from the Gene Ontology (GO) (Ashburner et al, 2000), where available, is provided in Supplementary Table I. (See Supplementary Information for further details on bait selection and disease associations.)
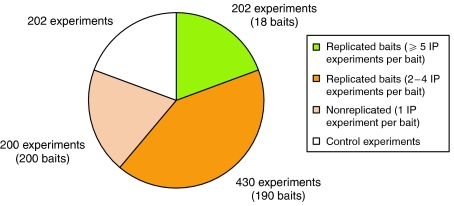
Analytical processing and mass spectrometry were carried out as described in Materials and methods. In total, 1034 individual immunoprecipitation experiments were resolved by SDS–PAGE, and proteins visualized by colloidal Coomassie stain. Processing of the corresponding gel lanes yielded 16 321 gel bands that were processed by mass spectrometry generating over 400 000 MS/MS spectra that matched a peptide sequence in the database. For over half of the baits, replicated immunoprecipitation experiments were performed. Figure 1 shows a breakdown of the total set of experiments by type.
Figure 1.
Data processing summary. Pie chart showing categorization of all immunoprecipitation experiments by type.
Prey identification, scoring and filtering
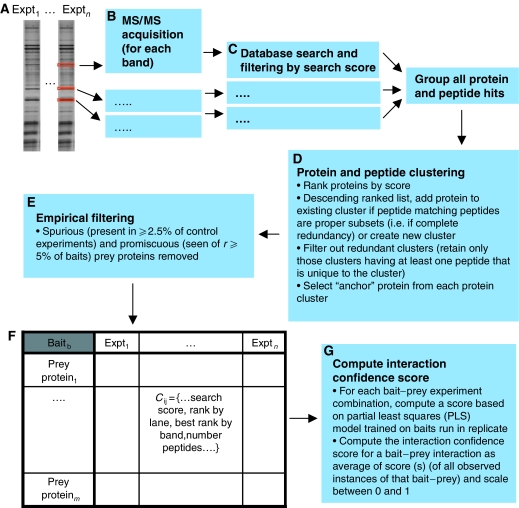
As shown in Figure 1, our data set consists of both replicated and single-pass immunoprecipitation experiments. An additional level of redundancy arises from the fact that prey proteins may or may not be restricted to single-gel bands in a given lane. We, therefore, devised a data-processing pipeline that would consolidate, organize and remove redundancy and provide us with an accurate master list of the prey proteins identified for each bait. Figure 2 provides an overview of this process. Having extracted each gel band and acquired the MS/MS data (Figure 2A), each data file was searched using the Mascot (www.matrixscience.com) search engine. Low-quality peptide and protein hits were then removed by applying score threshold rules (see Materials and methods). All protein and peptide hits corresponding to gel bands from the same bait were combined and clustered using the algorithm outlined in Figure 2D. The algorithm collapses proteins into clusters if their respective sets of matching peptides are proper subsets of one another (i.e., if one set of peptides is completely redundant with respect to the other). A representative protein (termed the ‘anchor' of the cluster) can then be selected from the cluster. The anchor is selected by ranking the proteins within a cluster by score or number of peptides and then choosing the top-ranked protein. Ties may be broken by consideration of other attributes, such as quantity and quality of annotation for each protein. This process removes redundancy at the level of the peptide matches and protein sequence; each of the anchor proteins is guaranteed to be non-redundant with respect to its complement of matching peptides. A small proportion (0.5%) of the reported interactions are, however, redundant at the level of the gene locus, that is, multiple reported prey proteins map to the same gene name. These may represent instances of protein isoforms or variants and are accordingly left in the data set.
Figure 2.
IP-HTMS data analysis pipeline. (A–B) All bands from the lane(s) corresponding to a bait are extracted and MS/MS data acquired. (C) Data from each MS/MS acquisition are searched against a non-redundant human protein sequence database using the Mascot search engine. (D) Data from all bands corresponding to each bait are merged and protein and peptides clustered to generate a non-redundant list of protein identifications. (E) Spurious proteins and promiscuous binding proteins are removed. (F) A data table is produced for each bait protein with all of the scoring information, including scores and ranks by band and experiment. This data table contains all data required for the estimation of bait–prey interaction probability. (G) An interaction confidence score is calculated based upon a partial least squares model trained on the replicated subset of the data.
Filtering out spurious and nonspecific proteins
In order to minimize the number of false-positive interactions, we applied an empirical filtering process to remove spurious/contaminant proteins and nonspecifically interacting proteins (Figure 2E). Three filtering steps were applied to the interaction network. These steps are summarized in Table I. First was the identification of the bait protein itself (bait–bait interactions were removed from the network; 97% of baits were identified at least once). Second, 207 interactions corresponding to instances of spill over from one gel lane to the next were removed. Finally, we built a database of spuriously occurring proteins and contaminants based on 202 control (vector only) immunoprecipitation experiments. Those proteins occurring in ⩾2.5% of control experiments were removed from the data set as were proteins interacting with ⩾5% of baits. This combined set of proteins includes many common contaminants of mass-spectrometry experiments (such as human keratins) as well as proteins observed to bind nonspecifically, and includes protein families such as tubulins, ribosomal proteins and heat-shock proteins.
Table 1.
Summary of IP-HTMS interactome network filtering
| Filtering step | Baits | Unique proteins | Interactions |
|---|---|---|---|
| Unfiltered interaction network | 407 | 2826 | 24 540 |
| Remove bait–bait interactionsa | 407 | 2826 | 24 211 |
| Remove spill-over interactionsb | 407 | 2826 | 24 005 |
| Remove frequent binders and control experiment proteinsc | 338 | 2235 | 6463 |
Those instances where the bait protein was identified in the mass spectrometry experiment.
Observations of apparent spill-over from one gel lane to another; detected by manual examination of gels and peptide/protein identification data.
Frequent binders defined as prey proteins identified for ⩾5% of baits; control proteins are those ‘prey' proteins identified in ⩾2.5% of control experiments.
Interaction confidence scores
As many peptide and protein identification metrics (scores, expect values, number of peptides, peptide coverage, etc.) can be used to assess the overall confidence of a prey identification, we sought to combine several of these metrics and generate an overall measure of the confidence of each prey observation (Figure 2G). Data corresponding to the set of 18 well-replicated (⩾5 immunoprecipitation experiments) baits (see Figure 1) were used as a training set to build a partial least squares (PLS)-based regression model of prey protein reproducibility, whereby the reproducibility was the dependent variable and six predictor variables were selected to build the model. The six predictor variables were the Mascot score for the prey in the lane, the total number of peptides observed for the prey in the lane, the rank of the prey protein in the lane, a binary value indicating whether the prey is in fact the bait protein itself, the maximum Mascot score for the prey across all of the bands in which it was observed in the lane and the best rank for the prey protein across all bands in the lane. The model was trained on the replicated set of baits and then applied to the remaining data set. Where multiple experiments were performed for a bait, an averaged predicted reproducibility value was calculated across all of the observations of the given prey protein. Finally, this value was normalized to between 0 and 1 and reported as the interaction confidence score. In a small number of cases (approximately 5% of the reported interactions), an interaction confidence score was not calculated, because one of the predictor variables was not available. For example, in some cases, a protein identified as present in a given lane may not be judged as present in any of the individual bands in that lane (peptides corresponding to the protein may be present in different bands, none of which scores highly enough for the protein to be considered as present); in these cases, the predictor variable corresponding to the best rank for the protein across all bands in the lane is not calculated. For 18% of the interactions in the accompanying data set, the interaction confidence score is reported as 0. These prey protein observations should still be interpreted as valid as they meet the required search engine score thresholds.
We validated our interaction scoring metric in several ways, demonstrating its utility as a measure of interaction confidence. First, using our training data set of reproduced baits, we performed a 10-fold cross-validation of the model, and measured the ability of our model to estimate prey reproducibility (see Supplementary Information). We found good correlation (r=0.66) between the observed reproducibility and predicted reproducibility across our training set. Second, by analyzing the subset of known interactions in the data set (see subsequent section), we observed that the interaction confidence scores assigned to the set of known interactions were significantly higher than those scores assigned to previously unknown interactions; the set of known interactions has a mean interaction confidence score of 0.43, whereas the mean of the entire set of interaction confidence scores is 0.21, a statistically significant difference (Wilcoxon rank sum test; P≪0.0001). Third, we analyzed the set of reciprocal interactions in the data set. In our study, no explicit effort was made to test bait–prey interactions reciprocally (i.e., to use the observed prey proteins as baits and see whether the original bait proteins are identified). A small number of interactions (21) were, however, observed reciprocally in the data set. The interaction confidence scores of these 21 reciprocally observed interactions (mean=0.43) were significantly higher (Wilcoxon rank sum test; P≪0.0001) than the set of interactions for which a reciprocal interaction was not observed (mean=0.25) or indeed the whole data set (mean=0.21). These observations show that the interaction confidence score is a useful means of ranking the interactions for subsequent data mining. To facilitate more in-depth analysis of such a large data set, we focused our in-depth interpretation of the interactions primarily on interactions with score ⩾0.3, corresponding to approximately one-third (2251 interactions) of the data set. This threshold was chosen because most interactions between subunits of well-characterized protein complexes represented in the data set (the proteasome and eukaryotic translation initiation factors—see below) have scores ⩾0.3. In addition, for 85% of prey proteins with interaction score ⩾0.3, two or more distinct peptide sequences were identified, consistent with emerging guidelines for mass spectrometry-based protein identification (Bradshaw et al, 2006).
Computational assessment and validation
Other types of genomic information, when combined with protein–protein interactions, can provide stronger evidence of functional relationships between genes. Several methods of utilizing these orthogonal genomic data to computationally assess high-throughput protein–protein interaction data have been proposed, such as comparison with gene expression, analysis of paralogous interactions and utilization of functional and sub cellular localization information (Deane et al, 2002; von Mering et al, 2002; Rual et al, 2005). In this section, we present a computational assessment of the IP-HTMS data set by integrating three classes of genomic information: other human protein–protein interaction data sources, GO annotations and gene expression microarray data.
An important consideration when integrating other data types is how to count the protein–protein interactions (von Mering et al, 2002). Two paradigms for modeling protein–protein interaction data have been proposed: the ‘spoke' model, whereby each bait is assumed to interact with each of its observed prey proteins, and the ‘matrix' model, whereby the bait and all of the preys interact with each other (Bader and Hogue, 2002) We adopted the ‘spoke' model for all of our analyses (unless stated otherwise), as the ‘matrix' model has been shown to produce higher rates of false positives (Bader and Hogue, 2002). We recognize, however, the limitations of the ‘spoke' model, in particular that bait–prey interactions identified in immunoprecipitation experiments may not actually represent direct physical interactions between the bait and prey protein.
Comparison to other protein–protein interaction data sources
Previous reports have in general found relatively little overlap between protein–protein interaction data sets (Bader and Hogue, 2002). For example, a recent comparison of a comprehensive literature-curated catalog of yeast interactions to all available high-throughput yeast interactions showed only a 14% overlap (Reguly et al, 2006). As pointed out by the latter authors, however, it is important to distinguish between the absolute intersection of the two data sets (the number of interactions in common between the data sets being compared) and the intersection of ‘interaction space' covered by each data set. For the IP-HTMS platform, the interaction space is the space covered by the set of bait proteins. For example, in comparing the IP-HTMS data set to a Y2H data set, we identify the IP-HTMS space as those Y2H interactions for which one or more of the interactors correspond to an IP-HTMS bait. Performing the comparisons in this way allows for realistic estimates of how interactions are recapitulated across different studies and technology platforms.
We compared the IP-HTMS data set to three other sources of human protein–protein interactions: a collation of known interactions (Ramani et al, 2005), a set of interactions predicted from lower eukaryotic interactome maps (Lehner and Fraser, 2004) and a high-throughput Y2H study (Rual et al, 2005). The overlap between these data sets and the IP-HTMS data set are summarized in Table II. The overlap between the IP-HTMS data set and these three other sources ranges from 6 to 11%, broadly in line with observations of the overlap between the human Y2H data set and literature-curated interactions (2–8%) (Rual et al, 2005). By randomly permuting the IP-HTMS bait–prey interactions and re-computing the overlaps, we confirmed that the overlaps are significantly greater than would be expected by chance (P<0.0001). Similar comparisons in yeast between IP-HTMS interactions (Ho et al, 2002) and literature-curated and tandem affinity purification (Gavin et al, 2002) and literature-curated interactions show 20 and 30% overlaps, respectively (Reguly et al, 2006), suggesting that a much greater proportion of the yeast interactome has been cataloged than that the human interactome.
Table 2.
Comparison of IP-HTMS data set to other sources of human protein–protein interactions
| Protein–protein interaction data set |
|||
|---|---|---|---|
| Knowna | Predictedb | Experimental (Y2H)c | |
| Interactions | 31 183 | 20 469 | 6727 |
| IP-HTMS baits featured in data setd | 216 | 123 | 94 |
| Overlap with IP-HTMS spacee | 2332 | 668 | 366 |
| Intersection with IP-HTMS (number of interactions, percentage of total)f | 149, 6.4% | 78, 11.4% | 29, 7.9% |
| Randomly permuted intersection with IP-HTMS (min, mean, max)g | 7, 14.3, 25 | 3, 8.0, 14 | 0, 1.8, 7 |
| Statistical significance of intersection (fold-enrichment, P-value)h | ∼10-fold, P<0.0001 | ∼10 fold, P<0.0001 | ∼15 fold, P<0.0001 |
IP-HTMS baits (from total of 343) featuring in the data set.
Number of interactions in the data set featuring one or more IP-HTMS baits.
Number of shared interactions between data set and IP-HTMS.
Number of shared interactions between randomly permuted (1000 iterations) IP-HTMS and data set.
Fold enrichment of observed intersection over intersection expected by chance.
The sets of interactions in common between the human IP-HTMS interactions and each of the other three data sets are themselves overlapping; of the total of 256 overlapping interactions between IP-HTMS and the other three data sets, 82 are found in two or more of the overlapping sets. We also note that interactions in common between the IP-HTMS and other sources of human protein–protein interactions have in general significantly higher confidence scores. The mean confidence scores for the interactions in common between IP-HTMS and the known set, IP-HTMS and the predicted set, and IP-HTMS and the Y2H set are 0.43, 0.43 and 0.42, respectively, higher than expected by chance (P≪0.0001; Wilcoxon rank sum test) given the overall distribution of confidence scores.
As already mentioned, it is probable that some of the bait–prey interactions identified in IP-HTMS experiments may not actually represent direct physical interactions between the bait and prey protein, but instead interactions between preys. To explore this further, we first extended our comparisons by considering the matrix of all possible interactions in the IP-HTMS data set (i.e., including all possible prey–prey interactions for each bait). Of the matrix of ∼225K possible IP-HTMS interactions, 1678 are in common with the known set (statistically significantly greater than expected by chance, P<0.0001). Although the accuracy of considering the matrix of all interactions is expected to be lower than when only considering bait–prey interactions (Bader and Hogue, 2002), clearly many valid interactions remain to be discovered from this broader approach.
Second, we compared our IP-HTMS interactions to the literature using the Pathway Studio software (Ariadne Genomics). This software enables rapid annotation of protein–protein interactions with literature mined from various sources. Using this approach, 145 protein–protein interactions in our IP-HTMS data set were annotated as present in the literature. In order to identify those IP-HTMS interactions that represent indirect interactions between bait and prey, we mined the literature in the following way. Bait–prey pairs from our IP-HTMS experiments that have literature validation in the Pathway Studio database were selected. The interaction network was then expanded by extracting all known interactors from the literature that are within two edges of the prey. We then overlapped the experimental interactions with the expanded network such that for each bait we considered all paths of length two where the (bait, prey) and the (bait, interactor of prey) pairs are both in IP-HTMS, and hence, the (prey, interactor of prey) pair can be inferred. We did the same for paths of length three, and we enumerated all the distinct length-one pairs from the literature that were part of the overlapping paths. This allows us to significantly expand the validation of our data set using the literature by including not just bait–prey but also prey–prey interactions. With our additional analysis, the total number of observed interactions that are reinforced by the literature increases to 375. This represents a 2.6-fold increase in validation corresponding to 6% of all of our interactions. This set of interactions is provided in Supplementary Table IV. We have utilized this approach in a detailed way to extend networks for individual bait proteins. An example of this is provided in Supplementary Figure III. Only four direct interactors of VHL from our data set matched with the literature. Using our novel approach, we extended the interaction surrounding VHL within two literature edges. This increased the number of proteins seen in the VHL IP-HTMS experiment that are linked to VHL through the literature to 13 (three-fold increase). The nine new associations are indirect but are linked through known interactors of VHL.
Paralogous interactions
Evolutionary relationships between genes both across and within species have been proposed as sources for discovery and confirmation of protein–protein interactions (Matthews et al, 2001). In yeast, interactions between pairs of proteins have been shown to be of higher confidence if interactions also occur between paralogs of the interactors (Deane et al, 2002). The latter authors developed the paralogous verification method, and showed that in yeast the method was able to predict 40% of true interactions with a 1% false-positive rate (Deane et al, 2002).
We explored the utility of this method for assessment of the IP-HTMS data set by first collating a set of 1999 groups of human paralogs (representing 6023 human genes) from the inparanoid database (O'Brien et al, 2005). Cross-referencing to the IP-HTMS data set identified 834 interactions for which both bait and prey could be assigned one or more paralogs. Overall, 154 of these 834 interactions (18%) had one or more paralogous interactions. The set of 154 paralogous interactions are provided as Supplementary Information (Supplementary Table III).
In many cases, these paralogous interactions are comprised of a single bait interacting with two or more related (paralogous) prey proteins. We also wished to test the rate at which paralogous baits identify the same or related prey proteins. The IP-HTMS data set provides an opportunity to do this, because for 16 of the IP-HTMS baits, one or more paralogs have also been used as baits. These 16 baits correspond to 157 interactions for which paralogs were assigned, and 57 of these interactions are paralogous (36%). One caveat to analyzing the IP-HTMS data in this way is that it is not possible to distinguish between independent interactions of paralogous baits with the same or related prey proteins and the scenario whereby paralogous baits interact with each other (e.g., heterodimers) and that complex then identifies the same set of preys regardless of which bait is used. The set of 16 paralogous baits includes three members of the 14-3-3 protein family, YWHAB, YWHAQ and YWHAZ. These proteins are known to form homo- and heterodimers in vivo (Jones et al, 1995) and together contribute 35 of the 57 interactions from paralogous baits. Nevertheless, this is a useful demonstration of the reproducibility of paralogous baits; the three 14-3-3 baits identify 117 prey proteins in total, 33 of which are identified by more than one of the baits. Finally, we note that interactions supported by a paralogous interaction have significantly higher interaction confidence scores; the set of 154 paralogous interactions have a mean score of 0.33, as compared to 0.21 across the whole data set (Wilcoxon rank sum test; P≪0.0001). As pointed out by Deane et al (2002), the paralogous verification method is useful only where paralogs can be identified. This is only possible for a relatively small fraction (834 out of 6463 interactions) of the IP-HTMS data set. Nevertheless, we believe that this first preliminary analysis of paralogous interactions in the human interactome illustrates the potential for further in-depth studies as our ability to assign paralogs improves and our knowledge of the human interactome increases.
Biological process and pathway enrichment
To gain an overview of the classes of proteins identified as preys for each of the baits, we used the GO (slim subsets) to analyze biological process and cellular component category representation. In both cases, the distribution of prey proteins among the categories is similar to the distribution of categories among bait proteins; the most well-represented bait biological process protein categories—protein modification, protein biosynthesis, cell cycle, transcription and signal transduction, are also the most well-represented prey protein categories.
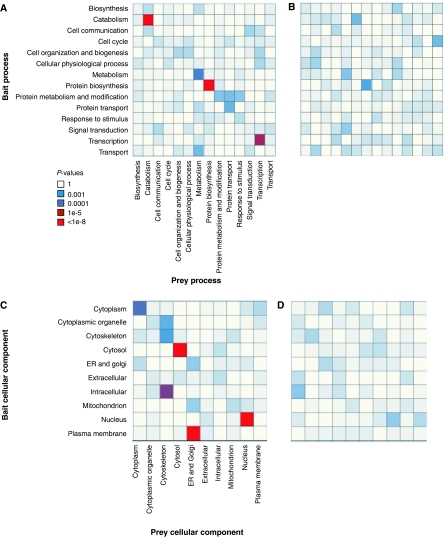
We used the GO annotation to analyze the degree to which bait and prey interactors share the same or related GO categories. For high-throughput yeast data, the fractions of interactions for which both interactors have the same high-level biological process or cellular component categories have been estimated at 20 and 27%, respectively (Reguly et al, 2006). For our human IP-HTMS data, these fractions are 12 and 20%, respectively. To illustrate these associations in more detail, we generated bait–prey coincidence maps (Figure 3) in which the association between each combination of bait and prey GO categories is tested using a contingency table and statistical test (Fisher exact test). Each combination of bait GO category, i, and prey GO category j, is represented as a cell in the matrix, and the color of the cell represents the statistical significance of the association between bait category i and prey category j. We also implemented a permutation procedure to characterize the distribution of P-values derived from random associations (see Materials and methods). The permutation-based P-value for each bait–prey category combination was calculated as the fraction of times the Fisher exact test P-value was less than the observed ‘real' P-value. On this basis, the bait–prey category combinations with P-values less than or equal to 0.0001 are all judged to be highly significant; smaller P-values for each of these category combinations were not observed across 1000 independent random permutations of the bait–prey categories.
Figure 3.
GO coincidence maps. Coincidence maps showing enrichment of bait–prey GO category combinations. Each bait–prey category combination is represented by a square in the matrix and colored according to the P-value from a pairwise statistical test (Fisher exact test) of association. (A) Bait–prey biological processes. (B) Randomly permuted bait–prey biological processes. (C) Cellular component categories. (D) Randomly permuted bait–prey cellular component categories.
This analysis revealed a significant tendency of baits to interact with prey proteins implicated in the same or similar biological process (Figure 3A and B). For example, the most significant bait–prey biological process category combinations were protein biosynthesis/protein biosynthesis (P=1.7e−09) and catabolism/catabolism (P=2.3e−08). These correspond to two highly connected clusters of interacting proteins representing known macromolecular complexes—translation initiation and elongation factors and the proteasome (both discussed in more detail below). Similar results were obtained for the cellular component categories (Figure 3C and D), except that significant off-diagonal associations were also seen. Most notably, a significant enrichment is seen between baits assigned to the plasma membrane baits and endoplasmic reticulum/Golgi preys. This enrichment is largely due to two members of the tumor necrosis factor receptor super-family baits (TNFRSF14 and TNFRSF5) interacting with several endoplasmic reticulum prey proteins. These two baits interact with an overlapping set of endoplasmic reticulum-associated proteins including several components of the microsomal signal peptidase complex and endoplasmic reticulum-associated protein disulfide isomerase family members. Although it is not clear what the actual biological explanation might be, we believe that these are not spurious observations as this group of prey proteins is also identified using the TRAF6 bait (TNF receptor-associated factor), a known mediator of signaling from TNFRSF5.
Integrated analysis of the IP-HTMS and GO categories also facilitated discovery of some very specific but potentially biomedically important interactions. Relatively few proteins in the IP-HTMS data set are assigned to the peroxisome (17 interactions involve a peroxosomal bait or prey). Of these interactions, a single interaction was observed between a peroxisomal bait and a peroxisomal prey: PHYH (phytanoyl-CoA 2-hydroxylase) bait identified ABCD3 (ATP-binding cassette, subfamily D) as a prey. Defects in the functioning of both PHYH and ABCD3 are implicated in Zellweger's syndrome and other peroxisomal biogenesis disorders, a set of potentially severe (fatal) inherited diseases (Moser, 1999; Steinberg et al, 2006). In addition, several studies have shown interactions between ABCD proteins and peroxisomal biogenesis factors (PEX proteins) and between PHYH and PEX proteins (Liu et al, 1999; Gloeckner et al, 2000). To our knowledge, our observation is the first indication of a protein–protein interaction between PHYH and ABDC3.
Cross-referencing gene expression information
Increased similarity of gene expression profiles for genes encoding interacting proteins has been demonstrated in yeast (Ge et al, 2001). Preliminary evidence that this may also be the case in higher eukaryotes has been reported for Caenorhabditis elegans (Li et al, 2004) and in humans (Hahn et al, 2005; Rual et al, 2005). In the latter case, enrichment for higher gene expression correlation was seen for both literature-derived interactions and, albeit at a lower level, for the experimentally derived data set (Rual et al, 2005). One of the principal issues in attempting to measure whether a relationship exists between gene expression and protein interaction data sets is the incompleteness and arbitrary nature of selecting appropriate human gene expression data. Rather than select individual data sets over which co expression could be measured, we made use of a compendium of co-expression measurements generated from 3924 microarrays from 60 different human studies (Lee et al, 2004). Co-expression links in this study are defined as positive or negative based upon their position within the extremes of the distributions of correlation for each study (Lee et al, 2004). Figure 4 shows the respective fractions of positive and negative co-expression links for several sets of interaction data. For the complete set of approximately 9 million co-expression measurements, a slight bias towards positive measurements was observed (Lee et al, 2004). We first confirmed that the ratio of positive to negative co-expression counts for measurements within the IP-HTMS space (i.e., where one or more of the pair of coexpressed genes corresponded to an IP-HTMS bait) was broadly similar to the bias observed in the complete data set (respective positive to negative ratios are 1.25 and 1.32). We then observed that elevated positive to negative ratios were observed for both the IP-HTMS data set and the human Y2H data set (Rual et al, 2005) and for the set of known interactions (Ramani et al, 2005), suggesting that human gene pairs encoding interacting proteins are also more likely to be coexpressed. The magnitudes of the positive to negative ratios for the IP-HTMS and Y2H data sets are similar (2.33 and 2.50, respectively), whereas the ratio for the known set is significantly higher (4.75). We also confirmed that the ratio of positive to negative co-expression counts for the IP-HTMS data set is statistically significantly higher (P<1e−6, 1 million iterations) than expected by chance by randomly sampling sets (1028 co-expression pairs—the same size as the observed overlap) of co-expression pairs from the IP-HTMS space (mean ratio=1.32, maximum ratio=1.68).
Figure 4.
Comparison of interaction data sets to gene co-expression data. Red and green fractions of each bar correspond respectively to the proportions of positive and negative co-expression correlations for each data set. The numbers above each column represent the numbers of co-expression measurements overlapping the respective data set, and the numbers in parentheses represent the ratio of positive co-expression correlations to negative co-expression correlations. (1) The complete set of co-expression correlation measurements (Lee et al, 2004). (2) The set of co-expression gene pairs mapping to one or more IP-HTMS baits. (3) The set of IP-HTMS bait–prey pairs for which a co-expression measurement is available. (4) The set of Y2H (Rual et al, 2005) interactions for which a co-expression measurement is available. (5) The set of known (Ramani et al, 2005) interactions for which a co-expression measurement is available.
We have also used the integrated IP-HTMS and gene co-expression data for further in-depth discovery of functional relationships between genes. The LYAR (Ly-1 antibody reactive) protein was originally isolated from a mouse T-cell leukemia cell line and shown to encode a predominantly nucleolar-localized protein (Su et al, 1993). As an IP-HTMS bait, LYAR identified 79 prey proteins, and of these, 32 were also found as coexpressed genes in the co-expression database (Lee et al, 2004). Twelve of these co-expression links are classed as stringent (co-expression observed across three or more gene expression studies) (Lee et al, 2004), and are represented in Figure 5. All of the 12 co-expressors/interactors are positively coexpressed and are nonrandomly distributed within the distribution of all co-expression P-values for LYAR (see Figure 5). Indeed, two LYAR interactors, BRIX and DKC1, are the two most highly coexpressed genes for LYAR across the complete co-expression database. All of the 12 (except one hypothetical protein) coexpressing/interacting proteins have been documented as nucleolar proteins (see Figure 5). Overall, these coherent co-expression/interaction patterns are not uncommon in our data set; 32 IP-HTMS baits show stringent co-expression with two or more of their prey proteins.
Figure 5.
LYAR interactors also show strong gene co-expression with LYAR. Box plot showing distribution of P-values for all genes coexpressed (in three or more studies) with LYAR. Red points indicate co-expression P-values for 12 LYAR IP-HTMS interactors. Interactor descriptions include known subcellular localizations in square brackets where available.
Biological interpretation of the interaction network
Global visualization of the IP-HTMS data set
To aid interpretation of the IP-HTMS data set, we visualized the interaction network in two ways. First, to globally visualize the data set, we developed the bait–bait connectivity map (Figure 6A and B). This visualization reduces the complexity and highlights salient features of the data set by representing only bait proteins and the degree to which they share prey proteins. Second, we visualized selected fragments of the complete (baits and preys) interaction map (Figure 6C–F). The biological significance of two of these maps (the NIMA family kinase, Nek6 interactions and translation initiation and elongation) is discussed in more detail in subsequent sections.
Figure 6ab.
Global and focused views of human interaction map. (A) Complete bait–bait connectivity map for 323 human bait proteins. Baits are represented as nodes in the graph. The size of the node represents the number of prey proteins identified for the bait. The thickness of edges between nodes represents the proportion of preys in common between the baits. Nodes are colored according to a combined disease and biological process classification, and selected classes indicated in the legend. (B) Focused views of selected bait–bait subnetworks (cross-referenced by roman numerals to panel A).
Figure 6c.
(C–F) Complete interaction networks (representing both baits and preys) for selected groups of baits. Nodes are colored according to cellular component or biological process as indicated on each figure. Baits are shown as large, labeled oval shapes, preys as small, labeled oval shapes. Arrow direction indicates a bait–prey relationship and line thickness indicates the interaction confidence score (see legend in panel C). Preys are grouped according to the baits with which they were identified (except panel E where they are grouped according to interaction confidence score). (C) Proteasome baits (corresponds to bait–bait cluster B (panel iv)). (D) Sumoylation pathway (corresponds to bait-bait cluster B (panel vi)). (E) Nek6. (F) Translation initiation and elongation (corresponds to bait–bait cluster B (panel iii)).
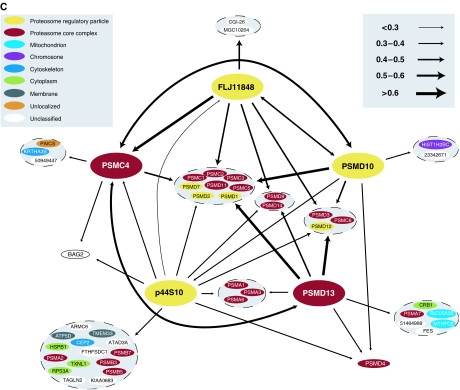
Several features of the data set are clear from the bait–bait map in Figure 6A. First, as shown in the lower part of the graph, many baits (approximately 30%) are poorly connected; that is, the set of prey proteins identified is quite distinct from the set of preys identified for any other bait. This is a consequence of both the empirical filtering that was applied to the data set (whereby frequent prey proteins and proteins observed in the control experiments that would otherwise tend to join all baits to one another were removed) and the fact that the baits selected for the study are proteins implicated in a wide variety of diseases, processes, pathways and complexes. Second, where data from multiple baits from the same complex and process are available, those baits are well connected to one another. Several of these interconnected sets of baits are indicated in Figure 6A and B (cross-referenced by roman numerals). For example, the five baits corresponding to the proteasome included in the study form a largely distinct, well-connected network as shown in Figure 6A and B, panel iv. The complete interaction map for these five baits is shown in Figure 6C. The identified prey proteins include many core and regulatory components of the proteasome. Other well interconnected sets of baits include spliceosome complex components (Figure 6A and B, panel i), chromatin remodeling components (Figure 6A and B, panel ii), the translation and elongation factor baits (Figure 6A and B, panel iii), the 14-3-3 protein baits (Figure 6A and B, panel v) and sumoylation pathway components (Figure 6A and B, panel vi). For several of these well-connected bait clusters, we have also represented the corresponding complete interaction maps (Figure 6A and B, panel iii corresponds to Figure 6F, Figure 6A and B, panel iv corresponds to Figure 6C, and Figure 6A and B, panel vi corresponds to Figure 6D). Each of the 14-3-3 baits (Figure 6A and B, panel v) identified a largely overlapping set of preys, an anticipated result given that these proteins form homo- and heterodimers in vivo (Jones et al, 1995). A subset of the experiments for the four 14-3-3 baits included in our study were previously reported and analyzed in-depth (Jin et al, 2004) (approximately 60% of the 14-3-3 prey proteins reported in the current study were reported by Jin et al (2004)). In addition, these authors analyzed the domain profiles of the identified prey proteins and validated the interaction with the Rho GTPase activator, AKAP13, an interaction identified in our study with two (YWHAB and YWHAG) of the four 14-3-3 baits.
Figure 6f.
(C–F) Complete interaction networks (representing both baits and preys) for selected groups of baits. Nodes are colored according to cellular component or biological process as indicated on each figure. Baits are shown as large, labeled oval shapes, preys as small, labeled oval shapes. Arrow direction indicates a bait–prey relationship and line thickness indicates the interaction confidence score (see legend in panel C). Preys are grouped according to the baits with which they were identified (except panel E where they are grouped according to interaction confidence score). (C) Proteasome baits (corresponds to bait–bait cluster B (panel iv)). (D) Sumoylation pathway (corresponds to bait-bait cluster B (panel vi)). (E) Nek6. (F) Translation initiation and elongation (corresponds to bait–bait cluster B (panel iii)).
Figure 6de.
(C–F) Complete interaction networks (representing both baits and preys) for selected groups of baits. Nodes are colored according to cellular component or biological process as indicated on each figure. Baits are shown as large, labeled oval shapes, preys as small, labeled oval shapes. Arrow direction indicates a bait–prey relationship and line thickness indicates the interaction confidence score (see legend in panel C). Preys are grouped according to the baits with which they were identified (except panel E where they are grouped according to interaction confidence score). (C) Proteasome baits (corresponds to bait–bait cluster B (panel iv)). (D) Sumoylation pathway (corresponds to bait-bait cluster B (panel vi)). (E) Nek6. (F) Translation initiation and elongation (corresponds to bait–bait cluster B (panel iii)).
NIMA family kinases and the mitotic cascade
The NIMA (never in mitosis gene a) was originally described in Aspergillus nidulans as a key regulator of entry into the mitotic cycle. Hence, families of NIMA-related kinases (Nek) have since been found to be widely distributed in eukaryotes with a conserved role in regulation of mitosis (Lu and Hunter, 1995; O'Connell et al, 2003). In humans, 11 members of the Nek family have been described. Nek6 was previously shown to be essential for mitotic progression in human cells, and was suggested to be particularly important for the metaphase–anaphase transition (Yin et al, 2003) and chromatin condensation (Hashimoto et al, 2002). Expression analysis also suggested an association of Nek family members with chromosome instability and cancer (Bowers and Boylan, 2004; Hayward and Fry, 2005). Nek6 bait was used in three IP-HTMS experiments, and 42 prey proteins were identified (see the interaction map in Figure 6E). Of particular interest in this set of Nek6 interacting proteins are those with roles in the mitotic cascade, chromosomal remodeling and regulation of the cell cycle. Both Nek7 and Nek9 were identified as interacting proteins with high confidence (Nek9 is the highest scoring prey for Nek6). In addition, these two prey proteins are quite specific; Nek7 was only identified in the Nek6 experiments, whereas Nek9 was identified with one other bait, GABARAPL2. Previous work showed that Nek9 activates Nek6 during mitosis and possibly regulates Nek7 as well (Belham et al, 2003). In addition, previous immunoprecipitation experiments showed that Nek6 binds Nek9 (Nercc1) (Roig et al, 2002). The Nek6 experiments also identified key components of the cohesin and condensin complexes (SMC1L1, SMC2L1, MTB, CNAP1, CSPG6 and MCM7) required for condensation, segregation and structural maintenance of chromosomes during mitosis in addition to several microtubule-associated proteins (EML2, EML3 and EML4). Although not well understood, the latter ‘echinoderm microtubule-associated' proteins are thought to play a role in regulation of microtubule dynamics during mitosis (Eichenmuller et al, 2002). In our study, these proteins were only identified with the Nek6 bait. Nek6 also identified several known cell-cycle regulators (WEE1, CDC37 and RBL1), although we note that the retinoblastoma-like protein, RBL1, was previously shown to bind MCM7 (Sterner et al, 1998), suggesting that Nek6 may not bind directly to both of these proteins. Besides the strong association of Nek6 with the mitotic cycle, Nek6 interacts with several proteins involved in nuclear import/export (TNPO3, IPO4, XPO5 and NUP93). Interestingly, NIMA kinase appears to be required for conformational changes to the nuclear pore complex during mitosis in Aspergillus (De Souza et al, 2004). Furthermore, a direct interaction between the nucleoporin 93 kDa (NUP93) and NIMA kinase has been shown and suggested to be required for nuclear accumulation of mitotic regulators (De Souza et al, 2003). Our data therefore suggest that the Nek family members are also required for nuclear pore complex regulation in higher eukaryotes.
Translation initiation and elongation factors
The molecular mechanisms underlying protein synthesis in eukaryotic organisms are complex and only partially understood (Kapp and Lorsch, 2004). The eukaryotic translation process can be divided into four steps: initiation—the assembly of the ribosome at the initiation codon, elongation—the positioning of aminoacyl tRNAs into the acceptor site, termination—occurring when a stop codon is encountered, and finally the recycling of the ribosomal machinery. As part of our protein interaction mapping, we selected six eukaryotic translation initiation factor (EIF) proteins as baits (EIF2B1, EIF3S10, EIF4A1, EIF4A2, EIF4EBP1 and GC20). A total of 222 interactions were identified for these six baits, primarily with GC20 (162 interactions) and EIF4A2 (42 interactions). Seventy-five interactions have an interaction confidence score greater than 0.3, and 60% of these interactions are with other eukaryotic initiation factor proteins or components of the translational machinery. We focus our discussion here on this subset of the interactions. Our results recapitulate many of the known complexes and steps involved in translation initiation and demonstrate both the specificity and sensitivity of the IP-HTMS approach. Figure 6F shows a bait–prey interaction map for the six initiation factor baits. All of the interactions are shown except for GC20 and EIF4A2 baits, for which only selected prey proteins are shown.
The first step of translation initiation is the formation of a ternary complex between GTP, Met-tRNA and EIF2 and binding of this complex and other EIFs to the 40S ribosomal complex to form the 43S preinitiation complex (Pestova et al, 2001). We observed several complexes that participate in this process. GC20 is a homolog of the yeast SUI1/EIF1 protein, known to be required for binding of the GTP/Met-tRNA/EIF2 complex to the 40S ribosome (Majumdar et al, 2003). In our experiments, the GC20 bait identified several components of the EIF2 complex.
EIF3 is also required for generation of a stable 40S pre-initiation complex. Our experiments with GC20 and EIF3S10 identified many of the EIF3 components (EIF1 (GC20 homolog) has previously been shown to interact with EIF3 (Fletcher et al, 1999)).
The EIF3S10 experiments demonstrate the specificity and sensitivity of the IP-HTMS approach; this bait identified eight prey proteins, seven of which are documented EIF3 subunits. Interestingly, the remaining prey protein, GA17, dendritic cell protein, contains a Proteasome/COP9/Initiation factor (PCI) domain, a domain of unknown function but which is seen in components of multi-subunit complexes, such as the proteasome, COP9 and EIF3. Our results support recent work suggesting that GA17 is an additional subunit of EIF3 (Unbehaun et al, 2004).
The next step in the process is mRNA binding to form the 43S pre-initiation complex. EIF4H is known to interact with EIF4A as part of this process and was observed in our experiments (Richter et al, 1999). Both EIF4A and EIF4H were observed in the raw data for the GC20 immunoprecipitation experiments, although EIF4A was removed based on our filtering criteria and EIF4H assigned a low interaction confidence score.
Eukaryotic messenger RNAs contain a modified guanosine, termed a cap, at their 5′ ends. For translation to proceed, binding of an initiation factor, EIF4E, to the cap structure is required (Richter and Sonenberg, 2005). EIF4B binds near the 5′-terminal cap of mRNA in the presence of EIF4F and ATP. EIF 4G1, 4G2, 4E and 4A are known components of the EIF4F multi-subunit complex, all of which were observed in our experiments with the EIF4 baits. EIF4E and protein translation as a whole are regulated in part by the EIF4E binding protein, EIF4EBP1 (Haghighat et al, 1995). In our experiments, the EIF4EBP1 bait identified a single prey protein, EIF4E. The PDCD4 (programmed cell death 4) protein was identified as a prey in both EIF4A1 and EIF4A2 experiments. The PDCD4 gene product has been reported to be a tumor and transformation suppressor and proposed as a target for cancer therapy (Lankat-Buttgereit and Goke, 2003). PDCD4 has also been shown to inhibit translation through its binding to EIF4A and EIF4G (Yang et al, 2004; Zakowicz et al, 2005). Our results support these reports and suggest that PDCD4 interacts very specifically with the translation machinery; PDCD4 was seen only with the EIF4A1 and EIF4A2 baits.
Finally, EIF2B functions to recycle the EIF2–GDP complex and recreate EIF2–GTP, which is then ready for a subsequent round of initiation. Immunoprecipitation using EIF2B1 identified six prey proteins, two of which (EIF2B3 and EIF2B5) are documented EIF2B components.
IP-HTMS has provided us with a snapshot of the interactions occurring during the complex process of eukaryotic translation initiation. With six bait proteins covering the major processes of initiation, we are able to identify many relevant interacting proteins and provide a rich data set for further discovery.
Future prospects
This study presents the first high-throughput analysis of native protein complexes by IP-HTMS in a human cell line. As illustrated in this report, our data set provides for both recapitulation of known complexes and discovery of new interactions and complexes. Although our data set maps interactions for proteins implicated in a broad range of pathways and processes, we anticipate that future, focused applications of the IP-HTMS approach will begin to probe in greater depth the impact of disease states and drug treatments on human protein–protein interactions.
Materials and methods
Cloning of the bait cDNAs and construction of entry clones
Full-length cDNAs encoding the genes for the respective protein baits were either purchased from Invitrogen (www.invitrogen.com) and the Kazusa project (www.kazusa.or.jp) or cloned in-house. Established polymerase chain reaction (PCR) methodologies were used to amplify the bait cDNAs from the corresponding parent plasmid DNAs. The oligonucleotide primers used for PCR (four required for each unique bait gene; two 5′-terminal primers, with Kozak code or not, and two 3′-terminal primers, with or without a stop codon) were designed to be complementary to the 5′ and 3′ ends of the bait coding region and to introduce an additional nucleotide sequence (29 bp), corresponding to Gateway attB recombination sites (Invitrogen), onto the ends of the PCR product. To create Gateway entry vectors, a portion of the purified PCR reaction product was added to the BP Reaction mixture, which contains a donor vector (encoding attP sites) and the BP CLONASE mix of recombination proteins. The recombination results in the oriented integration of the attB flanked PCR product into the attP sites of the donor vector, generating the Entry Clone in which the bait gene coding region is now flanked by attL sites (required for the LR Reaction, see below). A portion of the BP Reaction was used to transform competent Escherichia coli DH5α cells and the Entry Clone plasmid DNA was purified from selected transformants (antibiotic selection) using routine plasmid miniprep protocols (Sigma-Aldrich, www.sigmaaldrich.com). The integrity of each Entry Clone was verified by PCR amplification using gene-specific primers and DNA sequencing.
Construction of destination vectors
Two Destination Vectors, DV1 and DV2, were constructed based on a vector backbone using standard recombinant DNA methodologies. The Entry Clone and Destination Vector were subjected to the GATEWAY LR Reaction, which contains the LR CLONASE mix of recombination proteins. The LR Reaction results in the directional transfer of the bait gene coding region, flanked by the attL sites in the Entry Clone, to the Destination Vector (DV1 or DV2) through recombination with the attR flanked GATERC, generating the Expression Clone. A portion of the LR Reaction was used to transform competent DH5α cells and the Expression Clone plasmids were purified from selected transformants (antibiotic selection) using routine plasmid miniprep protocols. Following confirmation by PCR with gene-specific primers, milligram quantities of purified Expression Clones were prepared by standard protocols (Maxiprep; Sigma-Aldrich).
Cell culture
Anchorage-dependent human embryonic kidney 293 (HEK293) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and supplemented with 2 mM L-glutamine and 0.1 mM nonessential amino acids. Cells were grown in 10-cm-diameter or 24.5 × 24.5 cm2 tissue culture plates at 37°C in a 5% CO2 atmosphere. Cells were routinely tested for mycoplasma presence. A detailed protocol for the maintenance and passaging of cells is provided in Supplementary Information.
Transient transfection
A seed culture of HEK293 cells (at 70–80% confluence) was split and plated with fresh media the day before transfection and then grown to 30–40% confluency. Before performing transfection, cell plates were individually verified by microscopy. In particular, we verified that cells were healthy—no large vacuoles, no long extensions, not rounded up, no contamination was present (mould, yeast or bacteria) and less than 5% dead cells. We also confirmed that the plates were approximately 40% confluent. Any plates that did not meet the above criteria were discarded. Typically, approximately 1 × 107 cells were transiently transfected by adding 5 μg of DNA construct in the form of a calcium phosphate/DNA coprecipitation protocol. Briefly, a solution of calcium chloride and maxiprep Expression Clone plasmid DNA was diluted with an inorganic phosphate-containing buffer. The mixture was overlaid on the cells following a brief period to allow the calcium phosphate/DNA precipitate to develop. Cells were incubated at 37°C with the calcium phosphate DNA mixture for 12–16 h, the culture medium was replenished and the cells was cultured a further 24 h to ∼90% confluence before harvest. A similar procedure was used to culture HEK293 cells that were transiently transfected with the Destination Vector (no bait gene) in order to provide a negative-control sample. A detailed protocol for the transfection is provided in Supplementary Information.
Cell harvest and extract preparation
All methods used during the harvest procedure were performed at 4°C. Following the culture period described above (for each experimental and control culture), the media were removed from the plates by aspiration and the adherent HEK293 cells were washed thoroughly with Tris-buffered saline. Cells were then overlaid with a predetermined volume of detergent-containing lysis buffer (supplemented with a cocktail of protease inhibitors) and then scraped to concurrently dislodge and lyse the cells. Typically, cells were lysed by the addition of (1 ml) of lysis buffer (20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 10 μg/ml aprotinin, 0.2 mM AEBSF (Calbiochem)). The cell lysate was collected and then clarified by preparative centrifugation for 30 min at 20 000 g to yield a crude extract. In all cases, portions of the soluble and insoluble fractions from the centrifugation were separated by SDS–PAGE and immunoblotted with an anti-FLAG® (M2) monoclonal antibody (see below) to verify the bait's presence in the soluble extract fraction.
Immunoprecipitation of bait and bait-specific interacting proteins
The Flag-tagged bait proteins and their interacting partners were isolated from cell extracts by immunoprecipitation using M2-Agarose resin (Sigma-Aldrich). The M2-Agarose comprises the monoclonal anti-Flag M2 antibody immobilized onto an agarose resin and reacts specifically with fusion proteins possessing the Flag epitope at the N- or C-terminus. Briefly, the crude lysate were first incubated with 5 μg of agarose beads for 60 min at 4°C to remove nonspecific binders. The supernatant was then subjected to immunoprecipitation by adding 5 μg of anti-Flag monoclonal antibody covalently attached to crosslinked agarose beads (M2, Sigma). The mixture was gently agitated by inversion for 60 min at 4°C. Immunocomplexes associated with the insoluble fraction were recovered by centrifugation (1000 g for 2 min) and washed by three cycles of resuspension in lysis buffer followed by centrifugation as described above. Immunocomplexes were eluted from the beads by resuspension in 250 μl of 50 mM ammonium bicarbonate (prepared just before to use) containing 400 μM Flag peptide. Following a 30 min incubation, beads were removed by centrifugation and the supernatant containing Flag peptide as well as the eluted proteins was lyophilized.
Gel-based protein analysis
The dried immunopurified proteins were solubilized in a minimal volume of protein-loading buffer and subjected to SDS–PAGE. The immunopurified proteins were then separated by gel electrophoresis and detected by colloidal Coomassie staining. All gels were subjected to a visual appraisal before further processing; gel lanes that contained anomalies such as significant background across the entire lane or a large number of protein bands arising from nonspecific protein precipitation were rejected (approximately 40% of the gels were rejected based on these criteria). Band excision was automatically performed by a robotic system developed in-house and gel bands automatically transferred to a 96-well plate. Post-excision steps were carried out using commercially available automated robotic workstations (ProGest, Genomic Solutions). The proteins contained in the excised gel bands were treated with dithiothreitol (DTT) and the free sulfhydryl groups were alkylated using iodoacetamide. Proteins were then digested with trypsin and the resulting peptides were extracted from the gel slice using a series of wash steps. The extracted peptides were concentrated and analyzed directly by mass spectrometry.
Mass spectrometry
LC-ESI-MS/MS identification of proteins was performed as described previously (Figeys et al, 2001) using an automated network of mass spectrometers. Tryptic peptides recovered from individual gel bands were separated by reverse-phase chromatography on C18 resin and directly injected into a mass spectrometer. Ion trap mass spectrometers (LCQ Deca, Thermo Finnigan), operated in a data-dependent mode, which produces tandem MS spectra of all peptide species present above a programmed threshold, were used for these experiments.
Note: Additional detailed experimental protocols for cell transfection and passaging of cells are provided in Supplementary Information.
Data analysis
Data management
Laboratory data were managed using an in-house developed LIMS system that tracks all steps of immunoprecipitation, gel band excision as well as mass spectrometry acquisition names, annotated SDS–PAGE images and QC data. Mass spectrometry acquisition files were stored on a centralized network file system and processed using an automated analysis pipeline, including a cluster of Mascot nodes for peptide and protein identification.
Peptide and protein identification
All spectra were analyzed using Mascot version 1.9 (Matrix Sciences, www.matrixscience.com) searches against a non-redundant human protein sequence database (122 989 entries), constructed from all major sources of human protein sequences (GenBank, TrEMBL, SwissProt, IPI and Ensembl). Mascot was run in MS/MS Ion search mode with the following parameter settings: fixed modification (carbamidomethyl on cysteine), variable modification (oxidation on methionine), peptide mass tolerance 2 Da, fragment mass tolerance 0.4 Da, maximum missed cleavages two and enzyme trypsin. Peptide and protein identifications were included for further analysis according to the following criteria: for single peptide hit proteins, Mascot ionscore ⩾40; for proteins with multiple peptide hits, each Mascot peptide ionscore ⩾20. (The average Mascot recommended (P<0.05) ionscore for our data is ∼40.) Further assessment of the peptide and protein identification false-positive rates was made by searching a subset (500 gel bands; ∼3% of the data) against a randomized (each entry randomly shuffled) sequence database. Using Mascot ionscore thresholds as above, we estimate a protein false-positive rate of <7.5%. Mascot result files were parsed, proteins clustered and all data stored in a relational database. An in-house protein sequence index and annotation system was used to both provide the non-redundant sequence search database and to interpret and analyze the resulting protein hits. Spotfire (www.spotfire.com), cytoscape (www.cytoscape.org) softwares and Pathway Studio (Ariadne Genomics) were used extensively for data analysis and interaction map visualization respectively. The PLS regression analysis and generation of interaction confidence score was implemented in custom code using Python (www.python.org).
Comparisons to other data sets
Comparisons were made in general by cross-referencing NCBI Gene Ids where possible, or official HUGO gene symbols. For comparison to other protein interaction data sets, computation of statistical significance was carried out by repeatedly randomizing (1000 iterations) the IP-HTMS bait–prey associations and recalculating the interactions in common between the set of randomized interactions and the data set being compared. Minimum, mean and maximum counts of the interactions in common were then calculated from the 1000 trials. Cross-referencing to the inparanoid database (O'Brien et al, 2005) was performed by downloading all orthologous pairs for Homo sapiens and then forming paralogous groups of human genes in a simple, single-link fashion. Integration of the gene co-expression compendium (Lee et al, 2004) was performed by cross-referencing gene symbols.
GO analysis
GO-Slim versions of the Gene Ontology (www.geneontology.org/GO.slims.shtml) were used to map baits and preys to biological processes and cellular component categories (courtesy of Suparna Mundodi and Amelia Ireland, and MGI, www.spatial.maine.edu/~mdolan/MGI_GO_Slim.html), respectively. In addition, certain baits were ‘up-propagated' to parent categories where representation was low. Eighty percent of proteins in the interaction network were assigned biological process categories and 77% cellular component categories (55% of interactions were assigned biological process categories for both bait and prey, 33% of interactions were assigned cellular component categories for both bait and prey). Each combination of bait GO category and prey GO category was tested for association by constructing a 2 × 2 contingency table and using the Fisher exact test. Distributions of P-values from randomly permuted bait–prey categories were characterized as follows. Random permutation of bait–prey category associations (1000 trials) were performed, contingency tables for each bait–prey category combination constructed and the Fisher exact test P-value calculated. These distributions of 1000 P-values for each bait–prey category combination were then used to calculate the frequency with which a P-value less than or equal to the observed non-random P-value is seen by chance.
Supplementary Material
Supplementary Information
Supplementary Figure I
Supplementary Figure II
Supplementary Figure III
Supplementary Table I
Supplementary Table II
Supplementary Table III
Supplementary Table IV
Acknowledgments
DF acknowledges funding from the Canada Research Chair program, the Natural Sciences and Engineering Research Council of Canada, the Canadian Institutes of Health Research, la Fondation Jean-Louis Lévesque and MDS Inc. MM acknowledges funding from the Canada Research Chair Program. We also acknowledge past and present colleagues at MDS Proteomics/Protana who have contributed to this project.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW (2002) Analyzing yeast protein–protein interaction data obtained from different sources. Nat Biotechnol 20: 991–997 [DOI] [PubMed] [Google Scholar]
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL (2005) High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307: 1621–1625 [DOI] [PubMed] [Google Scholar]
- Belham C, Roig J, Caldwell JA, Aoyama Y, Kemp BE, Comb M, Avruch J (2003) A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J Biol Chem 278: 34897–34909 [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 [DOI] [PubMed] [Google Scholar]
- Bowers AJ, Boylan JF (2004) Nek8, a NIMA family kinase member, is overexpressed in primary human breast tumors. Gene 328: 135–142 [DOI] [PubMed] [Google Scholar]
- Bradshaw RA, Burlingame AL, Carr S, Aebersold R (2006) Reporting protein identification data: the next generation of guidelines. Mol Cell Proteomics 5: 787–788 [DOI] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S (1991) The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA 88: 9578–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Horn KP, Masker K, Osmani SA (2003) The SONB(NUP98) nucleoporin interacts with the NIMA kinase in Aspergillus nidulans. Genetics 165: 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Osmani AH, Hashmi SB, Osmani SA (2004) Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr Biol 14: 1973–1984 [DOI] [PubMed] [Google Scholar]
- Deane CM, Salwinski L, Xenarios I, Eisenberg D (2002) Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics 1: 349–356 [DOI] [PubMed] [Google Scholar]
- Eichenmuller B, Everley P, Palange J, Lepley D, Suprenant KA (2002) The human EMAP-like protein-70 (ELP70) is a microtubule destabilizer that localizes to the mitotic apparatus. J Biol Chem 277: 1301–1309 [DOI] [PubMed] [Google Scholar]
- Figeys D, McBroom LD, Moran MF (2001) Mass spectrometry for the study of protein–protein interactions. Methods 24: 230–239 [DOI] [PubMed] [Google Scholar]
- Fletcher CM, Pestova TV, Hellen CU, Wagner G (1999) Structure and interactions of the translation initiation factor eIF1. EMBO J 18: 2631–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Ge H, Liu Z, Church GM, Vidal M (2001) Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet 29: 482–486 [DOI] [PubMed] [Google Scholar]
- Gloeckner CJ, Mayerhofer PU, Landgraf P, Muntau AC, Holzinger A, Gerber JK, Kammerer S, Adamski J, Roscher AA (2000) Human adrenoleukodystrophy protein and related peroxisomal ABC transporters interact with the peroxisomal assembly protein PEX19p. Biochem Biophys Res Commun 271: 144–150 [DOI] [PubMed] [Google Scholar]
- Haghighat A, Mader S, Pause A, Sonenberg N (1995) Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J 14: 5701–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A, Rahnenfuhrer J, Talwar P, Lengauer T (2005) Confirmation of human protein interaction data by human expression data. BMC Bioinformatics 6: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Akita H, Hibino M, Kohri K, Nakanishi M (2002) Identification and characterization of Nek6 protein kinase, a potential human homolog of NIMA histone H3 kinase. Biochem Biophys Res Commun 293: 753–758 [DOI] [PubMed] [Google Scholar]
- Hayward DG, Fry AM (2005) Nek2 kinase in chromosome instability and cancer. Cancer Lett 2: 2. [DOI] [PubMed] [Google Scholar]
- Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C, Roechert B, Poux S, Jung E, Mersch H, Kersey P, Lappe M, Li Y, Zeng R, Rana D, Nikolski M, Husi H, Brun C, Shanker K, Grant SG, Sander C, Bork P, Zhu W, Pandey A, Brazma A, Jacq B, Vidal M, Sherman D, Legrain P, Cesareni G, Xenarios I, Eisenberg D, Steipe B, Hogue C, Apweiler R (2004) The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat Biotechnol 22: 177–183 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O'Donnell P, Taylor P, Taylor L, Zougman A, Woodgett JR, Langeberg LK, Scott JD, Pawson T (2004) Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol 14: 1436–1450 [DOI] [PubMed] [Google Scholar]
- Jones DH, Ley S, Aitken A (1995) Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett 368: 55–58 [DOI] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR (2004) The molecular mechanics of eukaryotic translation. Annu Rev Biochem 73: 657–704 [DOI] [PubMed] [Google Scholar]
- Lankat-Buttgereit B, Goke R (2003) Programmed cell death protein 4 (pdcd4): a novel target for antineoplastic therapy? Biol Cell 95: 515–519 [DOI] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P (2004) Co-expression analysis of human genes across many microarray data sets. Genome Res 14: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Fraser AG (2004) A first-draft human protein-interaction map. Genome Biol 5: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M (2004) A map of the interactome network of the metazoan C. elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LX, Janvier K, Berteaux-Lecellier V, Cartier N, Benarous R, Aubourg P (1999) Homo- and heterodimerization of peroxisomal ATP-binding cassette half-transporters. J Biol Chem 274: 32738–32743 [DOI] [PubMed] [Google Scholar]
- Lu KP, Hunter T (1995) Evidence for a NIMA-like mitotic pathway in vertebrate cells. Cell 81: 413–424 [DOI] [PubMed] [Google Scholar]
- Majumdar R, Bandyopadhyay A, Maitra U (2003) Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40 S preinitiation complex. J Biol Chem 278: 6580–6587 [DOI] [PubMed] [Google Scholar]
- Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, Vincent S, Vidal M (2001) Identification of potential interaction networks using sequence-based searches for conserved protein–protein interactions or ‘interologs'. Genome Res 11: 2120–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser HW (1999) Genotype–phenotype correlations in disorders of peroxisome biogenesis. Mol Genet Metab 68: 316–327 [DOI] [PubMed] [Google Scholar]
- O'Brien KP, Remm M, Sonnhammer ELL (2005) Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res 33: D476–D480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MJ, Krien MJ, Hunter T (2003) Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol 13: 221–228 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA 98: 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Bunescu RC, Mooney RJ, Marcotte EM (2005) Consolidating the set of known human protein–protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol 6: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguly T, Breitkreutz A, Boucher L, Breitkreutz B-J, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, Stark C, Ho Y, Botstein D, Andrews B, Boone C, Troyanskya OG, Ideker T, Dolinski K, Batada NN, Tyers M (2006) Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N (2005) Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480 [DOI] [PubMed] [Google Scholar]
- Richter NJ, Rogers GW Jr, Hensold JO, Merrick WC (1999) Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J Biol Chem 274: 35415–35424 [DOI] [PubMed] [Google Scholar]
- Roig J, Mikhailov A, Belham C, Avruch J (2002) Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev 16: 1640–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (2005) Towards a proteome-scale map of the human protein–protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW (2006) Peroxisome biogenesis disorders. Biochim Biophys Acta 1763: 1733–1748 [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE (2005) A human protein–protein interaction network: a resource for annotating the proteome. Cell 122: 957–968 [DOI] [PubMed] [Google Scholar]
- Sterner JM, Dew-Knight S, Musahl C, Kornbluth S, Horowitz JM (1998) Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol Cell Biol 18: 2748–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Hershberger RJ, Weissman IL (1993) LYAR, a novel nucleolar protein with zinc finger DNA-binding motifs, is involved in cell growth regulation. Genes Dev 7: 735–748 [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV (2004) Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 18: 3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P (2002) Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417: 399–403 [DOI] [PubMed] [Google Scholar]
- Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH (2004) A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol Cell Biol 24: 3894–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin MJ, Shao L, Voehringer D, Smeal T, Jallal B (2003) The serine/threonine kinase Nek6 is required for cell cycle progression through mitosis. J Biol Chem 278: 52454–52460 [DOI] [PubMed] [Google Scholar]
- Zakowicz H, Yang HS, Stark C, Wlodawer A, Laronde-Leblanc N, Colburn NH (2005) Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI. RNA 11: 261–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure I
Supplementary Figure II
Supplementary Figure III
Supplementary Table I
Supplementary Table II
Supplementary Table III
Supplementary Table IV