Abstract
Two genes encoding beta-1,3 glucanase activity were cloned from the gram-negative soil bacterium Cellvibrio mixtus. The two clones, designated cwd (cell wall degradation) and lam (laminarin degradation), had distinct endonuclease restriction patterns and encoded enzymes with distinct substrate specificities. The 3.7-kilobase cwd insert encoded an enzyme which degraded yeast cell walls as well as the soluble beta-1,3 glucan laminarin and the insoluble beta-1,3 glucans zymosan and pachyman. The 1.8-kilobase lam insert encoded an enzyme which degraded laminarin only. Both enzymes degraded laminarin in an endohydrolytic manner to yield laminarobiose, laminarotriose, and laminarotetraose as major end products. Radiolabeled translation products of the cwd and lam transcripts were identified.
Full text
PDF

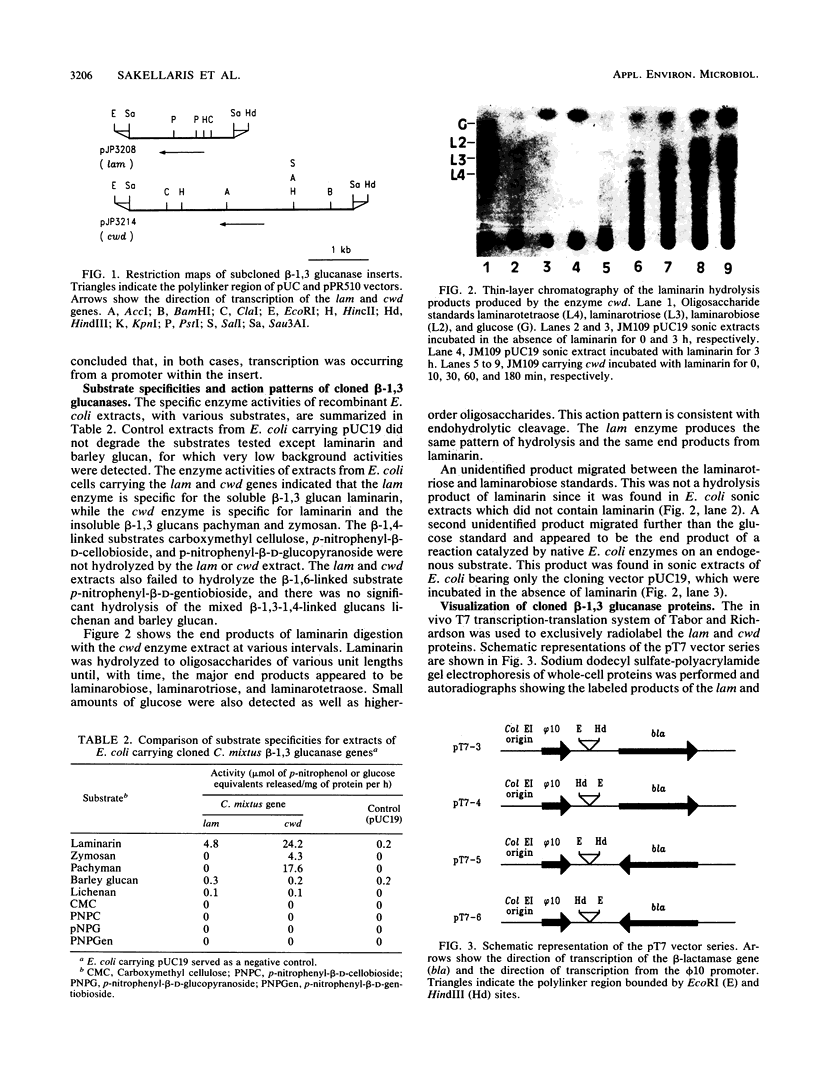


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barras D. R., Stone B. A. Beta-1,3-glucan hydrolases from Euglena gracilis. I. The nature of the hydrolases. Biochim Biophys Acta. 1969 Nov 4;191(2):329–341. doi: 10.1016/0005-2744(69)90252-6. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block M. D., Botterman J., Vandewiele M., Dockx J., Thoen C., Gosselé V., Movva N. R., Thompson C., Montagu M. V., Leemans J. Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 1987 Sep;6(9):2513–2518. doi: 10.1002/j.1460-2075.1987.tb02537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chung C. T., Niemela S. L., Miller R. H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande M. V., Eriksson K. E., Pettersson L. G. An assay for selective determination of exo-1,4,-beta-glucanases in a mixture of cellulolytic enzymes. Anal Biochem. 1984 May 1;138(2):481–487. doi: 10.1016/0003-2697(84)90843-1. [DOI] [PubMed] [Google Scholar]
- Doi K., Doi A. Cloning and expression in Escherichia coli of the gene for an Arthrobacter beta-(1----3)-glucanase. J Bacteriol. 1986 Dec;168(3):1272–1276. doi: 10.1128/jb.168.3.1272-1276.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardi G. L. Characterization of nonfermentative nonfastidious gram negative bacteria encountered in medical bacteriology. J Appl Bacteriol. 1971 Sep;34(3):623–644. doi: 10.1111/j.1365-2672.1971.tb02326.x. [DOI] [PubMed] [Google Scholar]
- Gobius K. S., Pemberton J. M. Molecular cloning, characterization, and nucleotide sequence of an extracellular amylase gene from Aeromonas hydrophila. J Bacteriol. 1988 Mar;170(3):1325–1332. doi: 10.1128/jb.170.3.1325-1332.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D., Grady K. L., Suslow T. V., Bedbrook J. R. Isolation and characterization of genes encoding two chitinase enzymes from Serratia marcescens. EMBO J. 1986 Mar;5(3):467–473. doi: 10.1002/j.1460-2075.1986.tb04235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen N. T., Yoshikawa M. beta-1,3-Endoglucanase from Soybean Releases Elicitor-Active Carbohydrates from Fungus Cell Walls. Plant Physiol. 1983 Mar;71(3):460–465. doi: 10.1104/pp.71.3.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mauch F., Hadwiger L. A., Boller T. Antifungal Hydrolases in Pea Tissue : I. Purification and Characterization of Two Chitinases and Two beta-1,3-Glucanases Differentially Regulated during Development and in Response to Fungal Infection. Plant Physiol. 1988 Jun;87(2):325–333. doi: 10.1104/pp.87.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch F., Mauch-Mani B., Boller T. Antifungal Hydrolases in Pea Tissue : II. Inhibition of Fungal Growth by Combinations of Chitinase and beta-1,3-Glucanase. Plant Physiol. 1988 Nov;88(3):936–942. doi: 10.1104/pp.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Quigley N. B., Reeves P. R. Chloramphenicol resistance cloning vector based on pUC9. Plasmid. 1987 Jan;17(1):54–57. doi: 10.1016/0147-619x(87)90008-4. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]