Abstract
The elucidation of cross-talk events between intersecting signaling pathways is one main challenge in biological research. The complexity of protein networks, composed of different pathways, requires novel strategies and techniques to reveal relevant interrelations. Here, we established a combinatorial RNAi strategy for systematic single, double, and triple knockdown, and we measured the residual mRNAs and proteins quantitatively by quantitative real-time PCR and reverse-phase protein arrays, respectively, as a prerequisite for data analysis. Our results show that the parallel knockdown of at least three different genes is feasible while keeping both untargeted silencing and cytotoxicity low. The technique was validated by investigating the interplay of tyrosine kinase receptor ErbB2 and its downstream targets Akt-1 and MEK1 in cell invasion. This experimental approach combines multiple gene knockdown with a subsequent quantitative validation of reduced protein expression and is a major advancement toward the analysis of signaling pathways in systems biology.
Keywords: combinatorial protein knockdown, reverse-phase protein arrays, signaling pathways, cross-talk, systems biology
Complex protein networks evoke an increasing demand for suitable methods to elucidate cross-talk between signaling pathways. For example, tumor invasion integrates three major pathways: the growth factor-signaling pathway, the integrin-mediated pathway, and the Rho family GTPases (1). Overall, these pathways comprise ≈130 proteins (2–4), and currently little is known about how cross-talk events between these pathways impact tumor invasion. A common strategy to obtain insight into protein activities would be to knock down each of these proteins and to measure the induced effects on cell invasion. However, synergistic information about pathway cross-talk should be better attained through analyzing several proteins in parallel, e.g., via RNAi by multiple knockdown. Xia et al. (5) established small-scale multiple knockdown via a tetracycline-regulated pol II promoter construct that allows for the simultaneous expression of up to three short hairpin RNAs (shRNAs). In a different approach, Huang et al. (6) used siRNAs directed against different variants of Rab5 to understand their contribution to clathrin-dependent endocytosis. This work offered insights into the use of multiple siRNAs and revealed that systematic approaches for the production of double or triple knockdown by siRNA technology might be promising for the analysis of complex cellular processes. However, this approach was restricted to the use of siRNAs against different variants of the same protein, and the protein knockdown was not quantified. The validation of gene knockdown is a critical parameter in RNAi experiments, and current technologies suffer from the poor correlation between RNA and protein levels of expressed genes [quantitative real-time PCR (qRT-PCR)] (7) or a low dynamic range (Western blot) (8).
Here, we describe a combinatorial RNAi strategy for a systematic knockdown of multiple genes by chemical siRNAs as a simple and highly reproducible method, and the quantitative measurement of gene silencing at the level of both mRNAs and proteins with help of qRT-PCR and reverse-phase protein arrays (RPPA), respectively. We validated this approach by producing single, double, and triple knockdown to reveal the interrelationship of tyrosine kinase receptor ErbB2 and its downstream targets Akt-1 and MEK1 in tumor cell invasion. The parallel knockdown of different proteins in combination with a sensitive and quantitative analysis of especially protein expression will be an essential tool to study cross-talk between different pathways in a systematic way and help elucidate the role of proteins in a defined network.
Results
Targeting Three Endogenously Expressed Genes in Parallel and Optimization of Transfection Conditions.
In an initial proof-of-concept study, HCC1954 cells (derived from human primary breast tumor) were transfected with a pool of ErbB2 siRNAs either alone (single transfection), in combination with either GFP siRNA or Lamin A/C siRNA (double transfection), or in combination with GFP and Lamin A/C siRNAs (triple transfection). Then, we investigated the level of ErbB2 knockdown in these transfection set-ups. The ErbB2 protein expression was efficiently reduced (>60%) in single, double, and triple knockdown [supporting information (SI) Fig. 5]. These results demonstrated the feasibility of applying multiple siRNAs without saturating the RNAi machinery (RNA-induced silencing complex).
Next, a parallel knockdown of the three proteins Akt-1, ErbB2, and MEK1 was performed, and the repression of each protein in single, double, and triple siRNA transfections was quantified. To define the siRNA concentration with respect to the optimal balance between side effects and high level of protein silencing, we tested three different concentrations (in pools of 20 nmol/liter, 40 nmol/liter, and 80 nmol/liter final concentration for each gene). The genes were silenced with pools of four different siRNAs and corresponding protein knockdowns were examined initially by Western blot (Fig. 1A). The ErbB2 knockdown was successful at all three siRNA concentrations.
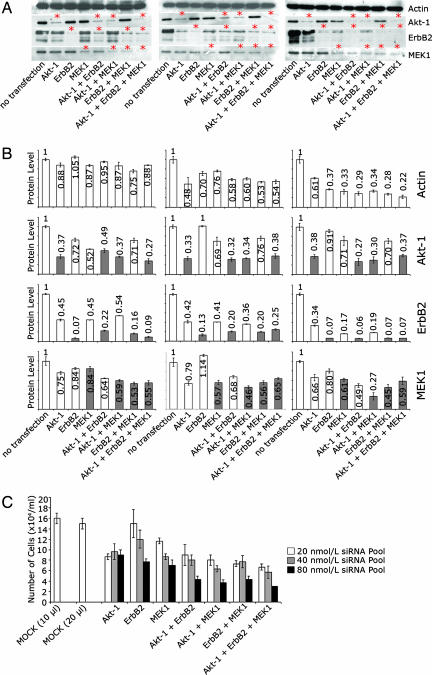
Fig. 1.
Simultaneous knockdown of three different proteins (Akt-1, ErbB2, and MEK1) using different siRNA concentrations. (A) Western blot showing the reduction at the protein level after RNAi (48 h) targeting Akt-1, ErbB2, and MEK1, alone or in the indicated combinations. For each gene, three different concentrations of siRNA pools [20 nmol/liter (Left), 40 nmol/liter (Center), and 80 nmol/liter (Right)] were tested. Red stars indicate samples where a reduction of protein was expected after transfection with the corresponding siRNAs. (B) Determination of protein knockdown after RNAi by RPPA. Signal intensities were normalized to 1 for the non-targeted control protein actin, or for the respective target proteins (ErbB2, Akt-1, or MEK1) in the case of nontransfected cells. For each gene, three different concentrations of siRNA pools [20 nmol/liter (Left), 40 nmol/liter (Center), and 80 nmol/liter (Right)] were tested. Gray bars indicate samples where a reduction of the protein level was expected whereas the bars in white show samples where no such reduction was presumed. (C) Cytotoxicity of siRNA transfections was determined for each concentration of siRNA by cell counting. MOCK (10 μl) is for transfections applied in 10 μl of transfection reagent, and MOCK (20 μl) is for the 20-μl set-up. Each sample was counted four times in three independent experiments, and the averages and standard deviations are shown.
RPPAs (Fig. 1B) showed that the knockdown was >75% for single, double and triple transfections, already for the lowest concentration. However, we also observed a reduction of ErbB2 protein in Akt-1 or/and MEK1 transfected samples, and this effect increased with higher siRNA concentrations. In the case of Akt-1 knockdown, the reduction was similar in single, double, and triple transfections for all siRNA concentrations. Compared with the ErbB2 knockdown, the repression of Akt-1 protein level was less (between 51% and 73%), and did not increase significantly with higher siRNA concentrations (Fig. 1B) or higher amount of transfection reagent (SI Fig. 6). Although we could clearly observe a reduced expression of MEK1 in single, double and triple transfections by Western blot (Fig. 1A), the microarray-based detection did not confirm these results. However, TaqMan analysis (SI Fig. 7) shows that MEK1 reduction was already efficient with 20 nmol/liter siRNA. Knockdown at the mRNA level was >70% for ErbB2 and Akt-1 and >90% for the MEK1, both with siRNA pools and with individual siRNAs (SI Fig. 7 and Fig. 2). Overall, our data show that effective knockdown (>60%) at the protein level for each of the genes was obtained at 20 nmol/liter siRNA, and that this concentration led to lowest cytotoxic effects (Fig. 1C).
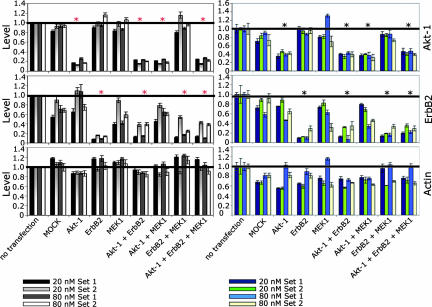
Fig. 2.
Evaluation of different siRNA sets on specific and untarget effects. Cells were transfected with 20 nmol/liter or 80 nmol/liter siRNAs of two different siRNA sets, respectively, each containing siRNAs against a single target region within the corresponding genes. The effects of both sets were measured on mRNA level (Left) by using qRT-PCR, as well as on protein level (Right) by RPPA. The stars on top of the bars show where the reduction was presumed for the given set-up.
Untarget effects, i.e., the repression of non-targeted proteins (7), increase with elevated concentrations of siRNA (9). Our results for actin and ErbB2 protein levels let us suppose that this increase of untarget effects is also true for the combinatorial RNAi strategy. The Western blot data suggested that the expression level of actin was not different in any of the transfection conditions, but the protein microarrays detected clear changes in the actin expression level. Whereas the actin signals varied between 5% and 25% for the transfections with 20 nmol/liter siRNA, this range increased to between 24% and 51% for 40 nmol/liter siRNA, and to between 39% and 78% for the 80 nmol/liter siRNA concentration (Fig. 1B). These data correlate with the results of the cytotoxicity assay (Fig. 1C).
Analysis of Different siRNAs in Combinations.
To validate the specificity of knockdown for the targeted proteins as well as the nonspecific effects on actin and ErbB2 expression changes, we next assembled two different siRNA sets from individual duplices (SI Materials and Methods and SI Fig. 8) for each gene and measured the efficiency of down regulation on mRNA and protein levels (Fig. 2). For Akt-1, an efficient down-regulation on mRNA level (80%) and protein level (60%) was measured for both sets with 20 nmol/liter and 80 nmol/liter siRNA. Down-regulation of ErbB2 was more efficient with set 1, especially in double knockdowns. As already observed with siRNA pools, with both siRNA sets the knockdown of Akt-1 and MEK1 reduced the expression of the non-targeted ErbB2, and this effect was strongest with 80 nmol/liter siRNA. It was also stronger for set 1, especially on mRNA level. Unexpectedly, we detected low fluctuations of the actin mRNA levels for both sets and at both concentrations, whereas we determined considerable heterogeneity at the protein level. In contrast to siRNA pools, we determined a substantial reduction of actin protein levels with both siRNA sets at 20 nmol/liter siRNA, whereas at 80 nmol/liter only little effect was seen. Obviously, siRNA pools dilute the untarget effects of different siRNAs on actin, but not on ErbB2.
Measurement of Untarget Effects.
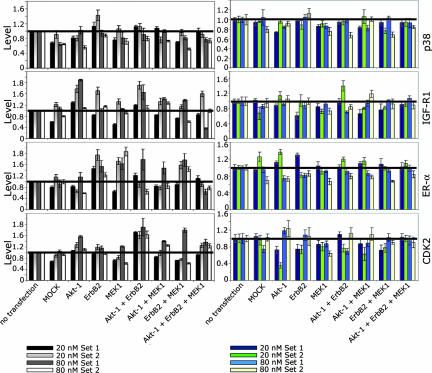
To examine untarget effects more thoroughly, we selected four additional non-targeted proteins and quantified the corresponding mRNAs and proteins after single, double, and triple transfections using siRNA sets at 20 nmol/liter and 80 nmol/liter. The stress-activated p38 MAPK is not related to the growth factor activated ErbB2-signaling pathway (SI Fig. 9), and we did not detect any significant changes in mRNA or protein levels at any condition (Fig. 3). The insulin-like growth factor 1 receptor (IGF-1R) and the estrogen receptor α (ER-α) are related to the ErbB2-signaling pathway. IGF-1R activates Akt-1 and MEK1, and the ER-α is a downstream substrate of both Akt-1 and MEK1 (SI Fig. 9). The TaqMan data for both genes show a high, nonsystematic fluctuation of mRNA levels. However, we could not detect significant changes on protein level (Fig. 3). For ER-α, we measured a slight reduction for both siRNA sets in the 80 nmol/liter transfections, but the level of reduction did not exceed 20%. It has been shown that inhibition of the Akt-1 pathway through ErbB2 and phosphatidylinositol 3-kinase (PI3-kinase) results in down-regulation of CDK2 at the transcriptional level (10). We detected this effect also when we measured the protein expression of untargeted CDK2 after down-regulation of Akt-1 or ErbB2 (Fig. 3) with 20 nmol/liter siRNAs, but not at the 80 nmol/liter concentrations (Fig. 3). In double transfections, this effect was only seen with siRNA set 2 at 20 nmol/liter. However, we could not confirm these specific effects by TaqMan analysis. Conversely, the TaqMan result for the double transfection Akt-1 and ErbB2 siRNAs suggests an up-regulation of CDK2.
Fig. 3.
Evaluation of untarget effects. Cells were transfected with 20 nmol/liter or 80 nmol/liter siRNAs of two different siRNA sets against Akt-1, ErbB2, and MEK1, respectively. Potential effects were measured for both sets on p38, IGF-1R, ER-α, and CDK2 expression, as well as on mRNA level (Left) and on protein level (Right).
Systematic Knockdown of Proteins as Suitable Tool to Reveal Their Role in Cell Invasion.
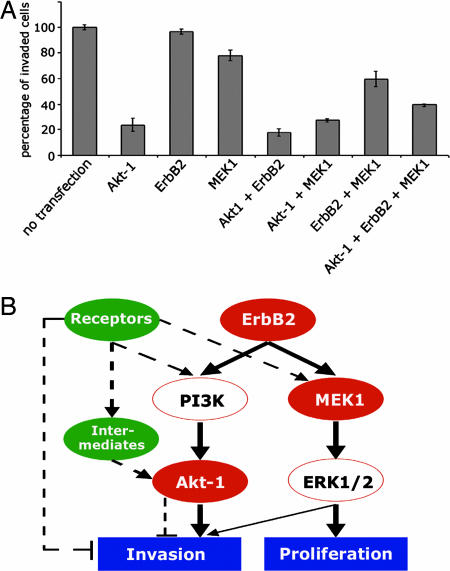
We next tested the combinatorial RNAi strategy for its applicability to analyze a protein network and to gain knowledge on the interconnections of ErbB2, Akt-1 and MEK1 proteins during cell invasion (Fig. 4). To this end, we tested whether the combinatorial knockdown of proteins was suited to induce effects on the invasive phenotype of the highly invasive HCC1954 cell line. Any changes in the number of invaded cells would directly reflect the role of each protein alone or in combination with the others on cell invasion. Loss of Akt-1 protein indeed resulted in a strong reduction in the number of invading cells, demonstrating that 60% depletion of Akt-1 protein (Figs. 1B and 2) was sufficient to induce this phenotypic effect. In contrast, repression of the ErbB2 protein alone did not affect the invasive capacity of HCC1954 cells, even though knockdown was >90%. Similarly, down-regulation of MEK1 resulted only in a slight reduction of invaded cells. These results indicate that the Akt-1 pathway is dominant for the invasive phenotype (Fig. 4B). However, double-transfection with ErbB2 and MEK1 siRNAs induced a significant decrease of invasion potential in HCC1954 cells, when compared with single knockdown of either protein. Knockdown of Akt-1 together with ErbB2 or MEK1 led to the same effect as knockdown of Akt-1 alone. Unexpectedly, the triple knockdown induced a lower reduction of invasiveness.
Fig. 4.
Systematic pathway analysis by combinatorial use of siRNAs. (A) Effect of protein knockdown by combinatorial RNAi strategy on the invasion of HCC1954 cells. Cells were transfected with 20 nmol/liter siRNA pools for single, double, and triple knockdown of the respective proteins (Akt-1, ErbB2, MEK1) in 10 μl of transfection reagent. The cells that had invaded were detached from the membrane by trypsinization, centrifuged, and then resuspended in PBS. The number of invaded cells for each condition was determined by flow cytometry. Error bars were calculated from triplicate experiments. (B) Scheme of the ERK1/2 and PI3-kinase pathway (red boxes), and assumed cross-talk effects via other pathways (green boxes, intermittent arrows). ErbB2 activates two pathways in parallel: the Akt-1 pathway influences cell invasion, whereas the ERK1/2 pathway regulates proliferation. For the HCC1954 cells, we showed that, indeed, the Akt-1 pathway is dominant for modulating the invasion capacity as compared with the ERK1/2 pathway. Our result for ErbB2 knockdown suggests that other receptors activate Akt-1 because the invasion capacity is not reduced. The double knockdown of ErbB2 and MEK1 implies that the activation of Akt-1 through other receptors results in a reduction of cell invasion. Our data for the triple knockdown reveal parallel mechanisms that attenuate the invasion potential of HCC1954 cells when both the PI3-kinase and ERK1/2 are blocked.
Discussion
In the present study, we developed and optimized a combinatorial RNAi strategy, allowing a fast, efficient, and systematic investigation of pathway interrelations. We knocked down three proteins, Akt-1, ErbB2, and MEK1 alone and in combinations and quantified the expression level of each mRNA and protein. With our approach we aimed to keep the untarget effects and cytotoxicity at the lowest possible level. The phenotypic effects of single, double, and triple transfections were determined by means of a cell invasion assay.
The measurement of knockdown was done quantitatively by RPPA and was compared with qRT-PCR data. In contrast to established protein analysis procedures like Western blot (8) and ELISA, RPPAs allow for a quantitative analysis of protein expression providing a linear relationship between protein concentration and antibody signal over a higher range of protein concentrations. In addition, protein expression profiling can be performed on a large scale for a high number of different target proteins (11). Employing the RPPA approach, we showed that the endogenous protein level of ErbB2 protein was reduced by 77–93%, and for Akt-1, the reduction was between 51–73%. However, we could not quantify the MEK1 protein with RPPA, indicating that the availability of specific and sensitive antibodies is still a pressing issue.
As the RNAi mechanism targets specific mRNAs, it is well established to use TaqMan analysis to monitor untarget effects, although it is rather the reduced level of the targeted protein that is responsible for phenotypic changes. Furthermore, down-regulation of mRNA is not always correlated with the amount of residual protein (7, 9). Our data suggest that high fluctuations on mRNA level occur especially for genes which are related to the pathways under consideration, but these fluctuations do not necessarily reproduce on protein level. Turnover rates of target proteins and buffering systems within the cell may contribute to this observation. Our results for ER-α, CDK2, and ErbB2 as well as the cytotoxicity test demonstrate that untarget effects can be minimized by using siRNA at low concentrations. With respect to our results on actin, we conclude that using siRNA pools could dilute untarget effects that were observed with single siRNA sets.
To test the combinatorial RNAi strategy for the study of a cellular process, we examined the effects of ErbB2, Akt-1, and MEK1 knockdown on invasion. Gene amplification and overexpression of ErbB2 are linked with increased risk of metastasis (12–14). Receptor activation is mediated by heterodimerization with other family members (EGFR, ErbB3, ErbB4) and potentiates tumor cell motility and invasion. Activated ErbB2 simultaneously initiates stimulation of linear signaling cascades, such as the Akt, MAPK, and the protein kinase C pathways (15). Compared with other breast cancer cell lines, ErbB2 expression is up-regulated in HCC1954 cells (SI Fig. 10), which correlates with their high invasive potential (Fig. 4A). Unexpectedly, we did not measure any reduced invasive capacity of these cells when the ErbB2 level had been reduced by >90% in single knockdown. Even if basal levels of ErbB2 were still present, a reduction of cell invasion should be expected if this protein was a dominant inducer. Most likely, other receptors than ErbB2 can compensate the signaling through Akt-1 and MEK1. For the HCC1954 cells, the EGF receptor could be a candidate for this buffering effect, as this protein is highly up-regulated in this cell line (SI Fig. 10). Furthermore, the IGF-1R is expressed in these cells (SI Fig. 10) and another candidate to test. The 80% reduction of invasion capacity for the Akt-1 single knockdown indicates a dominant role for Akt-1 in cell invasion as compared with MEK1 single knockdown (Fig. 4 A and B). When both ErbB2 and Akt-1 are down-regulated, signaling through MEK1 could still occur via other growth factor receptors. This cross-talk, however, does not increase the cell-invasion potential, as the effect of ErbB2/Akt-1 double knockdown is the same as for Akt-1 alone. When both, ErbB2 and MEK1, are down regulated, the invasion capacity of HCC1954 cells is lower as compared with MEK1 single knockdown. Apparently, the activation of Akt-1 through other growth factor receptors or intermediates inhibits cell invasion, although Akt-1 knockdown almost abolishes invasion. The simultaneous knockdown of Akt-1 and MEK1 reduces cell invasion by ≈75%, indicating that ErbB2 signaling through other substrates, e.g., PKC, cannot revert the inhibition of invasion once Akt-1 and MEK-1 are down-regulated. In the triple knockdown, ErbB2 signaling is also abolished and other receptors could activate neither Akt-1 nor MEK1. Nevertheless, we detected a reduction of invasion by a mere 60%, meaning that the invasive potential was not as efficiently lowered as in any single or double knockdown involving Akt-1. This reduction was still higher as compared with the ErbB2/MEK1 knockdown. We reason that other pathways could attenuate the inhibiting effect of Akt-1 knockdown in the HCC1954 cells once both the Erk1/2 and the PI3-kinase pathways are blocked, and also ErbB2 is not expressed. These alternative pathways could, for instance, be the integrin- or the Wnt-signaling pathways.
Our results clearly demonstrate that the use of combinatorial RNAi does not only reveal interrelations between the proteins under consideration, but also provides information about the key points where cross-talk with other pathways may occur (Fig. 4B). It should be noted, however, that these data reflect the situation in the cell line we analyzed, whereas the network and the relation of signaling pathways could well be different in other cell lines.
Individual signaling systems are believed to contribute to a phenotype in a defined and characteristic manner. However, each system is interconnected with others and therefore cannot be viewed in isolation. Currently, there is no technique available which would allow determining cross-talk between pathways in complex protein networks. Applying the presented approach to the systematic investigation of complex protein networks will pave the way toward a new level of understanding in biology.
Materials and Methods
Cell Culture.
HCC1954 (ATCC; CRL-2338) cells were cultured in RPMI 1640 Modified Medium (American Type Culture Collection). The medium was supplemented with 50 units/ml penicillin, 50 μg/ml streptomycin sulfate, 1% nonessential amino acids, and 10% FBS (all from GIBCO/BRL, Gaithersburg, MD). Cells were split two to three times per week in a 1:3 ratio and cultured until a maximum passage number of 25.
siRNA Sequences.
ErbB2, Akt-1, and MEK1 siRNAs were designed by Dharmacon (Lafayette, CO). GFP and Lamin A/C siRNAs were designed and obtained from Qiagen (Hilden, Germany). All siRNA sequences are given in SI Materials and Methods.
siRNA Transfections.
HCC1954 cells were seeded at a number of 1.5 × 105 per well in antibiotic-free medium in a six-well format (NUNC, Roskilde, Denmark) before transfection. The confluency of the cells was 50–60% at the day of transfection. The indicated amounts of siRNA and Lipofectamine 2000 (Invitrogen, Carlsbad, CA) were diluted separately in reduced-serum medium OptiMEM (GIBCO/BRL) and incubated for 5 min at room temperature. The two solutions were mixed and incubated for 20 min at room temperature. siRNA-Lipofectamine 2000 mixture was then added to the cells, and the plate was mixed by gentle rocking. MOCK transfected cells were treated with Lipofectamine 2000 but without siRNA. Transfected cells were incubated at 37°C and 5% CO2 for 48 h.
Antibodies and Western Blot.
Antibodies against Akt-1, ErbB2, and MEK1 were obtained from BD Biosciences (Heidelberg, Germany), NeoMarkers (Fremont, CA), and Cell Signaling Technology (Danvers, MA), respectively. After a 48-h incubation of siRNA transfected cells, cell extracts were prepared with M-PER lysis buffer (Pierce, Rockford, IL) containing a mixture of protease inhibitors (Complete Mini, EDTA-free; Roche Diagnostics, Mannheim, Germany). Protein concentration was determined with a Micro BCA Protein Assay Reagent Kit (Pierce). For Western blotting, proteins were denatured with 4× Roti Load (Roth, Karlsruhe, Germany) at 95°C for 5 min, and 60 μg of protein were loaded in every lane of a 12% SDS gel. Proteins were electroblotted to PVDF membranes (Hybond-P; Amersham Biosciences, Buckinghamshire, U.K.) and exposed to primary antibodies as indicated. All antibodies were used at a dilution of 1:500. An anti-actin (20–33) antibody (Sigma, Steinheim, Germany) was used at a dilution of 1:750 for normalization. Horseradish peroxidase-conjugated mouse or rabbit antibodies (Amersham Biosciences) were used as secondary antibodies, and signals were detected by enhanced chemiluminescence (Amersham Biosciences).
Protein Quantification and RPPA.
siRNA transfected samples were lysed with the mammalian lysis buffer M-PER (Pierce). The total protein concentration was measured with the BCA Protein Assay Kit (Pierce). Protein concentrations were adjusted to the concentration of the sample having the lowest concentration. Then, samples were diluted 1:2 with spotting buffer (Whatman, Brentford, U.K.) and spotted onto nitrocellulose coated FAST-slides (Whatman) with a Biochip Arrayer (PerkinElmer, Shelton, CT) or a Nanoplotter NP2 (GeSiM, Dresden, Germany). Each dot corresponded to 1 nl of lysate. For each target protein (Akt-1, ErbB2, and MEK1) selected for knockdown, two microarray-chips were incubated with a specific antibody that had been previously validated by Western blot. All three antibodies were applied at a 1:250 dilution except for the anti-actin (20–33) antibody (Sigma), which was used at a 1:500 dilution. Antibodies against untarget proteins p38 MAPK, ER-α, IGF-1R, and CDK2 were used at a dilution of 1:500, 1:150, 1:200, and 1:250. ER-α and IGF-1R antibodies were obtained from Upstate (Temecula, CA), and CDK2 antibody was obtained from BD Biosciences. p38 MAPK antibody (sc-7149) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Detection was performed with the near infrared dye-coupled secondary antibody described above, and signal intensities were analyzed with the LI-COR Odyssey Scanner System and Software (LI-Cor, Bad Homburg, Germany). All samples were spotted in at least two subarrays and four independent replicates, resulting in eight data points per sample. Normalization was by setting the antibody signal for nontransfected cells of each experiment to 1.
qRT-PCR (TaqMan).
Total RNA of the cell lines was extracted by using the Invisorb Spin cell RNA mini kit (Invitek GmbH, Berlin, Germany), and single-stranded cDNA was transcribed by using RevertAid H Minus First Strand cDNA Synthesis kit (Fermentas, St. Leon-Rot, Germany). Ten nanograms of total RNA was used for each reaction. qRT-PCR for target genes Akt-1, ErbB2, and MEK1 and untarget control genes ACTB, p38 MAPK, ER-α, IGF-1R, and CDK2 was performed with the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Weiterstadt, Germany) by using probes of the Universal Probe Library (Roche). Sequences of primers, and the respective UPL probes are given in SI Materials and Methods.
Cell Counting.
After an incubation of siRNA-transfected cells for 48 h at 37°C and 5% CO2, the cells were washed with D-PBS (GIBCO/BRL) and trypsinized (Sigma). The same amount of medium was added to each sample to inhibit the trypsin. Ten microliters of four replicate aliquots were taken from each sample and counted in a Neubauer chamber.
Invasion Assay.
The BioCoat tumor invasion system (BD Biosciences) was used for the invasion assay. HCC1954 cells were grown to 50–60% confluency in six-well plates and transfected with siRNAs. After 24 h, the medium was removed, and cells were washed with PBS and were then serum starved in medium containing 1% FBS for 24 h. After rehydrating the BioCoat Invasion plates with PBS, medium with 10% FBS (chemoattractant) was added to the bottom wells. Cells from the six-well plates were trypsinized, counted, and resuspended in serum-free medium. Triplicates of 1 × 105 cells for each transfection condition were seeded into separate wells of the top chamber and incubated for 72 h at 37°C and 5% CO2. Invaded cells were detached by trypsinization from the bottom side, centrifuged, and then resuspended in PBS. The number of invaded cells for each condition was determined by flow cytometry (FACSCalibur; BD Biosciences).
Supplementary Material
Acknowledgments
We thank Jens Mattern, Özlem Sener Sahin, Christian Schmidt, Ute Ernst, and Daniele Harmon for excellent technical assistance. This project was supported by the German Federal Ministry of Education and Research (BMBF) within National Genome Research Network Program Grant 01GR0420–SMP-Cell and BMBF Grant 0313336, and within EU FP6 Transfog Project (LSHC-CT-2004-503438).
Abbreviations
- RPPA
reverse-phase protein array
- PI3-kinase
phosphatidylinositol 3-kinase
- IGF-1R
insulin-like growth factor 1 receptor
- ER-α
estrogen receptor α;
- qRT-PCR
quantitative real-time PCR.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606827104/DC1.
References
- 1.Wang GK, Zhang W. Histol Histopathol. 2005;20:593–602. doi: 10.14670/HH-20.593. [DOI] [PubMed] [Google Scholar]
- 2.Gilcrease MZ. Cancer Lett. 2007;247:1–25. doi: 10.1016/j.canlet.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 3.Pollak MN, Schernhammer ES, Hankinson SE. Nat Rev Cancer. 2004;4:505–518. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 4.Karnoub AE, Symons M, Campbell SL, Der CJ. Breast Cancer Res Treat. 2004;84:61–71. doi: 10.1023/B:BREA.0000018427.84929.5c. [DOI] [PubMed] [Google Scholar]
- 5.Xia XG, Zhou H, Xu Z. BioTechniques. 2006;41:64–68. doi: 10.2144/000112198. [DOI] [PubMed] [Google Scholar]
- 6.Huang F, Khvorova A, Marshall W, Sorkin A. J Biol Chem. 2004;279:16657–16661. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
- 7.Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, Collins FS. Proc Natl Acad Sci USA. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koller A, Watzig H. Electrophoresis. 2005;26:2470–2475. doi: 10.1002/elps.200500024. [DOI] [PubMed] [Google Scholar]
- 9.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le XF, Bedrosian I, Mao W, Murray M, Lu Z, Keyomarsi K, Lee MH, Zhao J, Bast RC., Jr Cell Cycle. 2006;5:1654–1661. doi: 10.4161/cc.5.15.3007. [DOI] [PubMed] [Google Scholar]
- 11.Loebke C, Sueltmann H, Schmidt C, Henjes F, Wiemann S, Poustka A, Karf U. Proteomics. 2007;4:558–564. doi: 10.1002/pmic.200600757. [DOI] [PubMed] [Google Scholar]
- 12.Braun S, Schlimok G, Heumos I, Schaller G, Riethdorf L, Riethmuller G, Pantel K. Cancer Res. 2001;61:1890–1895. [PubMed] [Google Scholar]
- 13.Jing X, Kakudo K, Murakami M, Nakamura Y, Nakamura M, Yokoi T, Yang Q, Oura S, Sakurai T. Cancer. 1999;86:439–448. [PubMed] [Google Scholar]
- 14.Seton-Rogers SE, Lu Y, Hines LM, Koundinya M, LaBaer J, Muthuswamy SK, Brugge JS. Proc Natl Acad Sci USA. 2004;101:1257–1262. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yarden Y, Sliwkowski MX. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.