Abstract
Prokaryotic class I release factors (RFs) respond to mRNA stop codons and terminate protein synthesis. They interact with the ribosomal decoding site and the peptidyl-transferase centre bridging these 75 Å distant ribosomal centres. For this an elongated RF conformation, with partially unfolded core domains II·III·IV is required, which contrasts the known compact RF crystal structures. The crystal structure of Thermus thermophilus RF2 was determined and compared with solution structure of T. thermophilus and Escherichia coli RF2 by microcalorimetry, circular dichroism spectroscopy and small angle X-ray scattering. The structure of T. thermophilus RF2 in solution at 20°C is predominantly compact like the crystal structure. Thermodynamic analysis point to an initial melting of domain I, which is independent from the melting of the core. The core domains II·III·IV melt cooperatively at the respective physiological temperatures for T. thermophilus and E. coli. Thermodynamic analyses and the X-ray scattering results for T. thermophilus RF2 in solution suggest that the compact conformation of RF2 resembles a physiological state in absence of the ribosome.
INTRODUCTION
Remarkable advance in understanding the structure of ribosomes was achieved over the last decade (1,2). However, this structural information alone is not sufficient for detailed understanding of the mechanism of translation since a number of conformational changes on ribosomes are characteristic for this process (3–5). In addition, conformation of aminoacyl-tRNA affects the fidelity of decoding (6) and mobile ribosomal elements like the L7/L12 stalk seem to actively participate in substrate and factor recruitment (7).
After initiation and elongation is termination the last step in protein biosynthesis and leads to peptide release and recycling of ribosomes (8,9). This step is unique in translation since here, instead of aminoacyl-tRNAs, which read the codons during initiation and elongation, proteins, the release factors (RFs), act as adaptors for the decoding of stop codons. In prokaryotes two class I factors, RF1 and RF2, decode UAG/UAA and UGA/UAA codons, respectively, while the class II factor, the GTPase RF3, assists the removal of RFs from the ribosome (10). Finally RRF, leads together with EF-G to subunit dissociation (11). In eukaryotes one RF (eRF1), acting in complex with the GTPase eRF3 (12), decodes all three stop codons. The same is true for the archeal translation apparatus, while mitochondrial biosynthesis relies on two prokaryotic-type RFs (9). Site directed mutagenesis, biochemical probing and crosslinking provide evidence that class I RFs act as ribosomal A-site binders (9,13,14).
With respect to the stop codon recognition important differences between the prokaryotic and eukaryotic termination exist (15–17). In eukaryotes biochemical and genetic studies link the N-terminal domain of eRF1 with the NIKS and YxCxxxF consensus sequences to the decoding of stop signals (15–18) and a sequence comparison of eRF1s indicate the involvement of a complex 3D network of several amino acids (19). In contrast to this are in prokaryotes the tripeptide motifs PxT and SP(F/Y) for RF1 and RF2, respectively, associated with the recognition (13,20). However, similar to eRF1, more amino acids then a tripeptide are required for stop codon recognition, since domain swaps of at least 16 amino acids are needed to transplant the decoding specificity for UGA–RF1 or UAG–RF2 (21).
In contrast to stop codon recognition, there is a single conserved GGQ motif found in all class I RFs which is located at a position where the CCA-end of a tRNA in the ribosomal cocrystal structures was found (22). This GGQ motif is therefore assumed to participate in the hydrolysis of the peptidyl-residue from peptidyl-tRNA in the peptidyl transferase centre of the 50S subunit (23). The position of the GGQ-motif in the 3-D structure of eRF1 relative to the decoding-associated motif NIKS seems to be properly oriented to allow for bridging the decoding and peptidyl-transferase centres of the ribosome. However, the observed distance between these two regions in eRF1 (∼100 Å) exceeds the distance of 75 Å expected from the prokaryotic ribosome structures (24). In prokaryotic RF crystal structures, the conserved GGQ- and SP(F/Y)-motifs are only ∼20 Å apart (17,25). Cryo-EM and recent X-ray structure analysis of Escherichia coli and Thermus thermophilus ribosomes in complex with the corresponding class I RFs resolved this conflict by identification of elongated RFs bound to terminating ribosomes (22,26,27). This model of conformational adaptation of RFs upon ribosomal binding is supported by a molecular dynamic study (28).
However, it remained unanswered if the compact crystal structure of prokaryotic RFs is functionally relevant and if it exists under physiological conditions since the recent small angle X-ray scattering investigations (SAXS) on E. coli RF1 (29) indicated the existence of the elongated conformation also at ambient temperatures similar to human eRF1 (30).
In this work we present the crystal structure of RF2 from T. thermophilus, compare it to the crystal structure of E. coli RF2 and study the solution structures of both proteins by spectroscopic, small angle X-ray scattering and calorimetric analyses.
MATERIALS AND METHODS
Materials
Chemicals and biochemicals were from Merck (Darmstadt, Germany), with the following exceptions: acrylamide and 8-anilino-1-naphtalenesulphonic acid (ANS) was purchased from Sigma (Munich, Germany), sodium cacodylate from Fluka (Seelze, Germany), Ni-NTA agarose from Qiagen (Hilden, Germany), Q-SepharoseFF and FPLC-columns from GE-Healthcare/Amersham-Bioscience (Freiburg, Germany), DNase I, lysozyme and restriction enzymes from Roche (Mannheim, Germany), Taq polymerase from MBI fermentas (Vilnius, Lithuania), isopropyl-β-d-thiogalactoside (IPTG), ampicilline, kanamycine and chloramphenicol from Gerbu (Gaiberg, Germany), oligonucleotides, dNTPs and ready-to-use acrylamide solutions for SDS-gelelektrophoresis from Roth (Karlsruhe, Germany), screening solutions for crystallography from Hampton Research (Aliso Viejo, USA).
Bacterial strains, plasmids and protein overexpression
Escherichia coli strain DH5α [supE44 ΔlacU169 Φ80(lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA] was used for cloning, BL21(DE3)pLysS [hsdS gal(λclts857 ind1 Sam7 nin5 lacUV5-T7 gene1) (pLysS Camr)] for overproduction of RFs.
The T. thermophilus RF2 gene was amplified by PCR from T. thermophilus genomic DNA, cloned in pET30 and used for overexpression after transformation in E. coli BL21(DE 3)pLysS and addition of 4 mM IPTG. Four hours after induction, cells were harvested by centrifugation at 1000 g. The cells were stored at −20°C.
Escherichia coli K12 RF2(T246A) cloned in pET11a was a gift of Dr M. Ehrenberg (Uppsala University, Sweden) and used directly for expression or after subcloning into pET28c, to introduce a N-terminal His-tag. Overproduction was as described for the T. thermophilus protein. The T246A mutant of the K12 strain corresponds to the RF2 wild type of the B21 and MRE-600 strains (31) and provides a functional termination factor used in previous structural studies (17).
Purification of RFs
The S100 supernatant containing T. thermophilus RF2 was prepared as described previously (32), dialyzed against buffer containing 50 mM Tris–HCl, pH 7.5, 10 mM magnesium chloride and 1 mM 2-mercaptoethanol and applied to a Q-Sepharose FF column (5 × 50 cm). After washing the column with 2 l buffer, the proteins were eluted with a 4 l linear salt gradient from 0 to 500 mM potassium chloride in the same buffer. Fractions containing RF2, as judged by SDS–PAGE, were pooled and incubated for 20 min at 65°C, cooled to at 0°C for 30 min and centrifuged at 20 000 g to remove precipitated E. coli proteins. Proteins in the supernatant were precipitated by ammonium sulphate up to 70% (w/v) saturation, collected by centrifugation, dissolved in 30 ml 10 mM potassium phosphate pH 6.8 and dialyzed against the same buffer. Proteins from this step were separated on a hydroxyapatite column (0.5 × 12 cm) using 400 ml linear gradient from 10 to 500 mM potassium phosphate, pH 6.8. Fractions containing RF2 were pooled. After precipitation with ammonium sulphate, they were dissolved in 100 mM Tris–HCl pH 7.5, 100 mM potassium phosphate, 20 mM magnesium chloride, 2 mM 2-mercaptoethanol, 5% glycerol and dialyzed against the same buffer. Glycerol was added up to 50% v/v and the protein solution stored at −20°C. For crystallization the protein was kept precipitated and dialyzed just prior to the experiment.
For preparation of His-tagged E. coli RF2 the corresponding S100 supernatant was dialyzed against 50 mM sodium phosphate pH 7.0, 300 mM sodium chloride, 5% glycerol, 10 mM imidazol and applied to a Ni-NTA-agarose column (2.6 × 15 cm). After washing with 500 ml of the same buffer, bound proteins were step-wise eluted by 20 and 250 mM at imidazol pH 8.0, 50 ml each. The protein containing fractions were dialyzed against 50 mM sodium phosphate pH 7.0, 300 mM sodium chloride, 5% glycerol and precipitated with ammonium sulphate. Further purification by chromatography on hydroxyapatite followed the procedure described above for T. thermophilus RF2.
Proteins were analysed by SDS–PAGE. Purity was greater than 95%. Mass of T. thermophilus RF2 was confirmed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) and corresponded with 41.318 to the theoretically calculated value of 41.308 kDa.
Protein crystallization, X-ray data collection and structure determination
Thermus thermophilus RF2 was crystallized using the hanging drop vapour diffusion method at 18°C. Crystals were grown from 1.8 M ammonium formiate, 150 mM sodium thiocyanate and 100 mM sodium acetate at pH 4.3. The integrity of proteins obtained from dissolved crystals was analysed by SDS–PAGE using silver staining and by MALDI-TOF.
Crystals were tested for their diffraction quality on an in-house X-ray facility consisting of a rotating Cu-anode (Nonius FR 591, Bruker AXS, Karlsruhe, Germany) coupled to an image plate detector (mar345dtb, marresearch, Hamburg, Germany). Best crystals displayed a diffraction power to 3.4 Å and up to 2.6 Å (data collected at 100 K) at the ESRF synchrotron beamline ID 14-2 (Grenoble, France). All datasets were processed and scaled using XDS (33).
The structure determination was performed using Patterson search techniques. The search model comprised a truncated poly-Ala chain derived from the E. coli RF2 structure (PDB accession code 1GQE) and the best replacement solution was obtained with the program AMoRe (34). Model building was done with MAIN (35) and structure refinement was performed with CNS (36). The refined model displays good stereochemical values (Table 1). Some parts of domain I and the loop region around the GGQ-motif were ill-defined in the electron density and were modelled in analogy to the E. coli RF2 structure.
Table 1.
Crystallographic data for T. thermophilus RF2
| Dataset | RF2 |
|---|---|
| Total/unique refl. | 142 323/20 499 |
| Rsa | 0.068 (0.554) |
| Resolution (Å) | 20–2.50 (2.7–2.5) |
| Completeness (%) | 99.6 (100.0) |
| (I)/(σI) | 14.6 (3.3) |
| Model R/Rfree-factor (%)b | 26.5/31.2 |
| Rms deviation from ideal geometry | |
| Bonds (Å) | 0.0138 |
| Angles (°) | 2.0 |
| Ramachandran statistics (%) | |
| Most favored/additional/generously allowed/disallowed regions | 81.3/17.4/1.3/0.0 |
The values given in parentheses are for the highest resolution shell.
The Ramachandran statistics are given as defined by PROCHECK (51).
aRs = ∑h∑i|Ii(h) − 〈I(h)〉|/∑h∑i Ii(h); where i are the independent observations of reflection h.
bThe free R-factor was calculated from 5% of the data, which were removed at random before the refinement was carried out.
Differential scanning calorimetry
Differential scanning calorimetry (DSC) measurements were performed on a VP-DSC instrument (Microcal, Northampton, MA, USA) following general guidelines for calorimetry (37,38). The protein concentrations were determined with a Beckman DU spectrophotometer (Beckman Coulter, Krefeld, Germany) using the extinction coefficients at 280 nm of 43 215 M−1 cm−1 and 54 815 M−1 cm−1 for E. coli and T. thermophilus RF2, respectively (39). RFs were dialyzed at 4°C against 20 mM sodium cacodylate, pH 7.2, 300 mM sodium chloride and 1 mM 2-mercapthoethanol. Several other solvent compositions were tested, including different buffers (phosphate, cacodylate, HEPES), concentrations of sodium chloride (0–300 mM), concentration of 2-mercapthoethanol (0–10 mM) and concentration of EDTA (0–50 mM) in order to assure maximal reversibility of thermal transitions. Protein concentrations (8–50 μM) and scan rates (15–90 K/min) were varied as well. After DSC measurements, the protein integrity was verified by SDS–PAGE. Enthalpies of thermal transitions (ΔHcal) were obtained after numerical integration of the excess heat capacity. Alternatively, DSC data were analysed and deconvoluted by ORIGIN 6.0 software (OriginLabs, Northampton, MA, USA) with multiple non-two-state equations and Levenberg-Marquardt iterations for fitting. This analysis was only performed when the system under consideration was in equilibrium during denaturation.
Circular dichroism
Circular dichroism (CD) measurements were performed on a JASCO J-600 instrument (Jasco, Gross-Umstadt, Germany). Proteins were dialyzed either against 20 mM cacodylate buffer, pH 7.2, 300 mM sodium chloride, 1 mM 2-mercapthoethanol or 2 mM sodium phosphate buffer, pH 7.2. The latter was used for measurements below 200 nm. The final concentrations of T. thermophilus and E. coli RF2 were 1.4 and 2.95 μM, respectively. CD spectra represent the average of 12 consecutive scans. The measurements in the far-UV region were performed with a rectangular, 1 mm path length cuvette, with a bandwidth of 0.2 nm, response time of 0.5 s, and scan rate of 50 nm/min at 20°C. The mean residue ellipticity [Θ] was calculated according to Equation 1:
| 1 |
where M is the molecular mass (43.384 kDa for E. coli and 41.308 kDa for T. thermophilus RF2), N the number of residues (385 for E. coli and 365 for T. thermophilus RF2), d the path length in cm, c the concentration in mg/ml and Θobs the measured ellipticity in mdeg. Thermal transitions were followed by changes in the ellipticity at 220 nm with a scanning slope of 90 K/h.
Fluorescence and fluorescence quenching
Fluorescence measurements were performed at 20°C with a Perkin Elmer LS-50B spectrofluorometer (Rodgau-Jügesheim, Germany). Intrinsic tryptophan fluorescence at 345 nm after excitation at 295 nm and accumulation of four scan was recorded.
To study the rate of thermal-induced irreversibility proteins were dialyzed into 20 mM sodium cacodylate, 300 mM sodium chloride, 1 mM 2-mercapthoethanol at pH 7.2 and incubated at 60 and 90°C for the indicated time periods in tighty sealed PCR tubes at a final concentration of 2.95 and 1.4 μM for the E. coli and T. thermophilus RFs, respectively. The reactions were stopped by quick cooling on ice. Fluorescence emission was recorded between 400 and 600 nm upon selective excitation at 375 nm of the added ANS.
Isothermal guanidinum hydrochloride induced unfolding
Guanidium hydrochloride (GdmCl) -induced unfolding was monitored by measurement of the CD in the far-UV region at 220 nm in a 1 mm path length cuvette. Samples were preincubated for 12 h at 4°C in 20 mM cacodylate buffer, pH 7.2, 300 mM sodium chloride, 1 mM 2-mercapthoethanol and different concentrations of GdmCl. Measurements were performed at 20°C and analysed according to a three state unfolding mechanism (2):
| 2 |
where N is native, I intermediate and the U unfolded state. The intermediate state represents a structure with unfolded domain I and folded core composed of domains II·III·IV. Fully unfolded protein refolds upon dilution of GdmCl very rapidly, i.e. within the dead-time of manual mixing. A similar result was obtained for the protein unfolding. These experiments showed that folding/unfolding transitions of the secondary structure are fast, reversible and can be analysed by equilibrium thermodynamics. The unfolding curves showed a complex behavior: (i) pre-transition and post-transition dependence of protein ellipticities are non-linear functions; (ii) both curves showed a shoulder at low denaturant concentrations, indicating two independent unfolding events.
The observed protein ellipticity at 220 nm, Θ220, was fitted by the three-state Equation 3:
| 3 |
where , , and are the ellipticities of the native, intermediate and unfolded states, respectively. CD signals and denaturant dependences of the native and unfolded states can be obtained from fitting the post-transition and pre-transition baselines. The signal of the intermediate is not known. This parameter was therefore relaxed only upon the first rough curve fitting procedure, where only four parameters , , mNI and mNU, were used while the others were kept constant. In a second step, all parameters were relaxed. Chi-square was minimized to a value of 10−4 using Levenberg-Marquardt iterations and direct weighting of the residuals. Stability of the fit was verified by the introduction of small perturbation (±20%) of the initial parameters. Convergences of the fitting parameters into the original values indicate stability. Editing of equation, curve fitting and other mathematical procedure were performed using the ORIGIN 6.0 software (MicroCal, Inc.). Values obtained from the fitting of isothermal curves need to be handled with care as their accuracy might be affected by the parameters used in fitting procedure as well as by poorly resolved transitions.
Small angle X-ray scattering measurements
The synchrotron radiation X-ray scattering data were collected on the X33 camera of the EMBL on the storage ring DORIS III (DESY, Hamburg, Germany). Measurements were performed at 20°C in both 20 mM sodium cacodylate, pH 7.2, 300 mM sodium chloride, and 1 mM 2-mercaptoethanol as well as in 20 mM HEPES, pH 7.5, supplemented with 100 mM sodium chloride. Prior to the measurement, the solutions of T. thermophilus RF2 were adjusted to different protein concentrations ranging from 40 to 140 μM. Data were recorded using a MAR345 2D image plate detector at the sample-detector distance of 2.7 m and a wavelength λ = 1.5 Å, covering the range of momentum transfer 0.12 < s < 4.5 nm−1 (s = 4π sinθ/λ, where 2θ is the scattering angle). The data were averaged after normalization to the intensity of the incident beam and the scattering of the buffer was subtracted. The difference data were extrapolated to zero solute concentration following standard procedures. All data manipulations were performed using the program package PRIMUS (40). The forward scattering I(0) and the radius of gyration (Rg) were evaluated using the Guinier approximation assuming that at very small angles (s < 1.3/Rg) the intensity is represented as I(s) = I(0) exp[−(sRg)2/3]. These parameters were also computed from the entire scattering patterns using the indirect transform package GNOM (41). The molecular masses of the solute were evaluated by comparison of the forward scattering with that from reference solutions of BSA. The theoretical SAXS patterns from the existing crystallographic models of isolated RFs (PDB codes 1GQE, 1ZBT and 1RQ0) as well as from the X-ray and cryo-EM structures of RFs bound to ribosome and representing the open conformation (PDB codes 2B9M and 1ML5) were calculated using the program CRYSOL (42) and fitted to the experimental SAXS data. The quality of the fit is described by the discrepancy χ, with values closer to unity indicating better agreement between experimental and calculated scattering curves. The scattering from mixtures of different RF2 conformations was computed using the program OLIGOMER (40).
Protein Data Bank accession code
Atomic coordinates have been deposited in the Protein Data Bank (PDB) with the accession code 2IHR.
RESULTS
Cloning, overexpression and purification of T. thermophilus RF2
Cloning of the prfB gene into the pET30 expression vector and overexpression of T. thermophilus RF2 in E. coli followed standard techniques. Overexpression was strong, with no indication of interference with protein synthesis or general toxicity to the cell. Thermus thermophilus RF2 was found in the S100 fraction, could be bound tightly to a cation exchanger as expected for a protein with a calculated pI of 5.1 and was not denatured by a incubation at 65°C. The mobility of T. thermophilus RF2 in an SDS–PAGE was slightly higher than of the E. coli ortholog an effect observed previously for other thermostable proteins (32).
Crystal structure of T. thermophilus RF2
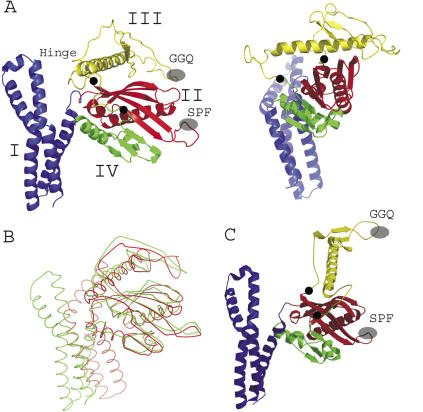
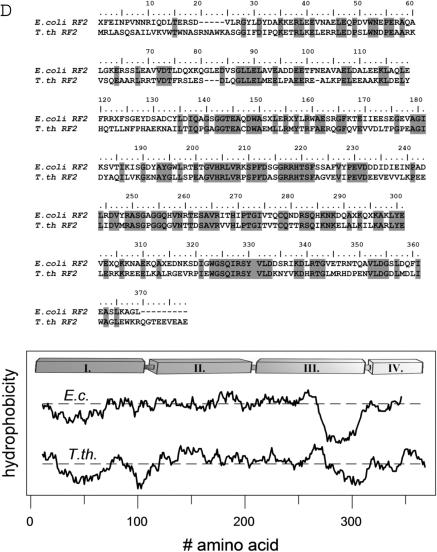
The crystal structure of T. thermophilus RF2 has a four domain architecture (Figure 1A). The N-terminal domain I (residues 5–105) consists of four α-helices and forms a bundle, which protrudes from the main body consisting of domains II·III·IV. A hinge region contains a short loop (residues 120–124) that connects domain I with domain II (residues 125–225) whose main structural element is a five-stranded antiparallel β-sheet with the SP(F/Y) motif in a loop connecting two of the β-strands. The antiparallel β-sheet covers a large part of a central α-helix (residues 120–143) running approximately parallel to the β-strands (Figure 1A). Domain III (residues 226–317) is dominated by long α-helix (α7; residues 262–294) but also contains an extended loop region which harbours the GGQ motif. The α-helix has a mixed hydrophilic/hydrophobic character and sticks out from the otherwise compact architecture of domains II·III·IV (Figure 1B). The N-terminal hydrophobic part of the helix interacts with a hydrophobic patch of the β-sheet of domain II, while its surface exposed C-terminal half is dominated by charged residues. This dual character of helix α7 is seen in the hydrophobic plots for E. coli and T. thermophilus RF2s, which overall possess a similar distribution of hydrophobicity/hydrophilicity along the polypeptide chains (Figure 1D). The C-terminal domain IV (residues 318–365) extends the antiparallel β-sheet of domain II and ends with two short α-helices (Figure 1A). The overall spatial arrangement places the GGQ and SP(F/Y) motifs in a distance of ∼20–25 Å.
Figure 1.
(A) Structure model of Thermus thermophilus RF2 in two orientations with the colour coded domains I (blue), II (red), III (yellow) and IV (green). Tripeptide motifs are highlighted [GGQ and SP(F/Y); grey ovals] as well as the possible hinge regions for movement of domain III (black circles). (B) Overlay of the ribbon structures of E. coli (red) and T. thermophilus (green) RF2 structure models. (C) Model of an elongated, ribosomal bound E. coli RF2 as determined by cryo-electron microscopy (26,27). (D) Alignment of primary structures and hydropathy plot of E. coli and T. thermophilus RF2s.
The structure of T. thermophilus RF2 is similar to the reported structures of E. coli RF2 (17), Thermotoga maritima RF1 (25) and a yet to be published structure of Streptococcus mutans RF1 (PDB accession code 1ZBT), all of which showing the unexpected short distance between the GGQ and SP(F/Y)/PxT motives. The relative orientation of domain I to the core of the structure build up by domains II·III·IV shows the largest variations between the T. thermophilus and E. coli RF2 structures (Figure 1C).
RF2 from T. thermophilus possesses structural homologies to the some ribosomal proteins as noted earlier for E. coli RF2 (17,25). Structural homologies exist also to several other proteins including aminoacyl-tRNA synthetases and other RNA-binding proteins (43). The homology is particularly evident for the central domains II·IV of RF2.
Structures of E. coli and T. thermophilus RF2 in solution
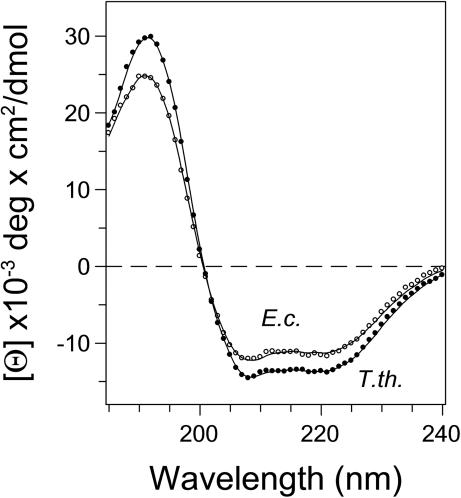
The secondary structures of E. coli and T. thermophilus RF2 in solution were studied by CD spectroscopy in the far-UV region. In accordance with the X-ray crystal structure indicate the CD spectra for both proteins a high content of α-helical structures, characterized by minima at 208 and 222 nm typical for α-helices (Figure 2). Analysis of the CD spectra by several algorithms (44) are in good agreement with the crystal structures. (i) For E. coli RF2 the highest scoring algorithm CDSSTR (45) provides 38% α-helices 17% β-strands, 19% turns and 27% unordered structures as compared with the crystal structure of E. coli RF2 (17) with 44.66 α-helix and 18.63% β-strands. (ii) In case of T. thermophilus RF2 the calculation from the CD spectra provided 46% α-helices 3% β-strands, 15% turns and 26% unordered structures. The content of secondary structure from the 3D model of T. thermophilus RF2 is 45% α-helices and 20% β-strands. A decrease in mean residue ellipticity of the E. coli RF2 by ∼1500 deg cm2/dmol at 220 nm might reflect subtle differences in secondary structure and/or higher structural dynamics (see Supplementary Data) compared with the T. thermophilus ortholog.
Figure 2.
Circular dichroism (CD) spectra of RF2 from E. coli and T. thermophilus in the far-UV region. CD spectra were measured in 2 mM sodium phosphate, pH 7.2, in a 1 mm path length cuvette. Concentrations of RF2 from E. coli (empty circles) and T. thermophilus (solid circles) were 2.95 and 1.4 μM, respectively. The fitted curves are shown by solid lines.
Isothermal denaturation of RF2 by GdmCl
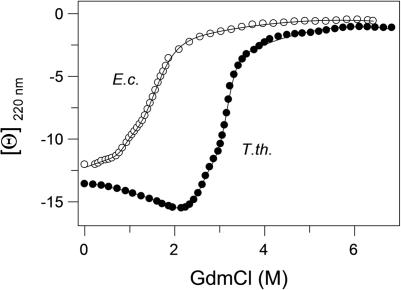
To compare the stability of E. coli and T. thermophilus RF2, isothermal GdmCl-induced unfolding was monitored by a change of ellipticity at 220 nm (Figure 3). Chemical unfolding of both proteins was fast and reversible (data not shown), which allowed to apply an equilibrium thermodynamic analysis. Apparent midpoints of the transitions for E. coli and T. thermophilus proteins were 1.5 and 3.2 M, respectively. The ellipticity dependences were fitted according Equation 3, resulting in a set of thermodynamic parameters for E. coli RF2 (mNI = 10.7 ± 0.3 kJ/mol/l, ΔGNI(H2O, 20°C) = 14 ± 1 kJ/mol, mND = 38 ± 6 kJ/mol/l, ΔGND(H2O, 20°C) = 32 ± 3 kJ/mol) and for T. thermophilus RF2 (mNI = 17 ± 2 kJ/mol/l, ΔGNI(H2O, 20°C) = 44 ± 4 kJ/mol, and mND = 26 ± 4 kJ/mol/l and ΔGND(H2O, 20°C) = 80 ± 10 kJ/mol). From this, the calculated free energies at 20°C indicate a 48 ± 13 kJ/mol higher stability of the T. thermophilus RF2 native state as compared with E. coli RF2. It was previously demonstrated that m-values correlate with the solvent accessible surface areas (ASAs) and thus have structural meaning (46). Additionally, the m-values of the unfolded states of E. coli RF2 and T. thermophilus RF2 correspond to changes in the ΔASA from 36 300 and 24 500 Å2, respectively. ΔASA for the E. coli protein (36 300 Å2) is in relatively good agreement with the calculated value based on the number of amino acid residues (32 900 Å2). The rather large difference in ΔASA values observed for the thermostable protein (measured 24 500 Å2, calculated 33 900 Å2) might indicate incomplete unfolding. The isothermal denaturation by GdmCl demonstrate that both RFs are well structured and stable (Figure 3).
Figure 3.
Guanidinium chloride induced unfolding of RF2 as monitored by the ellipticity at 220 nm. RF2 from E. coli (open circles) and T. thermophilus (solid circles) were measured at 1.5 μM protein concentration in 20 mM sodium cacodylate, pH 7.2 containing 300 mM NaCl and 1 mM β-mercaptoethanol at 20°C. Curves were generated by fitting the experimental data to Equation 3.
In an effort to assign the observed two transitions to unfolding of a particular structural units, we performed analysis of ASAs of the native and unfolded states of domain I and of the compact core consisting of domains II·III·IV using Surface Racer 4.0 (47). The data are in a good agreement with the experimentally obtained m-values (Figure 3) and the calculated ΔASA values based on amino acid composition (Table A). This indicates that the shoulder observed during the isothermal denaturation represents unfolding of domain I followed by cooperative unfolding of the compact core.
Thermal denaturation of RF2
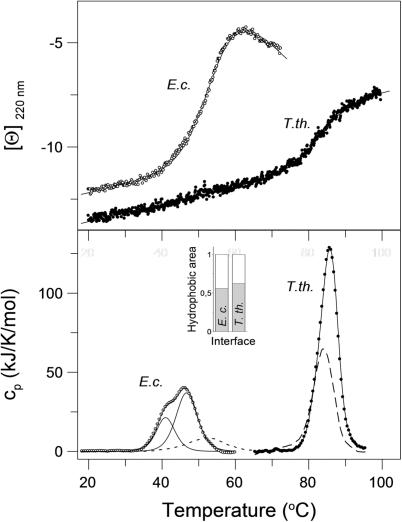
Thermal denaturation monitored by the ellipticity at 220 nm showed that the secondary structures of both RF2s denatured apparently cooperatively (Figure 4, upper panel). Midpoints of thermal transitions were 53.0 and 83.0°C for E. coli and T. thermophilus RF2, respectively. Detailed analysis using the second derivation of smoothed curves revealed a three-state transition of E. coli RF2 with transition temperatures at 48.5 and 53.7°C and a two-state transition of T. thermophilus RF2 at 83.0°C. The temperature-induced denaturations of secondary structures were completely reversible for both RFs.
Figure 4.
Temperature induced denaturation of E. coli and T. thermophilus RF2 monitored by CD and calorimetry. Upper panel: protein ellipticity in the far-UV region of CD at 220 nm. Scanning rate was 90 K/h, proteins concentration 1.5 μM in 20 mM sodium cacodylate, pH 7.2, 300 mM NaCl, and 1 mM β-mercapthoethanol. Ellipticity [Θ] is given in 10−3 deg cm2/dmol units. Lower panel: calorimetric scans of RFs (thick lines, E. coli and T. thermophilus) obtained upon subtraction of baselines. Second heating scans are shown as dashed (T. thermophilus) and dotted lines (E. coli). The scan rate was 90 K/min, the protein concentrations 24 μM. Curve fits of thermal denaturation based on the single and multiple two state processes are shown as solid lines. Inset: relative composition of ΔASA of interfaces between domain I and domains II·III·IV of RF2s from E. coli and T. thermophilus, respectively—polar amino acids (white box), nonpolar amino acids (grey box).
Thermal denaturation of E. coli RF2 as monitored by DSC resulted in a complex profile composed of two peaks (Figure 4, lower panel). The biphasic calorimetric trace was deconvoluted as two independent two-state transitions with transition temperatures of 45.6 and 51.3°C. The repeated scan using thermally denatured and cooled E. coli RF2 showed only one transition, shifted to higher temperature with Ttrs of 53°C. This indicates that one transition is partially (at least to 30%) reversible, while the shift to higher temperature points to a release of interface interactions that destabilized the more stable domain (48). Based on the structure, the amino acids distribution in the domains and the enthalpies of the transitions, it is reasonable to conclude that the first transition at 45.6°C corresponds to melting of domain I and the second transition at 51.3°C to the rest, namely, to the core. This is in accordance with results from CD-monitored isothermal denaturation.
Thermal denaturation of RF2 from T. thermophilus showed a single transition with Ttrs of 85.5°C and the reversibility of the transition obtained from the repeated heating scan was relatively high (∼60%). Even higher reversibility (70–80%) was achieved when the DSC scan was interrupted at half transition. From this it can be concluded that the denaturation of the T. thermophilus RF2 is controlled by a reversible melting.
Analysis of the calorimetric scans resulted in a calorimetric enthalpy of 840 ± 5 kJ/mol and a van't Hoff enthalpy of 656 ± 5 kJ/mol. The ratio of van't Hoff to calorimetric enthalpy r = 0.78 is significantly lower than 1.0 expected for a two state transition. This indicates an intramolecular process affecting the thermal transition (49). Apparently, the single observed peak in DSC may correspond to two non-resolved thermal transitions. Analysis of the interface between domain I and the core composed of domains II·III·IV identifies a peptide stretch of higher hydrophobicity for T. thermophilus RF2 (Figure 4, inset) as compared to its ortholog from E. coli. Strengthening the hydrophobic interactions at high temperatures, with the maximum at 70–80°C, explains this cooperative unfolding. A relatively large enthalpy and a shift of the transition temperature in the rescan indicate that, similarly to E. coli RF2, only one structural unit, most probably the core of T. thermophilus RF2, was partially reversible (see Supplementary Data).
Small angle X-ray scattering of RF2 from T. thermophilus
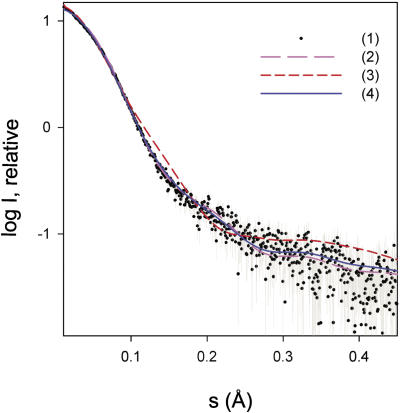
The conformation of RF2 in solution was studied by small angle X-ray scattering (SAXS), since the differences between the compact and the open conformation of the tertiary structure are expected to be sufficiently large. In agreement with Vestergaard et al. (29), aggregation in the concentrated solutions of E. coli RF2 was observed at 20°C preventing a SAXS analysis. In contrast, T. thermophilus RF2 remained under identical conditions monodisperse even at high concentrations. The molecular mass of the solute estimated from the obtained scattering curve (Figure 5) was 45 ± 5 kDa, which is in good agreement with the expected value calculated from the amino acid sequence. The radius of gyration (Rg) was 27.7 ± 1 Å at 20°C, which is somewhat smaller than the value of 29 Å reported for E. coli RF1 in solution (29).
Figure 5.
Small-angle X-ray scattering analysis of purified RF2 from T. thermophilus. (i) processed experimental SAXS pattern, dots with error bars; (ii) and (iii) scattering curves calculated from the closed X-ray structure of T. thermophilus RF2 determined and presented in this study and from the open X-ray structure of T. thermophilus RF2 when bound to a ribosome (PDB code 2B9M), respectively; (iv) scattering from a 78:22 mixture of the two conformations. The plot displays the logarithm of the relative scattering intensity as a function of the momentum transfer.
To perform a more detailed analysis, theoretical scattering curves were computed from the compact crystal structures of E. coli, T. thermophilus and S. mutans RF2, T. maritima RF1 as well as of the X-ray and the cryo-EM model of the open, ribosome bound T. thermophilus and E. coli RF2. These simulations were compared with the experimental SAXS data collected for T. thermophilus RF2. The theoretical SAXS curves derived from the open conformation models fit the experimental curve poorly, with the X-ray structure model of the open T. thermophilus RF2 being the best with a discrepancy χ of 2.6 (Figure 5, curve 3) and the cryo-EM model of the open E. coli RF2 being the worst with χ of 3.8 (curve not shown). In contrast, the theoretical curves derived from the compact X-ray structures fit the experimental data significantly better, with T. thermophilus RF2 being by far the best with χ of 1.5 (Figure 5, curve 2), followed by S. mutans and E. coli RF2 with discrepancies of 1.68 and 1.70, respectively (curves not shown). An even better fit, however, was obtained if a mixture of open and closed conformations was assumed. Using the program OLIGOMER and forming a weighted linear combination of the curves 2 and 3, a mixture containing 78% of closed and 22% of open conformation was found to provide the best fit to the experimental data with χ of 1.4 (Figure 5, curve 4).
DISCUSSION
As shown in this work the crystal structure of T. thermophilus RF2 is similar to the compact structures of E. coli RF2 (17) and RF1 from T.maritima (25). Minor deviations arise from different orientations of domain I in the molecules. These compact structures differ significantly from the structure of E. coli RF2 in complex with the ribosome (26,27). The main difference concerns the orientation of the large helix α7 and the adjacent loop forming domain III, which are either docked onto the β-sheet region of domain II·IV in the compact structure or extend to make contact to the peptidyl-transferase centre on the ribosome in the elongated structure (Figure 1). The recent 5.9 and 6.7 Å crystal structures of RFs bound to the 70S ribosome (22) together with the previous biochemical experiments (20,50) show beyond doubt, that the function of RFs on the ribosome requires the elongated RF contacting at the same time the decoding and the peptidyl-transferase sites. This parallels the situation of eRF1, which crystallized as isolated protein in an elongated form (24).
It remains striking that all isolated prokaryotic class I RFs, despite large differences in stability, structural dynamics, sequence, purification strategy, crystallization conditions, crystal symmetry and packing, crystallize in a similar compact form. For example, the RF2 crystals from E. coli and T. thermophilus have a P 1 21 1 symmetry with a very dense packing of molecules, while S. mutans and T. maritima RF1 with a tetragonal P 43 21 2 and a monoclinic P1 21 1 symmetry are much more dispersed. The latter contains actually three RF1 molecules in the asymmetric unit. All of this argues against the possibility that the compact structure of class I RFs is a crystal packing artifact. In contrary, this is an argument in support of the assumption that the compact state represents a natural conformation which prokaryotic RFs can adopt as well in solution. Otherwise it would not have occurred with such a high reproducibility. However, variations among the structures of prokaryotic RFs do exist, as can be seen in the different orientations of domain I, which generally has little contact to the remaining molecule. Its orientation is rather influenced by the inherent mobility and crystal packing contacts to neighbours. Flexibility in this domain is probably essential for interaction with the ribosomal GTPase centre and protein L11 (22). The orientation of domain I also differs between E. coli RF1 and RF2 when bound to the ribosome.
Domain III plays a central role in the function of bacterial class I RFs. It carries the GGQ consensus sequence, which is involved in the hydrolysis of the peptidyl residue from P-site bound peptidyl-tRNA. In all crystal structures of prokaryotic class I RFs the domain III is embedded in a compact structural unit formed by domains II·IV. Based on the B-factors of these structures, domains I and III in both E. coli and T. thermophilus RF2 possess increased dynamics, a fact also seen for T. maritima RF1, which contains three RF molecules in the asymmetric unit (25). This is in agreement with the relatively independent location of the domain I with respect to the rest of the molecule and with the required dynamics of domain III to allow ribosomal binding.
The expectation that the functionally essential movement of domain III can be monitored in unfolding experiments was not confirmed. However, the thermodynamic analyses of both thermal as well as isothermal denaturant-induced unfolding resulted in several important observations: (i) An initial and independent unfolding of domain I occurred for both RFs. (ii) Domains II·III·IV unfold in a cooperative manner at higher temperatures as well as higher denaturant concentration compared to domain I. This behavior agrees with the general temperature dependence of hydrophobic interactions that stabilize the core domains II·III·IV. This cooperativity has functional importance and was analogously observed in eRF1 (51). (iii) A partial unfolding of class I RFs at their respective physiological temperatures may be required for the interaction with the ribosomal A site. Moreover, it reflects inherent flexibility rather than stability.
Recent X-ray scattering experiments with RF1 from E. coli and human eRF1 measured larger radii of gyration than expected from models derived from prokaryotic RF crystal structures. This was taken as an indication for an elongated conformation of these proteins at room temperature. In contrary, the X-ray scattering analysis of T. thermophilus RF2 gave at 20°C a scattering profile more consistent with the compact form of the protein. The radius of gyration (2.8 nm) is larger than the simulated value of ∼2.5 nm for the X-ray models representing the closed form of RF2. This could be attributed to a slightly different orientation of domain I as discussed above rather than to a comparable large conformational change involving domains II·III·IV. However, a reasonable fit of the SAXS data was also obtained for a population of about 80% closed and 20% open RF2. Two conclusions can therefore be drawn: (i) the SAXS data confirms the postulate that the closed form is not a crystallographic artifact and reflects a structure RF2 can adopt in solution; (ii) it suggests inherent flexibility and a possible equilibrium of different conformational states. In as much a mixture of open and closed structures or a distribution of proteins with a varying degree of closure are present in solutions remains to be seen. While the SAXS data was measured at 20°C the thermodynamic analysis spans a wide range of temperatures up to the physiological temperature. This offers an indirect approach to access the conformational states of T. thermophilus RF2. The pre-transitional calorimetric baselines show no enthalpy associated process, which would correspond to an opening of the compact structure. Rather it reveals a highly cooperative conformational transition in absence of stable intermediates. In other words, a partially unfolded structure, which would correspond to an open conformation, is not stable below physiological temperatures in the absence of ribosomes.
The open structure is required for ribosomal binding and function, thus, the compact structure may have a physiological significance as well. As shown recently, the structure of E. coli RF1 adopts a closed conformation in complex with the modifying enzyme PrmC (52). This finding strengthens the biological importance of the closed form, as PrmC binds likely to the most stable solution conformation of the protein. The ratio of open to closed conformations is likely thermodynamically controlled to fit particular functions in the cell. Opening of the compact core exposes conserved hydrophobic residues at the interface between domains III and II·IV and increases the water accessible surface by 15% (27). It was suggested that the conformational transition of RF2 between free and ribosome-bound states are controlled by protonation of conserved histidines (28). Indeed, it can be expected that the overall negative charge of the ribosome will enhance the histidine protonation of RF2. It was, however, not possible to test this hypothesis since lowering the pH below pH 6 was accompanied by aggregation of RF2, which could be already taken as an indication of the proposed exposure of hydrophobic areas.
RF2 from T. thermophilus has with a Ttrs of 83°C a more than 30°C higher transition temperature and possesses with a free energy of 80 ± 10 kJ/mol at 20°C a 48 ± 13 kJ/mol higher stability as compared with E. coli RF2. Generally, the stability of proteins from thermophiles is attributed to an increased hydrophobicity, efficient atom packing, deletion or shortening of loops, avoidance of cavities, increase of surface area which is buried upon oligomerization, residue substitution, increased occurrence of proline residues in loops, decreased occurrence of thermolabile amino acid side chains, increased helical content, increased polar surface area, better hydrogen bonding, and last but not least stable salt bridge formation (53,54). The work presented here indicates that a combination of these factors is operating for RF2. With nearly the same number of total residues, the T. thermophilus RF2 has an increased number of Arg, Leu, Pro, while a significantly decreased content of Asn in comparison with its E. coli ortholog. The number of Gly residues, a typical residue replaced in thermostable proteins, remains unaffected. The high number of hydrophobic amino acids and the redistribution of charged amino acids in T. thermophilus as compared to E. coli RF2 reveal a strengthening of hydrophobic- and optimization of ionic interactions in T. thermophilus RF2. This is in accord with the generally accepted rules for protein stabilization and thermodynamic analysis of the stability of hydrophobic interactions, which reach their maximum around 75°C (55), the optimum growth temperature of T. thermophilus. Strengthening the hydrophobic interface at high temperatures and increasing the nonpolar area between the domains is most likely the basis for the tight coupling between structural domains in the thermostable RF2. This results in a single thermal transition. In addition, the cooperativity of thermal denaturation stabilizes labile domains in T. thermophilus RF2, making their partial exposure less favourable.
In conclusion, the compact structure reflects one physiological state of T. thermophilus and E. coli RF2 in absence of ribosomes, which require conformational changes to reach the ribosome-bound state. Class I RFs appear at their physiological conditions structurally destabilized facilitating these adaptations. Moreover, domains II·III·IV are closely interacting in the compact form and unfold cooperatively. Recent structural investigations indicate, that decoding of stop codons requires more contact sites than a simple peptide anticodon (22).
A binding model for the eukaryotic eRF1 based on photochemical crosslinking suggests a two state process: the first step is independent of the codon and relies on ribosomal proteins or rRNA, while the second isomerisation step takes only place if a UPuPu codon is present in the A-site (56). From the point of thermodynamic stability the elongated structure is clearly disadvantageous in solution as compared to the compact form. Dominating hydrophobic interactions at the domain II·III·IV interface have to be counterbalanced by ribosomal interactions to prevent formation of aggregates as observed during the calorimetric and SAXS measurements. On the other hand, an always elongated and active form of RF might result in constant binding competition with elongator-tRNA, which leads directly to premature termination as there are no preventive control mechanisms analogous to those operating during tRNA selection: aminoacyl-tRNA·EF-Tu·GTP ternary complex binding, GTP-hydrolysis and expulsion of non-coding aminoacyl-tRNA. Comparing the number of missense, frameshift and read-through events with premature termination shows that the latter occur much more infrequently. For eukaryotes it was found recently that complex formation between eRF1 and eRF3 prior to termination may stabilize an open conformation of eRF1, ensure rapid and efficient hydrolysis of peptidyl-tRNA (12) and may act in such way in proofreading to enhance the fidelity of termination (16).
Both the existence of a compact class I RF structure in which ribosomal binding elements are shielded and the controlled conformational change into an active form could therefore be of significant physiological importance in bacteria.
SUPPLEMENTARY DATA
Supplementary Data are available NAR Online.
Acknowledgments
We thank Dr Mans Ehrenberg for the clone of E. coli RF2(T246A), Dr Irina Dieser for crystallization and Norbert Grillenbeck for technical assistance. Financial support of the DFG (Sp 243/12-1) and the Slovak Grant Agency VEGA (1/3252/06) is gratefully acknowledged. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ogle J.M., Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 2.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 3.Gao H., Sengupta J., Valle M., Korostelev A., Eswar N., Stagg S.M., Van Roey P., Agrawal R.K., Harvey S.C., Sali A., et al. Study of the structural dynamics of the Escherichia coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 4.Frank J., Sengupta J., Gao H., Li W., Valle M., Zavialov A., Ehrenberg M. The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett. 2005;579:959–962. doi: 10.1016/j.febslet.2004.10.105. [DOI] [PubMed] [Google Scholar]
- 5.Schmeing T.M., Huang K.S., Strobel S.A., Steitz T.A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 6.Cochella L., Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaconu M., Kothe U., Schlunzen F., Fischer N., Harms J.M., Tonevitsky A.G., Stark H., Rodnina M.V., Wahl M.C. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Wilson D.N. Termination and ribosome recycling. In: Nierhaus K.H., Wilson D.N., editors. Protein Synthesis and Ribosome Structure. Germany: Wiley-VCH, Weinheim; 2004. pp. 367–395. [Google Scholar]
- 9.Kisselev L., Ehrenberg M., Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zavialov A.V., Mora L., Buckingham R.H., Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- 11.Hirokawa G., Kiel M.C., Muto A., Selmer M., Raj V.S., Liljas A., Igarashi K., Kaji H., Kaji A. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 2002;21:2272–2281. doi: 10.1093/emboj/21.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 13.Brown C.M., Tate W.P. Direct recognition of mRNA stop signals by Escherichia coli polypeptide chain release factor two. J. Biol. Chem. 1994;269:33164–33170. [PubMed] [Google Scholar]
- 14.Scarlett D.J., McCaughan K.K., Wilson D.N., Tate W.P. Mapping functionally important motifs SPF and GGQ of the decoding release factor RF2 to the Escherichia coli ribosome by hydroxyl radical footprinting. J. Biol. Chem. 2003;278:15095–15104. doi: 10.1074/jbc.M211024200. [DOI] [PubMed] [Google Scholar]
- 15.Seit-Nebi A., Frolova L., Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salas-Marco J., Bedwell D.M. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vestergaard B., Van L.B., Andersen G.R., Nyborg J., Buckingham R.H., Kjeldgaard M. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell. 2001;8:1375–1382. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- 18.Chavatte L., Seit-Nebi A., Dubovaya V., Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolosov P., Frolova L., Seit-Nebi A., Dubovaya V., Kononenko A., Oparina N., Justesen J., Efimov A., Kisselev L. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res. 2005;33:6418–6425. doi: 10.1093/nar/gki927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito K., Uno M., Nakamura Y. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 21.Mora L., Heurgue-Hamard V., Champ S., Ehrenberg M., Kisselev L.L., Buckingham R.H. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol. Microbiol. 2003;47:267–275. doi: 10.1046/j.1365-2958.2003.03301.x. [DOI] [PubMed] [Google Scholar]
- 22.Petry S., Brodersen D.E., Murphy F.V., Dunham C.M., Selmer M., Tarry M.J., Kelley A.C., Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 23.Seit-Nebi A., Frolova L., Justesen J., Kisselev L. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 2001;29:3982–3987. doi: 10.1093/nar/29.19.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song H., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F., Hemmings B.A., Barford D. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 25.Shin D.H., Brandsen J., Jancarik J., Yokota H., Kim R., Kim S.H. Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J. Mol. Biol. 2004;341:227–239. doi: 10.1016/j.jmb.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 26.Klaholz B.P., Pape T., Zavialov A.V., Myasnikov A.G., Orlova E.V., Vestergaard B., Ehrenberg M., van Heel M. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 27.Rawat U.B., Zavialov A.V., Sengupta J., Valle M., Grassucci R.A., Linde J., Vestergaard B., Ehrenberg M., Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 28.Ma B., Nussinov R. Release factors eRF1 and RF2: a universal mechanism controls the large conformational changes. J. Biol. Chem. 2004;279:53875–53885. doi: 10.1074/jbc.M407412200. [DOI] [PubMed] [Google Scholar]
- 29.Vestergaard B., Sanyal S., Roessle M., Mora L., Buckingham R.H., Kastrup J.S., Gajhede M., Svergun D.I., Ehrenberg M. The SAXS solution structure of RF1 differs from its crystal structure and is similar to its ribosome bound cryo-EM structure. Mol. Cell. 2005;20:929–938. doi: 10.1016/j.molcel.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 30.Kononenko A.V., Dembo K.A., Kiselev L.L., Volkov V.V. Molecular morphology of eukaryotic class I translation termination factor eRF1 in solution. Mol. Biol. (Mosk) 2004;38:303–311. [PubMed] [Google Scholar]
- 31.Dincbas-Renqvist V., Engstrom A., Mora L., Heurgue-Hamard V., Buckingham R., Ehrenberg M. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blank J., Grillenbeck N.W., Kreutzer R., Sprinzl M. Overexpression and purification of Thermus thermophilus elongation factors G, Tu, and Ts from Escherichia coli. Protein Expr. Purif. 1995;6:637–645. doi: 10.1006/prep.1995.1084. [DOI] [PubMed] [Google Scholar]
- 33.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 34.Navaza J. Amore—an automated package for molecular replacement. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- 35.Turk D. 1992. Weiterentwicklung eines Programms fuer Molekuelgraphik und Elektrondichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklaerungen. Technische Universität München. [Google Scholar]
- 36.Brunger A.T., Adams P.D., Clore G.M., Delano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Freire E. Thermal denaturation methods in the study of protein folding. Methods Enzymol. 1995;259:144–168. doi: 10.1016/0076-6879(95)59042-0. [DOI] [PubMed] [Google Scholar]
- 38.Lopez M.M., Makhatadze G.I. Differential scanning calorimetry. Methods Mol. Biol. 2002;173:113–119. doi: 10.1385/1-59259-184-1:113. [DOI] [PubMed] [Google Scholar]
- 39.Pace C.N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: a windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 41.Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 42.Svergun D., Barberato C., Koch M.H.J. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallography. 1995;28:768–773. [Google Scholar]
- 43.Holm L., Sander C. Protein-structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 44.Whitmore L., Wallace B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manavalan P., Johnson W.C., Jr Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 1987;167:76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- 46.Myers J.K., Pace C.N., Scholtz J.M. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsodikov O.V., Record M.T., Jr, Sergeev Y.V. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 48.Brandts J.F., Hu C.Q., Lin L.N., Mos M.T. A simple model for proteins with interacting domains. Applications to scanning calorimetry data. Biochemistry. 1989;28:8588–8596. doi: 10.1021/bi00447a048. [DOI] [PubMed] [Google Scholar]
- 49.Privalov P.L. Stability of proteins. Proteins which do not present a single cooperative system. Adv. Protein Chem. 1982;35:1–104. [PubMed] [Google Scholar]
- 50.Wilson K.S., Ito K., Noller H.F., Nakamura Y. Functional sites of interaction between release factor RF1 and the ribosome. Nature Struct. Biol. 2000;7:866–870. doi: 10.1038/82818. [DOI] [PubMed] [Google Scholar]
- 51.Mitkevich V.A., Kononenko A.V., Oparina N.Y., Kolosov P.M., Makarov A.A., Kisselev L.L. Thermal denaturation of eukaryotic class-1 translation termination factor eRF1. Relationship between stability of the eRF1 molecule and alteration of functional activity of its mutants. Mol. Biol. 2006;40:100–110. [PubMed] [Google Scholar]
- 52.Graille M., Heurgue-Hamard V., Champ S., Mora L., Scrima N., Ulryck N., van T.H., Buckingham R.H. Molecular basis for bacterial class I release factor methylation by PrmC. Mol. Cell. 2005;20:917–927. doi: 10.1016/j.molcel.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 53.Kumar S., Tsai C.J., Nussinov R. Factors enhancing protein thermostability. Protein Eng. 2000;13:179–191. doi: 10.1093/protein/13.3.179. [DOI] [PubMed] [Google Scholar]
- 54.Petsko G.A. Structural basis of thermostability in hyperthermophilic proteins, or ‘there's more than one way to skin a cat’. Methods Enzymol. 2001;334:469–478. doi: 10.1016/s0076-6879(01)34486-5. [DOI] [PubMed] [Google Scholar]
- 55.Makhatadze G.I., Privalov P.L. Energetics of protein structure. Adv. Protein Chem. 1995;47:307–425. doi: 10.1016/s0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- 56.Chavatte L., Frolova L., Laugaa P., Kisselev L., Favre A. Stop codons and UGG promote efficient binding of the polypeptide release factor eRF1 to the ribosomal A site. J. Mol. Biol. 2003;331:745–758. doi: 10.1016/s0022-2836(03)00813-1. [DOI] [PubMed] [Google Scholar]