Abstract
This study developed a method to boost the expression of recombinant proteins in a cell-free protein synthesis system without leaving additional amino acid residues. It was found that the nucleotide sequences of the signal peptides serve as an efficient downstream box to stimulate protein synthesis when they were fused upstream of the target genes. The extent of stimulation was critically affected by the identity of the second codons of the signal sequences. Moreover, the yield of the synthesized protein was enhanced by as much as 10 times in the presence of an optimal second codon. The signal peptides were in situ cleaved and the target proteins were produced in their native sizes by carrying out the cell-free synthesis reactions in the presence of Triton X-100, most likely through the activation of signal peptidase in the S30 extract. The amplification of the template DNA and the addition of the signal sequences were accomplished by PCR. Hence, elevated levels of recombinant proteins were generated within several hours.
INTRODUCTION
The cultivation of transformed cells has long been used as a standard route for preparing recombinant proteins. However, the recent explosion in the number of the newly identified open reading frames (ORFs) is now demanding a high throughput of protein expression that cannot be readily covered by the present in vivo expression technology.
Cell-free protein synthesis is attracting renewed attention as an alternative technique for overcoming the limited throughput of in vivo expression (1–4). While the conventional protocols of cell-free protein synthesis have been unable to provide sufficient quantities of proteins, recent improvements in the understanding of the key factors involved in cell-free synthesis have allowed the development of more efficient and robust protocols. Among the many attempts to enhance the accumulation of protein products in cell-free synthesis reactions, major improvements have been made through the stable supply of energy and substrates; ATP and amino acids (5–8). Kim et al. reported that the stable maintenance of the magnesium concentration against accumulating phosphate is also essential for prolonging the reaction period of cell-free protein synthesis (9).
However, the efficiency of gene expression is affected by the nature of the sequence elements as well as by environmental factors. Therefore, providing the optimal biochemical conditions is not sufficient for obtaining a high protein yield when the identity and arrangement of the nucleotide sequences have not been optimized for translation. In addition to the initiation codon and the Shine–Dalgarno (SD) sequence, several other sequence elements surrounding the start codon affect the efficiency of translation initiation significantly (10–14). In particular, the sequence of the first few nucleotides in close proximity to the start codon (downstream box) has a profound effect on the translation efficiency of many transcripts.
In this study, it was found that many of the naturally occurring signal peptide sequences stimulate protein synthesis in a cell-free protein synthesis system derived from Escherichia coli. For example, when the nucleotide sequence of the OmpA signal peptide (ompAss) was fused with the target genes, the amounts of the cell-free synthesized proteins were enhanced 10 times. The effectiveness of the different signal sequences were compared in a rapid and parallel manner through the PCR-based generation of the DNA templates for cell-free protein synthesis. As a result, it was found that the stimulatory effect of a signal peptide sequence was dependent on the identity of its second codon, and that most of the effective nucleotide sequences have an AAA triplet at the +2 position.
Finally, the ‘open’ nature of the cell-free protein synthesis enabled the in situ removal of the signal peptide from the expressed proteins. All the expression products were found at the native size of the target protein using the cell-free protein synthesis reactions in the presence of Triton X-100, most likely through the activation of the endogenous signal peptidase.
The method presented in this report enables the parallel expression of authentic proteins at elevated levels, and will provide a valuable platform for the large-scale translation of genomic information into protein molecules.
MATERIALS AND METHODS
Materials
ATP, GTP, UTP, CTP, creatine phosphate, creatine kinase and the E.coli total tRNA mixture were purchased from Roche Applied Science (Indianapolis, IN). The L-[U-14C]leucine (11.9 GBq/mmol) was obtained from Amersham Biosciences (Uppsala, Sweden). The detergents were obtained from Pierce Biotechnology (Rockford, IL). All the other reagents were purchased from Sigma (St. Louis, MO). The E.coli strain BL21-Star™(DE3) was purchased from Invitrogen (Carlsbad, CA). The S30 extract was prepared from the strain BL21-Star™(DE3) according to the method reported elsewhere (15,16).
Statistical analysis of nucleotide sequences
The full-length genome sequences of E.coli (strain K-12) were downloaded from the GeneBank database (NC_000913). The nucleotide sequences encoding the E.coli signal peptides were obtained from a signal peptide database (release 3.2) (17).
Construction of plasmid expression templates
Seven signal peptide sequences were fused to the structural gene of human erythropoietin (hEPO) using conventional PCR methods. The sequences were subcloned into the pK7 vector (18) between the nucleotide sequences of the T7 promoter and T7 terminator. The plasmid constructs were sequenced and purified using the Maxiprep kit (Qiagen, Valencia, CA) before being used as templates for cell-free protein synthesis reactions (Table 1 and Figure 1).
Table 1.
Strains and plasmids used in this study
| Bacterial strains | ||
| JM109 | e14–(McrA–) recA1 endA1 gyrA96 thi-1 hsdR17 ( ) supE44 relA1 Δ(lac-proAB) [F' traD36 proAB lacIqZΔM15] | Laboratory stock |
| BL21 star™(DE3) | F– ompT hsdSB( ) gal dcm rne131 | Invitrogen |
| Plasmidsa | ||
| pIVEX2.3d | Ampicillin resistant, C-terminal His-tag | Roche Applied Sciences |
| pK7hEPO | EPO (501 bp) cloned into NdeI/SalI sites | (18) |
| pK7pelBss-EPO | pK7EPO derivative containing the signal peptide sequence of pelB (66 bp) | This study |
| pK7ompAss-EPO | pK7EPO derivative containing the signal peptide sequence of ompA (63 bp) | This study |
| pK7phoAss-EPO | pK7EPO derivative containing the signal peptide sequence of phoA (63 bp) | This study |
| pK7malEss-EPO | pK7EPO derivative containing the signal peptide sequence of malE (78 bp) | This study |
| pK7ompCss-EPO | pK7EPO derivative containing the signal peptide sequence of ompC (63 bp) | This study |
| pK7ompTss-EPO | pK7EPO derivative containing the signal peptide sequence of ompT (60 bp) | This study |
| pK7heposs-EPO | pK7EPO derivative containing the signal peptide sequence of human EPO (81 bp) | This study |
aAll the pK7 plasmids have kanamycin resistance and the same 5′-UTR sequences including T7 promoter and RBS.
Figure 1.
Nucleotide and amino acid sequences of the N-terminal regions of the hEPO fused with the signal peptides.
Construction of linear expression templates
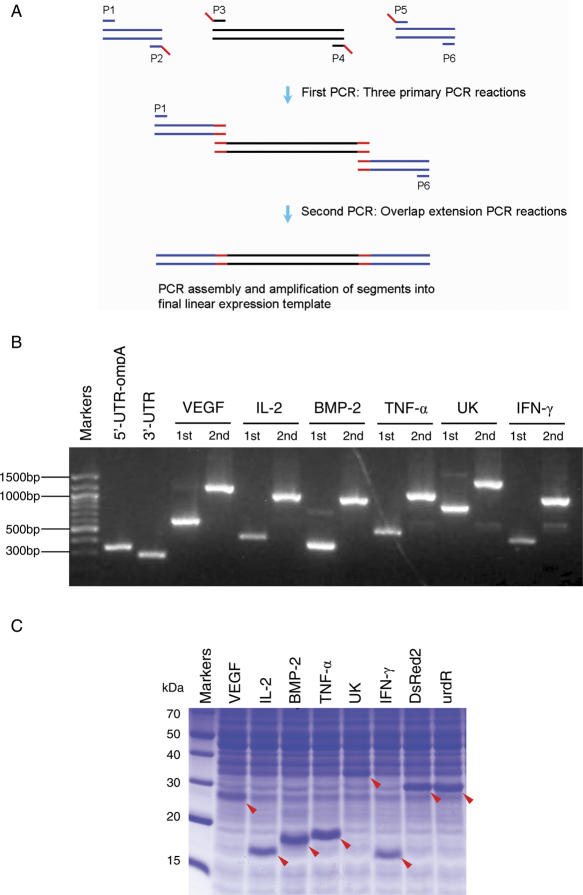
For the parallel expression of different ORFs, the PCR-amplified DNAs were used directly as templates for the cell-free synthesis reactions. The ompA fused-gene constructs were prepared using overlap extension PCR techniques. In the first-round PCR, three of the primary PCRs were carried out in separate reactions as follows: the T7 regulatory elements and ompA sequence were amplified from pK7ompAss-EPO using the primers, P1 (5′-TGGCACGACAGGTTTCCCGA-3′) and P2 (5′-GGCCTGCGCTACGGTAGCGA-3′). The target ORFs were amplified using the primers, P3 [5′-TCGCTACCGTAGCGCAGGCC-(20 nt sequence for target gene)-3′] and P4 [5′-TGATGATGAGAACCCCCCCC-(complementary 20 nt sequence for target gene)-3′] from the cloned plasmid or genomic DNA. The 3′-UTR region including the T7 terminator was amplified from the pIVEX2.3d plasmid (Roche Applied Science) using the primer P5 (5′-GGGGGGGGTTCTCATCATCA-3′) and P6 (5′-CCCAGTCCTGCTCGCTTCGC-3′). The 1st PCR products were purified by gel extraction and used for the second-round PCR, in which the full-length expression templates were synthesized using the primers P1 and P6. After amplification, the PCR products were purified using a PCR clean-up kit (Promega) before being used in the cell-free protein synthesis reactions.
Cell-free protein synthesis and analysis of the expressed proteins
The standard reaction mixture for cell-free protein synthesis consisted of the following components at a final volume of 15 μl: 57 mM Hepes–KOH (pH 8.2), 1.2 mM ATP, 0.85 mM each of CTP, GTP and UTP, 2 mM DTT, 0.17 mg/ml E.coli total tRNA mixture (from strain MRE600), 0.64 mM cAMP, 90 mM potassium glutamate, 80 mM ammonium acetate, 12 mM magnesium acetate, 34 μg/ml l-5-formyl-5,6,7,8-tetrahydrofolic acid (folinic acid), 1.0 mM each of 20 amino acids, 2% polyethylene glycol (PEG) 8000, 67 mM creatine phosphate (CP), 3.2 μg/ml creatine kinase (CK), 0.01 mM L-[U-14C]leucine (11.9 GBq/mmol, Amersham Biosciences), 6.7 μg/ml DNA, 4 μl of S30 extract. The cell-free synthesized protein was quantified by measuring the TCA-precipitated radioactivity using a liquid scintillation counter (WALLAC 1410), as described elsewhere (15). The size of the cell-free synthesized protein was analyzed by western blotting after running the reaction samples on a 13% Tricine-SDS-polyacrylamide gel, as described by Schagger and von Jagow (19).
Removal of the signal peptide from the expressed fusion proteins
The signal peptides were removed in situ by carrying out cell-free protein synthesis in the presence of different concentrations of Triton X-100 (0.01%–0.5%, w/v) from the start of incubation in order for the N-terminal signal peptide to be cleaved-off immediately after its translation in the reaction mixture.
RESULTS
Optimized biochemical conditions are not sufficient for the efficient cell-free expression of many ORFs
It was previously reported that the efficiency of cell-free protein synthesis is improved remarkably when the reaction mixture is supplied with sufficient amounts of energy and substrates (9). For example, in reactions utilizing creatine phosphate for the continuous regeneration of ATP, the use of an excess amount of creatine phosphate enabled prolonged protein synthesis provided that the reaction mixture was supplied periodically with fresh magnesium ions to compensate for their loss through the formation of insoluble magnesium phosphates. Using this approach, as much as 1.2 mg/ml of chloramphenicol acetyltransferase (CAT) could be produced from a single batch reaction of cell-free protein synthesis. An increase in the productivity of many other proteins was also shown after improving the supply of ATP and amino acids (9).
However, the provision of optimal biochemical conditions does not always enhance the productivity of cell-free protein synthesis. For example, the expression level of hEPO was not affected significantly in the presence of the elevated levels of ATP and amino acids, and remained at <100 μg/ml. Since all of our ORFs were routinely cloned in the same expression vectors (pK7 and pIVEX2.3d) (9,18), such a discrepancy would be due to the nature of the nucleotide sequences of the structural genes.
Signal peptide sequences-mediated stimulation of protein expression
With respect to the effect of the sequence elements on the efficiency of gene expression, many recent reports strongly suggest that the translational efficiency of a gene is determined primarily by the nature of the initial nucleotide sequences. For example, Stenström and Isaksson recently demonstrated that the translational efficiency of the lacZ reporter gene was critically affected by the identity of the first +2 to +5 codons (13). In particular, the expression level was dramatically changed according to which codon was placed at the +2 position. While the expression level of the reporter gene was strongly stimulated when the AAA codon was located at +2 position (11), the placement of the NGG codons (CGG, AGG, GGG and UGG) at the same position was associated with remarkably lower expression efficiency (12,20).
Based on their results, it was assumed that the yields of otherwise poorly expressed genes might be stimulated by placing the optimal nucleotide sequences beforehand. It was also expected that such a stimulatory sequence could be used as a universal translation enhancer to increase the productivity of recombinant proteins. In search of the nucleotide sequences that possibly enhance the expression of their fusion partners, the python script was used to analyze the codon usage in the entire E.coli genome. Interestingly, it was found that the nucleotide sequences of many signal peptides had strong bias for AAA as the second codon. Approximately 40% of signal peptides had the AAA triplet as the second codon (Table 2).
Table 2.
Codon biases of the nucleotide sequence of the E.coli signal peptides
| Sequence data | Frequency of AAA codon at position +2 |
|---|---|
| Whole E.coli K12 genome | 440/4241 (10.3%) |
| Unverified E.coli signal sequences | 22/45 (48.9%) |
| Verified E.coli signal sequences | 16/54 (29.6%) |
E.coli signal sequence data set were obtained from a signal peptide database (http://proline.bic.nus.edu.sg/spdb/). This database currently contains 99 entries of E.coli signal sequences: 54 of the entries are experimentally verified signal sequences (the N-terminal amino acid residues were sequenced), and 45 are computationally predicted sequences (17).
According to the analysis reported by Sato et al. (21), >10% of the E.coli genes use AAA as their second codon whereas the frequency of AAA triplet in the entire E.coli genome is 3.3%. It was also suggested that the higher frequency of the AAA triplet at the +2 position is related to the translation efficiency of a gene. Therefore, from the exceptionally high frequency of the AAA triplet as the second codon, it was assumed that the signal sequences might work as a translation-enhancing element.
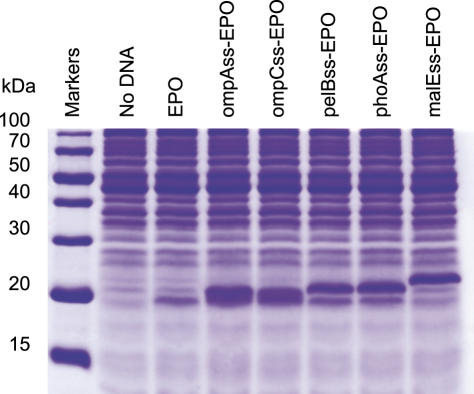
Therefore, the genes of hEPO with different signal sequences were fused and experiments were carried out to determine if their presence affects the efficiency of protein synthesis in our cell-free synthesis system. The nucleotide sequences of the seven different signal peptides (pelBss, ompAss, phoAss, malEss, ompCss, ompTss, heposs) were fused to the upstream of hEPO sequence in the plasmid pK7hEPO, and the resulting constructs were expressed in the reaction mixture for cell-free protein synthesis. As summarized in Table 3, the five signal sequences except for ompTss and heposs dramatically enhanced the expression level, yielding 9–10 times higher amounts of the target proteins (Figure 2). For example, the expression level of hEPO was increased from 55 to 588 μg/ml through its fusion with the ompA sequence.
Table 3.
Expression level of the signal peptide-EPO fusion constructs
| Plasmid | Signal peptide | Second codon | Expression levela(μg/ml) | Relative level of expressionb |
|---|---|---|---|---|
| pK7pelBss-EPO | pelB | AAA | 569 ± 34 | 10.3 |
| pK7ompAss-EPO | ompA | AAA | 588 ± 28 | 10.7 |
| pK7phoAss-EPO | phoA | AAA | 515 ± 31 | 9.4 |
| pK7malEss-EPO | malE | AAA | 524 ± 13 | 9.5 |
| pK7ompCss-EPO | ompC | AAA | 522 ± 24 | 9.5 |
| pK7ompTss-EPO | ompT | CGG | 107 ± 17 | 1.9 |
| pK7heposs-EPO | heposs | GGG | 101 ± 13 | 1.8 |
aData from three independent experiments.
bExpression level as compared with the expression of the wild-type EPO (EPO expression level = 1; EPO expression level = 55 μg/ml)
Figure 2.
SDS–PAGE analysis of the cell-free synthesized hEPOs fused with different signal peptides. The reaction mixture was incubated at 37°C for 2 h. The reaction mixture (2 μl) was loaded on a 13% Tricine-SDS–PAGE gel and stained with Coomassie Brilliant Blue.
Effect of the identity of the second codon on the translation efficiency
However, not all the signal sequences were effective in enhancing the expression of the downstream ORF. For example, the nucleotide sequence of the OmpT signal sequence (ompTss) failed to stimulate the synthesis of hEPO with the amount of the accumulated protein being similar to the wild-type sequence. Similarly, the addition of the natural signal sequence of hEPO (heposs) did not affect the expression level of hEPO.
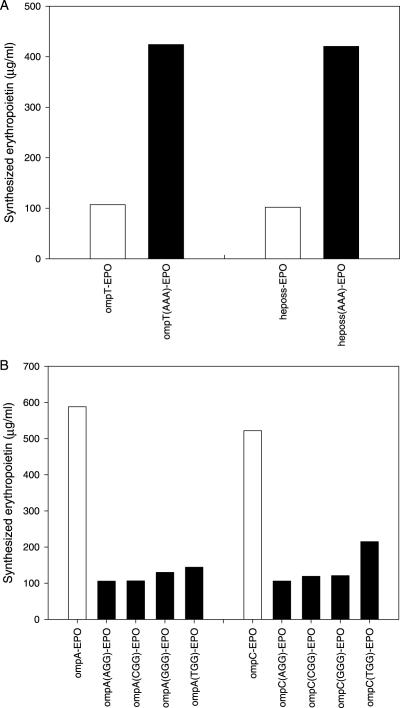
Interestingly, when the second codons of ompTss and heposs were switched to AAA (from CGG and GGG, respectively), there was an ∼4-fold increase in protein synthesis (Figure 3A). In contrast, switching the second codons of ompAss and ompCss from AAA to NGG (AGG, TGG, GGG and CGG) almost completely abolished its stimulatory effect (Figure 3B). This suggests that the identity of the second codon critically affects the stimulatory effect of the signal sequences.
Figure 3.
Effect of the expression level of the fused proteins according to the identity of the second codon. The efficiency of cell-free protein synthesis was enhanced by switching the second codon of ompTss and heposs into AAA (A). In contrast, protein synthesis was repressed when the second codons (AAA) of ompAss and ompCss were mutated into NGG codons (AGG, TGG, GGG and CGG) (B). After 2 h incubation, 15 μl of the reaction samples were withdrawn from the reaction mixture, and the [14C]leucine-labeled radioactivity was measured as described in Materials and Methods.
PCR-based addition of the stimulatory signal sequence and the direct expression of the amplified DNAs
Encouraged by the dramatic increase in hEPO expression in the presence of the signal sequences, this study examined whether or not the signal sequence-assisted stimulation of protein synthesis is generally applicable to the expression of other proteins. Sixteen different ORFs that normally show a very low expression level in our cell-free protein synthesis system were selected. Instead of cloning each of the fusion constructs in the expression vector, the PCR-amplified DNA with or without the ompAss were used directly as the templates for cell-free protein synthesis, thereby eliminating the time- and labor-intensive cloning steps (Figure 4). As summarized in Table 4, the presence of ompAss remarkably enhanced the expression of the target proteins in all the cases examined, which suggests that the proposed strategy can be employed as a general method for boosting the expression of the recombinant proteins in a cell-free protein synthesis system. However, the extent of stimulation showed significant variations among different proteins implying that the expression of protein can also be substantially affected by the nucleotide sequences of the target ORFs.
Figure 4.
Construction and expression of the linear expression templates by PCR. (A) Schematic diagram of the PCR-based generation of the ompA fusion constructs. Three primary PCR products with defined overlapping ends were synthesized by the first PCR. These three fragments were joined in the second PCR, overlap extension PCR, which also simultaneously introduced the regulatory elements and the OmpA signal peptide sequence to the target genes. The red bar indicates the overlapping region. The blue and black bar indicate UTR and ORF, respectively. (B) Agarose gel electrophoresis of the PCR products. Second PCR products have an additional 600 bp as a result of the fusion with 5′-UTR and 3′-UTR. (C) Cell-free expression of ompA-fused PCR products. The reaction mixture for cell-free protein synthesis was prepared as described in Materials and Methods. After 2 h of incubation, 2 μl of the reaction mixture was loaded on a 13% Tricine-SDS–PAGE gel and stained with Coomassie Brilliant Blue. Cell-free expressed proteins are indicated by arrows.
Table 4.
Cell-free expression of the ompAss fused genes
| Protein | Wild-type (μg/ml) | ompA fusion (μg/ml) | Relative level of expressiona | GenBank ID/references | Molecular weight of protein (kDa) | Origin |
|---|---|---|---|---|---|---|
| IL-2 | 91 | 545 | 6.0 | S77834 | 15.5 | Homo sapiens |
| TNF-a | 226 | 513 | 2.3 | X01394 | 17.5 | Homo sapiens |
| DsRed2b | 98 | 665 | 6.8 | — | 25.9 | Synthetic gene |
| BMP2 | 128 | 431 | 3.4 | M22489 | 13.0 | Homo sapiens |
| UK | 83 | 387 | 4.7 | NM_008873 | 30.5 | Mus musculus |
| VEGF | n.d. | 489 | — | X62568 | 22.3 | Homo sapiens |
| IFN-γ | 45 | 460 | 10.2 | K00083 | 16.0 | Mus musculus |
| ϖ-TACc | 125 | 487 | 3.9 | AAK25105 | 47.8 | Caulobacter crescentus |
| ϖ-TAMl | 206 | 537 | 2.6 | BAB48961 | 48.2 | Mesorhozobium loti |
| ϖ-TATr | 173 | 436 | 2.5 | AAL44116 | 49.0 | Agrobacterium tumefaciens |
| ϖ-TAVf | 237 | 539 | 2.3 | (32) | 50.3 | Vibrio fluvialis |
| ϖ-TAXc | 198 | 529 | 2.7 | AAM41635 | 48.4 | Xanthomonas axonopodis |
| smTG | 11 | 358 | 32.5 | AAS68222 | 45.7 | Streptomyces mobaraensis |
| oleV | 35 | 645 | 18.4 | AAD55451 | 53.1 | Streptomyces antibioticus |
| oleW | 26 | 689 | 26.5 | AAD55450 | 35.8 | Streptomyces antibioticus |
| urdR | n.d. | 702 | — | AAF72551 | 26.8 | Streptomyces fradiae |
aThe relative expression level was determined by dividing the amount of the ompA fused protein by the expressed wild-type protein.
bPlasmid pDsRed2 is obtained from Clontech.
n.d., not determined.
In situ cleavage of signal peptide in an E.coli cell-free protein synthesis system
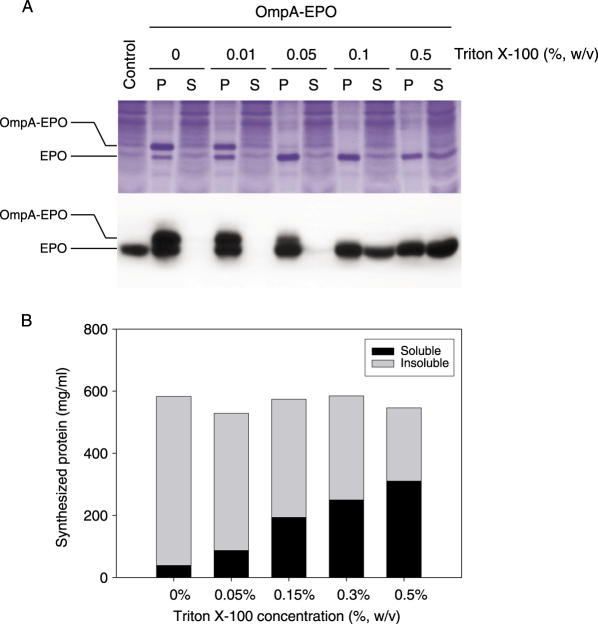
Although it was shown that the presence of ompAss dramatically enhances the expression of the recombinant proteins, it also adds 21 non-native amino acid residues to the target proteins that might interfere with the biological functions and proper folding of the expressed proteins. Therefore, it would be desirable if the signal peptides are removed during protein synthesis so that the resulting polypeptide of the target protein follows its natural folding pathway. Since the reactions of cell-free protein synthesis are carried out in a crude lysate of E.coli (S30 extract), it was presumed that most of the soluble E.coli proteins are present in the reaction mixture. In addition, the signal peptidase of E.coli retains its activity even when it is detached from the cellular membrane (22,23). Therefore, the added signal peptide should be removed in situ by the endogenous signal peptidase activity. As shown in Figure 5A, SDS–PAGE and western blot analyses confirmed that a significant proportion of the cell-free expressed protein had the native size without the signal peptide. This indicates that the S30 extract retains the signal peptidase activity under the reaction conditions for cell-free protein synthesis.
Figure 5.
In situ cleavage of the signal peptides during cell-free protein synthesis. (A) The indicated concentration of Triton X-100 was added to the reaction mixture from the start of the synthesis reaction, and examined to determine it could increase the level of signal peptide cleavage. After incubation, the reaction mixture was centrifuged at 10 000 g for 10 min, and the soluble and pellet fractions were analyzed by 13% Tricine-SDS–PAGE stained with Coomassie Brilliant Blue (upper panel) and western blot using the anti-EPO antibody (lower panel). P and S represent the insoluble and soluble fractions of the cell-free synthesized EPO, respectively. (B) [14C]leucine-labeled radioactivity of the expressed protein was measured as described under Materials and Methods.
Tschantz et al. reported that the catalytic activity of truncated E.coli signal peptidase, which lacks the membrane-anchoring domain, was stimulated in the presence of the detergent Triton X-100 (22,24). By taking advantage of the ‘open’ nature of cell-free protein synthesis, an attempt was made to express the ompAss-hEPO construct in the presence of Triton X-100 with the anticipation that the enhanced signal peptidase activity would increase the fraction of the correctly sized product. Indeed, the efficiency of the cleavage of signal peptide was improved remarkably upon the addition of Triton X-100, and virtually all the products were found at the correct molecular weight (Figure 5A). When tested at different concentrations, the presence of Triton X-100 >0.1% (w/v) effectively facilitated the removal of the OmpA sequence, and virtually all of the cell-free expressed products were found at the native size of EPO. At least up to the concentration of 0.5% (w/v), the presence of Triton X-100 did not affect the efficiency of protein synthesis (Figure 5B). Therefore, enhanced efficiency of the cleavage of the signal sequence was not due to a reduced yield of protein synthesis. In addition, solubility of the cell-free expressed products was substantially enhanced in the presence of Triton X-100. While the EPO molecules (regardless of the presence of signal sequences) were almost completely insoluble in the reaction mixture without any detergents, ∼70% of the expressed protein was found in the soluble fraction.
DISCUSSION
In this study, it was found that the nucleotide sequences of signal peptides could serve as efficient downstream boxes to stimulate protein synthesis. When analyzed in a cell-free protein synthesis system derived from E.coli, different target genes showed increases in protein expression of 2.3- to 32-fold through their fusion with the nucleotide sequences of the signal peptides. The stimulatory effect of the signal sequences appeared to be closely related to the presence of the AAA triplet at the +2 position.
Based on this discovery, a method was developed to rapidly generate authentic protein molecules at elevated levels. First, overlap extension PCR techniques were used to amplify the target genes in the fused form with the nucleotide sequence of signal peptide. The amplified DNA was then used to direct protein synthesis, and protein molecules were generated within several hours. In addition, by mimicking the in vivo process of the signal peptide cleavage, the target proteins were produced in their native amino acid sequences without additional signal peptides.
Although the detailed mechanism of the signal sequence-mediated stimulation of protein synthesis is not completely understood, it appears that the effect of the initial nucleotide sequences is not related to the relative abundance or stability of the mRNA species. Computational analyses of the mRNA structure did not indicate any notable difference caused by the presence of these signal sequences. In addition, among the gene constructs examined in this study, the relative level of mRNA with or without the signal sequence was similar (data not shown). Therefore, it appears that the initial nucleotide sequence affects the process of translation initiation and/or the early phase of elongation (12,20,25). In particular, Gonzalez de Valdivia and Isaksson demonstrated that the presence of NGG triplets in the early coding region causes inefficient translation through an accelerated drop-off of peptidyl-tRNA (12,20).
Although extensive evidence has been presented that entire codon context in the early coding region (from positions +2 to +5) influence gene expression, the presence of additional AAA triplet at the positions +3 to +5 of ompTss did not further increase the yield of protein synthesis in our experiments (data not shown). However, still, it might be of worth to note that some of the effective signal sequences have additional AAA triplet in the early coding region (e.g. malEss and ompCss have an additional AAA triplet at +4 position). It might be possible to further improve the effectiveness of the signal sequences by engineering the nucleotide sequences in this region. Apart from the precise mechanism of action, the use of signal sequences as a translation-enhancing downstream box offers distinctive advantages in cell-free protein synthesis. In addition to the remarkably enhanced expression level of the target proteins, the in situ cleavage of the signal peptides provides a simple way for removing the N-terminal methionine. The issue of removing the translation initiator, N-formyl-methionine or methionine, from a recombinant protein is often critical for obtaining active and stable recombinant proteins (26–30).
It is believed that these results can be further extended to controlling the expression level of genes. Using the signal sequences that have been engineered to stimulate protein synthesis at different efficiencies, a mixture of template DNA can be co-expressed to generate the proteins in the desired abundances. Such a system will provide an ideal platform for constructing an in vitro metabolic network. As demonstrated by Ku et al. (31), the in vitro reproduction of a metabolic pathway can be ideally used to further understand and optimize the production of metabolites.
Acknowledgments
Authors gratefully acknowledge the financial support from the Ministry of Commerce, Industry and Energy, Korea (grant No. 10021962). We wish to thank Mr. Joo-Hyun Seo for his kind help and advice in the computational analysis of the bias of codon usage in E.coli genome. We also thank Professor Byung-Gee Kim for providing the transaminase genes. Funding to pay the Open Access publication charges for this article was provided by Ministry of Commerce, Industry and Energy, Korea (grant No. 10021962).
Conflict of interest statement. None declared.
REFERENCES
- 1.Doi N., Takashima H., Kinjo M., Sakata K., Kawahashi Y., Oishi Y., Oyama R., Miyamoto-Sato E., Sawasaki T., Endo Y., Yanagawa H. Novel fluorescence labeling and high-throughput assay technologies for in vitro analysis of protein interactions. Genome Res. 2002;12:487–492. doi: 10.1101/gr.218802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawasaki T., Ogasawara T., Morishita R., Endo Y. A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl Acad. Sci. USA. 2002;99:14652–14657. doi: 10.1073/pnas.232580399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katzen F., Chang G., Kudlicki W. The past, present and future of cell-free protein synthesis. Trends Biotechnol. 2005;23:150–156. doi: 10.1016/j.tibtech.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Murthy T.V., Wu W., Qiu Q.Q., Shi Z., LaBaer J., Brizuela L. Bacterial cell-free system for high-throughput protein expression and a comparative analysis of Escherichia coli cell-free and whole cell expression systems. Protein Expr. Purif. 2004;36:217–225. doi: 10.1016/j.pep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Calhoun K.A., Swartz J.R. Energizing cell-free protein synthesis with glucose metabolism. Biotechnol. Bioeng. 2005;90:606–613. doi: 10.1002/bit.20449. [DOI] [PubMed] [Google Scholar]
- 6.Kim D.M., Swartz J.R. Prolonging cell-free protein synthesis by selective reagent additions. Biotechnol. Prog. 2000;16:385–390. doi: 10.1021/bp000031y. [DOI] [PubMed] [Google Scholar]
- 7.Kim D.M., Swartz J.R. Regeneration of adenosine triphosphate from glycolytic intermediates for cell-free protein synthesis. Biotechnol. Bioeng. 2001;74:309–316. [PubMed] [Google Scholar]
- 8.Kim D.M., Swartz J.R. Efficient production of a bioactive, multiple disulfide-bonded protein using modified extracts of Escherichia coli. Biotechnol. Bioeng. 2004;85:122–129. doi: 10.1002/bit.10865. [DOI] [PubMed] [Google Scholar]
- 9.Kim T.W., Kim D.M., Choi C.Y. Rapid production of milligram quantities of proteins in a batch cell-free protein synthesis system. J. Biotechnol. 2006;124:373–380. doi: 10.1016/j.jbiotec.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 10.Stenstrom C.M., Holmgren E., Isaksson L.A. Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene. 2001;273:259–265. doi: 10.1016/s0378-1119(01)00584-4. [DOI] [PubMed] [Google Scholar]
- 11.Stenstrom C.M., Jin H.N., Major L.L., Tate W.P., Isaksson L.A. Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene. 2001;263:273–284. doi: 10.1016/s0378-1119(00)00550-3. [DOI] [PubMed] [Google Scholar]
- 12.de Valdivia E.I.G., Isaksson L.A. A codon window in mRNA downstream of the initiation codon where NGG codons give strongly reduced gene expression in Escherichia coli. Nucleic Acids Res. 2004;32:5198–5205. doi: 10.1093/nar/gkh857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenstrom C.M., Isaksson L.A. Influences on translation initiation and early elongation by the messenger RNA region flanking the initiation codon at the 3' side. Gene. 2002;288:1–8. doi: 10.1016/s0378-1119(02)00501-2. [DOI] [PubMed] [Google Scholar]
- 14.Etchegaray J.P., Inouye M. Translational enhancement by an element downstream of the initiation codon in Escherichia coli. J. Biol. Chem. 1999;274:10079–10085. doi: 10.1074/jbc.274.15.10079. [DOI] [PubMed] [Google Scholar]
- 15.Kim D.M., Kigawa T., Choi C.Y., Yokoyama S. A highly efficient cell-free protein synthesis system from Escherichia coli. Eur. J. Biochem. 1996;239:881–886. doi: 10.1111/j.1432-1033.1996.0881u.x. [DOI] [PubMed] [Google Scholar]
- 16.Ahn J.H., Chu H.S., Kim T.W., Oh I.S., Choi C.Y., Hahn G.H., Park C.G., Kim D.M. Cell-free synthesis of recombinant proteins from PCR-amplified genes at a comparable productivity to that of plasmid-based reactions. Biochem. Biophys. Res. Commun. 2005;338:1346–1352. doi: 10.1016/j.bbrc.2005.10.094. [DOI] [PubMed] [Google Scholar]
- 17.Choo K.H., Tan T.W., Ranganathan S. SPdb—a signal peptide database. BMC Bioinformatics. 2005;6:249. doi: 10.1186/1471-2105-6-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahn J.H., Choi C.Y., Kim D.M. Effect of energy source on the efficiency of translational termination during cell-free protein synthesis. Biochem. Biophys. Res. Commun. 2005;337:325–329. doi: 10.1016/j.bbrc.2005.09.061. [DOI] [PubMed] [Google Scholar]
- 19.Schagger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 20.de Valdivia E.I.G., Isaksson L.A. Abortive translation caused by peptidyl-tRNA drop-off at NGG codons in the early coding region of mRNA. FEBS J. 2005;272:5306–5316. doi: 10.1111/j.1742-4658.2005.04926.x. [DOI] [PubMed] [Google Scholar]
- 21.Sato T., Terabe M., Watanabe H., Gojobori T., Hori-Takemoto C., Miura K. Codon and base biases after the initiation codon of the open reading frames in the Escherichia coli genome and their influence on the translation efficiency. J. Biochem. (Tokyo) 2001;129:851–860. doi: 10.1093/oxfordjournals.jbchem.a002929. [DOI] [PubMed] [Google Scholar]
- 22.Tschantz W.R., Paetzel M., Cao G., Suciu D., Inouye M., Dalbey R.E. Characterization of a soluble, catalytically active form of Escherichia coli leader peptidase: requirement of detergent or phospholipid for optimal activity. Biochemistry. 1995;34:3935–3941. doi: 10.1021/bi00012a010. [DOI] [PubMed] [Google Scholar]
- 23.Karla A., Lively M.O., Paetzel M., Dalbey R. The identification of residues that control signal peptidase cleavage fidelity and substrate specificity. J. Biol. Chem. 2005;280:6731–6741. doi: 10.1074/jbc.M413019200. [DOI] [PubMed] [Google Scholar]
- 24.Paetzel M., Dalbey R.E., Strynadka N.C. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature. 1998;396:186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 25.Jin H.N., Bjornsson A., Isaksson L.A. Cis control of gene expression in E.coli by ribosome queuing at an inefficient translational stop signal. EMBO J. 2002;21:4357–4367. doi: 10.1093/emboj/cdf424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao Y.D., Wang S.C., Leu Y.J., Wang C.F., Chang S.T., Hong Y.T., Pan Y.R., Chen C. The structural integrity exerted by N-terminal pyroglutamate is crucial for the cytotoxicity of frog ribonuclease from Rana pipiens. Nucleic Acids Res. 2003;31:5247–5255. doi: 10.1093/nar/gkg746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boix E., Wu Y., Vasandani V.M., Saxena S.K., Ardelt W., Ladner J., Youle R.J. Role of the N terminus in RNase A homologues: differences in catalytic activity, ribonuclease inhibitor interaction and cytotoxicity. J. Mol. Biol. 1996;257:992–1007. doi: 10.1006/jmbi.1996.0218. [DOI] [PubMed] [Google Scholar]
- 28.Adachi K., Yamaguchi T., Yang Y., Konitzer P.T., Pang J., Reddy K.S., Ivanova M., Ferrone F., Surrey S. Expression of functional soluble human alpha-globin chains of hemoglobin in bacteria. Protein Expr. Purif. 2000;20:37–44. doi: 10.1006/prep.2000.1277. [DOI] [PubMed] [Google Scholar]
- 29.Endo S., Yamamoto Y., Sugawara T., Nishimura O., Fujino M. The additional methionine residue at the N-terminus of bacterially expressed human interleukin-2 affects the interaction between the N- and C-termini. Biochemistry. 2001;40:914–919. doi: 10.1021/bi001170r. [DOI] [PubMed] [Google Scholar]
- 30.Mine S., Ueda T., Hashimoto Y., Imoto T. Improvement of the refolding yield and solubility of hen egg-white lysozyme by altering the Met residue attached to its N-terminus to Ser. Protein Eng. 1997;10:1333–1338. doi: 10.1093/protein/10.11.1333. [DOI] [PubMed] [Google Scholar]
- 31.Ku B., Jeong J.C., Mijts B.N., Schmidt-Dannert C., Dordick J.S. Preparation, characterization, and optimization of an in vitro C30 carotenoid pathway. Appl. Environ. Microbiol. 2005;71:6578–6583. doi: 10.1128/AEM.71.11.6578-6583.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin J.S., Yun H., Jang J.W., Park I., Kim B.G. Purification, characterization, and molecular cloning of a novel amine:pyruvate transaminase from Vibrio fluvialis JS17. Appl. Microbiol. Biotechnol. 2003;61:463–471. doi: 10.1007/s00253-003-1250-6. [DOI] [PubMed] [Google Scholar]