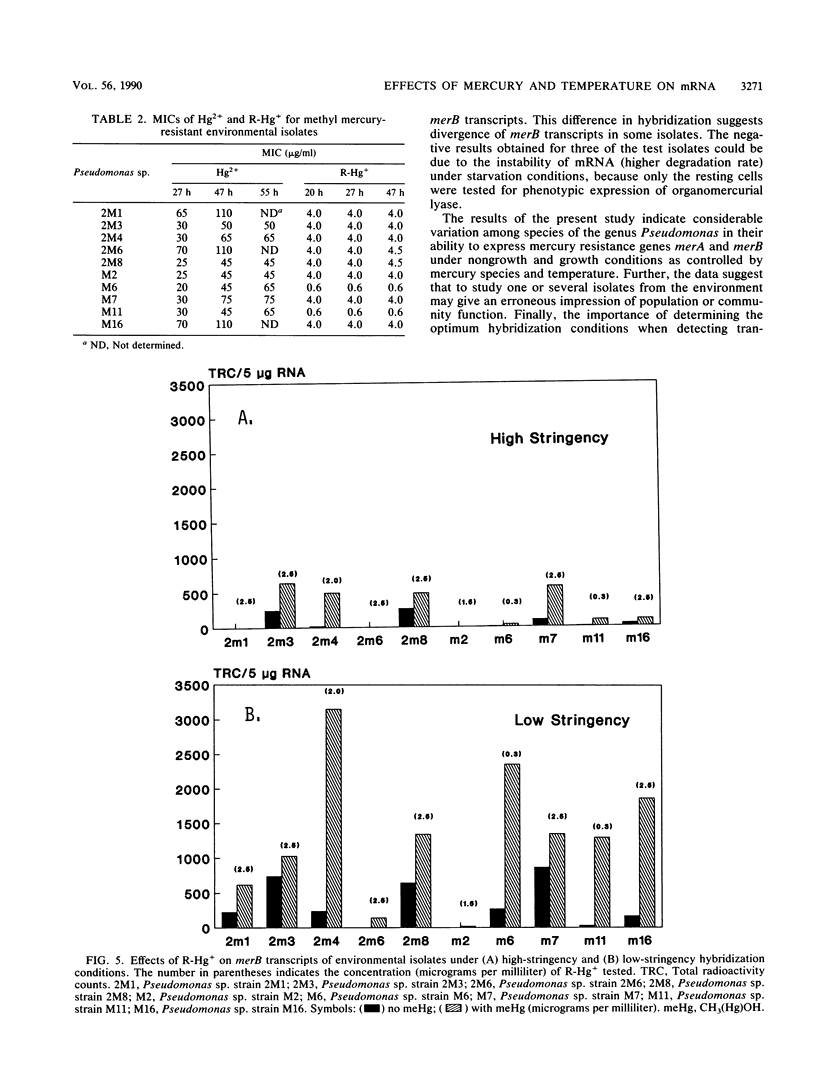
Abstract
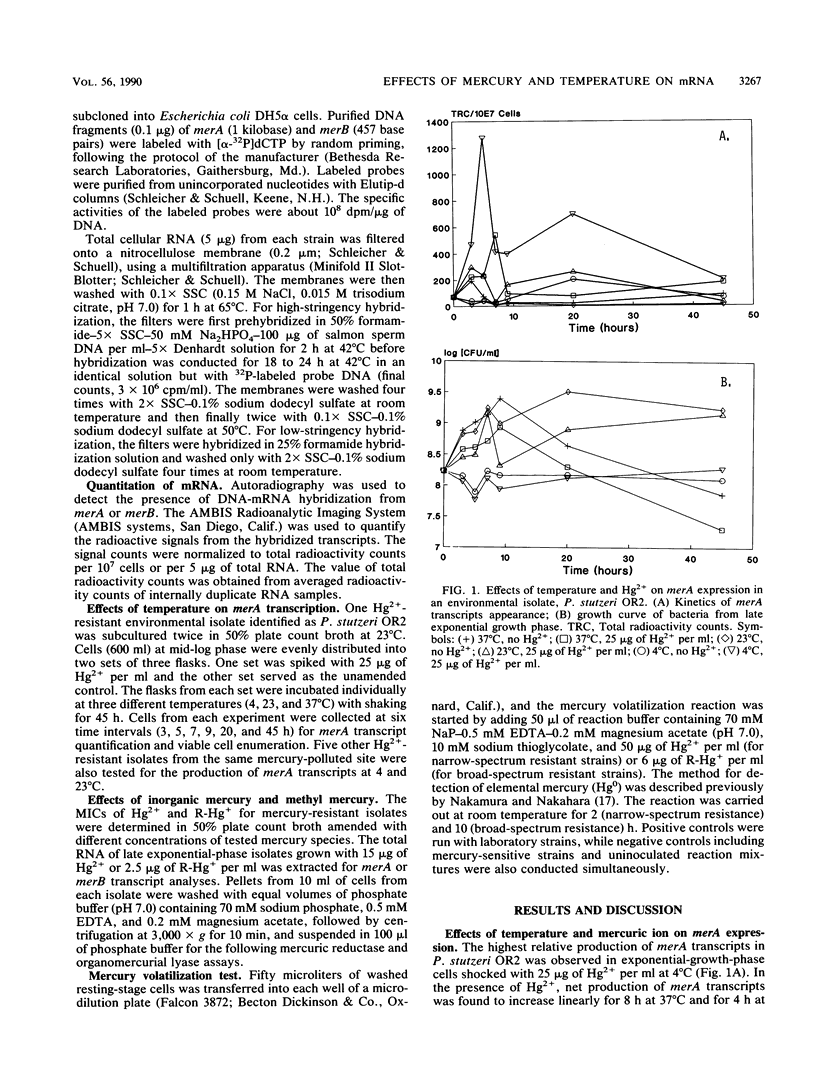
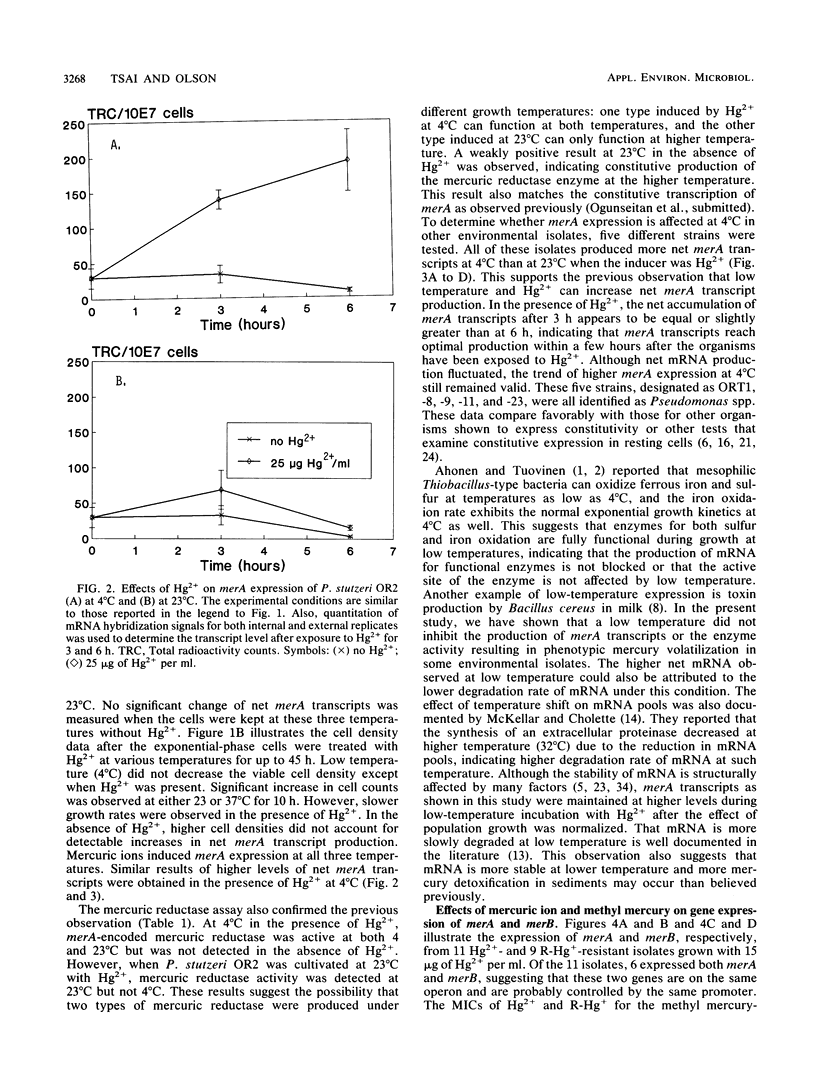
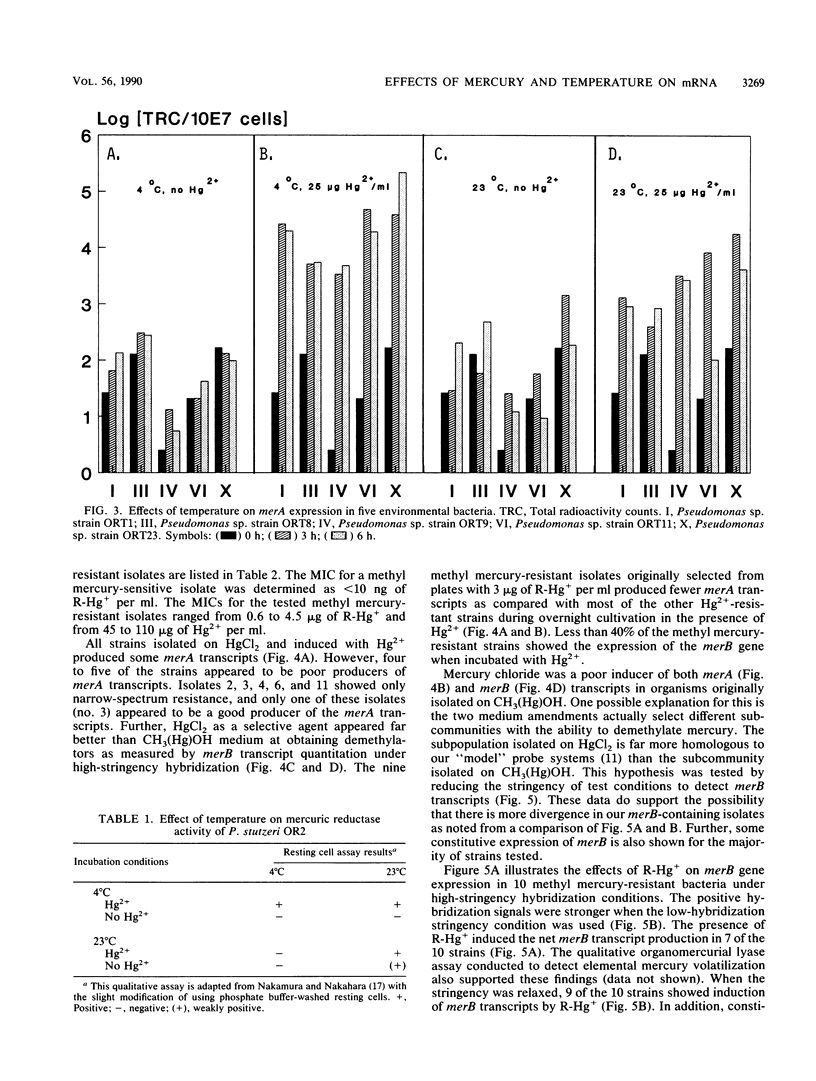
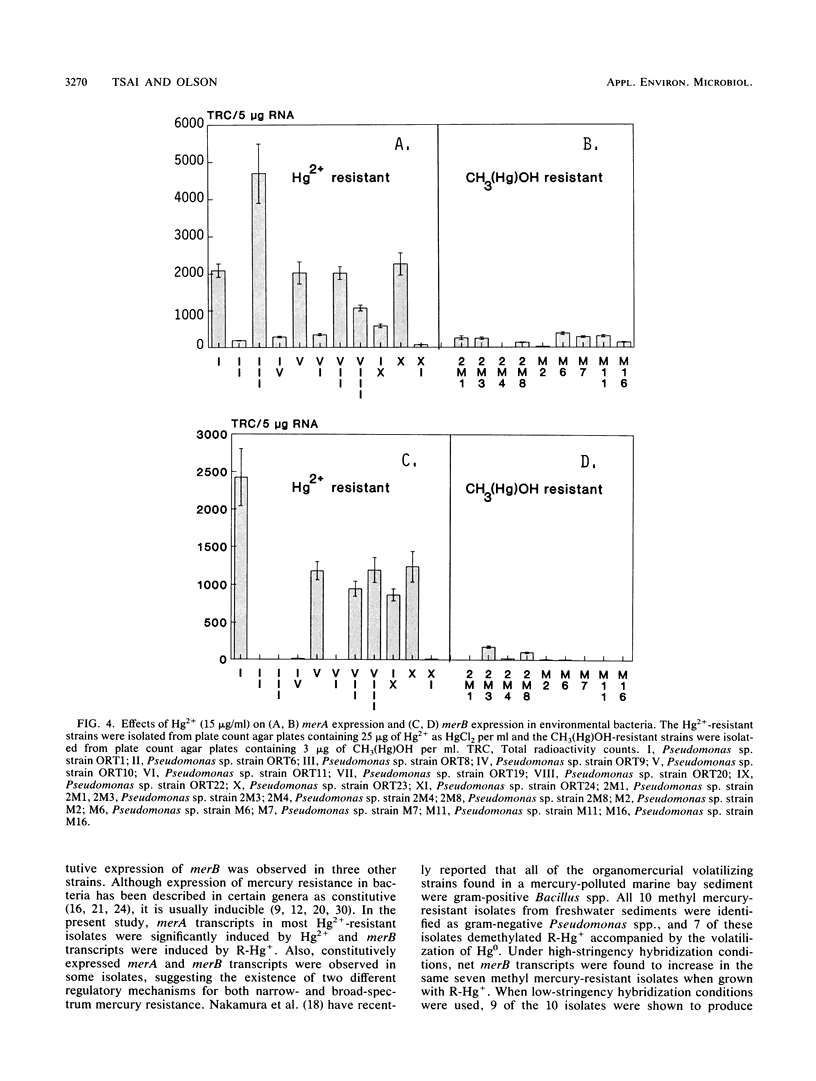
Twenty different bacterial isolates obtained from a mercury-contaminated site in Oak Ridge, Tenn., were grown on plate count agar amended with 25 μg of Hg2+ or 3 μg of CH3-Hg+ (R-Hg+) per ml. The total cellular RNA was extracted from each isolate by an acid-guanidine-thiocyanate-phenol-chloroform method. The transcripts of merA and merB were detected and quantitated by Northern (RNA) hybridization. A qualitative assay of mercuric reductase was used to confirm the enzyme activity. Low temperature (4°C) with the presence of Hg2+ (25 μg/ml) significantly increased the net merA transcripts of mid-log-phase cells of six environmental isolates. The net merA transcript production by 18 of the isolates increased when they were grown on 50% plate count broth with 15 μg of Hg2+ per ml, but only 8 isolates showed increased production of merB transcripts. The MICs of Hg2+ and R-Hg+ for 10 methyl mercury-resistant isolates ranged from 45 to 110 μg of Hg2+ and 0.6 to 4.5 μg of R-Hg+ per ml. R-Hg+ was able to induce the expression of merB in 70% of methyl mercury-resistant strains.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahonen L., Tuovinen O. H. Kinetics of sulfur oxidation at suboptimal temperatures. Appl Environ Microbiol. 1990 Feb;56(2):560–562. doi: 10.1128/aem.56.2.560-562.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahonen L., Tuovinen O. H. Microbiological oxidation of ferrous iron at low temperatures. Appl Environ Microbiol. 1989 Feb;55(2):312–316. doi: 10.1128/aem.55.2.312-316.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Liebert C., Gillman M. Environmental significance of the potential for mer(Tn21)-mediated reduction of Hg2+ to Hg0 in natural waters. Appl Environ Microbiol. 1989 May;55(5):1196–1202. doi: 10.1128/aem.55.5.1196-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Olson B. H. Phenotypic and genotypic adaptation of aerobic heterotrophic sediment bacterial communities to mercury stress. Appl Environ Microbiol. 1986 Aug;52(2):403–406. doi: 10.1128/aem.52.2.403-406.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Nilsson G., von Gabain A., Cohen S. N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986 Jul 18;46(2):245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Christiansson A., Naidu A. S., Nilsson I., Wadström T., Pettersson H. E. Toxin production by Bacillus cereus dairy isolates in milk at low temperatures. Appl Environ Microbiol. 1989 Oct;55(10):2595–2600. doi: 10.1128/aem.55.10.2595-2600.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J. The genetics and biochemistry of mercury resistance. Crit Rev Microbiol. 1987;15(2):117–140. doi: 10.3109/10408418709104455. [DOI] [PubMed] [Google Scholar]
- Fox B., Walsh C. T. Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J Biol Chem. 1982 Mar 10;257(5):2498–2503. [PubMed] [Google Scholar]
- Griffin H. G., Foster T. J., Silver S., Misra T. K. Cloning and DNA sequence of the mercuric- and organomercurial-resistance determinants of plasmid pDU1358. Proc Natl Acad Sci U S A. 1987 May;84(10):3112–3116. doi: 10.1073/pnas.84.10.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Wang Y., Mahler I., Walsh C. T. Homologous metalloregulatory proteins from both gram-positive and gram-negative bacteria control transcription of mercury resistance operons. J Bacteriol. 1989 Jan;171(1):222–229. doi: 10.1128/jb.171.1.222-229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennell D., Bicknell I. Decay of messenger ribonucleic acid from the lactose operon of Escherichia coli as a function of growth temperature. J Mol Biol. 1973 Feb 15;74(1):21–31. doi: 10.1016/0022-2836(73)90351-3. [DOI] [PubMed] [Google Scholar]
- McKellar R. C., Cholette H. Effect of temperature shifts on extracellular proteinase-specific mRNA pools in Pseudomonas fluorescens B52. Appl Environ Microbiol. 1987 Aug;53(8):1973–1976. doi: 10.1128/aem.53.8.1973-1976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munjaal R. P., Conneely O. M., O'Malley B. W. In situ detection of progesterone receptor mRNA in the chicken oviduct using probe-on slides. Biotechniques. 1989 Nov-Dec;7(10):1104–1108. [PubMed] [Google Scholar]
- Nakahara H., Schottel J. L., Yamada T., Miyakawa Y., Asakawa M., Harville J., Silver S. Mercuric reductase enzymes from Streptomyces species and group B Streptococcus. J Gen Microbiol. 1985 May;131(5):1053–1059. doi: 10.1099/00221287-131-5-1053. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Nakahara H. Simplified X-ray film method for detection of bacterial volatilization of mercury chloride by Escherichia coli. Appl Environ Microbiol. 1988 Nov;54(11):2871–2873. doi: 10.1128/aem.54.11.2871-2873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Sakamoto M., Uchiyama H., Yagi O. Organomercurial-volatilizing bacteria in the mercury-polluted sediment of Minamata Bay, Japan. Appl Environ Microbiol. 1990 Jan;56(1):304–305. doi: 10.1128/aem.56.1.304-305.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Halloran T. V., Frantz B., Shin M. K., Ralston D. M., Wright J. G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989 Jan 13;56(1):119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- Olson G. J., Porter F. D., Rubinstein J., Silver S. Mercuric reductase enzyme from a mercury-volatilizing strain of Thiobacillus ferrooxidans. J Bacteriol. 1982 Sep;151(3):1230–1236. doi: 10.1128/jb.151.3.1230-1236.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinderle S. J., Booth J. E., Williams J. W. Mercuric reductase from R-plasmid NR1: characterization and mechanistic study. Biochemistry. 1983 Feb 15;22(4):869–876. doi: 10.1021/bi00273a025. [DOI] [PubMed] [Google Scholar]
- Ross J. The turnover of messenger RNA. Sci Am. 1989 Apr;260(4):48–55. doi: 10.1038/scientificamerican0489-48. [DOI] [PubMed] [Google Scholar]
- Rudrik J. T., Bawdon R. E., Guss S. P. Determination of mercury and organomercurial resistance in obligate anaerobic bacteria. Can J Microbiol. 1985 Mar;31(3):276–281. doi: 10.1139/m85-051. [DOI] [PubMed] [Google Scholar]
- Sahlman L., Lambeir A. M., Lindskog S., Dunford H. B. The reaction between NADPH and mercuric reductase from Pseudomonas aeruginosa. J Biol Chem. 1984 Oct 25;259(20):12403–12408. [PubMed] [Google Scholar]
- Schottel J. L. The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J Biol Chem. 1978 Jun 25;253(12):4341–4349. [PubMed] [Google Scholar]
- Silver S., Misra T. K. Plasmid-mediated heavy metal resistances. Annu Rev Microbiol. 1988;42:717–743. doi: 10.1146/annurev.mi.42.100188.003441. [DOI] [PubMed] [Google Scholar]
- Spangler W. J., Spigarelli J. L., Rose J. M., Miller H. M. Methylmercury: bacterial degradation in lake sediments. Science. 1973 Apr 13;180(4082):192–193. doi: 10.1126/science.180.4082.192. [DOI] [PubMed] [Google Scholar]
- Summers A. O., Lewis E. Volatilization of mercuric chloride by mercury-resistant plasmid-bearing strains of Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa. J Bacteriol. 1973 Feb;113(2):1070–1072. doi: 10.1128/jb.113.2.1070-1072.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O. Organization, expression, and evolution of genes for mercury resistance. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- Tezuka T., Tonomura K. Purification and properties of an enzyme catalyzing the splitting of carbon-mercury linkages from mercury-resistant Pseudomonas K-62 strain. I. Splitting enzyme 1. J Biochem. 1976 Jul;80(1):79–87. doi: 10.1093/oxfordjournals.jbchem.a131261. [DOI] [PubMed] [Google Scholar]
- Weiss A. A., Murphy S. D., Silver S. Mercury and organomercurial resistances determined by plasmids in Staphylococcus aureus. J Bacteriol. 1977 Oct;132(1):197–208. doi: 10.1128/jb.132.1.197-208.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H. C., Chang S. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc Natl Acad Sci U S A. 1986 May;83(10):3233–3237. doi: 10.1073/pnas.83.10.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. S., Kaplan S. Effects of light, oxygen, and substrates on steady-state levels of mRNA coding for ribulose-1,5-bisphosphate carboxylase and light-harvesting and reaction center polypeptides in Rhodopseudomonas sphaeroides. J Bacteriol. 1985 Jun;162(3):925–932. doi: 10.1128/jb.162.3.925-932.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]


