Abstract
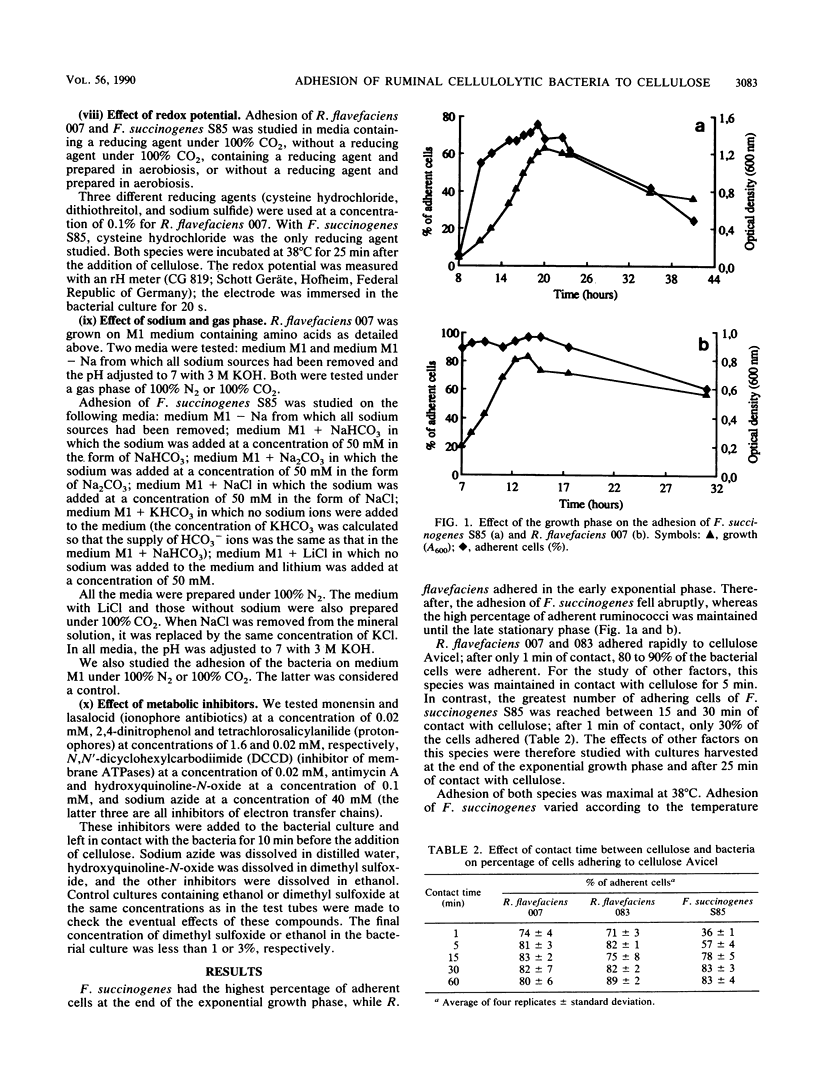
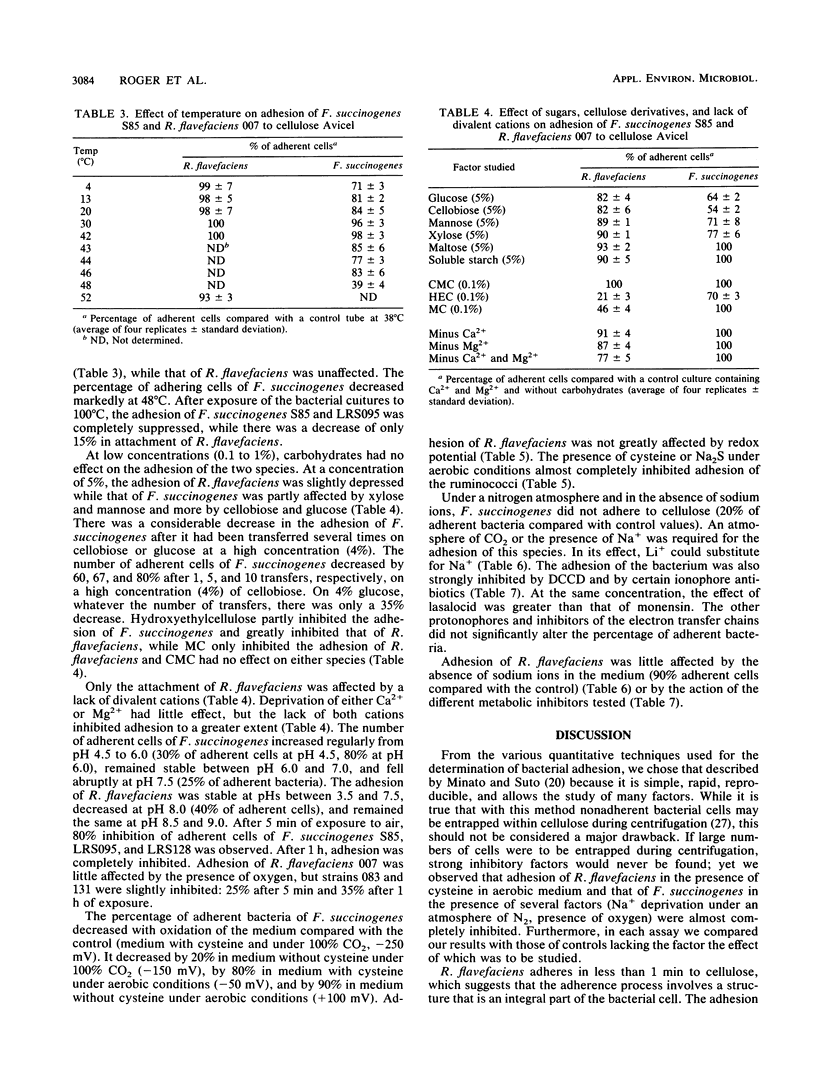
Ruminococcus flavefaciens adhered instantly to cellulose, while Fibrobacter succinogenes had the highest percentage of adherent cells after about 25 min of contact between bacteria and cellulose. Adhesion of R. flavefaciens was unaffected by high concentrations of sugars (5%), temperature, pH, oxygen, metabolic inhibitors, and lack of Na+. In contrast, the attachment was affected by the removal of divalent cations (Mg2+ and Ca2+), the presence of cellulose derivatives (methylcellulose and hydroxyethylcellulose), and cystine. Adhesion of F. succinogenes was sensitive to low and high temperatures, high concentrations of glucose and cellobiose (5%), hydroxyethylcellulose (0.1%), redox potential, pH, lack of monovalent cations, and the presence of an inhibitor of membrane ATPases or lasalocid and monensin. Cells of F. succinogenes heated at 100°C no longer were adherent. On the other hand, adhesion was insensitive to the lack of divalent cations (Mg2+ and Ca2+), the presence of 2,4-dinitrophenol, tetrachlorosalicylanilide, or inhibitors of the electron transfer chains. Adhesion of F. succinogenes seems to be related to the metabolic functions of the cell. External proteins and/or cellulases themselves might play a part in the attachment process. Several mechanisms are probably involved in the adhesion of R. flavefaciens, the main one being the interaction between the large glycocalyx and the divalent cations Ca2+ and Mg2+. Hydrophobic bonds and enzymes may also be involved.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akin D. E., Barton F. E., 2nd Rumen microbial attachment and degradation of plant cell walls. Fed Proc. 1983 Jan;42(1):114–121. [PubMed] [Google Scholar]
- Akin D. E. Evaluation by electron microscopy and anaerobic culture of types of rumen bacteria associated with digestion of forage cell walls. Appl Environ Microbiol. 1980 Jan;39(1):242–252. doi: 10.1128/aem.39.1.242-252.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akin D. E. Microscopic evaluation of forage digestion by rumen microorganisms--a review. J Anim Sci. 1979 Mar;48(3):701–710. doi: 10.2527/jas1979.483701x. [DOI] [PubMed] [Google Scholar]
- Akin D. E. Ultrastructure of rumen bacterial attachment to forage cell walls. Appl Environ Microbiol. 1976 Apr;31(4):562–568. doi: 10.1128/aem.31.4.562-568.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRYANT M. P., ROBINSON I. M. Some nutritional characteristics of predominant culturable ruminal bacteria. J Bacteriol. 1962 Oct;84:605–614. doi: 10.1128/jb.84.4.605-614.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale D., Morris E. J., Bacon J. S. Electron microscopy of the microbial populations present and their modes of attack on various cellulosic substrates undergoing digestion in the sheep rumen. Appl Environ Microbiol. 1978 Jul;36(1):160–168. doi: 10.1128/aem.36.1.160-168.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg C. W., Beveridge T. J., Hellstrom A. Cellulase and Xylanase Release from Bacteroides succinogenes and Its Importance in the Rumen Environment. Appl Environ Microbiol. 1981 Nov;42(5):886–896. doi: 10.1128/aem.42.5.886-896.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklund C. V., Glass T. L. Glucose uptake by the cellulolytic ruminal anaerobe Bacteroides succinogenes. J Bacteriol. 1987 Feb;169(2):500–506. doi: 10.1128/jb.169.2.500-506.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Forsberg C. W. Factors affecting adhesion of Fibrobacter succinogenes subsp. succinogenes S85 and adherence-defective mutants to cellulose. Appl Environ Microbiol. 1989 Dec;55(12):3039–3044. doi: 10.1128/aem.55.12.3039-3044.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groleau D., Forsberg C. W. Cellulolytic activity of the rumen bacterium Bacteroides succinogenes. Can J Microbiol. 1981 May;27(5):517–530. doi: 10.1139/m81-077. [DOI] [PubMed] [Google Scholar]
- Kudo H., Cheng K. J., Costerton J. W. Electron microscopic study of the methylcellulose-mediated detachment of cellulolytic rumen bacteria from cellulose fibers. Can J Microbiol. 1987 Mar;33(3):267–272. doi: 10.1139/m87-045. [DOI] [PubMed] [Google Scholar]
- Lamed R., Naimark J., Morgenstern E., Bayer E. A. Specialized cell surface structures in cellulolytic bacteria. J Bacteriol. 1987 Aug;169(8):3792–3800. doi: 10.1128/jb.169.8.3792-3800.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham M. J., Brooker B. E., Pettipher G. L., Harris P. J. Ruminococcus flavefaciens Cell Coat and Adhesion to Cotton Cellulose and to Cell Walls in Leaves of Perennial Ryegrass (Lolium perenne). Appl Environ Microbiol. 1978 Jan;35(1):156–165. doi: 10.1128/aem.35.1.156-165.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M. A., Hespell R. B., White B. A., Bothast R. J. Inhibitory Effects of Methylcellulose on Cellulose Degradation by Ruminococcus flavefaciens. Appl Environ Microbiol. 1988 Apr;54(4):890–897. doi: 10.1128/aem.54.4.890-897.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M. A., White B. A., Hespell R. B. Improved assay for quantitating adherence of ruminal bacteria to cellulose. Appl Environ Microbiol. 1989 Aug;55(8):2089–2091. doi: 10.1128/aem.55.8.2089-2091.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B. A proposed mechanism of monensin action in inhibiting ruminal bacterial growth: effects on ion flux and protonmotive force. J Anim Sci. 1987 May;64(5):1519–1525. doi: 10.2527/jas1987.6451519x. [DOI] [PubMed] [Google Scholar]
- Russell J. B. Effect of extracellular pH on growth and proton motive force of Bacteroides succinogenes, a cellulolytic ruminal bacterium. Appl Environ Microbiol. 1987 Oct;53(10):2379–2383. doi: 10.1128/aem.53.10.2379-2383.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Strobel H. J. Effect of ionophores on ruminal fermentation. Appl Environ Microbiol. 1989 Jan;55(1):1–6. doi: 10.1128/aem.55.1.1-6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCOTT H. W., DEHORITY B. A. VITAMIN REQUIREMENTS OF SEVERAL CELLULOLYTIC RUMEN BACTERIA. J Bacteriol. 1965 May;89:1169–1175. doi: 10.1128/jb.89.5.1169-1175.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack R. J., Hungate R. E. Effect of 3-Phenylpropanoic Acid on Capsule and Cellulases of Ruminococcus albus 8. Appl Environ Microbiol. 1984 Jul;48(1):218–223. doi: 10.1128/aem.48.1.218-223.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K. A., Crosby B., McGavin M., Forsberg C. W., Thomas D. Y. Characteristics of the endoglucanase encoded by a cel gene from Bacteroides succinogenes expressed in Escherichia coli. Appl Environ Microbiol. 1987 Jan;53(1):41–46. doi: 10.1128/aem.53.1.41-46.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. A., Rasmussen M. A., Gardner R. M. Methylcellulose inhibition of exo-beta-1,4-glucanase A from Ruminococcus flavefaciens FD-1. Appl Environ Microbiol. 1988 Jun;54(6):1634–1636. doi: 10.1128/aem.54.6.1634-1636.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T. M., Wilson C. A., Stewart C. S. Preparation of the cellulase from the cellulolytic anaerobic rumen bacterium Ruminococcus albus and its release from the bacterial cell wall. Biochem J. 1982 Jul 1;205(1):129–137. doi: 10.1042/bj2050129. [DOI] [PMC free article] [PubMed] [Google Scholar]


