Abstract
Genetic analysis of the Drosophila Bloom's syndrome helicase homolog (mus309/DmBLM) indicates that DmBLM is required for the synthesis-dependent strand annealing (SDSA) pathway of homologous recombination. Here we report the first biochemical study of DmBLM. Recombinant, epitope-tagged DmBLM was expressed in Drosophila cell culture and highly purified protein was prepared from nuclear extracts. Purified DmBLM exists exclusively as a high molecular weight (∼1.17 MDa) species, is a DNA-dependent ATPase, has 3′→5′ DNA helicase activity, prefers forked substrate DNAs and anneals complementary DNAs. High-affinity DNA binding is ATP-dependent and low-affinity ATP-independent interactions contribute to forked substrate DNA binding and drive strand annealing. DmBLM combines DNA strand displacement with DNA strand annealing to catalyze the displacement of one DNA strand while annealing a second complementary DNA strand.
INTRODUCTION
Bloom's Syndrome is a rare autosomal recessive genetic disorder that results in cancer-prone individuals with short stature, immune deficiency and sensitivity to sunlight [reviewed in (1)]. The hallmark feature of cells from Bloom's syndrome patients is an elevated frequency of sister-chromatid exchanges (SCE's) (2), which is often interpreted to indicate hyper-recombination in the absence of the Bloom's syndrome protein (BLM). The gene mutated in Bloom's syndrome encodes a helicase belonging to the RecQ family of DNA helicases (3). The RecQ family derives its name from the Escherichia coli RecQ helicase, Saccharomyces cerevisiae contains a single RecQ homolog (SGS1); Drosophila contains three RecQ homologs (mus309/DmBLM, RecQ4, and RecQ5); humans have a total of five RecQ helicases (RecQ1, RecQ2/WRN, RecQ3/BLM, RecQ4, and RecQ5). The WRN and RECQ4 genes are mutated in the heritable diseases Werner's and Rothmund–Thomson syndromes, respectively (4,5).
Purified, recombinant human BLM exists as a multimeric ring complex with 4- or 6-fold symmetry (6). BLM has 3′→5′ DNA helicase activity that is most robust on substrates that have forked or non-complementary DNA ends as well as synthetic X-junctions and G-quadruplex DNA (7,8) [also reviewed in (1)]. Similar 3′→5′ DNA helicase activity has been shown for most RecQ helicase family members, with WRN protein in particular having a substrate preference that is comparable to BLM. Besides helicase activity, several recent studies showed enhanced complementary DNA strand annealing by BLM and other RecQ helicases. Such activity has been demonstrated for BLM (9), WRN (10,11), RecQ5β (12), and RecQ1 (13). Strand annealing occurs most efficiently in the absence of ATP and is inhibited by single-strand binding proteins (SSBs) such as Replication Protein A (RPA) and SSB. In addition, BLM and WRN combine strand displacement and strand pairing to promote strand exchange (10).
A possible role for the Bloom's helicase in the recombination pathway has been demonstrated by an in vitro Holliday junction resolution activity that resolves double-Holliday junctions without crossing-over. The double-Holliday junction dissolution reaction requires Topoisomerase3α (Top3α) and is stimulated by a recently identified protein termed BLAP75 (BLM associated protein, 75 kD) (14–17). BLM, Top3α, and BLAP75 form a stable complex in vivo (15,18). However, BLM likely has additional in vivo functions besides Holliday junction resolution. For example, BLM interacts and co-localizes with the recombination repair factor Rad51 (19,20); this interaction is stimulated by treatment with ionizing radiation, suggesting a DNA damage-induced association between these two proteins. Holliday junctions are unlikely to contain Rad51, suggesting that BLM may function to resolve Rad51-containing structures independently of Holliday junction resolution.
The Drosophila mus309 gene encodes a RecQ family helicase that is thought to be most closely related to BLM (21,22). In this paper we refer to the mus309 gene product as DmBLM. Mus309 was originally identified in a screen for mutagen sensitivity (23). Mutant flies have reduced fertility, increased non-disjunction and chromosome loss, as well as sensitivity to DNA damaging agents and P element excision (22,24). Studies examining DNA repair following P element-induced DNA double-strand breaks in mus309 mutant flies demonstrate an increased frequency of deletions at DNA break sites and a defect in homologous recombination repair via the synthesis-dependent strand annealing (SDSA) pathway (24–26). This result is striking since most BLM phenotypes are interpreted to suggest an increased frequency of recombination in the absence of BLM, whereas in Drosophila, DmBLM is required specifically to promote the SDSA recombination repair pathway. In mus309 mutant flies P element excision is accompanied by a 6-fold increase in deletions flanking the P element donor site. These flanking deletions are suppressed in mus309; rad51 double mutant flies (26), thus indicating that lack of DmBLM results in degradation of one or both of the broken DNA ends in a Rad51-dependent manner, and suggests that DmBLM functions after Rad51-dependent strand invasion.
The requirement of DmBLM for efficient SDSA in Drosophila (25), suggests that DmBLM has a role in resolving recombination intermediates other than Holliday junctions. In the SDSA pathway of recombination a single strand invasion event results in formation of a displacement-loop (D-loop) structure as the free broken DNA end anneals to a complementary template DNA strand, often the sister chromatid (Figure 1B). The 3′ end of the broken DNA strand is then used to initiate copying of the template DNA. The D-loop structure is subsequently resolved and the newly synthesized strand re-anneals with the adjacent broken DNA end to yield the repair product. Thus, SDSA does not involve Holliday junction formation and a role for DmBLM in this recombination pathway indicates that DmBLM acts to resolve an SDSA-specific recombination intermediate.
Figure 1.
DNA substrates and synthesis-dependent strand annealing (SDSA). (A) DNA oligonucleotides and small duplex substrate molecules used in this study. (B) Model for DNA double-strand break repair by SDSA. Top strand contains the DNA double-strand break, bottom strand is the template DNA, typically the sister chromatid.
In this study we have isolated highly purified DmBLM from Drosophila tissue culture cells. Biochemical analysis shows that DmBLM is similar to human BLM. In addition, DNA binding assays indicate that DmBLM prefers forked substrate DNAs due to an ATP-independent interaction that is not observed with single-stranded or partial duplex DNA. Strand annealing is also an ATP-independent reaction, suggesting that DmBLM may interact weakly with two or more single-stranded DNAs. In the presence of a 100-fold molar excess of competitor oligonucleotide DNA DmBLM duplex unwinding is severely inhibited. Surprisingly, a 100-fold molar excess of complementary oligonucleotide does not inhibit duplex unwinding and also results in strand-swapping. Several observations suggest that DmBLM interacts with both the duplex substrate and complementary single-stranded DNA simultaneously during the strand-swapping reaction.
MATERIALS AND METHODS
Cloning and expression of DmBLM
The Py-DmBLM expression vector was created as follows. A 5′ segment of the DmBLM coding sequence was amplified by PCR to introduce a 5′ XhoI site and a six amino acid Py epitope tag sequence (EYMPME) flanked by glycine residues and with a 5′ methionine. The following primers were used (B15) GGCGGTCTCGAGCGTGATGGGTGAGTACATGCCAATGGAGGGTATGTCCAAGAAGCCTGTCGCGCAAAGAAAA and (B25) GATCTTCCTCTCATTTTGCGTCACTTTC, the PCR product was cleaved with XhoI and BspEI. A 3′ segment of the coding sequence was amplified by PCR to introduce a 3′ BamHI site. The following primers were used (B13) GATGCAAGCCGTCCTGGACGAA and (B14) GGCGGTGGATCCTTATTTTGATCCTGGCAGTGGCATTAAATCG, the PCR product was cleaved with NotI and BamHI. A final fragment was generated by cleaving the DmBLM coding sequence with BspEI and NotI to yield a 3575 bp internal fragment. The two PCR products and the large internal fragment were ligated into the pUC-MT-Hyg (pMTH) expression vector that was prepared by cleaving with XhoI and BamHI. pMTH contains the CuSO4-inducible metallothionien promoter as well as a hygromycin resistance gene for selecting stably transfected cells. Ligation yielded full-length clones that were designated pMTH-Py-DmBLM. Transfection of Drosophila tissue culture S2 cells with the pMTH-Py-DmBLM construct yielded CuSO4-inducible expression of full-length Py-DmBLM. Immunoblot analysis with both polyclonal anti-DmBLM antibody [see (27)] and monoclonal anti-Py antibody showed that Py-DmBLM was comparable in size to endogenous DmBLM, was CuSO4-inducible, and contained the Py epitope tag (data not shown). Transfected cells were selected with 50 μg/ml hygromycin to generate the Py-DmBLM-1 cell line that was used to express and purify the Py-DmBLM used in this study.
Purification of DmBLM
Four liters of Py-DmBLM-1 cells were grown to ∼5 × 106 cells/ml and treated with 500 μM CuSO4 for 16–20 h to induce Py-DmBLM expression. Cells were collected by centrifugation at ∼1000 g and washed once with cold (4°C) phosphate-buffered saline. The cell pellet was resuspended in ice cold hypotonic buffer A (10 mM Hepes-KOH, pH 7.6, 15 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 0.5 mM DTT, 100 μM phenylmethlysulfonyl fluoride (PMSF)] to a volume of 50 ml. Cells were allowed to swell for 20 min. on ice and then were lysed by dounce homogenization with 5–15 strokes of the B pestle of a Bellco dounce homogenizer. To the lysed cells was added 1/10 volume ice cold buffer B (50 mM Hepes-KOH, pH 7.6, 1 M KCl, 30 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 0.5 mM DTT, 100 μM PMSF) and the nuclei were pelleted by centrifuging at 8000 rpm for 10 min. in a Sorvall SS34 rotor. The nuclear pellet was resuspended with 20 ml isotonic buffer (9:1 buffer A to buffer B) and resuspended by gentle homogenization with the A pestle. The nuclei were then lysed by addition of 1/10 volume saturated (NH4)2SO4 and rotated at 4°C for 30min. The nuclear lysate was centrifuged for 1 h at 4°C in a Beckman Ti45 rotor at 35 000 rpm in a Beckman ultracentrifuge. The nuclear supernatant was precipitated by addition of finely ground (NH4)2SO4 (0.3 g/ml of nuclear supernatant) and the precipitated proteins were pelleted by centrifugation (12 000 rpm in a Sorvall SS34 rotor). Ammonium sulfate precipitates were stored at −80°C until later use. Before immuno affinity purification the ammonium sulfate nuclear pellet was resuspended in 10 ml IP buffer (20 mM Tris–HCl, pH 7.5, 0.5 M NaCl, 0.1% TritonX-100, 10% Glycerol, 0.5 mM DTT, 100 μM PMSF) and dialyzed against the same buffer (2× 2L, 2h each). The nuclear extract was incubated with ∼100 μL anti-Py antibody resin for 12–16 h at 4°C and washed with ∼100 volumes of IP buffer in a disposable 10 ml chromatography column (Bio-rad). The resin was transferred to a microfuge tube and Py-DmBLM eluted by incubating the resin with 2 volumes of elution buffer [IP buffer + 200 μg/ml Py peptide (EYMPME)] for 1 h on ice. The elution was repeated 1 or 2 additional times and the fractions pooled. Pooled fractions were diluted to 200 mM NaCl and loaded onto a 1.2 ml POROS-heparin column using a SMART system (Pharmacia). The column was loaded and washed with 200 mM NaCl buffer (50 mM Tris–HCl, pH 7.5, 0.2 M NaCl, 1 mM EDTA, 10% glycerol, 1 mM DTT) and eluted with a linear gradient of NaCl from 0.2 to 1.0 M. Py-DmBLM eluted at ∼400 mM NaCl. Pooled peak fractions were analyzed by silver stain (Figure 2) and the concentration determined by comparing to known amounts of BSA on a Coomassie stained SDS–PAGE gel. Working stock solutions (100 nM) were prepared by diluting pooled fractions into storage buffer (20 mM Tris–HCl, pH 7.5, 250 mM NaCl, 20% glycerol, 0.05% TritonX-100, 1 mM DTT) and kept at −80°C. Stock solutions were thawed once and never refrozen for subsequent use.
Figure 2.
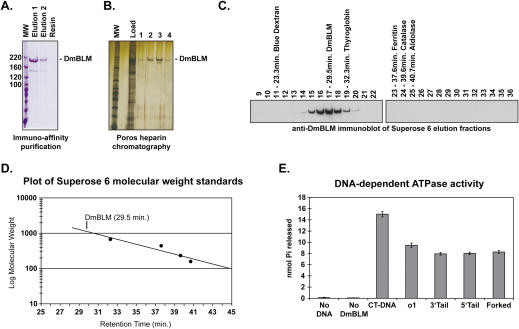
DmBLM is a large multimeric holoenzyme and DNA-dependent ATPase. (A) Immuno affinity purification of Py-DmBLM. The coomassie stained SDS–PAGE gel shows the first and second immuno affinity elution fractions and the material that remained bound to the anti-Py resin. (B) Silver stained SDS–PAGE gel of POROS-heparin fractions. The gel shows the pooled immuno affinity purified fractions (Load) and the POROS-heparin elution fractions (1–4). (C) Immunoblot of size exclusion chromatography fractions with polyclonal anti-DmBLM antibody. Py-DmBLM eluted as a single peak centered at fraction 17, the position and retention time of the protein molecular weight standards is shown above the fractions. (D) Plot of molecular weight standards used to calibrate the Superose 6 column. The position of the peak DmBLM elution at 29.5 min. is shown. (E) ATPase activity in the presence and absence of both DmBLM and DNA. CT-DNA is sonicated and denatured calf thymus DNA, all other DNA substrates are described in Figure 1A.
ATPase activity assay
ATPase activity was determined by orthophosphate detection using malachite green/phosphomolybdate. Assays were performed in helicase assay buffer (20 mM Tris–HCl pH 7.5, 4 mM MgCl2, 50 mM NaCl, 1 mM DTT) supplemented with 10 mM ATP, 5 nM DmBLM, and 100 ng of the indicated DNAs at 30°C for 60 min. Reactions (10 μL) were stopped by addition of 190 μL 100 mM EDTA and subsequently mixed with 750 μL of MGAM solution (1 part 4.2% ammonium molybdate in 4 N HCl, 3 parts 0.045% malachite green HCl in 0.1 N HCl). The reactions were allowed to develop for 5min. at room temperature before adding 100 μL 34% sodium citrate. Samples were then aliquoted into a 96 well plate in duplicate (250 μL) and the OD650 determined using a Molecular Devices Emax plate reader with SoftMax pro software. Orthophosphate concentration was determined by comparing the absorbance to known phosphate-containing standard reactions.
Size exclusion chromatography
Size exclusion chromatography was performed using a SMART system (Pharmacia) with a Superose 6 PC 3.2/30 column. The column was equilibrated with run buffer (50 mM Tris–HCl, pH 7.5, 500 mM NaCl, 0.1 mM EDTA, 10% glycerol, 1 mM DTT), all protein samples were applied to the column and eluted using the same buffer. The column was run at 40 μL/min. and 4°C. Pooled immuno affinity elution fractions of Py-DmBLM were loaded (∼40 μL/8 μg) and 50 μL fractions were collected. Protein standards used were, thyroglobin (669 kDa), ferritin (440 kDa), catalase (232 kDa), and aldolase (158 kDa). Void volume was determined by blue dextran. Plots of Log molecular weight versus retention time estimated the molecular weight of native Py-DmBLM to be 1170 kDa.
Preparation of radiolabeled DNA substrates
Purified oligonucleotide DNA was 5′ end labeled with [γ-32P]ATP (ICN crude, 167 mCi/ml, 7000 Ci/mmol). Labeled oligonucleotide was mixed with an equal amount of complementary unlabeled oligonucleotide, dH2O, and NaCl to give 100 mM NaCl final and was annealed by heating to 100°C and slow cooling to room temperature. Duplex, labeled DNAs were purified on a 12% native polyacrylamide gel run in TBE buffer at room temperature with a cooling fan. The duplex, annealed DNAs were cut from the gel and eluted by incubating with TEN-100 buffer (TE + 100 mM NaCl). Eluted DNAs were then concentrated and buffer exchanged on a microcon-10 micro-concentrator, using TEN-100 buffer to wash the DNAs. The concentration of purified duplex DNAs was determined by comparing 32P signal intensitiy between a known concentration of oligonucleotide from the initial labeling reaction and the purified duplex DNAs.
Helicase, strand-annealing, and strand-swapping assays
Assays were performed at 30°C in helicase assay buffer (20 mM Tris–HCl pH 7.5, 4mM MgCl2, 50 mM NaCl, 1 mM DTT) supplemented with 5 mM ATP, unless otherwise specified. Reactions were halted by the addition of an equal volume of 2× stop solution (100 mM EDTA, 1% SDS, 10% glycerol, and 0.1% bromophenol blue supplemented with 100 μg/ml proteinase K). Samples were resolved on 12% native polacrylamide gels run in TBE buffer (5 μL/lane) at 8mA at room temperature with a cooling fan. Gels were dried and visualized using a Fuji BAS-IIIs imaging screen and a Molecular Devices Typhoon 9400 scanner. The relative intensities of each DNA species were determined using ImageQuant software. For helicase assays the fraction of single-stranded DNA present at time = 0 was subtracted. SSB was purchased from Epicentre at 2 mg/ml concentration and was diluted to 600 nM in dilution buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA). Reactions that were compared to SSB-containing reactions contained an equal volume of dilution buffer without SSB.
DNA filter-binding assays
Filter-binding assays were performed using a Bio-Rad Bio-Dot SF apparatus as per the manufacturer's instructions. The slot-blot was assembled so that the samples first contacted a nitrocellulose (Bio-Rad) membrane to trap protein–DNA complexes and then a Hybond-N+ (Amersham) membrane to trap free DNA. Reactions (25 μL) were performed in helicase assay buffer (see above) in the presence or absence of 2 mM ATPγS for 30 min. at 30°C. Samples were then applied to the slot-blot apparatus and washed once with 500 μL wash buffer (20 mM Tris–HCl, pH 7.5, 4 mM MgCl2, 50 mM NaCl, 1 mM DTT). The membranes were then dried and signal intensities quantified as for helicase assay gels described above.
RESULTS
Purification and characterization of DmBLM
DmBLM was overexpressed in Drosophila tissue culture cells as an N-terminal polyoma epitope tag fusion protein (Py-DmBLM) and was highly purified from nuclear extracts by immuno affinity chromatography followed by POROS-heparin ion-exchange chromatography (Figures 2A and B). The POROS-heparin eluate is highly purified as evidenced by silver stain analysis (Figure 2B). Analytical gel filtration was used to determine the native molecular weight of Py-DmBLM. Immuno affinity purified Py-DmBLM applied to a superose 6 column elutes after the void volume and well before thyroglobin (669 kDa) (Figure 2C). By comparing the retention time of Py-DmBLM to the various protein standards shown in Figure 2C we predicted the molecular weight of Py-DmBLM to be ∼1170 kD, approximately seven times the size of monomeric Py-DmBLM (167 kDa) (Figure 2D). Py-DmBLM elutes from the Superose 6 column as a single protein species, indicating that the purified Py-DmBLM does not contain any other co-purifying proteins (Data not shown). Furthermore, we were unable to detect any monomeric DmBLM in both immuno affinity and POROS-heparin purified fractions (Figure 2C and data not shown, monomeric Py-DmBLM should elute in fractions near aldolase), indicating that the Py-DmBLM preparation exists exclusively as a large multimeric holoenzyme. These data are consistent with a previous study that showed human BLM, expressed in yeast, also forms a high molecular weight complex. Human BLM was additionally shown to form a ring-shaped structure with 4- or 6-fold symmetry, the similar elution profile of DmBLM from a superpose 6 column suggests that DmBLM likely forms a similar multimeric enzyme complex.
The ATPase activity of DmBLM was determined in the presence of sonicated, denatured calf thymus DNA (CT-DNA), a 50mer oligonucleotide (o1), and various oligonucleotide duplex substrates used in this study. Activity was highest in the presence of CT-DNA while single-strand oligonucleotide provided slightly better (∼19%) stimulation than an oligonucleotide duplexes (Figure 2E). Similar results were seen with a different oligonuleotide and oligonucleotide duplex pair (data not shown), indicating that single-stranded DNA stimulates ATPase activity to a slighter higher degree than partial duplex DNA, even when the duplex is an efficient substrate for DmBLM (see below).
DmBLM duplex unwinding
DmBLM helicase activity was compared using oligonucleotide substrates with a 3′ tail (3′ Tail) or a 5′ tail (5′ Tail). Robust helicase activity was observed with the 3′ Tail substrate while the 5′ Tail substrate was mostly unaffected by DmBLM (Figure 3A and B). Addition of SSB to the reaction resulted in a significant enhancement of helicase activity (Figure 3A and B). Interestingly, our DmBLM preparation showed an initial rate of unwinding (0–0.5 min.) that was 26.6 nM substrate/μM DmBLM/min. Addition of SSB to the reaction enhanced this rate to 38.5 nM substrate/μM DmBLM/min and drove the reaction further, to 72% unwinding as compared to 58% unwinding in the absence of SSB. These data clearly show that DmBLM, like human BLM, has 3′→5′ helicase activity that is stimulated by SSB. Stimulation by SSB may result from inhibiting the intrinsic strand-annealing activity of DmBLM, as shown below.
Figure 3.
DmBLM is a 3′→5′ DNA helicase (A) Duplex unwinding. Assays were performed with the indicated substrate DNAs. Reactions contained 1 nM substrate DNA and 20 nM DmBLM at 30°C and were started by the addition of ATP, 10 μL aliquots were removed at the indicated times and stopped by mixing with an equal volume of stop solution. (B) Plot of data from (A). (C) Duplex unwinding. Assays were performed with the indicated substrate DNAs. Reactions contained 1 nM substrate DNA and 2 nM DmBLM at 30°C and were started by the addition of radiolabeled substrate DNA, 10 μL aliquots were removed at the indicated times. (D) Plot of data from (C) and two additional identical experiments. (E) Plot of % radiolabeled DNA bound in filter-binding assays. Reactions contained the indicated substrate DNAs at 0.5 nM and increasing concentrations of DmBLM, 2 mM ATPγS was present, as indicated. Samples were applied first to a nitrocellulose membrane to recover protein-bound DNA then to a Hybond-N+ membrane to recover any unbound radiolabeled DNA that did not bind the nitrocellulose. The fraction bound (% Bound) was determined by taking the ratio of bound (nitrocellulose) to total (nitrocellulose and Hybond-N+) and subtracting the fraction bound at 0 nM DmBLM. (F) Summary of DNA binding at 10 nM DmBLM. Data from four independent filter-binding reactions is shown.
Purified human BLM prefers forked or bubble substrates to 3′ tail substrates, we compared DmBLM activity on the 3′ Tail substrate and an identical substrate with a 12 nt unpaired flap on the 5′ end of the unlabeled strand (Forked, see Figure 1A). DmBLM unwound the Forked substrate with a higher initial rate and to a greater extent than the 3′ Tail substrate (Figure 3C and D). The increased rate of unwinding with the forked substrate may occur because the unpaired ends destabilize the duplex and promote unwinding or because DmBLM has a greater affinity for the Forked substrate.
In order to explore the basis of this difference in substrate preference we used a DNA filter-binding assay to examine DmBLM-substrate affinity. DmBLM was incubated with Forked and 3′ Tail radiolabeled substrate DNAs in helicase assay buffer and assayed by slot-blotting. DmBLM failed to bind any of the 3′ Tail substrate in the absence of ATP, while a significant fraction was retained in the presence of non-hydrolysable ATPγS in a protein-dependent manner (Figure 3E). In contrast, DmBLM bound the Forked substrate in the absence of ATP and retained a greater fraction (∼10%) of the Forked substrate than the 3′ Tail substrate in the presence of ATPγS (Figure 3E). Although DmBLM showed greater affinity for the Forked DNA at all concentrations tested, the reproducibility of this difference was further examined at 10 nM DmBLM (Figure 3F). These data indicate that the difference in substrate specificity is due, at least in part, to the greater affinity of DmBLM for the Forked DNA substrate. In addition, DmBLM binds the Forked DNA substrate in the absence of ATP, indicating that the presence of a forked junction stabilizes the DmBLM–DNA interaction. Binding in the absence of ATP further suggests that DNA binding occurs independently of the DmBLM helicase domain.
DmBLM strand annealing
Annealing of the o1 and o3 oligonucleotides yields the Forked duplex substrate used in the previously described unwinding assays. Strand annealing reactions were performed with equimolar amounts (1 nM each) of o1 and o3 in the absence of DmBLM, in the presence of DmBLM, in the presence of DmBLM and ATP, and in the presence of DmBLM and SSB (Figure 4A and B). DmBLM stimulated the rate of annealing by complementary oligonucleatides 100-fold, indicating that DmBLM is able to bind two or more single-strand DNAs simultaneously. Addition of ATP to the reaction had little effect on the initial rate of strand annealing, yet resulted in a reduced amount (∼50%) of total duplex product (Figure 4A and B). The reduced yield in the presence of ATP is likely caused by unwinding of the annealed product by DmBLM. Addition of SSB completely blocks strand annealing, resulting in a rate of annealing that is less than that in the absence of DmBLM protein. This suggests that SSB inhibits annealing by preventing the pairing of complementary DNAs. These data further suggest that SSB stimulates unwinding of duplex substrates by DmBLM by preventing DmBLM-dependent strand re-annealing.
Figure 4.
DmBLM anneals complementary oligonucleotide DNAs (A) DNA strand annealing. Assays contained 1nM each of radiolabeled o1 and unlabeled o3 oligonucleotides and were incubated at 30°C with 10 nM DmBLM, 10 mM ATP and/or 60 nM SSB, as indicated. Reactions were started by the addition of radiolabeled DNA and 10 μL aliquots were removed at the indicated times. (B) Plot of data from A, including time points not shown in (A). (C) Plot of % radiolabeled DNA bound in filter-binding assays. Reactions contained radiolabeled o1 oligonucleotide at 0.5 nM, and were otherwise performed as in Figure 3.
DNA filter-binding assays were unable to detect an interaction between DmBLM and single-strand DNA in the absence of ATP (Figure 4C), suggesting that the DNA–DmBLM interaction that drives strand annealing is less stable than the ATP-dependent DNA–DmBLM interaction that occurs during strand unwinding. DmBLM bound single-strand DNA in the presence of ATPγS to a similar degree as 3′ Tail binding (compare Figure 3E to Figure 4C). Furthermore, single-stranded DNA promoted the ATPase activity of DmBLM ∼19% more efficiently than the 3′ Tail substrate (Figure 2E), thus indicating that DmBLM likely translocates on single-stranded DNAs in an ATP-dependent manner as well as on partial duplex DNAs.
Although DmBLM does not interact with single-stranded oligonucleotide DNA in a filter-binding assay, DmBLM can catalyze strand annealing and therefore must interact with free single stranded DNA, albeit weakly. The ATP-independent interaction between DmBLM and the Forked substrate suggests that multiple weak DmBLM–DNA interactions are sufficient to stabilize binding enough to detect this interaction by filter binding (Figure 3E). However, single-stranded DNAs must contain some complementary sequence in order to stabilize DmBLM binding, as no stable interaction is observed between DmBLM and non-complementary single-stranded DNAs in the absence of ATP.
DmBLM-dependent strand swapping
While examining the effect of unlabeled competitor DNAs on DmBLM helicase activity we found that a 100-fold molar excess of non-compementary oligonucleotide DNA completely inhibits strand unwinding (Figure 5A, 100× o6) while complementary oligonucleotide DNA does not (Figure 5A, 100× o1). Single-stranded DNA stimulates the ATPase activity of DmBLM as well as 3′ Tail duplex DNA (Figure 2E) and interacts with DmBLM as well as 3′ Tail duplex DNA (Figure 3E and 4C). Therefore, single-stranded oligonucleotide DNA competes with 3′ Tail partial duplex for DmBLM unwinding. Surprisingly, a 100-fold excess of complementary oligonucleotide DNA does not inhibit strand displacement (Figure 5A, 100× o1). One possible explanation for this observation is that the unlabeled complementary oligonucleotide is efficiently swapped with the labeled strand of the duplex substrate, thereby resulting in strand displacement at a similar rate and to a similar extent as unwinding in the absence of competitor DNA altogether. In order to test this hypothesis a 100-fold excess of o3 oligonucleotide DNA was included in an unwinding reaction (Figure 5A, 100× o3). The presence of complementary o3 oligonucleotide results in strand swapping between the unlabeled o2 oligonucleotide in the 3′ Tail substrate and the free o3 oligonucleotide to yield Forked duplex DNA product (o1 + o2 = 3′ Tail, o1 + o3 = Forked). While a 100-fold excess of o3 results in strand swapping exclusively, an equimolar amount of 3′ Tail and o3 oligonucleotide results in similar amounts of 3′ Tail, Forked, and single-stranded DNA species (Figure 5B). Since the Forked product is also a substrate for DmBLM it is likely that this reaction reaches equilibrium between the unwinding of substrate DNAs to yield single-stranded DNA and strand-swapping to yield Forked and 3′ Tail products. It is notable that unwinding and strand-swapping products form with similar kinetics in this reaction (Figure 5B) The presence of a 10-fold molar excess of o3 (10 nM) results mostly in strand-swapping while a 100-fold molar excess of o3 (100 nM) results in strand-swapping exclusively (Figure 5C). Strand-swapping reactions require both DmBLM and ATP, indicating that DmBLM-dependent strand unwinding is necessary to achieve strand swapping (Figure 5A and C).
Figure 5.
DmBLM combines duplex unwinding with strand annealing. (A) Complementary oligonucleotide DNA results in strand-swapping. Assays were performed as in a helicase reaction with the exception that various oligonucleotide DNAs were present at a 100-fold (100×) molar excess over substrate DNA. Reactions contained 1 nM 3′ Tail substrate DNA, 5 nM DmBLM and were started by the addition of ATP and oligonucleotide DNA (as indicated). Substrate and product DNAs and their constituent oligonucleotides are indicated. (B) Strand swapping in the presence of equimolar (1 nM each) 3′ Tail substrate and o3 oligonucleotide. (C) Strand-swapping is concentration-dependent and ATP-dependent. Assays were performed as in a helicase reaction with the exception that o3 oligonucleotide DNA was present at the indicated fold molar excess over substrate DNA. Reactions contained 1 nM 3′ Tail substrate DNA, 5 nM DmBLM and were started by the addition of ATP and o3 oligonucleotide (as indicated). (D) Non-complementary oligonucleotides inhibit duplex unwinding. Helicase assays were performed with 1 nM 3′ Tail substrate DNA and 5 nM DmBLM at 30°C and the indicated times. Reactions were started by the addition of ATP and the indicated non-complementary oligonucleotide DNAs (o6 or o7). (E) Complementary oligonucleotide DNA enhances strand displacement. Strand displacement may occur by duplex unwinding (helicase) to yield single-stranded DNA product or by strand swapping to yield forked duplex DNA product. The rate of these reactions was examined during the 0–2 min time interval of the experiments shown in parts C and D. 1×, 10×, and 100× indicates the fold molar excess of oligonucleotide relative to the 3′ Tail substrate DNA (1, 10 and 100 nM, respectively).
The most striking feature of the strand-swapping reaction is that it occurs efficiently at high concentrations of oligonucleotide DNA that otherwise inhibit DmBLM unwinding reactions. Since DmBLM presumably interacts with non-complementary oligonucleotide DNA as well as complementary oligonucleotide DNA then DmBLM likely interacts with the o3 oligonucleotide DNA and the 3′ Tail duplex DNA simultaneously during the strand-swapping reaction. As shown in Figure 5D, incubation with non-complementary oligonucleotide DNA inhibits DmBLM helicase activity. Inhibition was concentration-dependent with a ∼20-fold reduction in rate occurring between 1 and 100 nM competitor single-stranded DNA (Figure 5D and E). In contrast, the rate of strand swapping in the presence of a 10-fold molar excess of complementary single-stranded DNA was two to four times the rate of unwinding in the presence of 10-fold excess non-complementary DNA (Figure 5E). While a 100-fold excess complementary single-stranded DNA resulted in a rate of strand swapping that was 10-fold higher than the rate of unwinding in the presence of a 100-fold excess non-complementary single-stranded DNA (Figure 5E). These differences are not due to differences in the length of the single-stranded DNA used, since strand-swapping occurs with a 50mer oligonucleotide (o3), while competition occurs in the presence of 52mer oligonucleotides (o6 and o7). These data indicate that the presence of complementary single-stranded DNA stimulates strand displacement by DmBLM. Free complementary single-stranded DNA may stimulate strand displacement by an allosteric effect on DmBLM or by trapping product DNA during the unwinding reaction to drive the reaction forward.
The relative enhanced rate of strand-displacement in strand-swapping reactions indicates that DmBLM unwinding activity is stimulated in the presence of a single-stranded complementary DNA. This may occur because such triplex DNA structures are a favored substrate for DmBLM in vivo. In fact, a recent report indicates the mobile D-loops, generated by RecA-mediated strand-invasion and subsequently de-proteinated, are a preferred substrate for human BLM (28). Unwinding of a D-loop structure occurs with a complementary single-stranded DNA in close proximity to the displaced strand, therefore suggesting that the presence of the third unpaired strand may stimulate duplex unwinding, as in the strand-swapping reaction. These observations may also indicate that DmBLM is highly non-processive in the absence of a free complementary DNA strand. Therefore, the presence of complementary DNA stimulates DmBLM processivity enough to overcome the effects of a 100-fold excess of competitor DNA (Figure 5E).
DISCUSSION
Biochemical activities of DmBLM
In many respects DmBLM closely resembles human BLM; DmBLM exists as a large multimeric holoenzyme, is a DNA-dependent ATPase, is a 3′→5′ DNA helicase that prefers forked DNA substrates, and anneals complementary single-strand DNAs. DmBLM binds forked DNAs in the absence of ATP, suggesting a mechanism for the substrate specificity shown by DmBLM and other RecQ helicases. In addition, DmBLM performs strand swapping at a rate that is comparable to strand displacement and is able to drive strand unwinding in the presence of excess single-stranded DNA. It is difficult to determine whether the strand-annealing and strand-swapping activities of DmBLM in vitro are functionally significant in vivo. It is certainly possibly that this activity simply reflects an ability to bind two single-stranded DNAs simultaneously with the result that annealing is enhanced. Furthermore, the inhibitory effect of ATP and SSB on the strand annealing reaction suggests that this activity may not occur in vivo. However, the strand annealing activity surely indicates that DmBLM interacts with single-stranded DNA and that this interaction may be of specific functional relevance. Although strand annealing has been widely observed in RecQ-family helicases (see Introduction), this activity is not observed with E.coli UvrD helicase or viral NS3 helicase, both of which are 3′→5′ helicases (10). The extreme C-terminal domain of human BLM is required for strand annealing (amino acids 1267–1417), indicating that this activity involves a DNA-binding domain that is distinct from the conserved RecQ helicase domain (9). In addition, strand swapping occurs in the presence of ATP and with kinetics comparable to duplex unwinding. Therefore, the preference for forked end DNA substrates, the ability to anneal single-stranded DNAs, and the strand-swapping activity of DmBLM all suggest that DmBLM coordinates a free single-stranded DNA concurrently with ATP-dependent DNA binding and helicase activity.
Role of BLM in DNA recombination
The human BLM helicase and its yeast homolog Sgs1 are both thought to suppress recombination. This idea is based on the observation that loss of function in either of these proteins results in an increased frequency of recombination. In human cells this is seen as an increased frequency of SCEs (2) while in yeast there is an increased frequency of interchromosomal recombination, intrachromosomal excision recombination, and ectopic recombination (29). However, BLM and Sgs1 may not act to suppress recombination in general, but rather to prevent certain recombination products from forming. Sgs1 mutant yeast have an increased frequency of recombination that results in crossing-over between damaged and template DNA strands (30). Therefore the increased frequency of SCEs in Bloom's syndrome cells may result not from increased recombination, but rather from a failure to resolve recombination without crossing-over. Furthermore, DmBLM and Sgs1 are required for SDSA and heteroallelic recombination reactions respectively (25,31), indicating that these proteins also function to promote recombination. The resolution of Holliday junctions by human BLM to yield non-crossover products provides a mechanism whereby BLM may act in the recombination pathway to promote recombination that does not result in crossing-over. This proposed role for human BLM has enjoyed a wealth of supporting biochemical data including the dependence of the reaction on Top3α (14), the requirement of a functional HRDC domain (32), and the stimulation of the Holliday junction dissolution reaction by BLAP75 (16,17). However, a recent study showed that crossing-over in sgs1 mutants can be rescued by expression of helicase-defective Sgs1 (33). Therefore, Bloom's helicase activity is not required to suppress crossing-over in yeast and may be important for other aspects of DNA recombination.
DmBLM in synthesis-dependent strand annealing
The Holliday junction dissolution activity of Bloom's fails to account for a role of DmBLM in promoting recombination repair by SDSA, as Holliday junctions are not thought to form during recombination by SDSA. Analysis of SDSA in Drosophila suggests that repair may involve several rounds of strand invasion and strand displacement (34). In this process one or both ends of a DNA break invade the template DNA independently and are resolved from the template DNA independently. If the resolved strands fail to re-anneal and complete the repair process, then a subsequent round of strand invasion and synthesis occurs. Since mutation of spn-A/rad51 suppresses flanking deletions in mus309 mutants and DmBLM acts to promote recombination, DmBLM likely functions after strand invasion in the recombination pathway. Therefore, in the SDSA pathway, DmBLM may promote copying of the template DNA by unwinding ahead of the D-loop or DmBLM may act to displace the invading strand and resolve the D-loop (see Figure 1B). Alternatively, DmBLM could promote the strand-annealing step of the reaction, although this is unlikely since DmBLM is not required for repair by single-strand annealing in Drosophila ((35) and B. Weinert, unpublished data). Failure at any of these steps during SDSA could cause the observed defects in recombination repair and the appearance of flanking deletions at DNA break sites. However, only a failure to displace the invading strand and resolve chromosome exchanges is likely to result in the non-disjunction and chromosome loss seen in mus309 mutant flies.
BLM and displacement-loop resolution
The biochemical data presented here show that DmBLM interacts with free single-stranded DNA and that free complementary single-stranded DNA enhances DmBLM helicase activity and results in strand swapping. The strand-annealing and strand-swapping activities likely only occur in our in vitro reactions; however, these activities may indicate that DmBLM acts on three-stranded substrates such as D-loops in vivo. DmBLM could resolve a D-loop structure by combining strand-displacement and strand-annealing activities to displace the invading strand while insuring re-annealing of the template DNA. In addition, the three-stranded D-loop structure presents several forked DNA junctions that may provide a preferred binding site for DmBLM. This proposed function is consistent with genetic observations in mus309 mutant flies showing increased flanking deletions at DNA breaks [a failure to resolve the D-loop may result in an endonuclease cleaving the invading strand to yield a flanking deletion (26)] and the requirement of Rad51 in mus309 mutant flies for the formation of these deletions [suggesting that DmBLM functions after strand invasion (26)]. In addition, a recent study shows that mobile D-loops (generated by RecA-mediated strand invasion) are a preferred substrate for human BLM (28). Resolution of D-loops during SDSA repair is functionally equivalent to Holliday junction resolution in that the damaged DNA strand is resolved from the undamaged template DNA strand. It may be that DmBLM (and human BLM) functions in the resolution of both of these types of recombination intermediates with similar biological consequences. Therefore, the type of recombination intermediate acted on by BLM would depend on which recombination pathway is used to repair the DNA double-strand break. Mounting evidence, mainly from S.cerevisiae, suggests that recombination proceeds by distinct pathways with different repair outcomes, supporting the notion that recombination might proceed by both double-Holliday junction and SDSA-type mechanisms [reviewed in (36)].
Acknowledgments
We would like to thank members of the Rio lab for insightful discussion and technical assistance; in particular we thank Marco Blanchette who was always willing to discuss the primary data and troubleshoot technical challenges, Jerod Ptacin for discussion and the ATPase assay protocol, and Aurora Trapane for cell culture media. Special thanks to Stuart Linn for careful and critical reading of the manuscript. This research was supported by NIH grant #R01GM48862. Funding to pay the Open Access publication charges for this article was provided by NIH GM48862.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bachrati C.Z., Hickson I.D. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaganti R.S., Schonberg S., German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl Acad. Sci. USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 4.Yu C.E., Oshima J., Fu Y.H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 5.Kitao S., Shimamoto A., Goto M., Miller R.W., Smithson W.A., Lindor N.M., Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nature Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 6.Karow J.K., Newman R.H., Freemont P.S., Hickson I.D. Oligomeric ring structure of the Bloom's syndrome helicase. Curr. Biol. 1999;9:597–600. doi: 10.1016/s0960-9822(99)80264-4. [DOI] [PubMed] [Google Scholar]
- 7.Karow J.K., Chakraverty R.K., Hickson I.D. The Bloom's syndrome gene product is a 3′–5′ DNA helicase. J. Biol. Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 8.Mohaghegh P., Karow J.K., Brosh, R.M., Bohr V.A., Hickson I.D. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheok C.F., Wu L., Garcia P.L., Janscak P., Hickson I.D. The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machwe A., Xiao L., Groden J., Matson S.W., Orren D.K. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 11.Machwe A., Lozada E.M., Xiao L., Orren D.K. Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol Biol. 2006;7:1. doi: 10.1186/1471-2199-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia P.L., Liu Y., Jiricny J., West S.C., Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S., Sommers J.A., Choudhary S., Faulkner J.K., Cui S., Andreoli L., Muzzolini L., Vindigni A., Brosh R.M., Jr Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 14.Wu L., Hickson I.D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 15.Yin J., Sobeck A., Xu C., Meetei A.R., Hoatlin M., Li L., Wang W. BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L., Bachrati C.Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl Acad. Sci. USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raynard S., Bussen W., Sung P., Wu L., Bachrati C.Z., Ou J., Xu C., Yin J., Chang M., Wang W., et al. A double holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75 BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. J. Biol. Chem. 2006;4:4. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 18.Wu L., Davies S.L., North P.S., Goulaouic H., Riou J.F., Turley H., Gatter K.C., Hickson I.D. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 19.Wu L., Davies S.L., Levitt N.C., Hickson I.D. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 20.Bischof O., Kim S.H., Irving J., Beresten S., Ellis N.A., Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusano K., Berres M.E., Engels W.R. Evolution of the RECQ family of helicases: A drosophila homolog, Dmblm, is similar to the human bloom syndrome gene. Genetics. 1999;151:1027–1039. doi: 10.1093/genetics/151.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusano K., Johnson-Schlitz D.M., Engels W.R. Sterility of Drosophila with mutations in the Bloom syndrome gene—complementation by Ku70. Science. 2001;291:2600–2602. doi: 10.1126/science.291.5513.2600. [DOI] [PubMed] [Google Scholar]
- 23.Boyd J.B., Golino M.D., Shaw K.E., Osgood C.J., Green M.M. Third-chromosome mutagen-sensitive mutants of Drosophila melanogaster. Genetics. 1981;97:607–623. doi: 10.1093/genetics/97.3-4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beall E.L., Rio D.C. Drosophila IRBP/Ku p70 corresponds to the mutagen-sensitive mus309 gene and is involved in P-element excision in vivo. Genes Dev. 1996;10:921–933. doi: 10.1101/gad.10.8.921. [DOI] [PubMed] [Google Scholar]
- 25.Adams M.D., McVey M., Sekelsky J.J. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 26.McVey M., Larocque J.R., Adams M.D., Sekelsky J.J. Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl Acad. Sci. USA. 2004;101:15694–15699. doi: 10.1073/pnas.0406157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min B., Weinert B.T., Rio D.C. Interplay between Drosophila Bloom's syndrome helicase and Ku autoantigen during nonhomologous end joining repair of P element-induced DNA breaks. Proc. Natl Acad. Sci. USA. 2004;101:8906–8911. doi: 10.1073/pnas.0403000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachrati C.Z., Borts R.H., Hickson I.D. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watt P.M., Hickson I.D., Borts R.H., Louis E.J. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ira G., Malkova A., Liberi G., Foiani M., Haber J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gangloff S., Soustelle C., Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nature Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 32.Wu L., Chan K.L., Ralf C., Bernstein D.A., Garcia P.L., Bohr V.A., Vindigni A., Janscak P., Keck J.L., Hickson I.D. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo Y.C., Paffett K.S., Amit O., Clikeman J.A., Sterk R., Brenneman M.A., Nickoloff J.A. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol. Cell Biol. 2006;26:4086–4094. doi: 10.1128/MCB.00136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McVey M., Adams M., Staeva-Vieira E., Sekelsky J.J. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics. 2004;167:699–705. doi: 10.1534/genetics.103.025411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preston C.R., Engels W., Flores C. Efficient repair of DNA breaks in Drosophila: evidence for single-strand annealing and competition with other repair pathways. Genetics. 2002;161:711–720. doi: 10.1093/genetics/161.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haber J.E., Ira G., Malkova A., Sugawara N. Repairing a double-strand chromosome break by homologous recombination: revisiting Robin Holliday's model. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2004;359:79–86. doi: 10.1098/rstb.2003.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]