Abstract
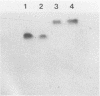
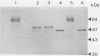
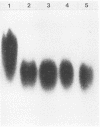
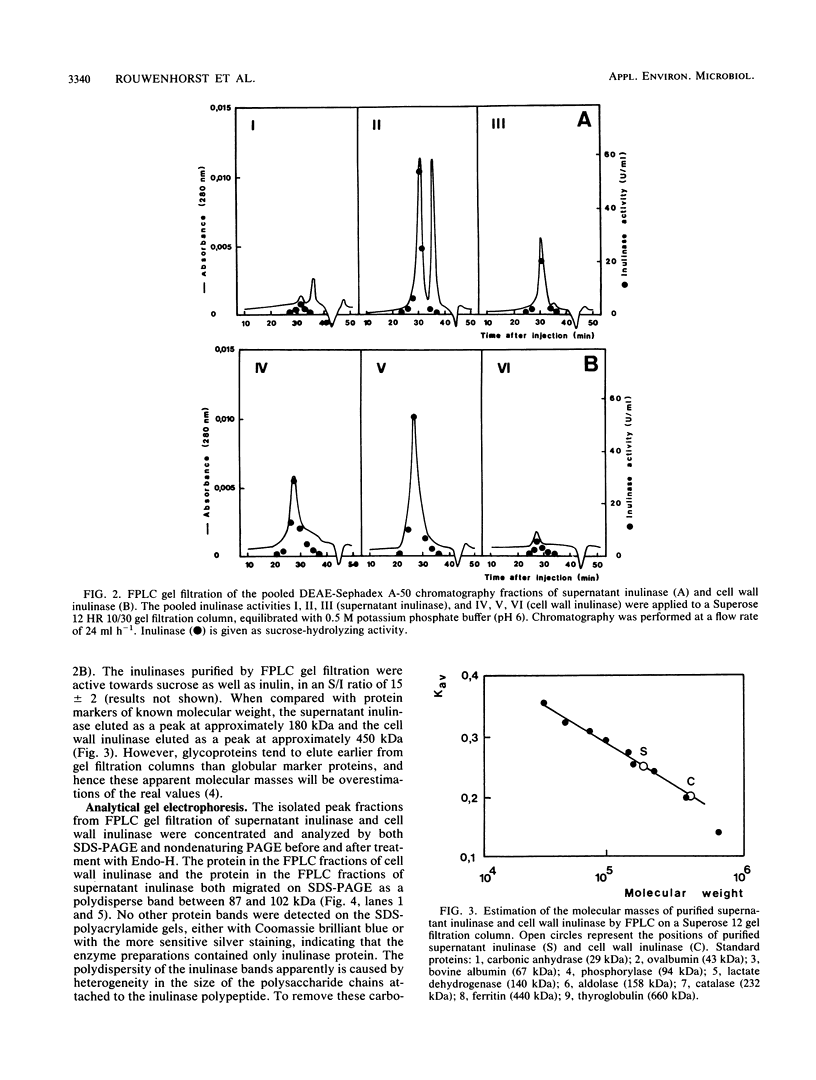
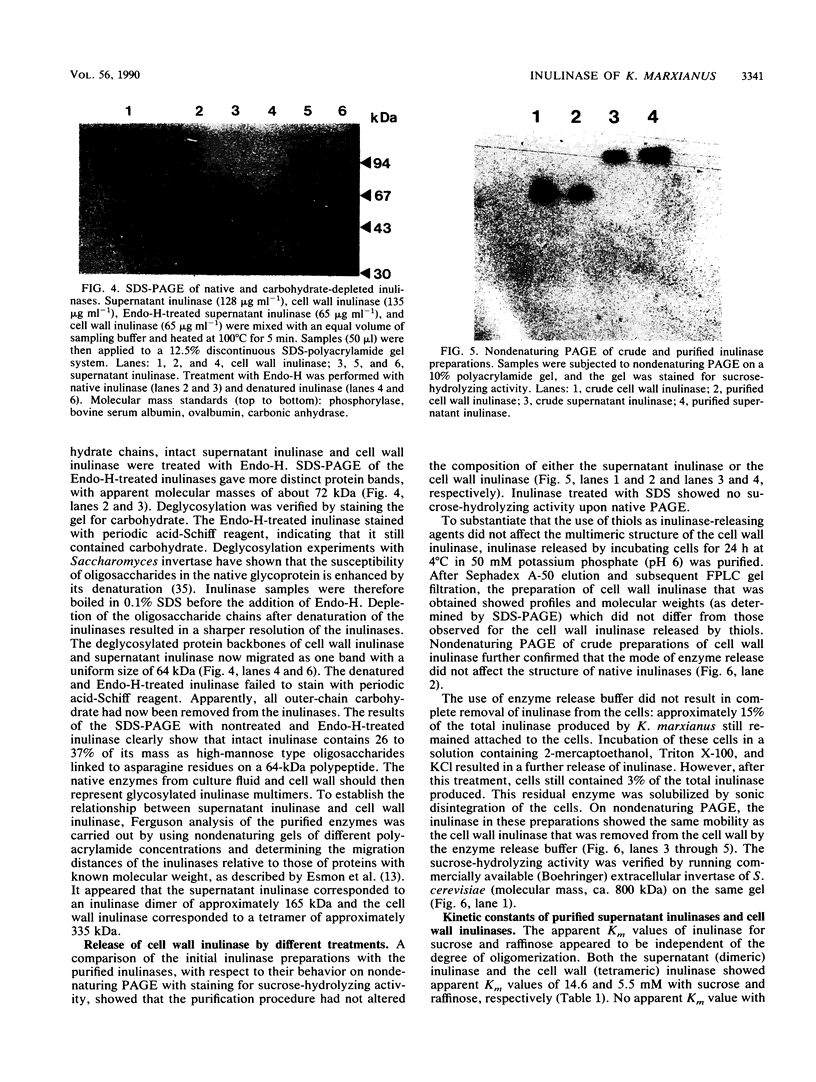
In the yeast Kluyveromyces marxianus two forms of inulinase were present, namely, an inulinase secreted into the culture fluid and an inulinase retained in the cell wall. Both forms were purified and analyzed by denaturing and nondenaturing polyacrylamide gel electrophoresis. With the use of endo-beta-N-acetyl-glucosaminidase H, it was established that the enzyme retained in the cell wall and the enzyme secreted into the culture fluid have similar subunits consisting of a 64-kDa polypeptide with varying amounts of carbohydrate (26 to 37% of the molecular mass). The two forms of inulinase differed in size because of their differences in subunit aggregation. The enzyme present in the culture fluid was a dimer, and the enzyme retained in the cell wall was a tetramer. The differences in oligomerization did not affect the apparent Km values towards the substrates sucrose and raffinose. These findings support the hypothesis that the retention of glycoproteins in the yeast cell wall may be caused by a permeability barrier towards larger glycoproteins. The amino-terminal end of inulinase was determined and compared with the amino terminus of the closely related invertase. The kinetic and structural evidence indicates that in yeasts two distinct beta-fructosidases exist, namely, invertase and inulinase.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Organization of the SUC gene family in Saccharomyces. Mol Cell Biol. 1983 Mar;3(3):351–359. doi: 10.1128/mcb.3.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Taussig R., Kustu S., Botstein D. The secreted form of invertase in Saccharomyces cerevisiae is synthesized from mRNA encoding a signal sequence. Mol Cell Biol. 1983 Mar;3(3):439–447. doi: 10.1128/mcb.3.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F. K., Watorek W., Maley F. Factors affecting the oligomeric structure of yeast external invertase. Arch Biochem Biophys. 1983 Jun;223(2):543–555. doi: 10.1016/0003-9861(83)90619-7. [DOI] [PubMed] [Google Scholar]
- Davies R., Wayman F. J. The effect of thiols on Saccharomyces fragilis. Antonie Van Leeuwenhoek. 1975;41(1):33–58. doi: 10.1007/BF02565035. [DOI] [PubMed] [Google Scholar]
- Edelman J., Jefford T. G. The metabolism of fructose polymers in plants. 4. Beta-fructofuranosidases of tubers of Helianthus tuberosus L. Biochem J. 1964 Oct;93(1):148–161. doi: 10.1042/bj0930148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmon B., Esmon P. C., Schekman R. Early steps in processing of yeast glycoproteins. J Biol Chem. 1984 Aug 25;259(16):10322–10327. [PubMed] [Google Scholar]
- Esmon P. C., Esmon B. E., Schauer I. E., Taylor A., Schekman R. Structure, assembly, and secretion of octameric invertase. J Biol Chem. 1987 Mar 25;262(9):4387–4394. [PubMed] [Google Scholar]
- Fuchs A., de Bruijn J. M., Niedeveld C. J. Bacteria and yeasts as possible candidates for the production of inulinases and levanases. Antonie Van Leeuwenhoek. 1985;51(3):333–343. doi: 10.1007/BF02439942. [DOI] [PubMed] [Google Scholar]
- Grossmann M. K., Zimmermann F. K. The structural genes of internal invertases in Saccharomyces cerevisiae. Mol Gen Genet. 1979 Sep;175(2):223–229. doi: 10.1007/BF00425540. [DOI] [PubMed] [Google Scholar]
- Kidby D. K., Davies R. Thiol induced release of invertase from cell walls of Saccharomyces fragilis. Biochim Biophys Acta. 1970 Feb 24;201(2):261–266. doi: 10.1016/0304-4165(70)90300-4. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehle L., Cohen R. E., Ballou C. E. Carbohydrate structure of yeast invertase. Demonstration of a form with only core oligosaccharides and a form with completed polysaccharide chains. J Biol Chem. 1979 Dec 10;254(23):12209–12218. [PubMed] [Google Scholar]
- Martin I., Débarbouillé M., Ferrari E., Klier A., Rapoport G. Characterization of the levanase gene of Bacillus subtilis which shows homology to yeast invertase. Mol Gen Genet. 1987 Jun;208(1-2):177–184. doi: 10.1007/BF00330439. [DOI] [PubMed] [Google Scholar]
- Neumann N. P., Lampen J. O. The glycoprotein structure of yeast invertase. Biochemistry. 1969 Sep;8(9):3552–3556. doi: 10.1021/bi00837a010. [DOI] [PubMed] [Google Scholar]
- Novick P., Ferro S., Schekman R. Order of events in the yeast secretory pathway. Cell. 1981 Aug;25(2):461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Rouwenhorst R. J., Ritmeester W. S., Scheffers W. A., Van Dijken J. P. Localization of inulinase and invertase in Kluyveromyces species. Appl Environ Microbiol. 1990 Nov;56(11):3329–3336. doi: 10.1128/aem.56.11.3329-3336.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouwenhorst R. J., Visser L. E., Van Der Baan A. A., Scheffers W. A., Van Dijken J. P. Production, Distribution, and Kinetic Properties of Inulinase in Continuous Cultures of Kluyveromyces marxianus CBS 6556. Appl Environ Microbiol. 1988 May;54(5):1131–1137. doi: 10.1128/aem.54.5.1131-1137.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarentino A. L., Plummer T. H., Jr, Maley F. The release of intact oligosaccharides from specific glycoproteins by endo-beta-N-acetylglucosaminidase H. J Biol Chem. 1974 Feb 10;249(3):818–824. [PubMed] [Google Scholar]
- Trimble R. B., Maley F. Subunit structure of external invertase from Saccharomyces cerevisiae. J Biol Chem. 1977 Jun 25;252(12):4409–4412. [PubMed] [Google Scholar]
- Vandamme E. J., Derycke D. G. Microbial inulinases: fermentation process, properties, and applications. Adv Appl Microbiol. 1983;29:139–176. doi: 10.1016/s0065-2164(08)70356-3. [DOI] [PubMed] [Google Scholar]
- Weimberg R., Orton W. L. Elution of exocellular enzymes from Saccharomyces fragilis and Saccharomyces cerevisiae. J Bacteriol. 1966 Jan;91(1):1–13. doi: 10.1128/jb.91.1.1-13.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman W. E., Day D. F. Purification and properties of the beta-fructofuranosidase from Kluyveromyces fragilis. FEBS Lett. 1983 Aug 22;160(1-2):16–20. doi: 10.1016/0014-5793(83)80927-2. [DOI] [PubMed] [Google Scholar]