Abstract
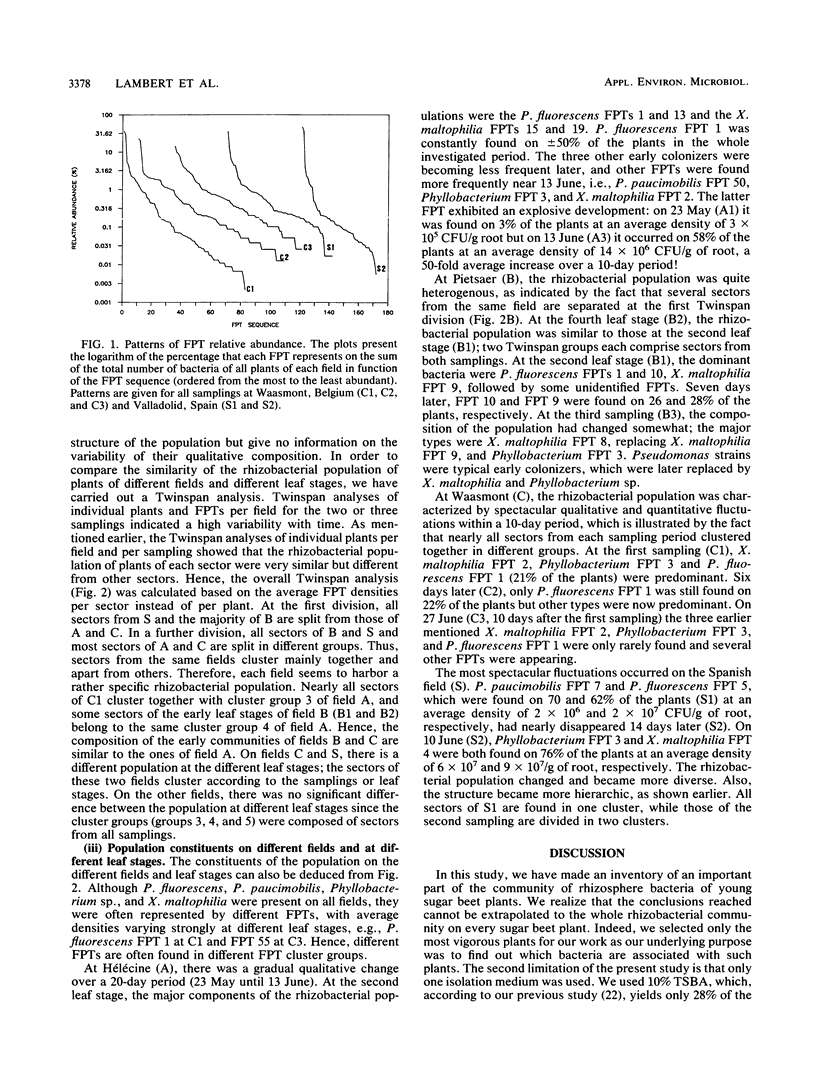
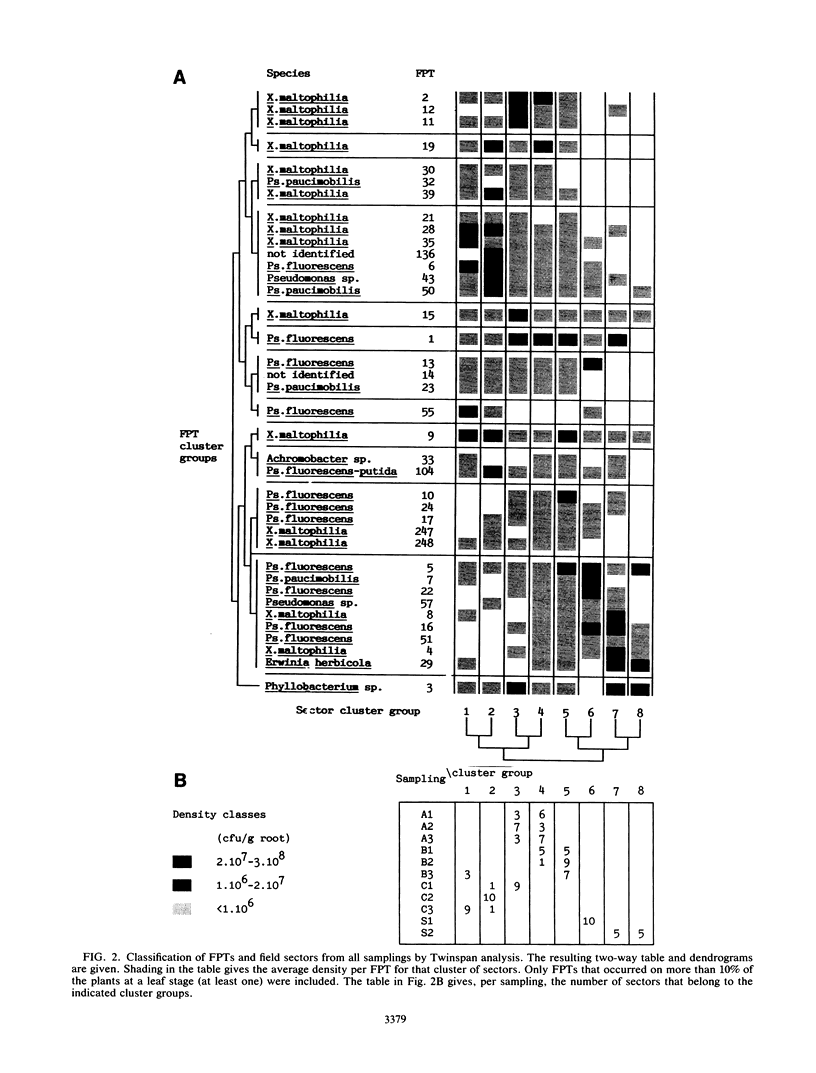
Fast-growing, aerobic, heterotrophic bacteria from the root surface of young sugar beet plants were inventoried. Isolation of the most abundant bacteria from the root surface of each of 1,100 plants between the second and tenth leaf stage yielded 5,600 isolates. These plants originated from different fields in Belgium and Spain. All isolates were characterized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of total cellular proteins. Comparison of protein fingerprints allowed us to inventory the bacteria of individual plants of different fields or leaf stages and to analyze the composition and variability of the rhizobacterial population of young sugar beet plants. Each field harbored a specific population of bacteria which showed a highly hierarchic structure. A small number of bacteria occurring frequently at high densities dominated in each field. The major bacteria were identified as Pseudomonas fluorescens, Xanthomonas maltophilia, Pseudomonas paucimobilis, and Phyllobacterium sp. The former three species showed a high genetic variability as they were represented by different protein fingerprint types on the same or different fields or leaf stages. Twinspan analysis and relative abundance plots showed that the structure and composition of the bacterial populations varied strongly over time. Pseudomonads were typically early colonizers which were later replaced by X. maltophilia or Phyllobacterium sp.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdel-Nasser M., Makawi A. A. Occurrence of certain physiological groups of soil micro-organisms in the rhizosphere and rhizoplane of watermelon, cucumber, and cowpea. Zentralbl Bakteriol Naturwiss. 1979;134(4):310–315. doi: 10.1016/s0323-6056(79)80003-8. [DOI] [PubMed] [Google Scholar]
- Debette J., Blondeau R. Présence de Pseudomonas maltophilia dans la rhizosphère de quelques plantes cultivées. Can J Microbiol. 1980 Apr;26(4):460–463. [PubMed] [Google Scholar]
- Kersters K., De Ley J. Identification and grouping of bacteria by numerical analysis of their electrophoretic protein patterns. J Gen Microbiol. 1975 Apr;87(2):333–342. doi: 10.1099/00221287-87-2-333. [DOI] [PubMed] [Google Scholar]
- Lambert B., Joos H., Dierickx S., Vantomme R., Swings J., Kersters K., Van Montagu M. Identification and Plant Interaction of a Phyllobacterium sp., a Predominant Rhizobacterium of Young Sugar Beet Plants. Appl Environ Microbiol. 1990 Apr;56(4):1093–1102. doi: 10.1128/aem.56.4.1093-1102.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert B., Leyns F., Van Rooyen L., Gosselé F., Papon Y., Swings J. Rhizobacteria of maize and their antifungal activities. Appl Environ Microbiol. 1987 Aug;53(8):1866–1871. doi: 10.1128/aem.53.8.1866-1871.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur J. V., Kovacic D. A., Smith M. H. Genetic diversity in natural populations of a soil bacterium across a landscape gradient. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9621–9624. doi: 10.1073/pnas.85.24.9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Castro F. A., Alexander M. Method for establishing a bacterial inoculum on corn roots. Appl Environ Microbiol. 1983 Jan;45(1):248–254. doi: 10.1128/aem.45.1.248-254.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. W. Use of Agrobacterium radiobacter in agricultural ecosystems. Microbiol Sci. 1988 Mar;5(3):92–95. [PubMed] [Google Scholar]
- Moore W. E., Hash D. E., Holdeman L. V., Cato E. P. Polyacrylamide slab gel electrophoresis of soluble proteins for studies of bacterial floras. Appl Environ Microbiol. 1980 Apr;39(4):900–907. doi: 10.1128/aem.39.4.900-907.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Evidence for clonal population structure in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jan;81(1):198–201. doi: 10.1073/pnas.81.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paau A. S. Improvement of Rhizobium inoculants. Appl Environ Microbiol. 1989 Apr;55(4):862–865. doi: 10.1128/aem.55.4.862-865.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira A. D., Sands D. C. Fluorescent pseudomonads--a residual component in the soil microflora? J Appl Bacteriol. 1971 Mar;34(1):253–259. doi: 10.1111/j.1365-2672.1971.tb02284.x. [DOI] [PubMed] [Google Scholar]
- Sands D. C., Rovira A. D. Pseudomonas fluorescens biotype G, the dominant fluorescent pseudomonad in South Australian soils and wheat rhizospheres. J Appl Bacteriol. 1971 Mar;34(1):261–275. doi: 10.1111/j.1365-2672.1971.tb02285.x. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancura V. Fluorescent pseudomonads in the rhizosphere of plants and their relation to root exudates. Folia Microbiol (Praha) 1980;25(2):168–173. doi: 10.1007/BF02933018. [DOI] [PubMed] [Google Scholar]
- Zambryski P., Tempe J., Schell J. Transfer and function of T-DNA genes from agrobacterium Ti and Ri plasmids in plants. Cell. 1989 Jan 27;56(2):193–201. doi: 10.1016/0092-8674(89)90892-1. [DOI] [PubMed] [Google Scholar]



