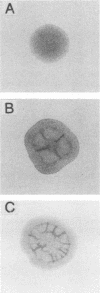
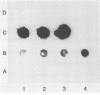
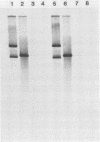
Abstract
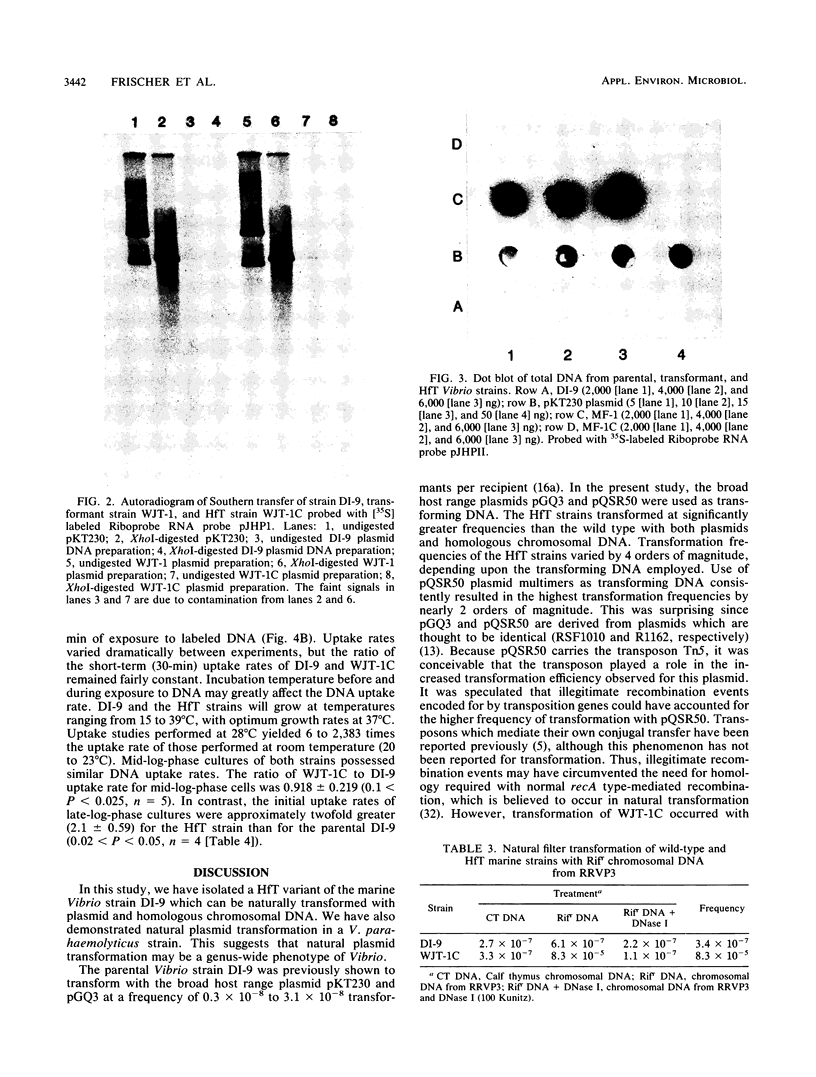
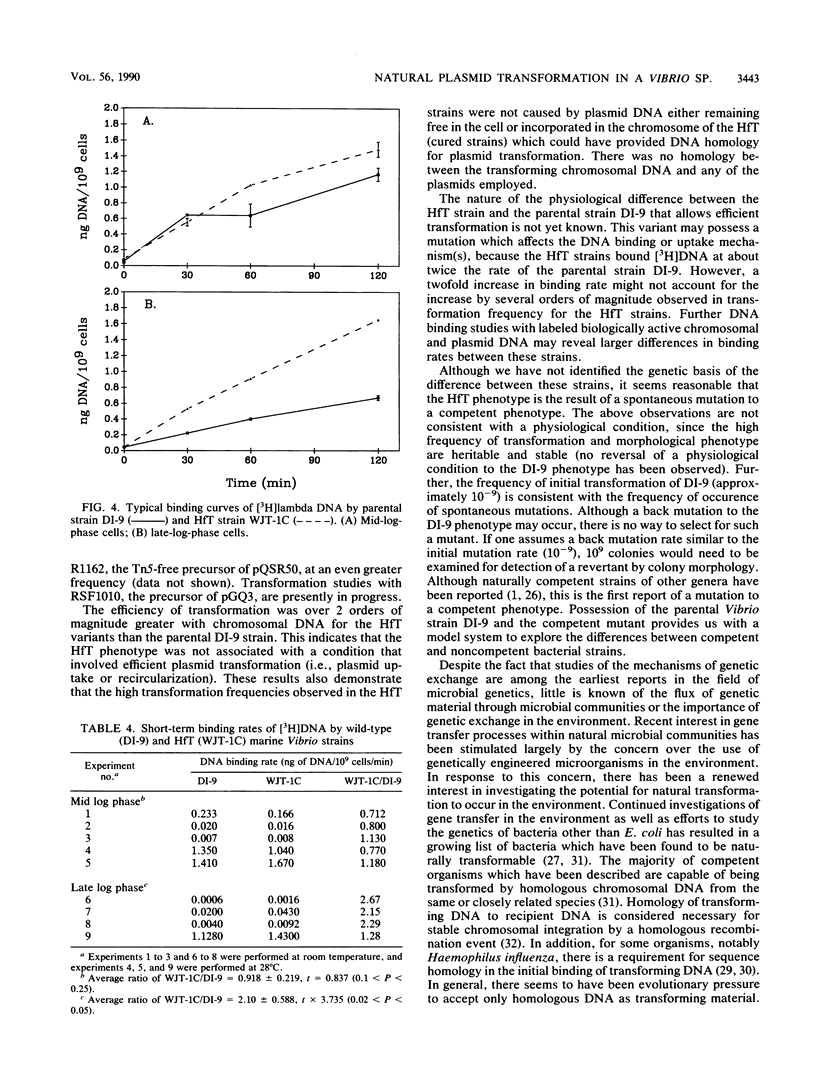
The estuarine bacterium Vibrio strain DI-9 has been shown to be naturally transformable with both broad host range plasmid multimers and homologous chromosomal DNA at average frequencies of 3.5 X 10(-9) and 3.4 X 10(-7) transformants per recipient, respectively. Growth of plasmid transformants in nonselective medium resulted in cured strains that transformed 6 to 42, 857 times more frequently than the parental strain, depending on the type of transforming DNA. These high-frequency-of-transformation (HfT) strains were transformed at frequencies ranging from 1.1 X 10(-8) to 1.3 X 10(-4) transformants per recipient with plasmid DNA and at an average frequency of 8.3 X 10(-5) transformants per recipient with homologous chromosomal DNA. The highest transformation frequencies were observed by using multimers of an R1162 derivative carrying the transposon Tn5 (pQSR50). Probing of total DNA preparations from one of the cured strains demonstrated that no plasmid DNA remained in the cured strains which may have provided homology to the transforming DNA. All transformants and cured strains could be differentiated from the parental strains by colony morphology. DNA binding studies indicated that late-log-phase HfT strains bound [3H]bacteriophage lambda DNA 2.1 times more rapidly than the parental strain. These results suggest that the original plasmid transformation event of strain DI-9 was the result of uptake and expression of plasmid DNA by a competent mutant (HfT strain). Additionally, it was found that a strain of Vibrio parahaemolyticus, USFS 3420, could be naturally transformed with plasmid DNA. Natural plasmid transformation by high-transforming mutants may be a means of plasmid acquisition by natural aquatic bacterial populations.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Buluwela L., Forster A., Boehm T., Rabbitts T. H. A rapid procedure for colony screening using nylon filters. Nucleic Acids Res. 1989 Jan 11;17(1):452–452. doi: 10.1093/nar/17.1.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C. A., Stewart G. J., Ingraham J. L. Thymidine salvage in Pseudomonas stutzeri and Pseudomonas aeruginosa provided by heterologous expression of Escherichia coli thymidine kinase gene. J Bacteriol. 1985 Jul;163(1):291–295. doi: 10.1128/jb.163.1.291-295.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughter J. P., Stewart G. J. Genetic exchange in the environment. Antonie Van Leeuwenhoek. 1989;55(1):15–22. doi: 10.1007/BF02309615. [DOI] [PubMed] [Google Scholar]
- Felsenstein K. M. A modified protocol for the PZ523 spin columns: a legitimate purification alternative for plasmid DNA. Biotechniques. 1988 Oct;6(9):847–848. [PubMed] [Google Scholar]
- Griffith O. M. Large-scale isolation of plasmid DNA using high speed centrifugation methods. Biotechniques. 1988 Sep;6(8):725–727. [PubMed] [Google Scholar]
- Jeffrey W. H., Paul J. H. Thymidine uptake, thymidine incorporation, and thymidine kinase activity in marine bacterium isolates. Appl Environ Microbiol. 1990 May;56(5):1367–1372. doi: 10.1128/aem.56.5.1367-1372.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R., Laux R., Boch G., Hinds M., Bayly R., Shapiro J. A. Broad-host-range IncP-4 plasmid R1162: effects of deletions and insertions on plasmid maintenance and host range. J Bacteriol. 1982 Oct;152(1):140–150. doi: 10.1128/jb.152.1.140-150.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michod R. E., Wojciechowski M. F., Hoelzer M. A. DNA repair and the evolution of transformation in the bacterium Bacillus subtilis. Genetics. 1988 Jan;118(1):31–39. doi: 10.1093/genetics/118.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Deflaun M. F., Jeffrey W. H. Mechanisms of DNA utilization by estuarine microbial populations. Appl Environ Microbiol. 1988 Jul;54(7):1682–1688. doi: 10.1128/aem.54.7.1682-1688.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of hoechst 33258. Appl Environ Microbiol. 1982 Jun;43(6):1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H. Use of hoechst dyes 33258 and 33342 for enumeration of attached and planktonic bacteria. Appl Environ Microbiol. 1982 Apr;43(4):939–944. doi: 10.1128/aem.43.4.939-944.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochelle P. A., Day M. J., Fry J. C. Occurrence, transfer and mobilization in epilithic strains of Acinetobacter of mercury-resistance plasmids capable of transformation. J Gen Microbiol. 1988 Nov;134(11):2933–2941. doi: 10.1099/00221287-134-11-2933. [DOI] [PubMed] [Google Scholar]
- Saye D. J., Ogunseitan O. A., Sayler G. S., Miller R. V. Transduction of linked chromosomal genes between Pseudomonas aeruginosa strains during incubation in situ in a freshwater habitat. Appl Environ Microbiol. 1990 Jan;56(1):140–145. doi: 10.1128/aem.56.1.140-145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisco K. L., Smith H. O. Sequence-specific DNA uptake in Haemophilus transformation. Proc Natl Acad Sci U S A. 1979 Feb;76(2):972–976. doi: 10.1073/pnas.76.2.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. J., Carlson C. A. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211–235. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]
- Stewart G. J., Sinigalliano C. D. Detection of horizontal gene transfer by natural transformation in native and introduced species of bacteria in marine and synthetic sediments. Appl Environ Microbiol. 1990 Jun;56(6):1818–1824. doi: 10.1128/aem.56.6.1818-1824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZINDER N. D., LEDERBERG J. Genetic exchange in Salmonella. J Bacteriol. 1952 Nov;64(5):679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervos P. H., Morris L. M., Hellwig R. J. A novel method for rapid isolation of plasmid DNA. Biotechniques. 1988 Mar;6(3):238–242. [PubMed] [Google Scholar]