Abstract
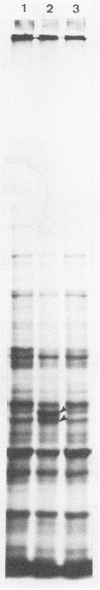
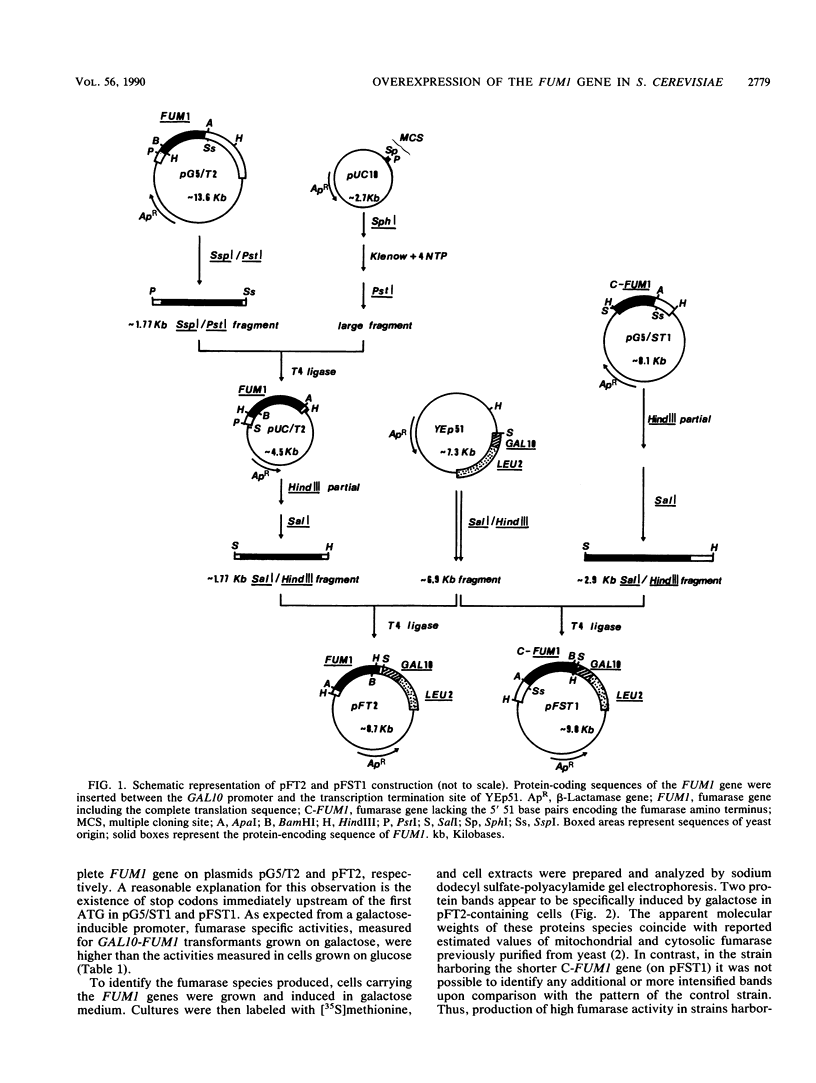
Cloning of the Saccharomyces cerevisiae FUM1 gene downstream of the strong GAL10 promoter resulted in inducible overexpression of fumarase in the yeast. The overproducing strain exhibited efficient bioconversion of fumaric acid to L-malic acid with an apparent conversion value of 88% and a conversion rate of 80.4 mmol of fumaric acid/h per g of cell wet weight, both of which are much higher than parameters known for industrial bacterial strains. The only product of the conversion reaction was L-malic acid, which was essentially free of the unwanted by-product succinic acid. The GAL10 promoter situated upstream of a promoterless FUM1 gene led to production and correct distribution of the two fumarase isoenzyme activities between cytosolic and mitochondrial subcellular fractions. The amino-terminal sequence of fumarase contains the mitochondrial signal sequence since (i) 92 of 463 amino acid residues from the amino terminus of fumarase are sufficient to localize fumarase-lacZ fusions to mitochondria and (ii) fumarase and fumarase-lacZ fusions lacking the amino-terminal sequence are localized exclusively in the cytosol. The possibility that both mitochondrial and cytosolic fumarases are derived from the same initial translation product is discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beeckmans S., Kanarek L. A new purification procedure for fumarase based of affinity chromatography. Isolation and characterization of pig-liver fumarase. Eur J Biochem. 1977 Sep;78(2):437–444. doi: 10.1111/j.1432-1033.1977.tb11756.x. [DOI] [PubMed] [Google Scholar]
- Boonyarat D., Doonan S. Purification and structural comparisons of the cytosolic and mitochondrial fumarases from baker's yeast. Int J Biochem. 1988;20(10):1125–1132. doi: 10.1016/0020-711x(88)90258-3. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Daum G., Böhni P. C., Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982 Nov 10;257(21):13028–13033. [PubMed] [Google Scholar]
- Goldberg I., Lonberg-Holm K., Bagley E. A., Stieglitz B. Improved conversion of fumarate to succinate by Escherichia coli strains amplified for fumarate reductase. Appl Environ Microbiol. 1983 Jun;45(6):1838–1847. doi: 10.1128/aem.45.6.1838-1847.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McAlister-Henn L., Thompson L. M. Isolation and expression of the gene encoding yeast mitochondrial malate dehydrogenase. J Bacteriol. 1987 Nov;169(11):5157–5166. doi: 10.1128/jb.169.11.5157-5166.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir A. The isolation of lambda transducing phages carrying the citG and gerA genes of Bacillus subtilis. J Gen Microbiol. 1983 Feb;129(2):303–310. doi: 10.1099/00221287-129-2-303. [DOI] [PubMed] [Google Scholar]
- Natsoulis G., Hilger F., Fink G. R. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell. 1986 Jul 18;46(2):235–243. doi: 10.1016/0092-8674(86)90740-3. [DOI] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. Distinct repressible mRNAs for cytoplasmic and secreted yeast invertase are encoded by a single gene. Cell. 1981 Aug;25(2):525–536. doi: 10.1016/0092-8674(81)90071-4. [DOI] [PubMed] [Google Scholar]
- Pines O., Lunn C. A., Inouye M. Defective Escherichia coli signal peptides function in yeast. Mol Microbiol. 1988 Mar;2(2):209–217. doi: 10.1111/j.1365-2958.1988.tb00022.x. [DOI] [PubMed] [Google Scholar]
- Woods S. A., Schwartzbach S. D., Guest J. R. Two biochemically distinct classes of fumarase in Escherichia coli. Biochim Biophys Acta. 1988 Apr 28;954(1):14–26. doi: 10.1016/0167-4838(88)90050-7. [DOI] [PubMed] [Google Scholar]
- Wu M., Tzagoloff A. Mitochondrial and cytoplasmic fumarases in Saccharomyces cerevisiae are encoded by a single nuclear gene FUM1. J Biol Chem. 1987 Sep 5;262(25):12275–12282. [PubMed] [Google Scholar]