Summary
Plasma-polymerized films deposited from AlAm, HxAm, NVP, NVFA, AA and FC were compared to TCPS and PS surfaces in supporting cellular attachment, viability, and proliferation in serum-based culture in vitro for extended periods of time (>7 d). Surface patterns were created using multi-step depositions with physical masks. Cell adhesion in the presence of serum was compared for (monocyte-) macrophage and fibroblast cell lines. Cellular response was tracked over time, reporting adhesive behavior, proliferative rates, and morphological changes as a function of surface chemistry. Micropatterned surfaces containing different surface chemistries and functional groups (e.g. –NH2, –COOH, –CF3) produced differential cell adhesive patterns for NIH 3T3 fibroblasts compared to J774A.1, RAW 264.7 or IC-21 (monocyte-) macrophage cell types. Significantly, macrophage adhesion is substantial on surfaces where fibroblasts do not adhere under identical culture conditions.
Keywords: cell culture, cell patterning, macrophage, plasma, surface chemistry
Introduction
Surface chemistry is well known to exert profound effects over interfacial biological processes in vitro relevant to biomedical device designs and applications.[1–4] Interfacial behaviors of both proteins and cell types are known to result from combined effects of surface chemical composition and physical topology.[5–10] Plasma polymerization (pp) is frequently employed to generate stable, sterile surfaces or coatings with varied chemical, topological and physical characteristics, facilitating such measurements.[11,12] Patterned surfaces representing regular arrangements of two or more chemically distinct materials can be readily generated, providing both spatial and chemical control of the interface useful for examining and producing differential biological responses.
Surface pattern deposition methods include soft (stamping) lithography, photolithography, ion milling and plasma patterning. Plasma polymerization techniques have been employed for numerous applications including deposition of Si-based materials for microelectronics applications,[13–15] hard coatings such as diamond-like carbon and carbon nitride films,[4,16,17] and organic polymer films for modifying substrates with complicated geometries.[18–21] Plasma polymerization techniques provide a facile preparation method for generation of diverse surface chemistries on a variety of substrates. The resultant surfaces are sterile, stable under ambient storage conditions and amenable to use with cultured cells.
Cell patterning studies to date have examined the effects of specific substrate chemistries and microfabrication techniques on the modulation of cell-substrate and cell-cell contacts.[22–26] Frequently, differential cell fidelity to underlying pattern chemistry is a determinant for distinguishing success of these approaches to elucidate cell-surface interaction mechanisms. Numerous methods for cellular micropatterning have been employed, with varied levels of success for specific substrate-substrate and substrate-cell combinations.[6,10,12,22–25] Many strategies have exploited unique combinations of surface chemistries or deposition techniques to achieve alternating cell adhesive and non-adhesive regions, often correlated with the presence or absence of surface-adsorbed cell adhesive matrix proteins,[24,26–29] thus modulating cell-substrate interactions.
Although many combinations of surfaces, patterns and cells have been tested to date, one important distinction is the ability to produce spontaneous cell pattern fidelity in serum protein-based culture versus that resulting from pre-adsorbed matrix protein (e.g., collagen, fibronectin) adsorption to surface patterns prior to cell seeding. Biased [e.g., pure mono-component extra-cellular matrix (ECM)] protein pre-adsorption to patterned chemistry readily produces protein patterns templated by the surface chemistry to which cells adhere. However, spontaneous cell pattern formation and long-term fidelity from serum media is less reported, understood or controlled because of difficulties in control of ECM protein surface selection from serum (a milieu of >500 proteins). Yet, this serum-based cell-surface scenario is much more relevant to cell response to implanted biomaterials than biased ECM protein adsorption templated control of cell patterning.
Numerous surface chemistries have been studied to date for cell patterning.[22–25,27] A broad general characterization of the conclusions from this exhaustive survey is that surface chemistries conducive to cell adhesion have specific chemical and physical parameters that endow them with a propensity to adsorb cell matrix proteins at sufficient density, and with sufficient adhesion strength, to support cell receptor and cell surface engagement successfully enough to produce cell adhesion, spreading and proliferative responses.[4,30] Frequently, this correlates with moderate surface polarity and mechanical modulus (neither extreme hydrophilicity nor extreme surface elasticity).[30]
Various cell types have been employed in cell patterning studies, including epithelial,[7] endothelial,[31] capillary endothelial,[32] monocyte-macrophage,[27,33] osteoblast,[24,34] fibroblast[35] and numerous others.[3] Interestingly, few studies report pattern responses of inflammatory cell types, including lymphocytes, or leukocytes such as monocytes and differentiated macrophages. This current contribution describes the use of plasma-patterned surface chemistry to contrast the adhesive and proliferative behavior of fibroblasts and macrophage cell lines cultured in serum-containing media. Few studies compare these distinct cell behaviors. This present study includes biologically relevant cell types of primary and secondary derivation for comparison: primary bone marrow-derived macrophages, three secondary (monocyte-) macro-phage cell lines (IC-21, RAW 264.7 and J774A.1), and one fibroblast cell line (NIH 3T3) frequently employed in patterning studies[35–37] (Figure 1). Results point to intrinsic differences in the responses of these cell types to identical chemistries and suggest important distinctions in phenotypic mechanisms for recognizing surfaces relevant to cell based drug testing in vitro (e.g., anti-fibrotic, anti-inflammatory drugs) and inflammatory processes with implanted biomaterials and surgical devices.
Figure 1.
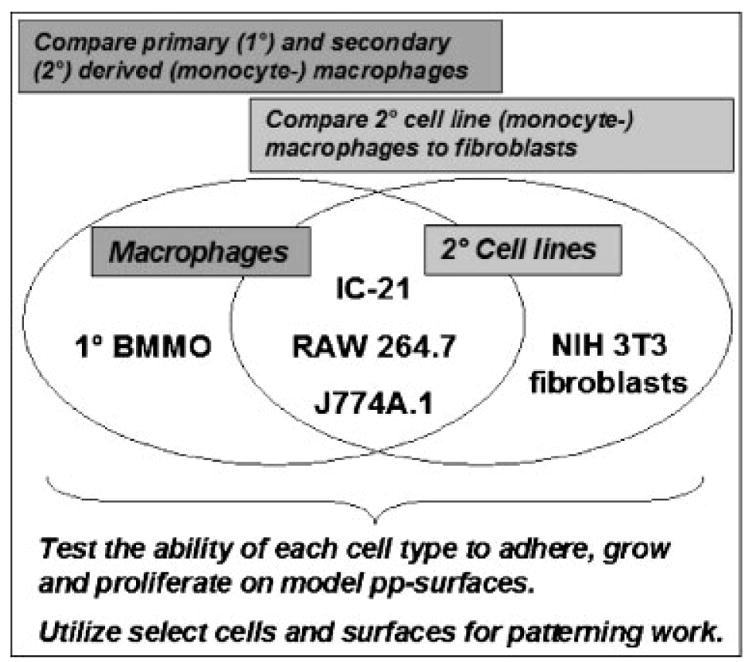
Schematic of relationships among cells utilized in this study.
Experimental Part
Preparation of Culture Surfaces Using Plasma Deposition
All pp-films were deposited as described previously.[38] Briefly, all films were deposited in our home-built inductively coupled radio frequency (RF) (13.56 MHz) plasma reactor, described previously.[39,40] The pulse duty cycle was varied using the internal pulse generator of an RF Power Products power supply. The peak applied RF pulse power (P) was kept constant at 300 W for fluorocarbon film depositions and between 16 and 150 W for other organic monomers. The duty cycle (d.c.) is the ratio of pulse on time to the total cycle time; thus, a pulse sequence of 10/190 ms has a 5% d.c. A 10 ms on time was used in all experiments; pulse off time was varied to achieve different d.c.’s, ranging from 5–50%. For all experiments, the gas flow was kept constant at 10.0 sccm (standard cubic centimeters per minute), resulting in a reactor pressure of ca. 200 mTorr.
The C3F8 (FC) (Air Products, 99%) was used without further purification. Acrylic acid (AA) (99%), N-vinyl-2-pyrrolidi-none (NVP) (99%), and N-vinylformamide (NVFA) (98%) were obtained from Aldrich Chemical Company. Allylamine (AlAm) (98%) and hexylamine (HxAm) (99%) were obtained from Acros Organics. All were used after undergoing several freeze-pump-thaw cycles to remove dissolved gases. Tissue culture polystyrene (TCPS, Falcon) and polystyrene (PS) suspension culture dishes (Corning), and glass coverslips (VWR Scientific) were used as substrates for pp deposition reactions. One macro solid disk mask (30 mm diameter) and copper TEM grids (Ted Pella, Inc.) of various dimensions and with distinctly different internal patterns (grids, circles and bullet shapes) were used as masters for pp-film patterning on TCPS, PS and glass substrates. All substrates were cleaned with methanol immediately before pp-deposition. Surface analysis was performed on patterned and unpatterned samples treated identically.
Optimized conditions for the various pp-films were determined previously.[38] Unless otherwise noted, all films were deposited 8 cm downstream from the coil region of the plasma using a 10% duty cycle. Specific conditions for each monomer deposition system include: C3F8 (FC) plasmas used a peak power of 300 W, for 20 min; AA plasmas had a 16% duty cycle (150 W peak power) for 5 min; NVP plasmas utilized a peak power of 100 W for 15 min; NVFA plasmas had a peak power of 30 W for 12 min; and AlAm and HxAm plasmas used a 150 W peak power for 10 min and 30 min, respectively. In some instances, deposition times were increased to 90 min to increase film thickness. Deposition rates varied from 11.6 Å · min−1 (HxAm) to 129.3 Å · min−1 (NVP). Water contact angles are reported below and adhesion tests revealed successful layering (i.e. no delamination) of pp-films on PS and PS coated with pp-C3F8. After deposition, samples were stored in petri dishes under ambient conditions until cell culture use (1–14 d).
Film aging has been a concern for many plasma-deposited materials. For all nitrogen-containing materials, we do observe some oxygen uptake in a matter of days following deposition, and a concomitant loss of nitrogen, most markedly at the surface, as determined from angle-resolved X-ray photoelectron spectroscopy (XPS) data. Over an 11-month period, the pp-AlAm film composition changed significantly, with the %O increasing from ≈4 to 20%, %N decreasing from 26 to 13%, and the N/C ratio increasing significantly. No significant changes in the overall N/C ratio in other films were observed (over a period of 6–9 d) although there are small decreases in nitrogen at the surface. For the patterned samples, some secondary ion mass spectrometry (SIMS) images were acquired using samples aged 14 months and still showed high fidelity of chemical composition. Thus, the patterned samples are extremely stable with respect to post-deposition aging.
Prior to use in cell studies, pp-surfaces were first thoroughly misted with 70% ethanol and then UVirradiated for 20 min in a biosafety cabinet. This process was previously shown to reliably produce sterility for cell culture with minimal changes to surface chemistry.[41] XPS analysis[38] performed on samples before and after this treatment demonstrated that the pp-deposited surface chemistry was not affected (data not shown).
Patterned Substrate Deposition by Plasma Polymerization
Patterned substrates comprising two different surface chemistries for cell culture were produced by a previously reported plasma deposition technique.[38] Plasma deposition for patterns occurred sequentially as described in Figure 2 and was then reversed in a separate preparation to create two analogous inverse macropatterned surfaces for each combination of surface chemistries employed (cf. Table 2).
Figure 2.
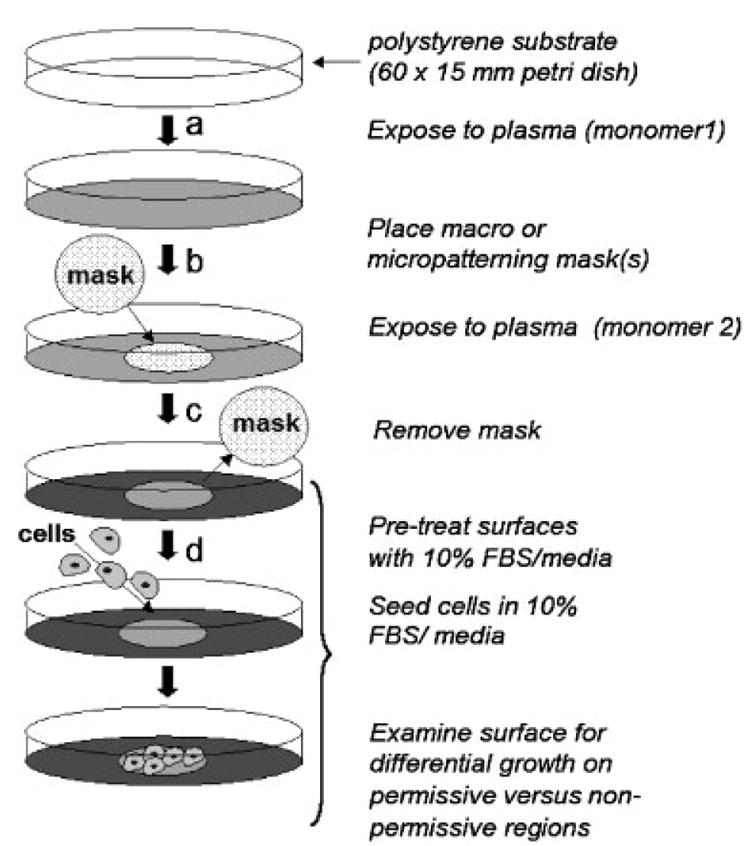
Schematic illustration of the patterning procedure using masks and sequential plasma polymerization techniques. (a) PS petri dishes were exposed to the first of two sequential plasma polymerization depositions. (b) One macro (30 mm diameter solid disk) or multiple micropatterning (3 mm diameter copper TEM grid) mask(s) of various patterns were placed onto the plasma-deposited substrates and the (c) second plasma deposition (different chemistry) was performed. The mask(s) were then removed. (d) Plates were treated with 70% ethanol and UV light for 20 min. Plates were subsequently exposed to 10% serum containing media for 3–24 h before cell seeding. Plates were seeded with NIH 3T3 fibroblasts and (monocyte-) macrophage cells at various densities as described in Materials and Methods.
Table 2.
Cell attachment and proliferation on plasma-macropatterned substrates.
| Cell pattern fidelity
|
|||||
|---|---|---|---|---|---|
| Surface descriptiona) | NIH 3T3 | RAW 264.7 | J774A.1 | IC-21 | dc) |
| NVP-FC | +/−b) | NDb) | ND | +/+ | 7 |
| FC-NVP | −/+ | ND | ND | +/+ | 7 |
| AlAm-FC | +/− | +/+ | ND | +/+ | 7 |
| FC-AlAm | −/+ | ND | +/+ | +/+ | 5 |
Abbreviations as used in text. See text for detailed description of patterning method.
+ (supportive of cell growth), − (no adherence or proliferation); order of ‘‘surface description’’ matches reported + or − order, respectively. ND (not determined). See ref.[37] for full surface analytical characterization. See text for a detailed description of successful combinations of surface chemistries for macropatterning based on cell line.
Cell pattern fidelity differences due to different seeding concentrations, doubling times, and affinity for surfaces.
Materials Surface Characterization
Surfaces of plasma-deposited polymeric materials were characterized as described previously using angle-resolved XPS, spectroscopic ellipsometry (SE) and measurement of static water contact angles.[38] Similarly, surface pattern fidelity was confirmed with scanning electron microscopy (SEM), scanning Auger microscopy (SAM), XPS imaging, and time-of-flight secondary ion mass spectroscopy (ToF-SIMS).[38] Aging studies were performed on nitrogen-containing materials by allowing exposure to atmosphere for 10–14 d, with subsequent reprobing of surfaces via XPS.[38]
Cell Culture
Murine fibroblast NIH 3T3, murine monocyte-macrophage RAW 264.7 and J774A.1, and murine macrophage IC-21 cell lines were obtained from the American Type Culture Collection (Manassas, VA). Cells were cultured in Dulbecco’s modification of Eagle’s medium (DMEM, Mediatech, for NIH 3T3 and J774A.1), or RPMI 1640 (Mediatech, for IC-21 and RAW 264.7) supplemented with 10% fetal bovine serum (FBS, HyClone, Inc.) and 1% penicillin-streptomycin (Life Technologies). Cultures were maintained in T-175 TCPS flasks (NuncTM) under standard conditions: incubation at 37 °C, 98% humidity and 5% CO2. Cells were dissociated from culture flasks by incubation with Ca2+– and Mg2+– free cell culture grade Hank’s balanced salt solution (HBSS, Life Technologies) (NIH 3T3, J774A.1 and RAW 264.7), or by scraping with a rubber policeman (IC-21).
Primary Cell Harvest
Bone marrow derived macrophages (BMMO) were prepared from bone marrow cells harvested from the femurs and tibias of C57BL/6 mice.[42] To differentiate bone marrow precursors into macrophages, bone marrow-extracted cells were cultured in DMEM supplemented with 10% L929 fibroblast-conditioned medium, 2 × 10−3 M L-glutamine, 0.01% HEPES [4-(2-hydroxyethyl)piperazineethanesulfonic acid], 1% penicillin- streptomycin, and 2 × 10−3 M non-essential amino acids (Sigma-Aldrich). Cells were grown under standard conditions (vida supra) with media changes every two days. This method has been shown to produce differentiated macrophages.[42]
Cell Culture on Patterned Surfaces
Cell seeding densities were adjusted, depending on observed cell-specific culture doubling times. Seeding densities were varied for each cell type to allow similar growth kinetics toward confluent density for comparison of cell types as follows: murine (monocyte-) macrophages cell lines were seeded at a density of 50 000 to 250 000 cells; NIH 3T3 murine fibroblasts at 150 000–200 000 cells; and primary-derived bone marrow macrophage cells at 6.2 × 106 cells; all per 15 × 60 mm culture surface. Cell line passage number ranged from 2–22 (beyond original stock subculture as obtained from the ATCC) and varied depending on the cell line employed. Cells counts were performed with a hemacytometer and viability was assessed using the standard Trypan blue dye exclusion test.[43] Surfaces were conditioned for up to 24 h with 10% FBS-containing media (RPMI 1640 or DMEM) prior to seeding. After seeding, surfaces were incubated at 37 °C, 98% humidity and 5% CO2 and media changes were performed as necessary. Cell attachment, growth and proliferation were studied by direct light (phase contrast) and fluorescent microscopy techniques.
Protein Pre-Adsorption and Cell Culture on pp-FC Surfaces
IC-21, NIH 3T3 or BMMO cells were seeded onto control PS suspension culture dishes and pp-FC surfaces that were treated for 24 h with one of the following solutions: 3 mg · ml−1 bovine serum albumin (BSA Fraction V, OmniPur®, Sigma) in sterile Dulbecco’s phosphate buffered saline (DPBS, HyClone), 100% FBS, or 10% FBS in DPBS. At 24 h, the protein solution or serum was removed by aspiration and cells were immediately seeded in an appropriate cell culture media containing 10% FBS. Culture conditions proceeded as described above.
Phase Contrast Microscopy
Images were obtained on either a Nikon Eclipse TS100 or a Nikon TMS inverted microscope using Nikon objectives. A Kodak DC290 camera was used to capture images that were subsequently processed in Adobe Photoshop 6.0 (Adobe Systems, Inc.).
Fluorescence Microscopy
Cells were stained with fluorescein diacetate (Molecular Probes). Briefly, culture media was aspirated from the surfaces, which were then rinsed once with sterile DPBS. A dilute fluorescein diacetate solution (0.01 mg · ml−1 in PBS) was added and surfaces with cells were allowed to stand for 15 min. Images were obtained on a Zeiss Axiomat® inverted microscope with Zeiss filter sets for fluorescein with a Dage 300 CCD camera (Zeiss Plan 2.5X objective, NA of 0.08, and 0.63 reducing lens). A DPS PVR digital recorder was used to digitize the video output of the camera.
Results and Discussion
Previously, a series of micropatterned substrates was created and characterized where the underlying substrate was first coated with one type of plasma polymer (most commonly a highly hydrophobic FC) and then patterned with a second type of plasma polymer (usually a relatively hydrophilic hydrocarbon).[38] This diverse group of polymer films was subsequently employed in patterning experiments using fibroblastic and (monocyte-) macrophage cell types, as reported here. Included in this group were the cell adhesion-promoting pp-AA films[44] containing carboxylic acid groups; the unique NVP monomer, a heterocyclic nitrogen-containing compound; the amide-containing NVFA monomer, an isomer of acrylamide that exhibits low toxicity; the extensively studied and employed nitrogen-containing AlAm; and the saturated amine monomer HxAm, thought to be promising for the production of films with high concentrations of primary amines.[38] Since material hydrophilicity is a critical factor in promoting protein adhesion and subsequent cellular adhesion, static water contact angle measurements were performed on non-patterned plasma polymerized films created on Si substrates[38] with the materials ranging from hydrophilic to very hydrophobic in nature: NVFA (43 ± 1°), AlAm (46 ± 1 °), NVP (53 ± 1 °), HxAm (77 ± 6 °), AA (74 ± 3 °), FC (114 ± 1 °).
Cell Attachment and Proliferation on Plasma-Polymerized Culture Substrates in Serum-Containing Media
First, each unpatterned pp-surface was assessed for its ability to support cell attachment and proliferation in vitro in 10% FBS using the following cell types: NIH 3T3 (fibroblast), RAW 264.7 and J774A.1 (monocyte-macrophage), IC-21 (macrophage) and primary-derived BMMO. Two dissimilar cell types were selected (fibroblast versus (monocyte-) macrophagea to determine differential surface responses based on cell type. On each unpatterned surface chemistry, at least one (monocyte-) macrophage cell line was compared to the NIH 3T3 fibroblast cell line. Results for control surfaces (PS and TCPS) and pp-film chemistries studied over 2–7 d culture times are shown in Table 1. Each surface tested supported the attachment and proliferation of all (monocyte-) macrophage cell lines, and all surfaces except the FC chemistry supported attachment and proliferation of NIH 3T3 fibroblasts, similar to fibroblast behavior observed on Teflon AF® fluorocarbon surfaces.[45,46] Representative phase contrast microscopy images for the IC-21 and NIH 3T3 cell lines are shown in Figure 3 and 4. Figure 3 shows results for control (TCPS) and three of the pp-surfaces tested: FC, AlAm and NVFA. Due to differences in doubling times, cell lines were seeded at varied densities on each substrate (as described in Materials and Methods) to facilitate comparative analysis over virtually equivalent culture time periods, based on predicted time to reach 100% confluence on TCPS. Results from Figure 3(B) and Figure 4(D–F) show minimal fibroblast attachment and survival, supporting previous assertions that albumin’s natural abundance in cell culture media prevents fibroblast-surface interactions required for attachment, growth and proliferation on FC surfaces.[31,45–47]
Table 1.
Cell attachment and proliferation on uniform plasma-deposited substrates.
| Duration
|
||||||
|---|---|---|---|---|---|---|
| Surface descriptiona) | NIH 3T3 | RAW 264.7 | J774A.1 | IC-21 | BMMO | dc) |
| FC | −b) | +b) | + | + | + | 2–5 |
| AA | + | + | NDb) | ND | ND | 2 |
| AlAm | + | + | + | + | ND | 4 |
| HxAm | + | ND | + | + | ND | 4 |
| NVFA | + | ND | ND | + | ND | 4 |
| NVP | + | + | + | + | ND | 4 |
| PS control | + | ND | + | + | + | 7 |
| TCPS control | + | + | + | + | + | 7 |
Abbreviations as used in text. See text for detailed description of patterning method.
+ (supportive of cell growth), − (no adherence or proliferation), ND (not determined). See ref.[37] for full surface analytical characterization.
Duration differences due to different seeding concentrations, doubling times, and affinity for surfaces.
Figure 3.
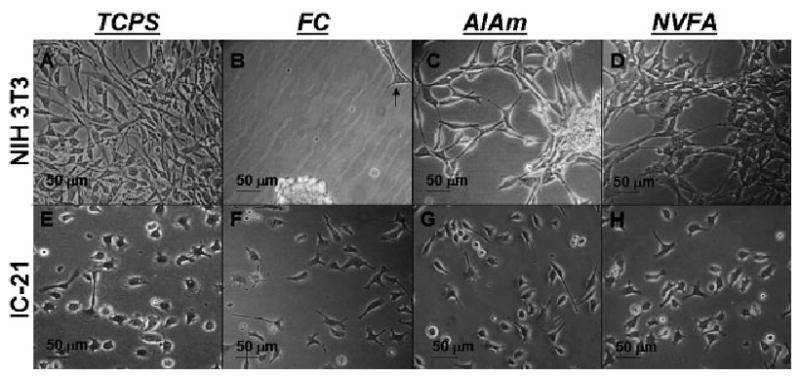
Phase contrast microscopy images of NIH 3T3 (A–D) and IC-21 (E–H) cell morphologies observed on control (TCPS) and FC, AlAm and NVFA pp-surfaces at sub-confluent culture conditions (time in culture varied based on cell type and substrate). Images shown are representative of results obtained for multiple fields (>5) and plates (≥2) for each surface and cell line. Arrow (B) indicates an anomalous adherent cell that has likely attached to a surface defect.
Figure 4.
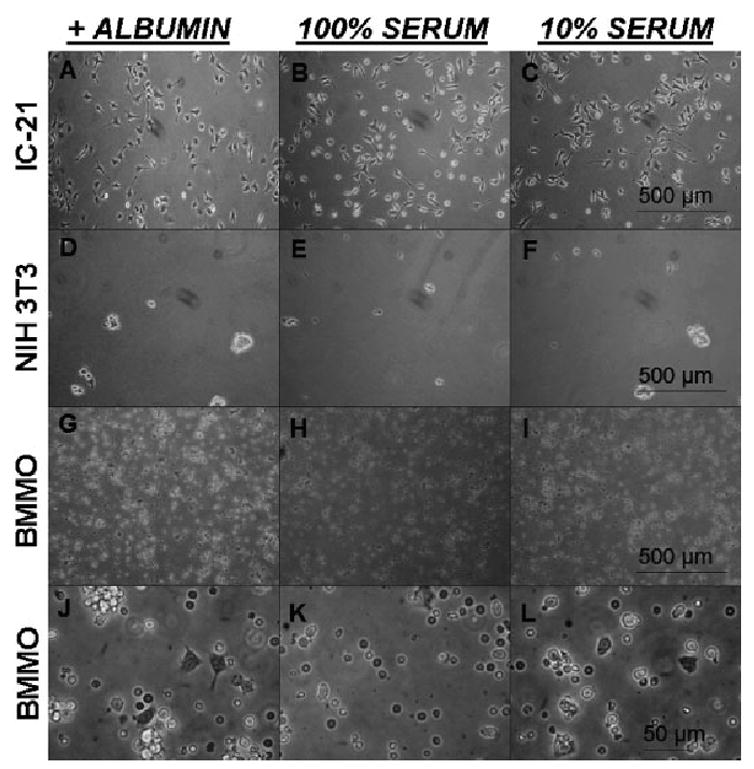
Phase contrast microscopy comparison of IC-21 macrophage, NIH 3T3 fibroblast and BMMO cell growth at 24 h post-seeding in 10% serum containing media on uniform FC surfaces pre-treated with pure (3 mg · ml−1) albumin (A, D, G and J), 100% (B, E, H and K) or 10% serum (C, F, I and L). NIH 3T3 fibroblasts fail to adhere to FC surfaces under any of the conditions tested (D–F). IC-21 (A–C) and BMMO (G–L) cells adhere and proliferate to nearly confluent cultures (data not shown) on FC surfaces under all test conditions. Images are representative of multiple fields (>5) and multiple plates (2) for each test condition. Since BMMO cells are significantly smaller than IC-21 and NIH 3T3 cells, BMMO images were further magnified (J–L) to allow visualization of differences in adherent (dark circular and angular with extensions) and non-adherent (light circular) cell morphologies.
To determine whether the noteworthy result of growth of (monocyte-) macrophage cell lines on FC surfaces was a cell line-dependent phenomenon, we performed limited experiments on FC surfaces with murine primary-derived BMMO. Significantly, BMMO cells readily adhered, grew and proliferated on FC surfaces (Figure 4(G–L)). Although at 24 h the majority of BMMO cells remained undifferentiated (monocytic) and either non-adherent or loosely adherent (indicated by a round shape), several cells with surface contacts and short filopodia can be seen even at this early time point (Figure 4(J–L)).
Surfaces prepared with AlAm (Figure 3(C,G)), HxAm (data not shown) and NVFA (Figure 3(D,H)) readily promoted cell adhesion and proliferation for fibroblast and (monocyte-) macrophage cell types. Cells elongated and spread at early time points suggestive of a motile phenotype, but as the cultures progressed, adherent cells adopted characteristic tightly packed cobblestone growth patterns associated with limited surface space (exemplified by cells grown on TCPS, Figure 3(A)). Fibroblasts exhibited normal morphology on AlAm (Figure 3(C)), HxAm (data not shown) and NVFA (Figure 3(D)) surfaces by comparison to TCPS (Figure 3(A)) with multiple attachment sites on these surfaces. Cultures progressed to 100% confluence if allowed (day 4), and cells remained adherent throughout the culture lifetime. No delamination of the cells was observed at any time.
On AlAm, HxAm and NVFA substrates, (monocyte-) macrophage cells also adopted typical adherent morphologies, which differed slightly based on the cell line. Representative data for IC-21 cells are shown in Figure 3(E–H). J774A.1 and RAW 264.7 cultures exhibited similar results (data not shown), with the appearance of characteristic features such as lamellipodia, filopodia, and membrane ruffles as the cells adhered to and spread on surfaces. These features have been previously reported for surfaces of varied chemical composition and hydrophilicity.[46] Surfaces comprising NVP were also observed to be generally cell-conducive for all cell lineages tested, with slightly slower proliferation rates compared to AlAm, HxAm and NVFA surfaces (unpublished observations). Limited culture performed on AA surfaces supported attachment and proliferation of both RAW 264.7 and NIH 3T3 cells, consistent with previous reports.[48,49]
Media Influences on Cell Proliferation on pp-FC and PS Surfaces Pre-Adsorbed With Proteins
Effects of serum protein-surface deposition from a pure albumin solution (at 10% of physiological concentration, 3 mg · ml−1, consistent with tissue culture conditions of 10% FBS) versus surface protein selection from complex cell culture milieu (10 or 100% FBS) on cellular attachment and proliferation, were assessed using NIH 3T3 fibroblasts, IC-21 macrophages and BMMO under the aforementioned culture conditions on moderately and very hydrophobic surfaces (PS and pp-FC, respectively). Surfaces were exposed to one of three protein pre-adsorption test conditions: 1) pure albumin solution (3 mg · ml−1 albumin in DPBS), 2) 100% FBS, or 3) 10% FBS in DPBS, for 24 h prior to seeding with either fibroblast or macrophage cells in an appropriate culture media also containing 10% FBS.
Murine NIH 3T3 fibroblasts failed to adhere and proliferate up to 48 h on FC surfaces under any of the pre-adsorbed test conditions. Instead, fibroblast cells displayed spherical morphologies characteristic of non-adherent/non-spreading phenotypes[3,41] or adhered to each other, forming cell clusters (Figure 4(D–F), 24 h after seeding). Significantly, IC-21 macrophages readily adhered to and proliferated on FC surfaces under all test conditions (Figure 4(A–C)) with no apparent differences in proliferative rate or morphological behavior as compared to growth on PS (data not shown). These cells progressed to near confluent conditions (data not shown) in culture and exhibited varied morphology including astral shapes, filopodia, lamellipodia and membrane ruffling, consistent with our previous observations of macrophage growth on Teflon AF® fluorocarbon surfaces.[45,46] BMMO cells also adhered, grew and proliferated regardless of test conditions (Figure 4(G–L)), and at later time points (data not shown) achieved cell morphologies similar to that observed for the IC-21 cell line on these surfaces under identical conditions. The morphologies observed for the BMMO at 24 h (Figure 4(J–L)) are typical for mixed cell population primary-derived BMMO cultures that have been recently collected. Light spherical cells are non-adherent (for BM-MO cultures, typically immature monocytic) cells. Dark spherical cells are assumed to be in proximity to the surface, and are likely establishing adhesive contacts. Cells with astral or elongated morphologies are adherent. Only a small percentage of the cell population exhibits filopodia at this early time point on this substrate.
Cell Attachment and Proliferation on Macropatterned Surfaces in Serum-Based Culture
Cell growth on macropatterned surfaces was tested using multiple combinations of two dissimilar plasma-deposited surfaces (Table 2); one surface generally observed to be cell-adhesive in serum-based culture (e.g., NVP, AlAm) was paired with another surface characterized as generally non-cell adhesive (e.g., FC). Plasma deposition of patterns occurred sequentially, and was then reversed in a separate preparation to create two analogous macro-patterned surfaces for each polymer combination studied (Figure 2). Representative results for macropattern cultures using IC-21 murine macrophages (Figure 5(A)) and NIH 3T3 murine fibroblasts (Figures 5(B) and 6(A)) are shown.
Figure 5.
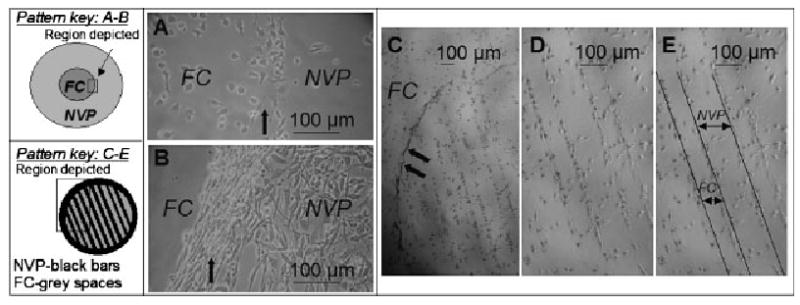
Patterning of IC-21 macrophages (A, C–E) and NIH 3T3 fibroblasts (B) on FC-NVP surfaces with macro (A, B) and micro (C–E, striped TEM grid) masks. Note: pattern cartoons are not to scale. Striped TEM grid (C–E) bars are 184 μm, spaces are 92 μm. A) IC-21 macrophages moderately and evenly populated the FC center of the pattern, while fewer cells attached to NVP regions (72 h). A small annulus (partially shown, bold arrow) was visible at the chemistry boundary. B) NIH 3T3 fibroblasts attached exclusively to the NVP region, with a ring forming at the margin between the NVP and FC chemistries (bold arrow, 72 h). Cells were loosely adherent at this boundary (an edge effect) with some cells floating above other adherent cells, attached on the NVP side only. C–E) Edge effect observed with IC-21 cells on NVP-FC micropatterns at 48 h (bold arrows). Macrophage cells grew preferentially at the interface of the surface chemistries at early time points. (D and E are the same image with E labeled for clarity).
Figure 6.
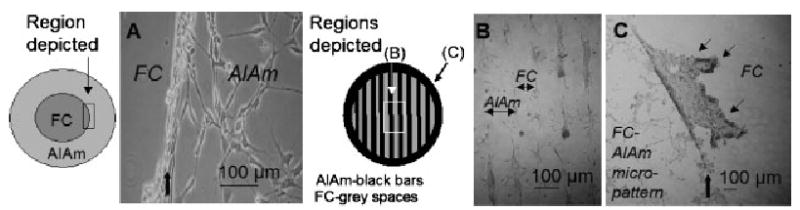
Patterning of NIH 3T3 fibroblasts on FC-AlAm surfaces with macro (A) and micro (B, C) masks. Striped TEM grid (B), bars are 184 μm, spaces are 92 μm. A) NIH 3T3 cells adhered to and proliferated on AlAm (right), but not on FC (left). Patterned growth fidelity appeared within 24 h and was maintained through day 7. An ‘‘edge effect’’ (see text for description, bold arrow) was noted at margins of masked regions. B) NIH 3T3 cell growth was confined to the region of the TEM grid (see also C, left), but not to the AlAm region exclusively (arrows indicate approximate widths of each polymer chemistry). C) NIH 3T3 cells did not grow on the larger FC background region (right) of the plates with micropatterns. Arrows indicate a large sheet of cells that are loosely adherent at the boundary (bold arrow) between the FC background and the FC-AlAm micropatterned region. Note: pattern cartoons are not to scale.
For either the NVP-FC or AlAm-FC macropatterns, NIH 3T3 fibroblasts adhered exclusively to and proliferated on the NVP (Figure 5(B)) or the AlAm (Figure 6(A)) regions. Cell patterns with fidelity to the anticipated surface chemistry pattern appeared within 24 h of seeding. The NIH 3T3 cells exhibited normal growth characteristics, kinetics and morphology in cell-supportive regions, and progressed to 100% confluence over the time course of the experiment, 7 d, in these regions. The boundary between the two co-patterned chemistries was also maintained for the duration of the culture experiment. At the boundary, cells would frequently grow parallel to the non-supportive chemistry boundary or extend toward and over the non-supportive (FC) region, surface-adherent only by cell contacts made in the cell-supportive (AlAm or NVP) region (Figure 5(B) and 6(A)). Many loosely adherent cells were observed at the pattern boundary. These cells were partially attached to the adherent NVP (or AlAm) chemistry zone, and floated above the non-adherent FC zone. Collectively, this behavior at the margin can be described as an ‘‘edge effect’’, attributed both to topological and chemical distinctions by cultured cells across the adherent/non-adherent pattern boundaries. The pp-film thickness for each deposition was calculated from the deposition rates and exposure times. For each experiment, deposited film thicknesses were: FC, 100 nm; NVP, 26 nm; AlAm, 90 nm; HxAm 47 nm.
By contrast, serum cultured IC-21 murine macrophages preferentially adhered to the FC region of the FC-NVP surfaces. These cells were evenly distributed over the FC substrate chemistry, suggesting that adherence was not due to a surface defect or topographical feature (Figure 5(A)). Initially upon seeding, cell morphology was markedly different on each chemistry; cells in the center of the FC pattern were more closely packed and exhibited primarily astral morphology, whereas the fewer cells scattered across the NVP region exhibited spreading behavior. Adherent macrophage cells exhibited characteristic membrane ruffling, lamellipodia and filopodia on both chemistries.[46] Although initial cell adherent densities indicated cell preference for FC regions, these cells progressed with a normal proliferative rate and eventually populated the entire surface (>7 d) with no discernible differences in cell behavior or morphology based on surface chemistry. However, over short time frames post-seeding (24–72 h), the IC-21 cells were observed to preferentially bind to the FC surfaces rather than the NVP, a behavior distinct from that previously documented in cultures of most other cell lines on similar FC or substantially hydrophobic surfaces,[31,41] but similar to that reported by Rich and Harris.[33]
Since the IC-21 macrophage represents the most physiologically relevant macrophage model within this study, use of the more monocytic RAW 264.7 and J774A.1 cell lines in macropatterning studies was limited. However, RAW 264.7 and J774A.1 behavior was examined on AlAm-FC and FC-AlAm, respectively. Both cell types attached to the FC and AlAm surface chemistries; neither demonstrated any discernible preference for either chemistry throughout the culture period, analogous to results obtained for the IC-21 cells (data not shown).
Cell Attachment and Proliferation on Micropatterned Surfaces
Cell behavior on micropatterned surfaces (see masks, Figure 7(A–C)) was tested using multiple combinations of five plasma-deposited polymers (Table 3), representing four adhesive (e.g., NVP, AlAm, HxAm, NVFA) and one non-adhesive (e.g., FC) cell culture surfaces. Plasma deposition was performed in a sequential manner (Figure 2) and all cell cultured micropatterning experiments were performed in media supplemented with 10% FBS.
Figure 7.
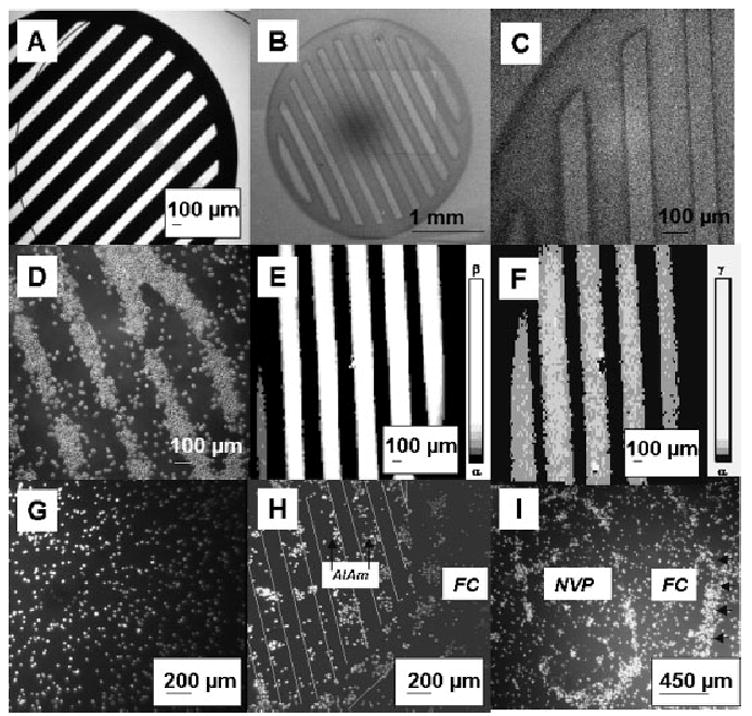
Micropatterning and control images. A) Phase contrast microscopy image of one type of (striped) TEM grid employed as a mask. Bars, 184 μm; spaces 92 μm. B, C) Electron microscopy images of the same grid. D) Phase contrast microscopy image of IC-21 cells grown on micropatterned HxAl-FC. Majority of cells shown are adhered to the HxAl bars. Patterns appeared by day 7 and were maintained through day 9, when the experiment was terminated. Cell seeding density was 100 000 cells per 15 × 60 mm2 plate. E, F) Scanning Auger maps of a pattern generated using a striped TEM grid identical to those used for preparing samples for use in cell culture experiments. Areas of most intensity (lightest) are strongest Auger signal. Intensity scales shown in E and F: α = 0, β = 281400, γ = 520633. E) Auger fluorine map (from the KLL signal) and F) Auger nitrogen map (from the KLL signal). G) Fluorescence microscopy of a non-patterned control FC surface showing an even and random distribution of IC-21 cells. H) Fluorescence microscopy of IC-21 cells on an AlAm-FC striped micropattern. Preferential growth was observed on the AlAm region (thicker bar) of the pattern at day 2, although cells also grew on the FC regions (thinner bar and background). Seeding density was 136 000 cells per 15 × 60 mm2 plate. I) Phase contrast images of IC-21 cells grown on a bullet shaped micropattern of NVP-FC, dimensions 1 × 2 mm2, 4 d post-seeding at a density of 250 000 cells per plate. Plate background is also NVP. Macrophage cells grew well on both the NVP and FC surfaces showing no preference for the NVP region. Edge effects were observed at the margins of the TEM grid patterns for both the central bullet shape and the outer ring, which represents the edge of the TEM grid (arrows).
Table 3.
Cell attachment and proliferation on plasma-micropatterned substrates.
| Cell pattern fidelity
|
|||
|---|---|---|---|
| Surface descriptiona) | NIH 3T3b) | IC-21b) | daysc) |
| HxAm-FC | +/− | +/+ | 9 |
| NVP-FC | +/− | +/+ | 9 |
| NVFA-FC | +/− | +/+ | 9 |
| AlAm-FC | +/− | +/+ | 7 |
| FC-AlAm | −/+ | +/+ | 7 |
Abbreviations as used in text. See text for detailed description of patterning method.
+ (supportive of cell growth), − (no adherence or proliferation); order of ‘‘surface description’’ matches reported + or − order, respectively. ND (not determined). See ref.[37] for full surface analytical characterization. See text for a detailed description of successful combinations of surface chemistries for micropatterning based on cell line.
Cell pattern fidelity differences due to different seeding concentrations, doubling times, and affinity for surfaces.
Micropattern cell pattern fidelity for the macrophage-derived IC-21 cell line was generally successful, although not absolute for cell exclusion versus adherence on the different chemistries where registration of patterned substrate chemistry and cell adhesion was directly observable in culture from 2–9 d (Figure 5(C–E) and 7(D,H,I)). Cell adhesive patterns became evident at different time points. Generally, at early time points, few cells adhered to both regions regardless of surface chemistry (illustrated in Figure 7(H) for cells grown on AlAm-FC chemistries day 2). At later time points, discernible preferences for one chemistry were observed on select surfaces (e.g., day 7 of culture for HxAm-FC, Figure 7(D)). Patterns were typically maintained for 2–5 d before the cells overgrew the pattern. Such fidelity to substrate micropatterned chemistry was less successful for NIH 3T3 fibroblasts. Although cell patterns developed on these surfaces (Figure 6(B)), as fibroblast cultures progressed, cellular delamination occurred readily, originating from the margins of the chemistry interfaces (Figure 6(C)). These results indicate that strong cell-cell contacts dominate over cell-surface interactions. As cells multiplied and adhesive space became limiting, strong cell-cell contacts allowed surface-bound cells to be pulled off of their patterns (during rinsing) by attached cells ‘‘hanging’’ off pattern edges bound only by cell-cell contacts. Additionally, after completely rinsing cells from the surface, cells repopulated the patterned region, but again could be rinsed away once space became limiting.
Representative results for micropatterning with IC-21 murine macrophages (Figure 5(C–E), Figure 7(D,H,I)) indicate that cell patterns develop for these cells on AlAm-FC (Figure 7(H)), NVP-FC (Figure 5(C–E), Figure 7(I)) and HxAm-FC (Figure 7(D)), but not on NVFA-FC (data not shown) where no preferential growth based on chemistries was observed. Phase contrast microscopy results for striped and bullet shaped TEM grid-patterns, HxAm-FC and NVP-FC respectively, are shown in Figure 7(D, I). Multiple examples of such pattern fidelity were detected on each plate. Additionally, at early culture times (48 h), an edge effect was observed for micropatterned IC-21 cells (Figure 5(C–E)), likely a response to topological features (roughness) at the margin of the two chemistries.[33]
Cellular response to various model materials has important fundamental significance to numerous bio-technological medical devices, diagnostic high-throughput cell-based screening and environmental applications. Specifically, surface interactions with leukocytes and lymphocytes are particularly important for understanding inflammatory responses to implanted device materials and soluble drug candidates. We have previously described the morphological and proliferative characteristics of a fibroblast and macrophage inclusive group cultured on model bio-materials surfaces.[45,46] This initial study has been extended here to a large group of previously characterized plasma-deposited co-patterned surfaces of distinct chemistries.[38]
Two important considerations distinguishing cell-surface behavior for in vitro-based cell adhesion studies are the presence of serum as opposed to biased pre-adsorption of ECM proteins, and the influence of cell phenotype.[3] Monocyte-macrophage and fibroblast cells have markedly different functions in vivo, and consequently their interactions with surfaces reflect these highly specialized phenotypic roles. Although typically considered non-adhesive in vivo unless activated by non-host materials (e.g., foreign bodies) macrophages, the sentinels of the immune system, are uniquely equipped to identify and respond to surfaces. The complex macrophage-mediated host response includes biomaterials attachment reactions,[50] interactions with lymphocytes[51] and cell-cell fusion to form foreign body giant cells.[52–54] Curiously, materials typically considered non-permissive to cell attachment and growth in culture unless pre-adsorbed with matrix proteins (e.g., hydrophobic surfaces), have been shown to promote macrophage attachment in serum cultures in vitro.[33,45,46,55] One major unresolved issue is the mechanism by which this occurs, compared to well-known integrin attachment mechanisms employed by most cell types.[56,57] Additionally, there is the question of how closely the frequently employed immortalized monocyte/macrophage-derived secondary cell lines accurately and faithfully represent phenotypic behavior of primary macrophage or monocytic cells under in vitro assay conditions. This has significant implications for in vitro pharmacological assays employing adherent cells as predictive indicators of inflammatory activation in vivo.
Given that macrophage adhesion to biomaterials plays a key role in mediating immune response to foreign materials,[50] adhesive behavior on surfaces designed to be non-fouling is of great interest. However, primary cells represent an expensive cell source with a short lifetime and uncertain phenotypic stability in culture. Therefore, we focused on the most physiologically relevant cell line, IC-21, as it shares many characteristics with normal peritoneal macrophages,[58] yet as an immortalized cell line it can be grown successfully over extended culture periods. In the case of our most interesting result, i.e. proficient growth on fluorocarbon surfaces, we compared the behavior of primary sourced and secondary-derived cell lines. Results for the (monocyte-) macrophage cell lines indicated similar behaviors for these cells on all pp-generated surfaces tested. The IC-21 and J774A.1 lines grew well on HxAm, and all three monocyte/macrophage cell lines successfully colonized FC, AlAm and NVP surfaces (Table 1).
Data demonstrate that IC-21, J774A.1 and RAW 264.7 secondary (monocyte-) macrophage cell lines tested in serum cultures adhered to and proliferated on all pp-surfaces, regardless of surface chemistry. In addition, limited tests with primary derived bone marrow macrophages (FC and PS controls) showed similar results, indicating that both the immortalized lineages and the primary macrophages behave consistently with respect to surface response within this select group of cells and surfaces.
Poor cell culture results on fluorocarbon surfaces have been previously reported for numerous cell lines and conditions.[3,27,31,33,41,59,60] Extremely hydrophobic per-fluorocarbon surfaces [e.g., poly(vinylidene fluoride), PVDF; poly(tetrafluoroethylene), PTFE] typically hinder cellular attachment from serum media.[3,47] This observed resistance to cell adhesion has been shown to be mediated by preferential albumin deposition onto these surfaces, blocking recognition by cell integrin receptors of trace matrix protein motifs on the surface required for cell adhesion.[47,60,61] Interestingly, our FC surfaces supported the attachment and proliferation of all (monocyte-) macrophage cell lines tested, but not the NIH 3T3 fibroblasts (Figure 3 and 4).[3,41] In addition, pre-treatment of surfaces for 24 h with either 3 mg · ml−1 albumin, 10 or 100% serum, to adsorb protein from a single biased or complex protein mixture, had no significant effect on the ability of cells to adhere to and proliferate on this surface (Figure 4). Despite little observable adhesion from other cell lines cultured on similar FC surfaces in serum-containing media,[3,31,41,59] murine macrophages were observed to grow well on various types of FC surfaces: all three macrophage-derived cell lines and primary-derived bone marrow macrophages adhered readily to FC patterns in the presence of serum. This distinct attachment capability from fibroblasts could result from: (1) different classes of cell adhesion receptors on macrophages compared to other cell types that typically rely on integrins and matrix proteins, (2) macrophage use of an integrin-independent surface attachment process that is not receptor-driven (i.e., protein-independent, electrostatic, membrane glycosylation-dependent), (3) much lower surface density requirements of adsorbed trace matrix proteins (e.g., fibronectin, collagens, vitronectin) for macrophages as compared to cells that attach to surfaces using their integrin classes, or (4) rapid up-regulation of endogenous matrix protein expression in these cells that facilitates surface attachment.
Grainger and coworkers recently showed that poly(tetra-fluoroethylene) fluorocarbon surfaces overwhelmingly adsorb excess albumin from serum[47] compared to other trace matrix proteins (e.g., fibronectin) essential for attachment of most cell types.[56,57] This result correlated with lack of observed probe antibody reactivity to several fibronectin-epitopes after serum exposure, as well as observed low endothelial cell adhesion. Biased surface selection of albumin seems to be a natural tendency of FC surfaces,[61] hindering cellular attachment in the presence of serum via an albumin blocking mechanism that either limits matrix protein deposition or masks its recognition by adhering cells. Therefore, cell-surface chemistry selection preference should result for specific surface chemistries patterned side-by-side. For example, this rationale applied to cell attachment to HxAm co-patterned with FC in serum predicts that albumin adsorption to the more apolar FC surface precludes cell adhesion to these areas, whereas increased matrix protein adsorbed abundance on HxAm promotes cell adhesion. This is in fact what is observed in studies reported here for macrophages and supported in several other analogous literature reports.[3,36,37] Typical cell pattern development from serum media relies on consistent cell adhesion preference for an attachment-permissive surface (e.g., HxAm,NVP, TCPS) adsorbing sufficient matrix proteins over non-permissive (e.g., FC) surfaces blocked by preferential albumin adsorption.
This predicted preferential behavior was evident in the NIH 3T3 fibroblast culture results, where cells bound exclusively and definitively to permissive regions of macro-patterned plates (Figure 5(B) and 6(A); analogous results obtained for inverse patterned chemistry, data not shown). These patterns were evident at early culture times (24 h) and maintained even if cells were allowed to progress to 100% confluence on the permissive substrate. In contrast, macrophages did not exhibit this distinct patterning behavior: they did not adhere exclusively to the predicted permissive regions. Although (monocyte-) macrophages consistently demonstrated the ability to adhere to and proliferate on all homogenous surface chemistries tested, they did exhibit preferences for specific chemistries within combinations of co-patterned substrates. Examples include the combination of FC and HxAm (Figure 7(D)) where IC-21 macrophages adhered preferentially to the HxAm over the FC, resulting in a pattern. This pattern evolved slowly with time, fully developed 7 d after cells were applied to the plates, and was maintained through day 9 when the experiment was terminated. By contrast, at early time points (24 h), IC-21 macrophages attached preferentially to FC regions of FC-NVP macropatterns (Figure 5(A)). In some cases no surface chemistry preference was discernible due to similar surface selection (e.g., J774A.1 and RAW 264.7 cultures on AlAm-FC and FC-AlAm, respectively, data not shown).
Numerous previous studies of cell adhesion to materials have utilized fibroblastic cell lines of diverse types.[3,33,36,37,41] The ability of fibroblasts to attach to and proliferate on, or in the presence of specific substrate chemistries in serum-containing milieu or pre-adsorbed with matrix proteins, is often used as a basic indicator of materials ‘‘biotolerance’’. Fibroblasts adhere to more hydrophobic substrate chemistries with difficulty[3,33,35,41,59,60] unless pre-adsorbed with purified matrix proteins prior to culture. Their attachment-dependent phenotype, adherent morphology and surface matrix protein requirements are distinct from monocyte/macrophage cells. In addition, the cell-cell contacts formed in cells grown on hydrophobic surfaces are stronger than the cell-surface contacts, exemplified by delamination of large intact sheets of cells (Figure 6(C)).
NIH 3T3 fibroblast cultures attached to and grew readily on all attachment-permissive surfaces tested, AlAm, HxAm, NVP and NVFA. This is consistent with previous reports[62–64] and leads to an interpretation that these adhesion-permissive surface chemistries (1) adsorb sufficient matrix proteins from serum to support cell integrin-dependent attachment, and (2) are not cytotoxic. In contrast to macrophage cultures, fibroblasts were non-adherent or poorly adherent to the FC surfaces in the presence of either serum (as previously observed by others)[3,41,59] or pure albumin solution pre-adsorption. Most cells were unable to adhere and remained free-floating in the media. Any observed fibroblast attachment was attributed to small surface coating defects in the plates (Figure 3(B), arrow). Ultimately, the NIH 3T3 fibroblast cells were not able to colonize the plates effectively, with only small, isolated areas of attachment, and no observed cell pattern development in any case, consistent over various cell-seeding densities. Limited fibroblast attachment and growth on non-permissive plasma-treated substrates was most frequently observed at the pattern edges (Figure 6(B)) where FC deposition was likely affected or disrupted by the pattern transition or walls of the plate.
Conclusion
Biomaterials design has become more sophisticated with numerous techniques currently available to develop surfaces with improved biocompatibility. Plasma-derived chemical functionalization of biomaterials is one commonly employed method for improving implant surface characteristics. We have examined a diverse group of pp-surfaces and the responses of two phenotypically distinct cell types to these surfaces in vitro.
NIH 3T3 fibroblast cell lineage adherence, growth and proliferation on pp-substrates in serum proceeds as expected based on known surface chemistry trends. Monocyte-macrophage cells derived from both primary and secondary cell sources show consistent adherence, growth and proliferation on all pp-surfaces tested, including fluorocarbons shown refractory to cell adhesion in many previous studies. Results suggest that these phenotypically distinct cell types have different requirements for successful surface adherence and proliferation. Cell-cell contacts on FC surfaces are stronger than cell-surface contacts made by fibroblast cultures, whereas (monocyte-) macrophage cells establish strong cell-surface contacts, even in serum on fluorocarbon.
The demonstrated ability of cultured, secondary-derived immortalized cells to attach to non-adhesive substrates such as FC is unusual. Equivalence with the more biologically significant primary-derived BMMO cells to adhere and proliferate on these surfaces suggests that the many elegant micropatterns designed for use in implantable devices and scaffolds may not be practically useful in vivo due to the attachment of immunogenic cells which not only elicit a foreign body inflammatory response, but also preclude the attachment and growth of desirable cell types. Additionally, use of apolar surfaces and albumin masking agents in common in vitro assays of anti-inflammatory drugs with cultured macrophages may not be sufficient to block cell-surface activation, thus confounding pharmacological assay results. Consequently, the interactions of macrophages as key cellular mediators must be carefully considered in an appropriate biologically relevant context.
Acknowledgments
We gratefully acknowledge Dr. I. T. Martin and Dr. G. Hagen for technical assistance and Dr. M. Gonzalez-Juarerro for assistance with primary BMMO harvests. This work was funded by NIH grant EB 000894.
Footnotes
The use of ‘‘(monocyte-) macrophage’’ is used to describe the secondary cell lines J774A.1, RAW 264.7 and IC-21 collectively despite presumed phenotypic differences, and may refer to any combination of the monocyte-macrophage cell lines J774A.1 and RAW 264.7 with each other and/or the more differentiated macrophage cell line IC-21.
References
- 1.Andrade JD. Med Instrum. 1973;7:110. [PubMed] [Google Scholar]
- 2.Hubbell JA. Biotechnology (NY) 1995;13:565. doi: 10.1038/nbt0695-565. [DOI] [PubMed] [Google Scholar]
- 3.van Kooten TG. Encyclopedia of Surface and Colloid Science. Marcel Dekker; New York: 2004. Growth of Cells on Polymer Surfaces; p. 1. [Google Scholar]
- 4.Liu WF, Chen CS. Mater Today. 2005;8:28. [Google Scholar]
- 5.Andrade JD. Surface and Interfacial Aspects of Biomedical Polymers: Protein Adsorption. Vol. 2. Plenum; New York: 1985. [Google Scholar]
- 6.Mrksich M, Whitesides GM. Annu Rev Biophys Biomol Struct. 1996;25:55. doi: 10.1146/annurev.bb.25.060196.000415. [DOI] [PubMed] [Google Scholar]
- 7.Dewez JL, Doren A, Schneider YJ, Rouxhet PG. Biomaterials. 1999;20:547. doi: 10.1016/s0142-9612(98)00207-5. [DOI] [PubMed] [Google Scholar]
- 8.Baszkin A, Lyman DJ. J Biomed Mater Res. 1980;14:393. doi: 10.1002/jbm.820140406. [DOI] [PubMed] [Google Scholar]
- 9.Sethuraman A, Han M, Kane RS, Belfort G. Langmuir. 2004;20:7779. doi: 10.1021/la049454q. [DOI] [PubMed] [Google Scholar]
- 10.Curtis A, Wilkinson C. Biomaterials. 1997;18:1573. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 11.Poncin-Epaillard F, Legeay G. J Biomater Sci, Polym Ed. 2003;14:1005. doi: 10.1163/156856203769231538. [DOI] [PubMed] [Google Scholar]
- 12.Schroder K, Meyer-Plath A, Keller D, Ohl A. Plasmas Polym. 2002;7:103. [Google Scholar]
- 13.Bouyer E, Schiller G, Muller M, Henne RH. Plasma Chem Plasma Process. 2001;21:532. [Google Scholar]
- 14.Meaudre R, Butte R, Vignoli S, Meaudre M, Saviot L, Marty O, Rocai I, Cabarrocas P. Eur Phys J Appl Phys. 2003;22:171. [Google Scholar]
- 15.Sopori B. J Electr Mater. 2005;34:564. [Google Scholar]
- 16.Cicala G, Bruno P, Losacco AM, Mattei G. Surf Coat Technol. 2004;180–181:222. [Google Scholar]
- 17.Mutsukura N, Handa Y. Plasma Chem Plasma Process. 2002;22:607. [Google Scholar]
- 18.Dhar R, Pedrow PD, Liddell KNC, Ming Q, Moeller TM, Osman MA. IEEE Trans Plasma Sci. 2005;33:138. [Google Scholar]
- 19.Luting Y, Wenjie S, Hezhuo M, Wei H, Zhigang G, Yikang P. Key Eng Mater. 2004;264–268:109. [Google Scholar]
- 20.Steen ML, Flory WC, Capps NE, Fisher ER. Chem Mater. 2001;13:2749. [Google Scholar]
- 21.Tang JX, Li YQ, Dong X, Wang SD, Lee CS, Hung LS, Lee ST. Appl Surf Sci. 2004;239:117. [Google Scholar]
- 22.Folch A, Toner M. Annu Rev Biomed Eng. 2000;2:227. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 23.Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, Ingber DE. Science. 1994;264:696. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 24.McFarland CD, Thomas CH, DeFilippis C, Steele JG, Healy KE. J Biomed Mater Res. 2000;49:200. doi: 10.1002/(sici)1097-4636(200002)49:2<200::aid-jbm7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell SA, Emmison N, Shard AG. Surf Interface Anal. 2002;33:742. [Google Scholar]
- 26.Sardella E, Gristina R, Senesi GS, d’Agostino R, Favia P. Plasma Process Polym. 2004;1:63. [Google Scholar]
- 27.Collier TO, Thomas CH, Anderson JM, Healy KE. J Biomed Mater Res. 2000;49:141. doi: 10.1002/(sici)1097-4636(200001)49:1<141::aid-jbm18>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 28.Thomas CH, McFarland CD, Jenkins ML, Rezania A, Steele JG, Healy KE. J Biomed Mater Res. 1997;37:81. doi: 10.1002/(sici)1097-4636(199710)37:1<81::aid-jbm10>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 29.Folch A, Jo BH, Hurtado O, Beebe DJ, Toner M. J Biomed Mater Res. 2000;52:346. doi: 10.1002/1097-4636(200011)52:2<346::aid-jbm14>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 30.Harbers GM, Grainger DW. In: An Introduction to Biomaterials. Guelcher SA, Hollinger JO, editors. Vol. 11. CRC Press; Boca Raton: 2006. p. 15. [Google Scholar]
- 31.Koenig AL, Gambillara V, Grainger DW. J Biomed Mater Res A. 2003;64:20. doi: 10.1002/jbm.a.10316. [DOI] [PubMed] [Google Scholar]
- 32.Mrksich M, Dike LE, Tien J, Ingber DE, Whitesides GM. Exp Cell Res. 1997;235:305. doi: 10.1006/excr.1997.3668. [DOI] [PubMed] [Google Scholar]
- 33.Rich A, Harris AK. J Cell Sci. 1981;50:1. doi: 10.1242/jcs.50.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Scotchford CA, Gilmore CP, Cooper E, Leggett GJ, Downes S. J Biomed Mater Res. 2002;59:84. doi: 10.1002/jbm.1220. [DOI] [PubMed] [Google Scholar]
- 35.Ruardy TG, Schakenraad JM, van der Mei HC, Busscher HJ. J Biomed Mater Res. 1995;29:1415. doi: 10.1002/jbm.820291113. [DOI] [PubMed] [Google Scholar]
- 36.Gallant ND, Capadona JR, Frazier AB, Collard DM, Garcia AJ. Langmuir. 2002;18:5579. [Google Scholar]
- 37.Tourovskaia A, Barber T, Wickes BT, Hirdes D, Grin B, Castner DG, Healy KE, Folch A. Langmuir. 2003;19:4754. [Google Scholar]
- 38.Malkov G, Martin IT, Schwisow WB, Chandler JP, Fisher ER. Chem Mater. submitted. [Google Scholar]
- 39.Bogart KHA, Dalleska NF, Bogart GR, Fisher ER. J Vac Sci Technol A. 1995;13:476. [Google Scholar]
- 40.Mackie NM, Dalleska NF, Castner DG, Fisher ER. Chem Mater. 1997;9:349. [Google Scholar]
- 41.McClary KB, Ugarova T, Grainger DW. J Biomed Mater Res. 2000;50:428. doi: 10.1002/(sici)1097-4636(20000605)50:3<428::aid-jbm18>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 42.Rhoades ER, Orme IM. Mech Ageing Dev. 1998;106:145. doi: 10.1016/s0047-6374(98)00113-4. [DOI] [PubMed] [Google Scholar]
- 43.Kaltenbach JP, Kaltenbach MH, Lyons WB. Exp Cell Res. 1958;15:112. doi: 10.1016/0014-4827(58)90067-3. [DOI] [PubMed] [Google Scholar]
- 44.Daw R, Candan S, Beck AJ, Devlin AJ, Brook IM, MacNeil S, Dawson RA, Short RD. Biomaterials. 1998;19:1717. doi: 10.1016/s0142-9612(98)00080-5. [DOI] [PubMed] [Google Scholar]
- 45.Godek ML, Duchsherer NL, McElwee Q, Grainger DW. Biomed Sci Instrum. 2004;40:7. [PubMed] [Google Scholar]
- 46.Godek ML, Michel R, Grainger DW. in preparation. [Google Scholar]
- 47.Grainger DW, Pavon-Djavid G, Migonney V, Josefowicz M. J Biomater Sci, Polym Ed. 2003;14:973. doi: 10.1163/156856203322381456. [DOI] [PubMed] [Google Scholar]
- 48.Haddow DB, France RM, Short RD, MacNeil S, Dawson RA, Leggett GJ, Cooper E. J Biomed Mater Res. 1999;47:379. doi: 10.1002/(sici)1097-4636(19991205)47:3<379::aid-jbm13>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 49.Haddow DB, Steele DA, Short RD, Dawson RA, Macneil S. J Biomed Mater Res A. 2003;64:80. doi: 10.1002/jbm.a.10356. [DOI] [PubMed] [Google Scholar]
- 50.Anderson JM. Cardiovasc Pathol. 1993;2:33S. [Google Scholar]
- 51.MacEwan MR, Brodbeck WG, Matsuda T, Anderson JM. J Biomed Mater Res A. 2005;74:285. doi: 10.1002/jbm.a.30316. [DOI] [PubMed] [Google Scholar]
- 52.Anderson JM. Curr Opin Hematol. 2000;7:40. doi: 10.1097/00062752-200001000-00008. [DOI] [PubMed] [Google Scholar]
- 53.Kyriakides TR, Foster MJ, Keeney GE, Tsai A, Giachelli CM, Clark-Lewis I, Rollins BJ, Bornstein P. Am J Pathol. 2004;165:2157. doi: 10.1016/S0002-9440(10)63265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNally AK, Anderson JM. Am J Pathol. 1995;147:1487. [PMC free article] [PubMed] [Google Scholar]
- 55.Davis GE. Exp Cell Res. 1992;200:242. doi: 10.1016/0014-4827(92)90170-d. [DOI] [PubMed] [Google Scholar]
- 56.Garcia AJ. Biomaterials. 2005;26:7525. doi: 10.1016/j.biomaterials.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 57.Hynes RO. Cell. 2002;110:673. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 58.Mauel J, Defendi V. J Exp Med. 1971;134:335. doi: 10.1084/jem.134.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schakenraad JM, Busscher HJ, Wildevuur CR, Arends J. J Biomed Mater Res. 1986;20:773. doi: 10.1002/jbm.820200609. [DOI] [PubMed] [Google Scholar]
- 60.Webb K, Hlady V, Tresco PA. J Biomed Mater Res. 1998;41:422. doi: 10.1002/(sici)1097-4636(19980905)41:3<422::aid-jbm12>3.0.co;2-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kiaei D, Hoffman AS, Horbett TA. J Biomater Sci, Polym Ed. 1992;4:35. [PubMed] [Google Scholar]
- 62.Griesser HJ, Chatelier RC, Gengenbach TR, Johnson G, Steele JG. J Biomater Sci, Polym Ed. 1994;5:531. doi: 10.1163/156856294x00194. [DOI] [PubMed] [Google Scholar]
- 63.Marchant RE, Johnson SD, Schneider BH, Agger MP, Anderson JM. J Biomed Mater Res. 1990;24:1521. doi: 10.1002/jbm.820241108. [DOI] [PubMed] [Google Scholar]
- 64.Morra M, Cassinelli C. Plasmas Polym. 2002;7:89. [Google Scholar]


