We want to see how, in some cases at least, the forms of living things, and of the parts of living things, can be explained by physical considerations.… D'Arcy Thompson (1917)
Proper organization of microtubule polymers is crucial to the form and function of all eukaryotic cells. Whether growing, dividing or polarizing, different cell types—be they neurons, plant cells or fungal cells—organize specialized microtubule patterns appropriate for their needs. Is it possible to identify the set of factors sufficient to organize microtubules in a specific cell, and to understand how the cell regulates those factors spatially and temporally such that they collectively sustain that particular cellular pattern of microtubules? In a recent issue of Cell, Marcel Janson and collaborators (Janson et al, 2007) use an elegant combination of quantitative microscopy, in vitro assays and computer simulations to try to elucidate the minimal set of components required to stably organize microtubule patterns in a unicellular eukaryote, the fission yeast Schizosaccharomyces pombe.
Microtubule organization in a given cell is partly dependent on the intrinsic turnover of each of its polymers. But it is the collective interaction of microtubules with microtubule-interacting proteins—nucleators determining polymer number and localization; stabilizing or destabilizing proteins regulating average polymer length; static or dynamical crosslinkers mediating polymer connectivity—that mostly determines the global spatial pattern and ‘systemic' cellular function of microtubules. Individually, every component of this ‘system' appears to behave independently; collectively, they generate an overall stable, organized and functional microtubule pattern.
In the fission yeast, the proper growth and form of each cell relies on the presence in its cytoplasm of ‘bipolar' bundles of antiparallel microtubules, organized with their more dynamic ‘plus' ends towards the cell tips and their ‘minus' ends overlapping at the cell centre (Drummond and Cross, 2000; Tran et al, 2001). It is generally thought that the organization of these bipolar microtubule arrays relies on two evolutionarily conserved microtubule interactors: the Prc1-related protein Ase1, which statically crosslinks or ‘bundles' microtubules (Loiodice et al, 2005; Yamashita et al, 2005); and the Kar3/Ncd-related, minus-end-directed kinesin motor Klp2, which mediates the transport (‘sliding') of short microtubules towards the cell centre along longer microtubules (Carazo-Salas et al, 2005). Recently, it has been suggested that these two interactors might suffice for bipolar microtubule arrays to ‘self-organize' without pre-existing organizational templates (Carazo-Salas and Nurse, 2006; Daga et al, 2006).
Janson et al (2007) set out to further develop this hypothesis. Using a sophisticated range of experimental methods, they first characterized the interaction of Klp2 and Ase1 with microtubules. By quantitative in vivo microscopy and statistical methods, they observed that Klp2 associated to the ‘plus' ends of short, sliding microtubules and was absent from microtubule ‘minus' ends. This suggested that Klp2 ‘pulls' microtubules only at their ‘plus' ends. They then analysed the interaction of bacterially expressed Ase1 protein with pure microtubules in vitro. They found that Ase1 forms oligomers that bind stably along the length of microtubules only when bound to multiple polymers, and that Ase1 preferentially bundles microtubules in a ‘biased' fashion with respect to microtubule polarity, leading to an antiparallel microtubular array.
With that information, they used computer simulations to test whether the collective interaction of microtubules with dynamic or static crosslinkers with properties similar to Klp2 and Ase1 could give rise to the organization of stable, bipolar microtubule arrays. In the simulations, neither ‘biased bundlers' alone, nor mixtures of ‘motors' and ‘unbiased bundlers', could efficiently organize focused bipolar microtubule arrays. However, when ‘motors' and biased ‘bundlers' were combined, bipolar microtubule arrays were formed with a higher degree of polarization, in agreement with what has been observed in vivo. These findings suggest that a fine-tuned antagonism between motor and bundling activities may be the ‘design principle' underlying the organization of microtubule arrays in fission yeast cells (Figure 1, simulations can also be downloaded from http://www.embl.de/ExternalInfo/nedelec/reprints and run live on Mac OS X and Windows platforms).
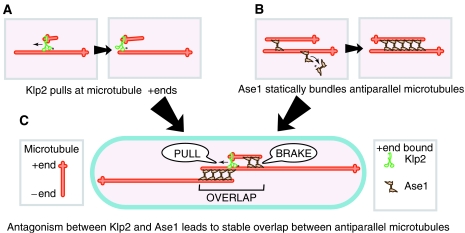
Figure 1.
Model for the formation of stable bipolar microtubule arrays in fission yeast. (A) The minus-end-directed kinesin Klp2 (green) mediates antiparallel microtubule sliding by ‘pulling' short microtubules at their plus end. (B) The bundling protein Ase1 (brown) preferentially crosslinks antiparallel microtubules (red). (C) A short, newly nucleated microtubule is ‘pulled' by Klp2 to the bundle midzone. Increasing levels of Ase1 bind to the microtubule as it grows, gradually breaking its motion. The antagonism between Klp2 and Ase1 leads to the formation of a stable overlap between antiparallel microtubules in the cell (blue).
The simulations make a number of assumptions about the biophysical properties of Klp2 and Ase1, which need to be tested experimentally to validate the model fully. It will also be interesting to further explore how this type of two-component system may have changed throughout evolution to generate different microtubule patterns in different cells. But altogether, this exciting piece of work underscores that a ‘systems' approach combining experimental and theoretical methodologies will be the right way forward to understand the ‘design principles' underlying the spatial organization and function of microtubular arrays in more complex eukaryotic cells (Karsenti et al, 2006), an endeavour that brings us closer to D'Arcy Thompson's objective of explaining the forms of living things in terms of physical considerations.
References
- Carazo-Salas RE, Antony C, Nurse P (2005) The kinesin Klp2 mediates polarization of interphase microtubules in fission yeast. Science 309: 297–300 [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Nurse P (2006) Self-organization of interphase microtubule arrays in fission yeast. Nat Cell Biol 8: 1102–1107 [DOI] [PubMed] [Google Scholar]
- Daga RR, Lee KG, Bratman S, Salas-Pino S, Chang F (2006) Self-organization of microtubule bundles in anucleate fission yeast cells. Nat Cell Biol 8: 1108–1113 [DOI] [PubMed] [Google Scholar]
- Drummond DR, Cross RA (2000) Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr Biol 10: 766–775 [DOI] [PubMed] [Google Scholar]
- Janson ME, Loughlin R, Loiodice I, Fu C, Brunner D, Nedelec FJ, Tran PT (2007) Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell 128: 357–368 [DOI] [PubMed] [Google Scholar]
- Karsenti E, Nedelec F, Surrey T (2006) Modelling microtubule patterns. Nat Cell Biol 8: 1204–1211 [DOI] [PubMed] [Google Scholar]
- Loiodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT (2005) Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell 16: 1756–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DAW (1917) On Growth and Form. Cambridge University Press: Cambridge [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoue S, Chang F (2001) A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol 153: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T (2005) The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell 16: 1378–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]